Abstract
A stable and robust trypsin-based biocatalytic system was developed and demonstrated for proteomic applications. The system utilizes polymer nanofibers coated with trypsin aggregates for immobilized protease digestions. After covalently attaching an initial layer of trypsin to the polymer nanofibers, highly concentrated trypsin molecules are crosslinked to the layered trypsin by way of a glutaraldehyde treatment. This process produced a 300-fold increase in trypsin activity compared with a conventional method for covalent trypsin immobilization, and proved to be robust in that it still maintained a high level of activity after a year of repeated recycling. This highly stable form of immobilized trypsin was resistant to autolysis, enabling repeated digestions of bovine serum albumin over 40 days and successful peptide identification by LC-MS/MS. This active and stable form of immobilized trypsin was successfully employed in the digestion of yeast proteome extract with high reproducibility and within shorter time than conventional protein digestion using solution phase trypsin. Finally, the immobilized trypsin was resistant to proteolysis when exposed to other enzymes (i.e. chymotrypsin), which makes it suitable for use in “real-world” proteomic applications. Overall, the biocatalytic nanofibers with trypsin aggregate coatings proved to be an effective approach for repeated and automated protein digestion in proteomic analyses.
Keywords: Enzyme aggregate coatings, Nanofibers, Protein digestion, Recyclable biocatalysts, Trypsin
1 Introduction
In bottom-up proteomics strategies, proteins are first digested into peptides and then separated prior to mass spectrometric analysis. Over the last decade, great advances have been achieved in the areas of peptide separation and mass spectrometric detection [1, 2]; however, developments to enhance protein digestion have lagged [3], considering most current protocols involve digestion incubation times on the order of 4–15 h. The most commonly used enzyme for protein digestion is trypsin, as it affords several advantages over other proteases; for example, trypsin hydrolyzes peptide bonds exclusively on the carboxyl side of lysine and arginine. Because these amino acid residues are present at approximately one residue per every 10–12 amino acids, the masses of the resulting peptides are ideal for mass spectrometric analysis. In addition, the positive charge on the lysine and arginine assists homogeneous fragmentation during collision induced dissociation in the mass spectrometer.
Protein digestion is commonly accomplished via one of two methods, i.e., either in gel or in-solution. In both cases, free soluble trypsin is used to perform the digestion, which can lead to autolytic digestion and subsequent loss of trypsin activity over time. To overcome these drawbacks, trypsin has been immobilized on solid supports such as agarose [4], polystyrene [5], porous silica [6, 7], sol-gel entrapment [8], magnetic particles [9] and also in a monolithic columns [10]. The use of a support also accelerates the reaction due to an increase in the enzyme to substrate ratio. Other means of improving digestion kinetics have included application of energy inputs, such as ultrasound [11], microwave radiation [12], and a combination of immobilized trypsin and irradiation [9]. Regardless the support used to enhance digestion rates, a reoccurring hurdle was the reusability/recyclability of the immobilized trypsin. In order to successfully be reused, immobilized trypsin must have high activity, high stability, and high resistance to autolysis and proteolysis by other proteases commonly found in proteomic samples. Such characteristics could enable nano-devices with automated digestion capabilities [13] that would consequently reduce total analysis time, as well as improve analytical efficiency.
In the work reported herein, we capitalized on the success of a biocatalytic system [14, 15] we developed earlier that uses nanostructures to stabilize enzymatic activity. Enzyme aggregate coatings display distinct features, such as higher stability and lower activation energies due to the multipoint covalent linkages and direct reaction with the substrate, respectively [14]. Compared to internal nanopores of mesoporous media, the well-exposed surface of nanofibers improve mass transfer of substrates and enable multilayered enzyme coatings in the open space between nanofibers. Electrospun nanofibers were used as support materials [14, 15], as they can be readily fabricated through a simple and versatile process of electrospinning. In addition, they are durable and can be easily recovered from the reaction solution. However, the earlier work of enzyme aggregate coatings on electrospun nanofibers used a small model substrate of less than 1 nm in size [14], and the successful application of enzyme coatings in proteomic analysis requires the evaluation of the activity of enzyme aggregate coatings in the digestion of large-sized protein molecules, which can possibly place a serious challenge in the mass transfer limitation of substrate proteins through the matrix of enzyme aggregate coatings. To modify the system for proteomics, we developed a process whereby polymer nanofibers are coated with trypsin aggregates for immobilized protease digestions. For the first time in this paper, we evaluated the trypsin-aggregate coated nanofibers in terms of protein digestion activity, long-term stability, and proteolytic resistance. Overall, the nanofibers proved to be active in protein digestion, incredibly stable over long periods of time, and resistant to proteolytic digestion, making their application in proteomic analyses very promising.
2 Materials and methods
2.1 Materials
Trypsin, α-chymotrypsin, glutaraldehyde (GA), Nα-Benzoyl-L-arginine 4-nitroanilide hydrochloride (L-BAPNA) and N,N-dimethylformamide (DMF) were purchased from Sigma (St Louis, MO, USA). Bovine serum albumin (BSA) was purchased from New England Biolab (Ipswich, MA, USA). Polystyrene (PS, Mw = 860,000) and poly (styrene-co-maleic anhydride) (PSMA, Mw=224,000; maleic anhydride content = 7 wt%) were purchased from Pressure Chemical Company (Pittsburgh, PA, USA) and Aldrich (Milwaukee, WI, USA), respectively. Tetrahydrofuran (THF; HPLC 99.9%) was purchased from Burdick and Jackson (Muskegon, MI, USA). All other reagents were purchased from Sigma and Aldrich in the highest grade commercially available.
2.2 Immobilization of trypsin on polymer nanofibers
Polymer nanofibers were prepared via electrospinning, as previously described [14] Briefly, a mixture of PS and PSMA in a 2:1 w/w ratio was dissolved in THF, and then loaded into a plastic syringe with a stainless steel needle. 7 kV was applied to the needle using a high voltage supply. The electrospun nanofibers were collected on clean aluminum foil, which was connected to the high voltage supply and also grounded. A non-woven mat form of polymer nanofibers were detached from the foil with tweezers, and stored at room temperature until later use. The PS-PSMA nanofibers (0.4 mg) were incubated in 1 mL of 10 mM sodium phosphate buffer (pH 7.9) containing 10 mg of trypsin. The glass vial containing nanofibers and trypsin was shaken at 200 rpm for 30 min, and then placed on the rocker (30 rpm) in a cold box (4 °C) for 2 h to covalently attach the trypsin onto the nanofibers (CA-TR). For the synthesis of enzyme-aggregate coatings (EC-TR), a GA solution (final GA concentration was 0.5% w/v) was added after 150 min incubation, and the mixture was shaken on the rocker (30 rpm) at 4 °C overnight. For the CA-TR, a buffer was added instead of the GA solution. After an overnight incubation, biocatalytic nanofibers (both CA-TR and EC-TR) were washed with 100 mM sodium phosphate, pH 7.8. To cap the un-reacted aldehyde groups, the biocatalytic nanofibers were incubated in 100 mM Tris-HCl (pH 7.8) for 30 min. Biocatalytic nanofibers were washed extensively with 10 mM sodium phosphate buffer (pH 7.9), and were stored in the same buffer at 4 °C until later use
2.3 Trypsin activity and stability of biocatalytic nanofibers
Trypsin activity of the biocatalytic nanofibers (CA-TR and EC-TR) was measured by the hydrolysis of L-BAPNA in a buffer solution (10 mM sodium phosphate, pH 7.9). In detail, biocatalytic nanofibers were transferred into a glass vial containing 4 mL of buffer and 40 µL of L-BAPNA (10 mg/mL in DMF), which initiates the enzymatic hydrolysis of L-BAPNA. The vials were shaken at 200 rpm and aliquots were removed every 10 min. The product of enzymatic catalysis in each aliquot was analyzed by using a spectrophotometer (Model Cary 5G UV-Vis-NIR spectrophotometer from Varian, Inc., Palo Alto, CA) to measure the absorbance at 400 nm (A400), and the initial rate was calculated from the slope of A400 over time. The stability of biocatalytic nanofibers was determined under iterative uses and continuous incubation in a buffer solution (10 mM sodium phosphate, pH 7.9) under rigorous shaking (200 rpm). The residual activity was measured at each time point as described above. Immediately after each activity measurement, the samples were washed three times with a fresh buffer solution to remove all residual substrates and products. The biocatalytic nanofibers were incubated at room temperature while shaking (200 rpm) until the next use. The relative activity was calculated as the ratio of residual activity over initial activity at each time point.
2.4 Stability of biocatalytic nanofibers in the presence of proteolytic hydrolysis
The stability of the biocatalytic nanofibers in the presence of a protease was evaluated using α-chymotrypsin as follows. Biocatalytic nanofibers were added to a vial containing 1 mg/mL α-chymotrypsin in a buffer solution (10 mM sodium phosphate, pH 7.9), and the mixture was incubated at room temperature while shaking (200 rpm). After incubation with chymotrypsin, the biocatalytic nanofibers were washed with fresh buffer to remove residual chymotrypsin, and then transferred to a new vial where the residual trypsin activity was measured. After the measurement of the residual activity, biocatalytic nanofibers were incubated with chymotrypsin again under the same conditions.
2.5 Protein digestion by biocatalytic nanofibers
In another set of experiments, we investigated the ability of the biocatalytic nanofibers to be used repeatedly as required in a “real world” proteomics application. For this purpose in-solution protein digestions were carried out. A 1 µM solution of BSA was used as a model protein substrate. First, BSA was reduced with 10 mM DL-dithiothreitol (DTT) in 25 mM ammonium bicarbonate (pH 8.25) at 37 °C for 1 h, after which iodoacetamide was added to a final concentration of 50 mM. The mixture was subsequently incubated at room temperature in darkness for 45 min to alkylate the free cysteines and then added to a glass vial containing biocatalytic nanofibers. The in-solution digestion was then carried out at room temperature while shaking (200 rpm) for 16 h. Finally, the supernatant was transferred to a new centrifuge tube and formic acid was added to quench the reaction. The samples were dried down in a Speedvac (Thermo-Fisher, San Jose, CA), and stored at −20 °C until further sample processing. The biocatalytic nanofibers were washed and stored at room temperature for future experiments.
The yeast proteome was prepared as described previously [16], and 3 aliquots of 50 µg each from the soluble fraction were dried down. Samples were resuspended in 50% TFE and sonicated for 3 min and reduced as described above. The samples were then diluted 10 times with 25 mM ammonium bicarbonate and either 1µg of trypsin was added or biocatalatic nanofibers from two different preparations were added to a 1.5 mL reaction tube. Digestions were carried out at 37 °C under shaking (1400 rpm) in a thermo-mixer for 6 hours. After digestion, supernatants were transferred to new 0.6 mL tubes, and dried down by centrifugal evaporation.
2.6 LC-MS/MS analysis
One picomol of the BSA digest was analyzed by LC-MS/MS. An Agilent HPLC-Chip system was coupled to a MSD Trap XCT Ultra ion trap (Agilent Technologies, Santa Clara, CA). The Agilent auto sampler was used to load the samples at 4 °C. Separations were performed on the chip, which contained a 40-nL enrichment column and a 43 mm × 75 µm analytical column. Both columns were packed with 5 µm ZORBAX 300SB C18 particles. A flow rate of 1 µL/min was employed for sample loading and enrichment, while elution was performed at a flow rate of 600 nL/min. Peptides were eluted using a 5 min gradient from 5% to 90% Solvent B (0.5% formic acid, 90% acetonitrile; Solvent A: 0.5% formic acid in water:acetonitrile of 97:3), with a separation window of ~2 min and total analysis time of 12 min. A blank was run between each sample to eliminate the possibility of cross contamination among different samples. The data were acquired in survey scans from 500 to 1600 amu (3 µscans) followed by five data dependent MS/MS scans, using an isolation window of 3 amu, a normalized collision energy of 35%, and a dynamic exclusion period of 2 min.
For the analyses of yeast digests, 5 µg of the digested peptides were analyzed as described previously [17]. Briefly, digests were analyzed using an in-house developed capillary LC system coupled online to a linear ion trap mass spectrometer (Thermo-Fisher, San Jose, CA) with an in-house ESI source. Data were collected in a data-dependent mode where the ten most intense ions from the survey scan were subjected to MS/MS fragmentation. A dynamic exclusion time was activated for 1 min and the normalized collision energy used was 35%.
Peptide identification from raw data was performed using Spectrum Mill software with the following constraints: tryptic cleavage, up to three missed cleavage sites, and tolerances ±2.5 Da for the precursor ions, and ±0.8 for the fragments. For database searches the swissprot.fasta protein database (October 10, 2003) was used. Spectra matching for BSA were manually identified. For the estimation of error rates in the yeast analyses, a reverse database was built. Score distributions for forward and reverse searches were constructed after normalization. Statistical analysis and curve fitting were performed using Microsoft Excel, histogram and solver functions. Score distributions were modeled assuming a Gaussian distribution. The mean and the standard deviation from the score distributions were determined by least squares fitting. False discovery rates (FDR) were calculated as proposed earlier.
2.7 SEM characterization
Fibers were coated with a thin layer of iridium (~ 20 Å) to make them conductive and prevent charging. Image characterization was performed using a JEOL 6700 F - FESEM (JEOL Ltd., Tokyo, Japan). All the samples were imaged at an acceleration voltage of 3 KV and an emission current of 20 µA.
3 Results and discussion
3.1 Preparation and characterization of trypsin-aggregate coated nanofibers
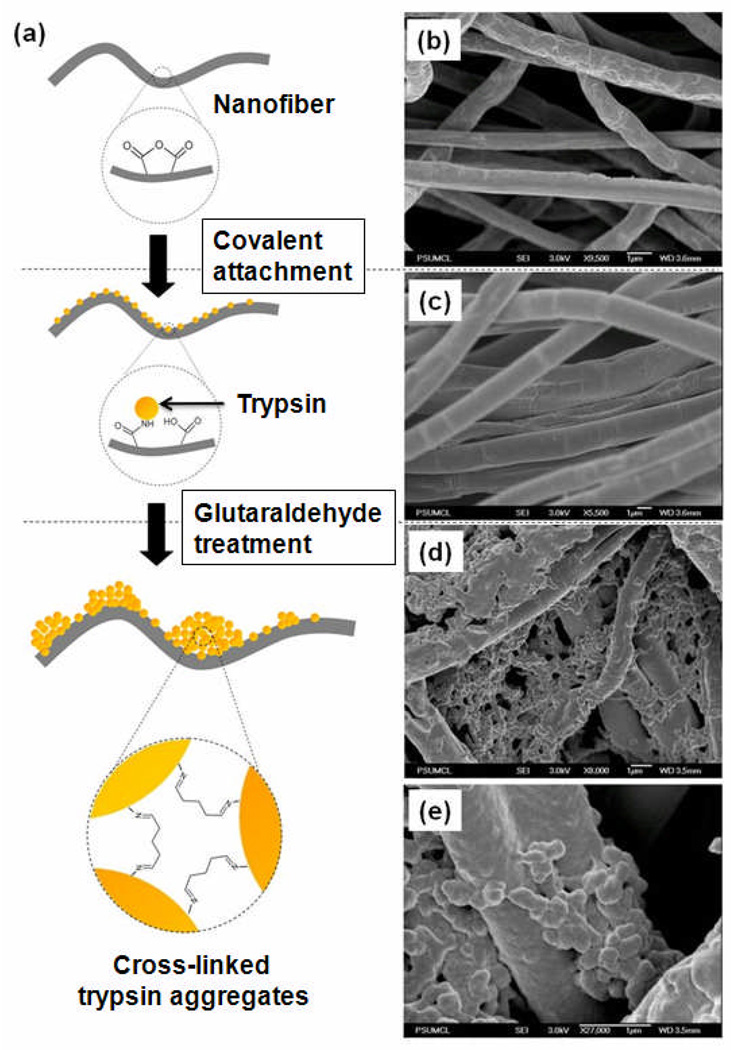
Figure 1a schematically depicts the stages involved in preparing the covalently-attached trypsin (CA-TR) and trypsin aggregate coatings (EC-TR) on polymer nanofibers. First, a mixture of PS and PSMA in a 2:1 w/w ratio is electrospun to build polymer nanofibers structures. Trypsin molecules are covalently attached to these polymer nanofibers (CA-TR) as a result of the reaction among the amino groups of trypsin molecules and the maleic anhydride groups of PSMA. After incubating CA-TR in a highly concentrated trypsin solution, the glutaraldehyde treatment is applied to crosslink the enzyme molecules, including the covalently attached trypsin, which results in the trypsin-aggregate coated nanofibers (EC-TR).
Figure 1.
(a) Schematic diagram for the preparation of covalently-attached trypsin (CA-TR) and trypsin-aggregate coated nanofibers (EC-TR) on the PS:PSMA nanofibers. SEM images of (b) bare nanofibers, (c) CA-TR, and (d) EC-TR. (e) A zoomed-in SEM image of EC-TR.
The SEM images of bare nanofibers, CA-TR, and EC-TR are shown in Figures 1b–1e. The surface morphologies of bare nanofibers and CA-TR (Figure 1b and 1c) appear similar, which suggests that the small size (~4 nm) of covalently-attached trypsin molecules make it difficult to visualize the monolayer. Conversely, SEM images of EC-TR (Figures 1d and 1e) vividly show thick coatings that represent crosslinked trypsin aggregates on the polymer nanofibers. The activities of CA-TR and EC-TR, measured by the hydrolysis of L-BAPNA, were 2.79 and 825 (M/min per mg fibers), respectively. In other words, the apparent activity of EC-TR is ~300 times higher than that of CA-TR as a result of the thick coating on EC-TR (Figures 1d and 1e) that significantly increases the enzyme-to-substrate ratio.
3.2 Stability of trypsin-aggregate coated nanofibers
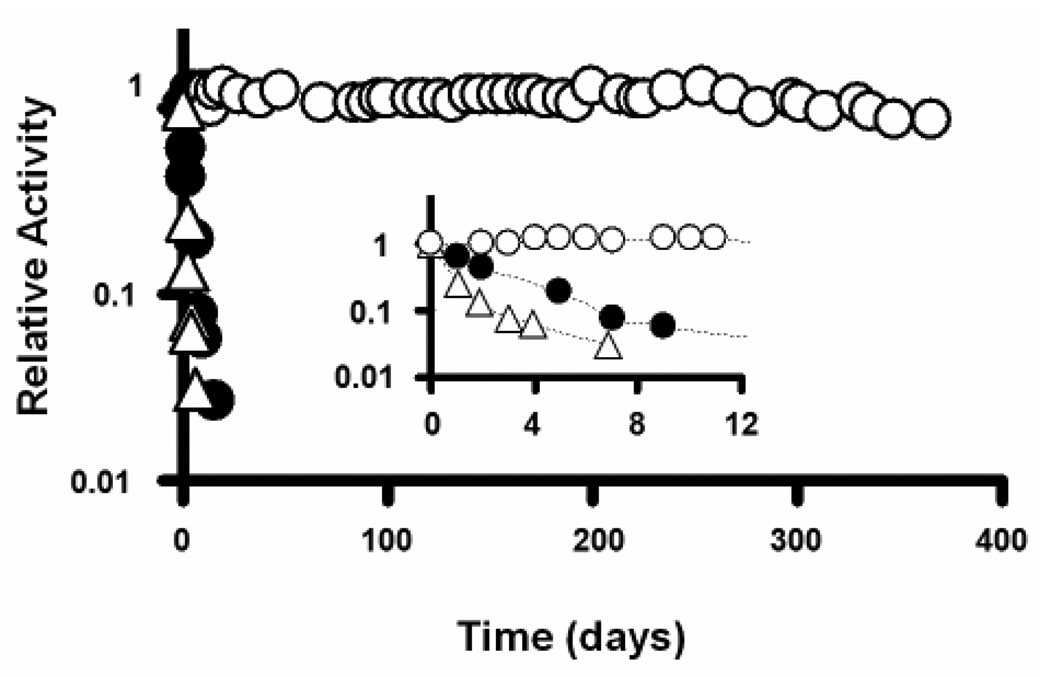
The stability of biocatalytic nanofibers was investigated by measuring the residual activity of each sample over time following continuous incubation in a buffer solution under rigorous shaking (200 rpm) at room temperature. Figure 2 shows the stabilities of free trypsin, CA-TR, and EC-TR. Note that both free trypsin and CA-TR exhibit rapid inactivation, possibly due to autolysis and denaturation under rigorous shaking. Conversely, EC-TR exhibits negligible loss of trypsin activity under iterative uses and continuous shaking over a period of one year. This unprecedented stabilization of enzyme activity is most likely the result of the multipoint linkages on the surface of enzyme molecules, which not only prevent autolysis, but also prevent enzyme denaturation. This long-term (one year) study is the first of its kind to obtain excellent enzyme stability results under iterative uses and continuous shaking.
Figure 2.
Enzyme stability of free trypsin (empty triangles), covalently-attached trypsin (CA-TR, filled circles), and trypsin-aggregate coatings (EC-TR, empty circles) under repeated use and rigorous shaking (200 rpm). Inserted figure shows the enzyme stability during the first 12 days.
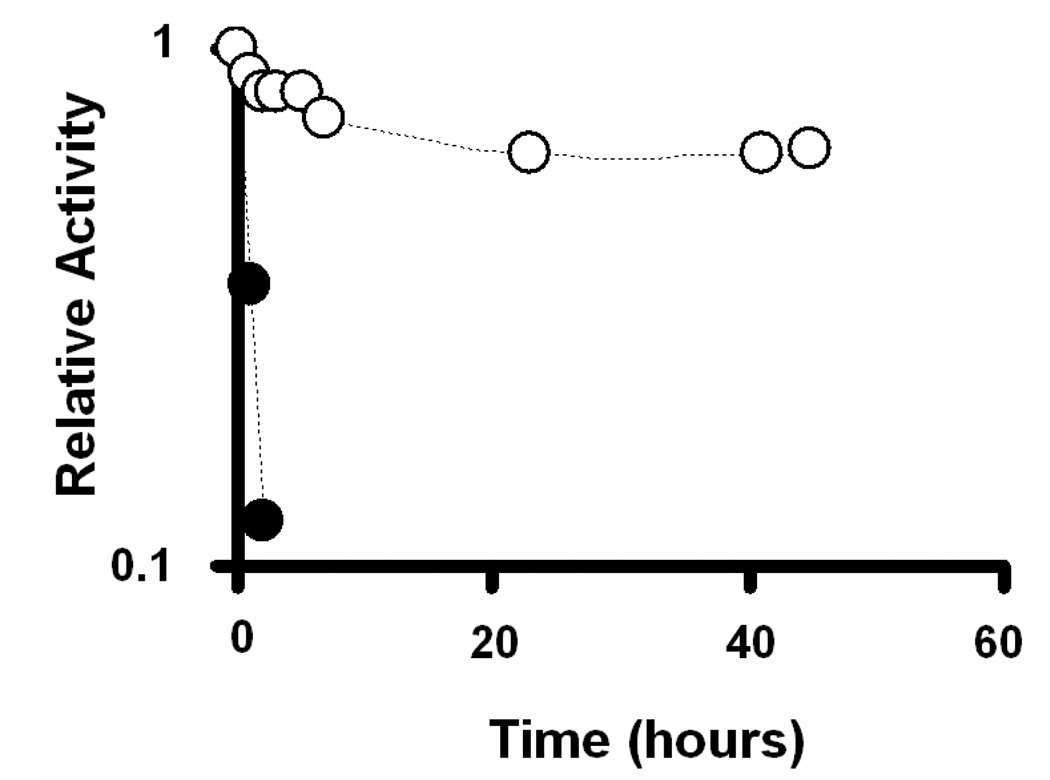
The resistance to undesired proteolytic hydrolysis is a valuable feature for practical applications using enzymes due to other ubiquitous proteases in complex cell protein extracts. To test the robustness of the trypsin-aggregate nanofibers in the presence of other proteases that may be present in a cell lysate sample, the stabilities of CA-TR and EC-TR were evaluated in the presence of chymotrypsin (Figure 3). After washing away all chymotrypsin, the activity of the biocatalytic nanofibers was measured at each time point by hydrolyzing L-BAPNA from the incubation solution. Figure 3 shows that the activity of CA-TR had dropped to < 85% after a 2 h incubation in the chymotrypsin solution. This rapid inactivation most likely resulted from the proteolytic hydrolysis of CA-TR by the free chymotrypsin. Conversely, EC-TR showed an excellent resistance to the proteolysis of chymotrypsin by maintaining 70% of its initial trypsin activity after 45 h incubation with the chymotrypsin. This observation suggests that the multipoint linkages among the trypsin molecules effectively reduce the potential for digestion by other proteases.
Figure 3.
Comparison of the enzyme stability of covalently-attached trypsin (CA-TR, filled circles) and trypsin-aggregate coatings (EC-TR, empty circles) in the presence of α-chymotrypsin.
3.3 Repeated protein digestion using biocatalytic nanofibers
Using BSA as the substrate, repeated digestions were carried out on the biocatalytic nanofibers to demonstrate their stability, reusability, and potential for high throughput proteomics applications. Following proteolysis, the protein digests were submitted for LC-MS/MS analysis using Chip-MS technology. The digested products were collected after each BSA digestion, and the biocatalytic nanofibers were washed with fresh buffer (10 mM sodium phosphate, pH 7.9) until residual protein digests were no longer observed. The biocatalytic nanofibers were stored at room temperature with shaking at 200 rpm until needed.
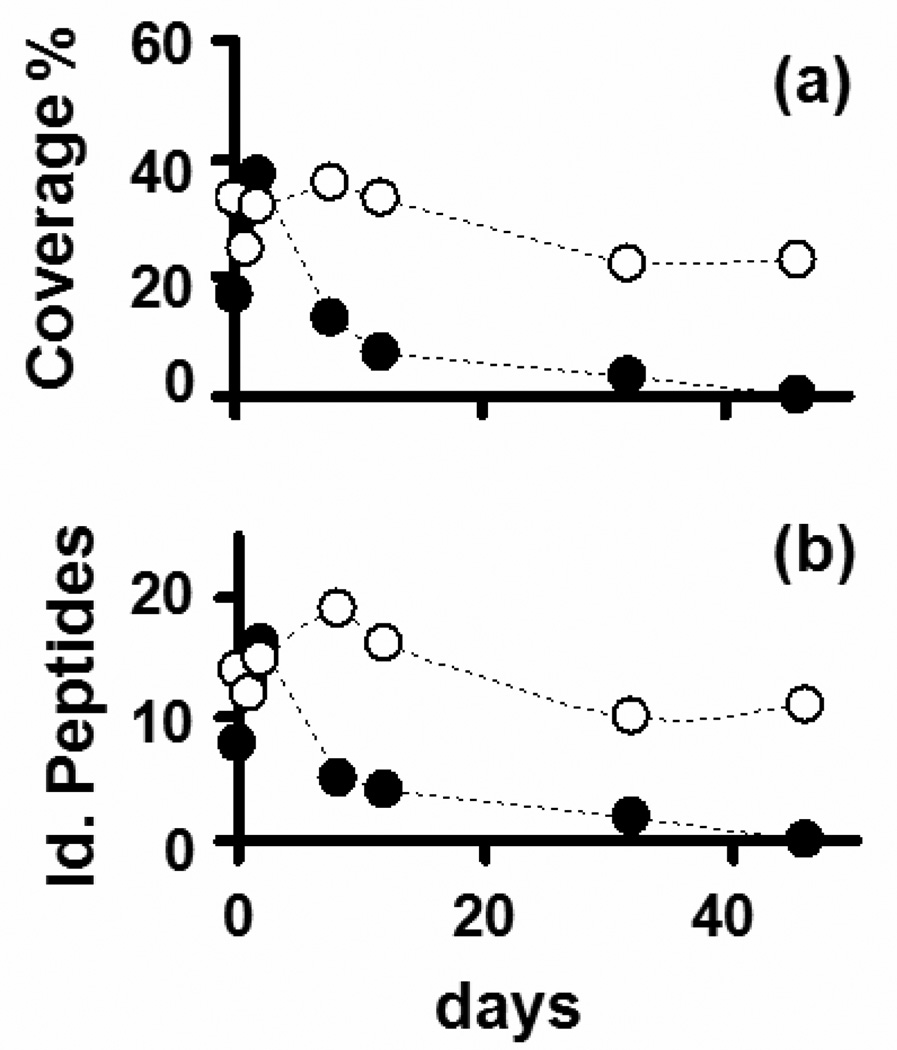
The LC-MS/MS analysis results of a BSA digestion after five repeated uses of the same biocatalytic nanofibers (CA-TR and EC-TR) are summarized in Figure 4. Note that the performance of CA-TR in terms of protein identification (represented by both the percentage of protein coverage and the number of identified BSA peptides) dropped rapidly after a few days of repeated use. Intact BSA could be observed at the end of the LC chromatograms, which suggests that BSA was not fully digested under this condition, possibly as a result of the poor stability exhibited by CA-TR (Figure 2). On the other hand, EC-TR maintained a high level of performance in terms of protein identification under repeated use, even after 46 days (Figure 4). Interestingly, the signal-to-noise ratio of the final time points was virtually the same as that of the first time point (data not shown).
Figure 4.
Time course analyses of different immobilization methods by iterative digestions of BSA across several days as a function of (a) protein sequence coverage; (b) number of identified BSA peptides using covalently-attached trypsin (CA-TR, filled circles) or trypsin-aggregate coatings (EC-TR, empty circles).
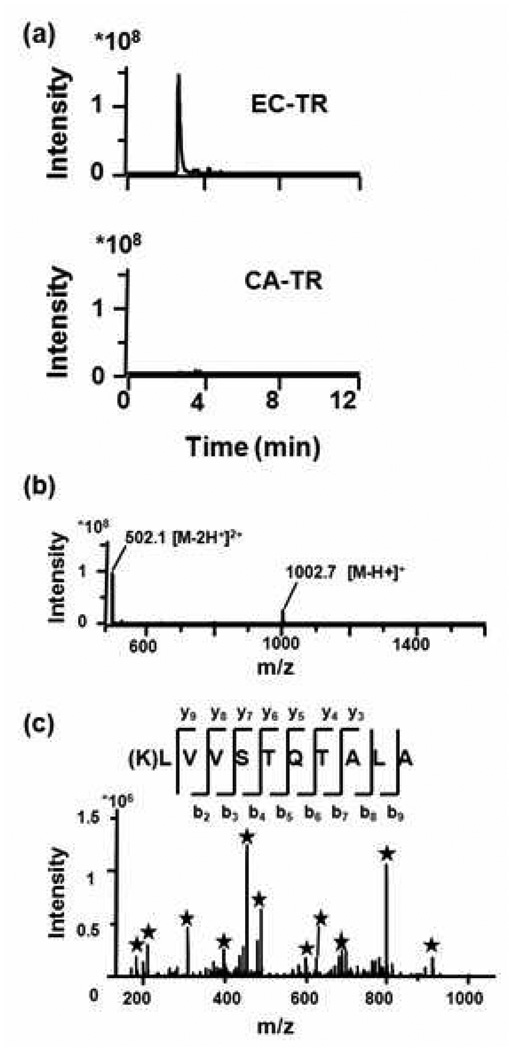
The difference between CA-TR and EC-TR is readily apparent in the base peak chromatograms of the BSA peptide LVVSTQTALA, which were obtained from the BSA digestions after 46-days (Figure 5a). The CA-TR nanofibers produced no detectable cleavage product, while the EC-TR nanofibers produced the highly intense +2 charge state peptide LVVSTQTALA. After repeated cycles of use for 46 days, quality MS and MS/MS spectra could still be obtained for EC-TR nanofibers (Figures 5b and 5c), that lead to confident BSA identification. Lastly, a BSA digestion was performed after the EC-TR nanofibers had been incubated in a chymotrypsin solution for 16 days. The protein coverage and number of unique identified peptides were 34% and 16, respectively, i.e., a further indication that EC-TR resisted proteolytic digestion by chymotrypsin.
Figure 5.
BSA digestion using covalently-attached trypsin (CA-TR) and trypsin-aggregate coatings (EC-TR) after 46 days of iterative use. (a) Comparison of CA-TR and EC-TR on the base peak chromatogram (trace of ions) corresponding to the charge +2 peptide LVVSTQTALA. (b) Mass spectrum showing the charge +1 and +2 of the peptide LVVSTQTALA from the BSA digestion using EC-TR. (c) MS/MS spectrum of +2 peptide LVVSTQTALA from the BSA digestion using EC-TR; asterisks mark the fragment peaks matching the theoretical ion masses from the y or b-series.
3.4 Application of biocatalytic nanofibers for protein digestion in high throughput proteomics
The soluble fraction of yeast proteome extract was used as a model to demonstrate, a) the applicability of the EC-TR-nanofibers to the digestion of a whole proteome sample, b) the digestion reproducibility of EC-TR-nanofibers, and c) the comparison of this novel digestion method with conventional trypsin digestion using solution phase trypsin. To accomplish these goals, three batches of digestion were carried out using two different samples of EC-TR-nanofibers and the solution phase trypsin.
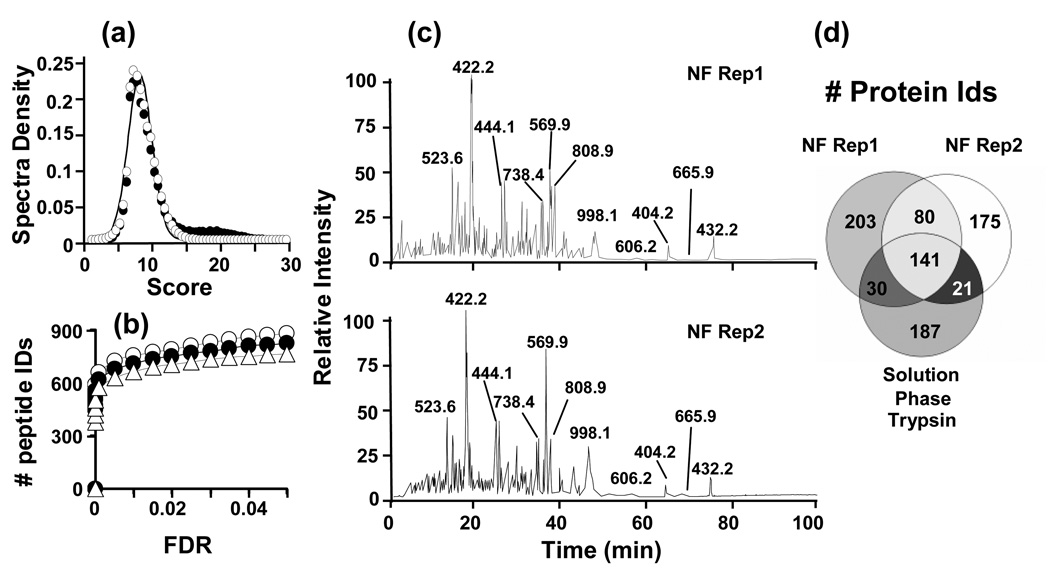
In order to evaluate the LC-MS/MS analyses, we had to develop an empirical method for differentiating between random and true peptide sequence assignations to the MS/MS spectra using Spectrum Mill™. This method was based on the distribution of the random scores in the reverse database search. As shown in Figure 6a, random scores either from the forward or reverse databases follow a Gaussian distribution and deviate from the true matches in the forward search. This finding allowed us to proceed in the same way as we have done before with other search engines such as SEQUEST [18, 19] and assign a formal error rate at the peptide identification level. By using this statistical method, we were able to identify at a 5% of FDR cut-off an average of ~850 unique peptides, corresponding to ~400 proteins for each dataset (Figure 6b). The number of identified peptides differed in less than 15% for both datasets, as well as for the dataset originated from the conventional trypsin digestion. These minimal differences can be explained as a consequence of the expected reproducibility in a typical shotgun proteomic experiment. Nevertheless, the visual comparison of chromatograms from the digestion using two different samples of EC-TR nanofibers reveals that both samples share most of intense peaks from each run (Figure 6c). A closer look into the identified proteins shows a very high overlap among the three datasets (Figure 6d). Remarkably, the overlap pair-to-pair comparison is greater than 50% in all cases in accordance with the expected levels of reproducibility for technical replicates in MS/MS shotgun experiments. Putting all together, we can summarize that not only the immobilization process generates consistent results, since the reproducibility in the digestion process is high for shotgun proteomic standards, but also comparable digestion efficiency as obtained with a free solution trypsin is achieved.
Figure 6.
Application of trypsin-aggregate coated nanofibers and solution phase trypsin to the digestion of a whole yeast protein extract. (a) Score distributions from the search of all datasets against swissprot.fasta database (filled circles) or against the inverted database (empty circles). Curves were generated with an assumption of a Gaussian distribution. (b) Comparison of the number of identified peptides at a given FDR following the Spectrum Mill analysis. EC-TR nanofibers batch 1 (filled circles), EC-TR nanofibers batch 2 (empty circles), solution phase trypsin (empty triangles). (c) Representative chromatograms for the replicates of yeast proteome digestion using two different batches of EC-TR nanofibers. (d) Overlap of the identified proteins.
4 Concluding remarks
The use of EC-TR biocatalytic nanofibers for protein digestion was demonstrated for the first time to be comparably better than either conventional in-solution or CA-TR trypsin digestions. The aggregated trypsin-containing nanofibers demonstrated excellent enzymatic activity (300 times higher than mono-layered nanofibers), long-term stability (i.e., negligible loss of activity over one year), and a high resistance to proteolysis by other proteases (such as α-chymotrypsin). In addition to being reusable, the enhanced activity and stability of EC-TR decreased the time involved to digest proteins and was applicable to the shotgun proteomic experiments. We anticipate that the chemistry employed for the trypsin-aggregate coatings can be implemented not only on nanofibers, but also on other solid supports (e.g.., silica beads, monolithic structures, magnetic beads, etc.). Furthermore, the process for coating nanofibers with aggregates is applicable to other enzymes that may be useful in proteomic research, as well as in other types of applications.
Acknowledgements
Portions of this work were supported by the National Center for Research Resources (RR18522), Battelle’s internal research and development funds, and by a Korea University Grant (Jungbae Kim). The research was performed in part at the W.R. Wiley Environmental Molecular Sciences Laboratory, a national scientific-user facility sponsored by the U. S. Department of Energy’s Office of Biological and Environmental Research, and located at the Pacific Northwest National Laboratory. The authors would like to acknowledge Dr. Tyler Heibeck, Penny Colton and Kim Hixson for their helpful assistance and suggestions during the writing of the manuscript.
Abbreviations
- 1D
one dimensional
- 2D
two dimensional
- Ambic
ammonium bicarbonate
- CA-TR
covalently attached trypsin onto the nanofibers
- EC-TR
trypsin-aggregate coatings onto the nanofibers
- GA
glutaraldehyde
- IAA
iodoacetamide
- L-BAPNA
Nα-Benzoyl-L-arginene 4 nitroanilide hydrochloride
- MeOH
methanol
- MW
molecular weight
- PS
polystyrene
- PSMA
poly (styrene-co-maleic anhydride)
Footnotes
The authors have declared no conflict of interest.
References
- 1.Smith RD. Future directions for electrospray ionization for biological analysis using mass spectrometry. Biotechniques. 2006;41:147–148. doi: 10.2144/000112217. [DOI] [PubMed] [Google Scholar]
- 2.Canas B, Lopez-Ferrer D, Ramos-Fernandez A, Camafeita E, Calvo E. Mass spectrometry technologies for proteomics. Brief. Funct. Genomic. Proteomic. 2006;4:295–320. doi: 10.1093/bfgp/eli002. [DOI] [PubMed] [Google Scholar]
- 3.Lopez-Ferrer D, Canas B, Vazquez J, Lodeiro C, et al. Sample treatment for protein identification by mass spectrometry-based techniques. Trac-Trends Anal. Chem. 2006;25:996–1005. [Google Scholar]
- 4. [last accessed January, 2008];Pierce, Immobilized TPCK Trypsin from Pierce: immobilized trypsin on agarose beads. http://www.piercenet.com/Products/Browse.cfm?fldID=02040710&WT.mc_id=go_Proteases_trypsin_brj&gclid=CKWdvPLDkpECFSgWiQodXwyqHQ.
- 5.ABI. [last accessed January, 2008];Porozyme (TM) from Applied Biosystems: immobilized trypsin on polystyrene beads. http://www3.appliedbiosystems.com/cms/groups/psm_support/documents/generaldocuments/cms_041677.pdf.
- 6.Krogh TN, Berg T, Hojrup P. Protein analysis using enzymes immobilized to paramagnetic beads. Anal. Biochem. 1999;274:153–162. doi: 10.1006/abio.1999.4254. [DOI] [PubMed] [Google Scholar]
- 7.Shui W, Fan J, Yang P, Liu C, et al. Nanopore-Based Proteolytic Reactor for Sensitive and Comprehensive Proteomic Analyses. Anal. Chem. 2006;78:4811–4819. doi: 10.1021/ac060116z. [DOI] [PubMed] [Google Scholar]
- 8.Sakai-Kato K, Kato M, Toyo'oka T. On-line trypsin-encapsulated enzyme reactor by the sol-gel method integrated into capillary electrophoresis. Anal. Chem. 2002;74:2943–2949. doi: 10.1021/ac0200421. [DOI] [PubMed] [Google Scholar]
- 9.Chen WY, Chen YC. Acceleration of microwave-assisted enzymatic digestion reactions by magnetite beads. Anal. Chem. 2007;79:2394–2401. doi: 10.1021/ac0614893. [DOI] [PubMed] [Google Scholar]
- 10.Peterson DS, Rohr T, Svec F, Frechet JMJ. Enzymatic microreactor-on-a-chip: Protein mapping using trypsin immobilized on porous polymer monoliths molded in channels of microfluidic devices. Anal. Chem. 2002;74:4081–4088. doi: 10.1021/ac020180q. [DOI] [PubMed] [Google Scholar]
- 11.Lopez-Ferrer D, Capelo JL, Vazquez J. Ultra fast trypsin digestion of proteins by high intensity focused ultrasound. J. Proteome Res. 2005;4:1569–1574. doi: 10.1021/pr050112v. [DOI] [PubMed] [Google Scholar]
- 12.Sun W, Gao S, Wang L, Chen Y, et al. Microwave-assisted protein preparation and enzymatic digestion in proteomics. Mol. Cell. Proteomics. 2006;5:S2–S2. doi: 10.1074/mcp.T500022-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Lazar IM, Ramsey RS, Ramsey JM. On-chip proteolytic digestion and analysis using "wrong-way-round" electrospray time-of-flight mass spectrometry. Anal. Chem. 2001;73:1733–1739. doi: 10.1021/ac001420+. [DOI] [PubMed] [Google Scholar]
- 14.Kim BC, Nair S, Kim J, Kwak JH, et al. Preparation of biocatalytic nanofibres with high activity and stability via enzyme aggregate coating on polymer nanofibres. Nanotechnology. 2005;16:S382–S388. doi: 10.1088/0957-4484/16/7/011. [DOI] [PubMed] [Google Scholar]
- 15.Jia HF, Zhu GY, Vugrinovich B, Kataphinan W, et al. Enzyme-carrying polymeric nanofibers prepared via electrospinning for use as unique biocatalysts. Biotechnol. Prog. 2002;18:1027–1032. doi: 10.1021/bp020042m. [DOI] [PubMed] [Google Scholar]
- 16.Shen Y, Hixson KK, Tolic N, Camp DG, et al. Mass spectrometry analysis of proteome-wide proteolytic post-translational degradation of proteins. Anal. Chem. 2008;80:5819–5828. doi: 10.1021/ac800077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Ferrer D, Heibeck TH, Petritis K, Hixson KK, et al. Rapid sample processing for LC-MS-based quantitative proteomics using high intensity focused ultrasound. J. Proteome. Res. 2008;7:3860–3867. doi: 10.1021/pr800161x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Ferrer D, Martinez-Bartolome S, Villar M, Campillos M, et al. Statistical model for large-scale peptide identification in databases from tandem mass spectra using SEQUEST. Anal. Chem. 2004;76:6853–6860. doi: 10.1021/ac049305c. [DOI] [PubMed] [Google Scholar]
- 19.Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]