SUMMARY
The mitochondrial antiviral signaling protein (MAVS; also known as IPS-1, VISA and CARDIF) is essential for innate immune response against RNA viruses. MAVS transduces signals from the cytosolic RIG-I-like receptors, which bind to viral RNAs, but how MAVS activates downstream transcription factors such as IRF3 to induce type-I interferons is not well understood. We have established a cell-free system in which mitochondria derived from virus-infected cells activates IRF3 in the cytosol. Fractionation of the cytosol led to the identification of Ubc5 as a ubiquitin-conjugating enzyme (E2) required for IRF3 activation. Using an inducible RNAi strategy, we demonstrate that catalytically active Ubc5 is required for IRF3 activation by viral infection. The activation of IRF3 also requires two ubiquitin-binding domains of NEMO. Furthermore, we show that replacement of endogenous ubiquitin with its K63R mutant abolishes viral activation of IRF3, demonstrating that K63 polyubiquitination plays a key role in IRF3 activation.
INTRODUCTION
A hallmark of the antiviral innate immune response is the induction of type-I interferons, including multiple subtypes of IFNα and a single IFNβ (Pichlmair and Reis e Sousa, 2007; Stetson and Medzhitov, 2006). Produced by a variety of cell types, type-I IFNs not only locally suppress viral infection and proliferation, but also facilitate effective adaptive immune responses to eliminate viral infection. Induction of type-I IFNs is initiated by detection of viral nucleic acids through receptors belonging to the pattern-recognition receptor family (PRRs). Viral RNAs in endosomal lumen are recognized by a subfamily of Toll-like receptors (TLRs), whereas cytosolic viral RNAs are sensed by RIG-I-like receptors (RLRs), including RIG-I, MDA5 and LGP2 (Yoneyama and Fujita, 2008; Yoneyama et al., 2004). The RLRs contain RNA helicase domains that bind to viral double-stranded RNA. In addition, RIG-I and LGP2 contain a C-terminal regulatory domain that binds to RNA bearing 5′-triphosphate (Cui et al., 2008; Hornung et al., 2006; Pichlmair et al., 2006). RIG-I and MDA5, but not LGP2, contain N-terminal tandem CARD domains which are important for triggering type-I IFN induction. Upon binding to RNA ligands, RIG-I and MDA5 activate mitochondrial antiviral signaling protein (MAVS; also known as IPS-1, VISA and CARDIF), probably through homotypic interaction between CARD domains of RIG-I and MAVS (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). MAVS then further activates downstream transcription factors such as NF-κB and IRF3, which converge on the induction of interferons (McWhirter et al., 2005; Seth et al., 2006).
MAVS activates NF-κB through the IKK complex, which consists of the catalytic subunits IKKα and IKKβ and the regulatory subunit NEMO (also known as IKKγ). The activation of IRF3 by MAVS requires the IKK-like kinase TBK1, and IKKε in some cells (Fitzgerald et al., 2003; McWhirter et al., 2005; Sharma et al., 2003). Several proteins including STING (also known as MITA or MPYS), NEMO, TANK, SINTBAD and NAP1, have been found to associate with TBK1 (Guo and Cheng, 2007; Ishikawa and Barber, 2008; Jin et al., 2008; Ryzhakov and Randow, 2007; Sasai et al., 2006; Zhao et al., 2007; Zhong et al., 2008). Phosphorylation of IRF3 leads to its dimerization and nuclear translocation, where it functions together with NF-κB to induce interferons. It has been demonstrated that recombinant TBK1 is sufficient to phosphorylate IRF3 at key serine residues required for its dimerization and activation (Panne et al., 2007); however, the mechanism by which TBK1 is activated by upstream signaling cascade following viral infection is poorly understood.
Recent studies have led to the discovery of several ubiquitination and deubiquitination enzymes that regulate the RIG-I pathway. For instance, TRIM25 is an E3 ubiquitin ligase that catalyzes K63 polyubiquitination of RIG-I (Gack et al., 2007). Deficiency of TRAF3, a member of TRAF ubiquitin ligase family, in mouse embryonic fibroblast (MEF) also results in a decrease of IFNα production (Hacker et al., 2005; Oganesyan et al., 2006). In addition, the deubiquitination enzymes CYLD and DUBA have been identified as negative regulators of IRF3 activation through deubiquitination of RIG-I and TRAF3, respectively (Friedman et al., 2008; Kayagaki et al., 2007; Zhang et al., 2008). These results suggest an important role of ubiquitination in the RIG-I pathway. However, direct evidence that K63 polyubiquitination is required for viral activation of IRF3 is still lacking. In addition, the biochemical mechanism by which ubiquitination regulates IRF3 activation remains largely unknown.
To dissect the biochemical mechanism of IRF3 activation by MAVS, we establish a cell-free system in which mitochondria derived from viral-infected cells activates IRF3 in the cytosol. Fractionation of the cytosolic extracts led to the identification of Ubc5 as an E2 that plays an important role in IRF3 activation. Using an inducible RNAi system, we also found that the catalytic activity of Ubc5 is required for viral activation of IRF3. Furthermore, by employing a strategy to replace endogenous ubiquitin with a K63R mutant of ubiquitin (Ub-K63R) in a human cell line, we demonstrated that K63 polyubiquitination is essential for IRF3 activation by viral infection. In addition, we found that activation of IRF3 requires two ubiquitin-binding domains of NEMO. Collectively, these results demonstrate the key role of Ubc5 and K63 polyubiquitination in viral activation of IRF3.
RESULTS
Establishment of a signal-dependent, cell-free IRF3 activation system
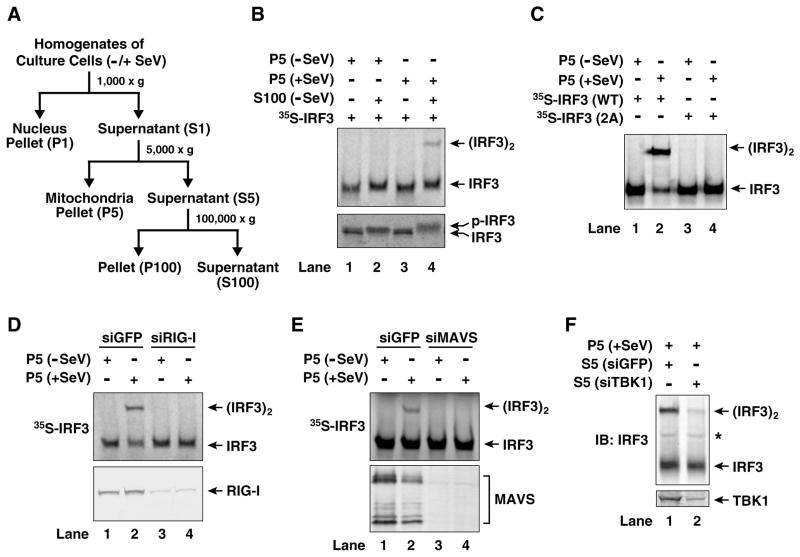
To understand how MAVS on the mitochondria activates IRF3 in the cytosol, we isolated mitochondria from HEK293T cells infected with Sendai virus, and incubated them with ATP-supplemented cytosolic extracts from uninfected cells (Fig. 1A). To facilitate detection of IRF3 activation, in vitro translated [35S]-labeled IRF3 was used as a substrate and its dimerization was analyzed by native gel electrophoresis. As shown in Figure 1B, addition of the crude mitochondrial fraction (P5) isolated from viral-infected cells, but not that from uninfected cells, to the cytosolic extracts (S100) led to the formation of a slower-migrating form of IRF3 on the native gel (Fig. 1B, upper panel). The mobility of this form of IRF3 was virtually identical to that of IRF3 from virus-infected cells (Supplementary Fig. S1A), which has been shown to be an active IRF3 dimer (Yoneyama et al., 1998). Indeed, the in vitro activated Flag-IRF3 and His8-IRF3 formed a complex that could be co-immunoprecipitated (Fig. S1B). Furthermore, the IRF3 isolated from an in vitro reaction containing virus-activated mitochondria (P5) eluted from a gel filtration column (Superdex-200) with an apparent molecular mass of ~300 kDa (Fig. S1C), which corresponds to a dimer of IRF3 based on analytical ultracentrifugation analysis (Panne et al., 2007). In contrast, in the absence of the virus-activated mitochondria, IRF3 eluted from Superdex-200 with an apparent molecular mass of 150 kDa (Fig. S1C), which corresponds to a monomer of IRF3. When the reaction products were analyzed by denaturing gel electrophoresis, a slower-migrating form of IRF3 indicative of its phosphorylation was apparent when viral-activated mitochondria were incubated with S100 (Fig. 1B; lower panel). The phosphorylated form of IRF3 was detected with an antibody specific for phospho-serine 396 (Supplementary Fig. S1A, bottom panel), a residue known to be important for viral-activation of IRF3 (Lin et al., 1998). Two other serines at the C-terminus of IRF3, S385 and S386, have also been shown to be phosphorylated in response to RNA virus infection and this phosphorylation is important for IRF3 dimerization (Yoneyama et al., 1998). Mutations of these two serine residues to alanine abolished the dimerization of IRF3 in vitro (Fig. 1C). Taken together, these results strongly suggest that the virus-activated mitochondria trigger a signaling cascade in the cell-free system that leads to the C-terminal phosphorylation and subsequent dimerization of IRF3.
Figure 1. Establishment of a virus-dependent, in vitro assay for IRF3 activation.
(A) Diagram of the fractionation procedure of cell homogenates. Culture cells infected with Sendai virus (+SeV) or mock treated (−SeV) were homogenized in hypotonic buffer, followed by sequential centrifugation to separate crude mitochondria (P5) from cytosolic supernatant (S5 and S100) as described in Experimental Procedures. (B) Viral activation of IRF3 in vitro. Mitochondrial fraction (P5) from mock- or Sendai virus-infected HEK293T cells was incubated with cytosolic extract (S100) from uninfected cells in the presence of ATP together with 35S-IRF3. Dimerization and phosphorylation of IRF3 were analyzed by native gel electrophoresis (upper panel) and SDS-PAGE (lower panel), respectively, followed by autoradiography. (C) Reactions were carried out as in (B) except that wild type (WT) 35S-IRF3 or its mutant (S385A/S386A; denoted as 2A) was used. (D & E) RIG-I and MAVS are required for activation of the mitochondrial fraction. HEK293T cells were transfected with siRNA against GFP, RIG-I (D) or MAVS (E) and then infected with Sendai virus or mock treated. Mitochondrial fractions (P5) from these cells were incubated with the cytosolic extracts (S5) from uninfected cells in the presence of ATP and 35S-IRF3. Dimerization of IRF3 was analyzed by native gel electrophoresis (upper panels), and the efficiency of RNAi was examined by immunoblotting (lower panels). (F) TBK1 in the cytosol is required for IRF3 activation in vitro. HEK293T cells were transfected with siRNA against GFP or TBK1. Cytosolic extracts (S5) from these cells were incubated with the mitochondrial fraction from Sendai virus-infected HEK293T cells in the presence of ATP. Dimerization of endogenous IRF3 was analyzed by native gel electrophoresis. The asterisk (*) indicates a non-specific band cross-reacting with the IRF3 antibody.
The activation of IRF3 by the mitochondrial fraction (P5) was entirely dependent on RIG-I and MAVS, because RNAi of either RIG-I or MAVS abolished the ability of P5 to induce IRF3 dimerization in vitro, whereas a control GFP RNAi had no effect (Fig. 1D and 1E). Overexpression of MAVS is known to activate IRF3 and induce type-I interferons (Seth et al., 2005). Consistently, the P5 fraction from HEK293T cells overexpressing MAVS potently activated IRF3 in the cytosol (Supplementary Fig. S1D). TBK1 was also required for IRF3 activation in this in vitro system, because the cytosolic extract (S5) derived from cells treated with siRNA against TBK1 failed to activate IRF3 in the presence of virus-activated mitochondria (Fig. 1F). Therefore, this cell-free system faithfully recapitulates IRF3 regulation in vivo in that its activation is dependent on RIG-I, MAVS and TBK1, as well as viral infection.
Purification of Ubc5 as an IRF3 activator
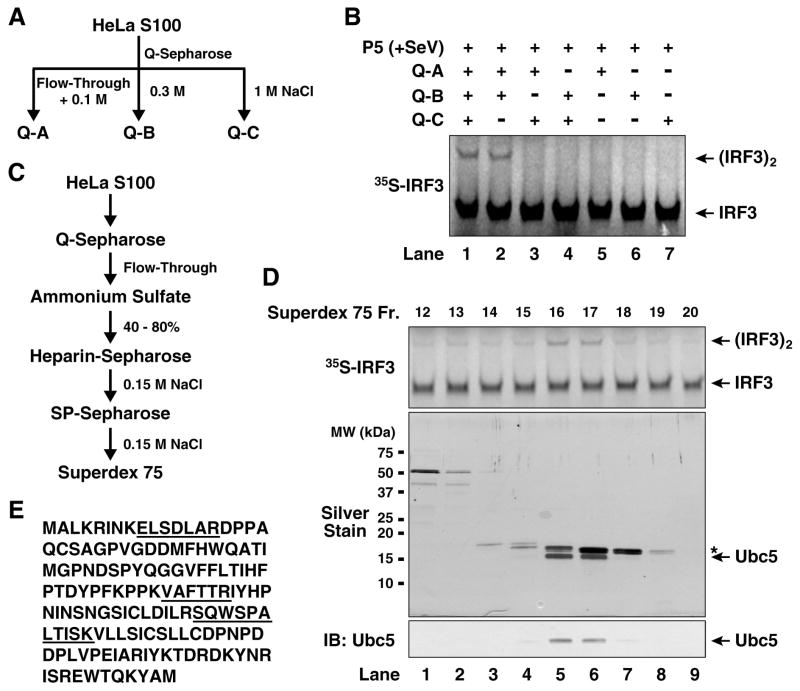
We separated HeLa S100 into three fractions using Q-Sepharose anion exchange chromatography. Q-A contained proteins unable to bind to the Q column at 0.1M NaCl, whereas Q-B and Q-C contained proteins eluted from the column with 0.3M and 1.0M NaCl, respectively (Fig. 2A). Activation of IRF3 by the virus-stimulated mitochondrial fraction required both Q-A and Q-B, but not Q-C (Fig. 2B). TBK1, the kinase that directly phosphorylates IRF3, was mainly present in Q-B fraction (data not shown). We further purified the activity in Q-A through five steps of conventional chromatography as shown in Figure 2C. Silver staining of the fractions from the last purification step (Superdex-75) revealed two major bands, the lower band co-purifying with the IRF3 dimerization activity (Fig. 2D). This band was excised for tandem mass spectrometry, which identified three peptides corresponding to Ubc5b and Ubc5c (Fig. 2E). Immunoblotting with a Ubc5 antibody confirmed that Ubc5 co-purified with the IRF3 dimerization activity (Fig. 2D, lower panel).
Figure 2. Identification of Ubc5 as an IRF3 activator.
(A) Diagram of initial fractionation of HeLa S100 on Q-Sepharose column. After loading the sample, the column was eluted step-wise with 0.1 M (Q-A; also contains the flow-through fraction), 0.3 M (Q-B) and 1.0 M NaCl (Q-C). (B) Reconstitution of IRF3 activation with Q-Sepharose fractions. In vitro assay for IRF3 activation was carried out as described in Figure 1, except that the cytosolic extracts were replaced with the indicated combinations of Q-A, Q-B and Q-C. (C) Scheme of biochemical fractionation of Q-A. (D) Identification of Ubc5 as the active component in Q-A. Aliquots of the fractions from Superdex-75 were tested for their ability to stimulate IRF3 dimerization in the presence of Q-B and virus-activated P5 (upper panel), and analyzed by silver staining (middle panel) and immunoblotting with a Ubc5 antibody (lower panel). The asterisk (*) in the silver-stained gel denotes a contaminating protein (cyclophilin A). (E) Amino acid sequence of Ubc5c. Peptide sequences identified by mass spectrometry are underlined.
Ubc5 is required for IRF3 activation
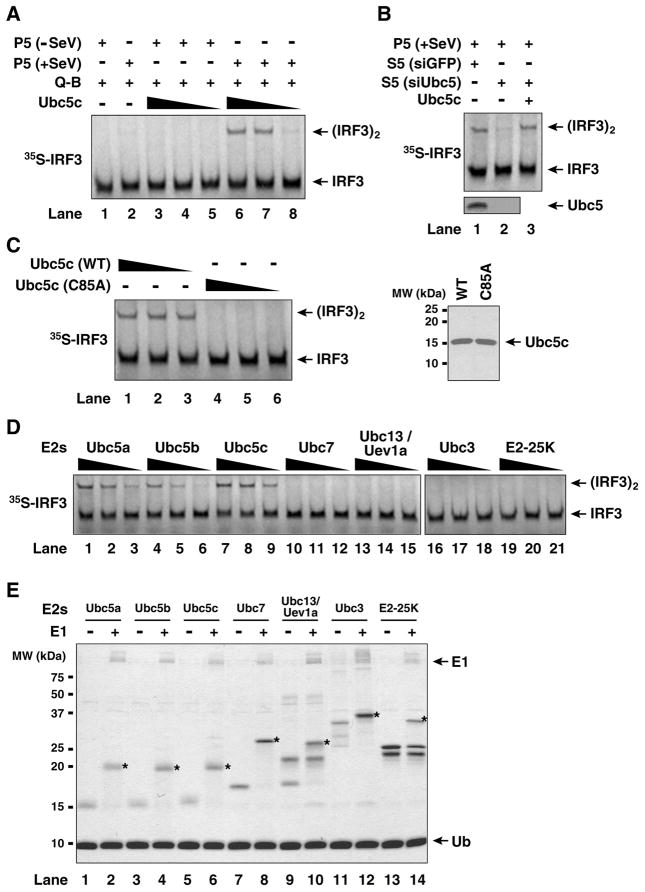
To determine if Ubc5 is indeed important for IRF3 activation in vitro, we substituted Q-A fraction with recombinant Ubc5c in the IRF3 dimerization assay as described in Figure 2B. As shown in Figure 3A, Ubc5c supported IRF3 dimerization in the presence of the mitochondrial fraction (P5) from viral infected cells, but not from mock-treated cells. Cytosolic extracts from HEK293T cells depleted of Ubc5 by RNAi failed to support IRF3 activation, but this activity was restored by adding back recombinant Ubc5c (Fig. 3B), indicating that Ubc5 is the E2 responsible for IRF3 activation in the cytosol. The catalytic activity of Ubc5c is essential, because a point mutation of the active site cysteine of Ubc5c (C85A) completely abrogated IRF3 dimerization (Fig. 3C). Among a panel of E2s tested, including Ubc5 (a, b & c), Ubc7, Ubc13/Uev1A, Ubc3 and E2-25K, only the Ubc5 isoforms could support IRF3 dimerization (Fig. 3D), although all of these E2s were active, as evidenced by the formation of thioesters with ubiquitin (Fig. 3E).
Figure 3. Specific requirement of Ubc5 in IRF3 activation in vitro.
(A) Recombinant Ubc5c activates IRF3. IRF3 dimerization assay was carried out as described in Figure 2B, except that Q-A was replaced by varying amounts of recombinant Ubc5c protein (0.1, 0.3 and 1.0 μM). (B) RNAi of Ubc5b and Ubc5c prevents IRF3 activation. HEK293T cells were transfected with siRNA oligos against both Ubc5b and Ubc5c, then cytosolic extracts (S5) were prepared for IRF3 dimerization assay. (C) Catalytic activity of Ubc5 is required for IRF3 activation. IRF3 dimerization assay was carried out in the presence of wild-type (WT) or Ubc5c (C85A) at varying concentrations (0.1, 0.3 and 1.0 μM). 2 g of the recombinant Ubc5c and the C85A mutant was analyzed by Coomassie blue staining (right panel). (D) Ubc5 isoforms, but not other E2s, support IRF3 activation. Varying concentrations (0.1, 0.3 and 1.0 μM) of indicated E2s were tested for their ability to support IRF3 dimerization. (E) Thioester assays of E2s. The same E2s used for in vitro IRF3 activation assay in (D) were analyzed for their ability to form thioester with ubiquitin in the absence or presence of E1. The asterisks (*) denote the positions of E2-ubiquitin thioesters visualized by Coomassie blue staining.
Next, we tested the effect of depleting Ubc5 on IRF3 activation by the kinase TBK1 or the mitochondria from MAVS-overexpressing cells (Supplementary Fig. S2A). The depletion of Ubc5 prevented IRF3 activation by MAVS, but not TBK1, indicating that Ubc5 functions at a step in the signaling cascade from MAVS to TBK1. Recent studies have identified STING as an IRF3-activating protein localized in the membrane of mitochondria or endoplasmic reticulum (ER) (Ishikawa and Barber, 2008; Zhong et al., 2008). Interestingly, we found that high-speed membrane pellets (P100) containing ER from STING-overexpressing cells could induce IRF3 dimerization in crude cell extracts depleted of Ubc5 (Fig. S2B). The reason for this Ubc5-independent activation of IRF3 is not clear, but it is possible that the overexpressed STING directly recruits TBK1 to phosphorylate IRF3 (Zhong et al., 2008). We have also found that the virus-stimulated mitochondria could activate IKK in crude cytosolic extracts, and that this activation was modestly impaired when Ubc5 was depleted (Fig. S2D). This in vitro system would be useful for future dissection of the IKK signaling cascade triggered by MAVS.
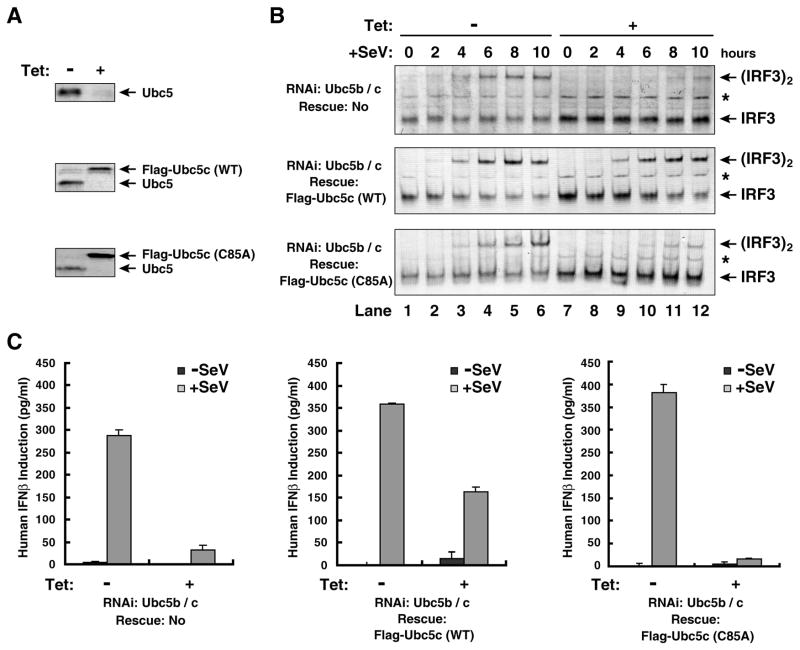
To determine if Ubc5 and its catalytic activity are required for IRF3 activation by viral infection, we used tetracycline-inducible short hairpin RNA (shRNA) to deplete endogenous Ubc5 in the human osteosarcoma cell line U2OS and simultaneously replaced it with wild type Ubc5c or the mutant Ubc5c (C85A), whose expression was also controlled by a tetracycline-inducible promoter. These cells were treated with tetracycline for seven days until endogenous Ubc5 was reduced to an undetectable level (Fig. 4A), then infected with Sendai virus for different lengths of time before cell lysates were harvested to detect endogenous IRF3 dimerization (Fig. 4B). RNAi of Ubc5 caused a significant delay in IRF3 dimerization, and this delay was rescued by wild type Ubc5c, but not Ubc5c (C85A). RNAi of Ubc5 also strongly inhibited the induction of IFNβ by Sendai virus (Fig. 4C). The interferon induction was rescued by the wild type Ubc5c, but not Ubc5c (C85A). Transient transfection of siRNA oligos against Ubc5b and Ubc5c in HEK293T cells alzso severely impaired Sendai virus-induced expression of luciferase reporters driven by interferon-stimulated response element (ISRE; Supplementary Fig. S3A) or by IFNβ promoter (Fig. S3B), further supporting the role of Ubc5 in IRF3 activation. The residual IRF3 activation in Ubc5b/c RNAi cells might be due to incomplete depletion of these Ubc5 isoforms or the presence of Ubc5a, which is resistant to silencing by the Ubc5b/c shRNA and siRNA oligo sequences. It is also possible that other E2s, such as Ubc13, may contribute to IRF3 activation by MAVS in vivo, although our biochemical experiments showed that Ubc13 could not support IRF3 activation by virus-activated mitochondria in vitro (Fig. 3D). On the other hand, it has been shown that ubiquitination of RIG-I is important for IRF3 activation by RNA viruses (Gack et al., 2007). Consistent with this notion, knockdown of Ubc5b/c in U2OS cells modestly delayed the activation of MAVS in the mitochondria (P5) by Sendai virus (Fig. S3C).
Figure 4. Viral activation of IRF3 depends on catalysis by Ubc5.
(A) RNAi knockdown and expression rescue of Ubc5. U2OS cells stably expressing tetracycline-inducible shRNA against Ubc5b and Ubc5c together with Flag-tagged Ubc5c (wild-type or C85A) were treated with tetracycline (Tet; 1 μg/ml) for 7 days, then expression of Ubc5 was examined by immunoblotting with an antibody against all Ubc5 isoforms. (B) Active Ubc5 is required for IRF3 activation by Sendai virus (SeV). U2OS cells were treated with tetracycline as described in (A), then infected with SeV for the indicated time. Dimerization of endogenous IRF3 was analyzed by native gel electrophoresis. The asterisk (*) indicates a non-specific band cross-reacting with the IRF3 antibody. (C) Active Ubc5 is required for viral induction of IFNβ. U2OS cells were treated with tetracycline for 6 days and then infected with Sendai virus for 16 hours. IFNβ in the culture medium was measured by ELISA. The error bars represent variation ranges of duplicate experiments.
Key role of K63 polyubiquitination in IRF3 activation
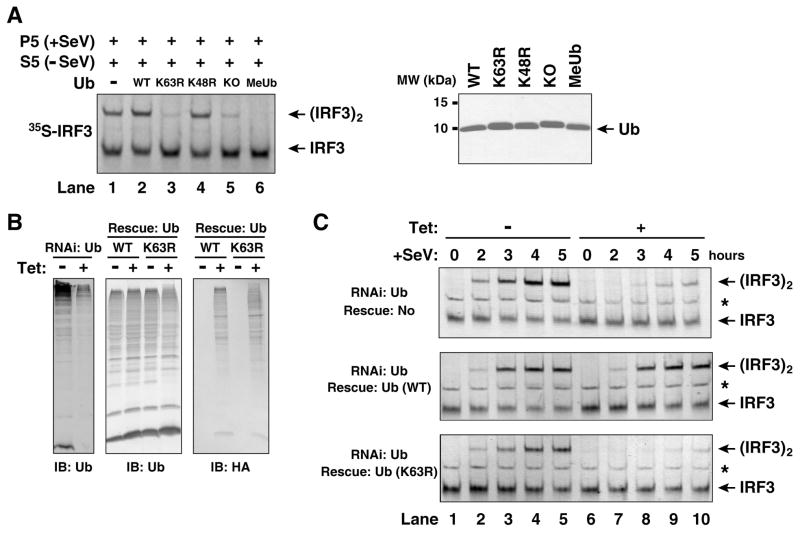
The requirement of the catalytic activity of Ubc5 in IRF3 activation implies a requirement of ubiquitination in this process. Previous studies have suggested a role of lysine 63 (K63)-linked polyubiquitination in IKK activation through a proteasome-independent mechanism (Deng et al., 2000; Wang et al., 2001). To determine if K63 of ubiquitin is involved in IRF3 activation by MAVS, we tested a panel of ubiquitin mutants for their ability to enhance or inhibit IRF3 activation in vitro (Fig. 5A). These mutants include those lacking any lysine (KO), containing methylated lysines (MeUb), or containing a point mutation at lysine 48 (K48R) or 63 (K63R). While wild type and K48R ubiquitin slightly enhanced IRF3 dimerization in the cytosolic extracts that contained endogenous ubiquitin, K63R, KO and MeUb inhibited IRF3 dimerization. In the absence of exogenous ubiquitin (lane 1 in Fig. 5A), IRF3 activation was observed because of endogenous ubiquitin present in the crude cell extracts as well as in rabbit reticulocyte lysates used for in vitro translation of IRF3 (Supplementary Fig. S4A). When affinity purified Flag-IRF3 and partially purified fractions from HeLa S100 were used in the assays, IRF3 activation was dependent on ubiquitin (Fig. S4B).
Figure 5. K63-linked polyubiquitination is essential for viral activation of IRF3.
(A) K63 of ubiquitin is required for IRF3 activation in vitro. In vitro assay for IRF3 activation was carried out as described in Figure 1B, except that 100 μM of recombinant ubiquitin and its lysine mutants were added to the assay mixtures. KO: lysine-less ubiquitin; MeUb: methylated ubiquitin. 2 μg of ubiquitin and its mutants were analyzed by Coomassie blue staining (right panel). (B) K63 of ubiquitin is required for IRF3 activation by Sendai virus. U2OS cells stably integrated with tetracycline-inducible shRNA against ubiquitin genes and an expression cassette for wild-type or the K63R mutant of ubiquitin were growing in the presence or absence of tetracycline for 3 (ubiquitin knockdown cells) or 4 days (ubiquitin rescue cells) before cells were infected with Sendai virus for the indicated time. The efficiency of knocking down endogenous ubiquitin and rescuing with HA-ubiquitin was examined by immunoblotting (B). Dimerization of endogenous IRF3 was analyzed by native gel electrophoresis (C). The asterisk (*) indicates a non-specific band cross-reacting with the IRF3 antibody.
Genetic studies of ubiquitin function in vivo have been hampered by the fact that there are four ubiquitin genes in yeast and mammals, two of which encode linear polyubiquitin (UBB and UBC) and the other two encode ubiquitin fused to ribosomal subunits (UBA52 and RPS27A) (Finley et al., 1987). To overcome the technical difficulty of deleting all four ubiquitin genes and the lethal effect of depleting ubiquitin in cells, we have recently devised a tetracycline-inducible “knock-down” and “knock-in” strategy to replace endogenous ubiquitin with its K63R mutant in U2OS cells (Xu, M., Skaug, B., Zeng, W., and Chen, Z., manuscript submitted). In this strategy, we identified two shRNA sequences that are complementary to all four ubiquitin genes and expressed them under the control of a tetracycline-inducible promoter in a stable U2OS cell line. We also constructed a tetracycline-inducible expression vector driving the expression of both UBA52 and RPS27, which carry silent mutations rendering them resistant to RNAi. These vectors, when introduced to the U2OS ubiquitin RNAi cells, rescued the expression of both ubiquitin and ribosomal subunits. By introducing the K63R mutation to both UBA52 and RPS27, we were able to “knock-in” K63R ubiquitin to replace endogenous ubiquitin in a tetracycline-dependent manner (Fig. 5B). As shown in Figure 5C, Sendai virus-induced dimerization of IRF3 was inhibited in the ubiquitin RNAi cells, but this was rescued when wild-type ubiquitin was expressed from the tetracycline-inducible promoter. Strikingly, replacement of endogenous ubiquitin with the K63R mutant completely blocked IRF3 dimerization, demonstrating the essential role of K63 polyubiquitination in IRF3 activation.
The ubiquitin-binding domains of NEMO are required for IRF3 activation
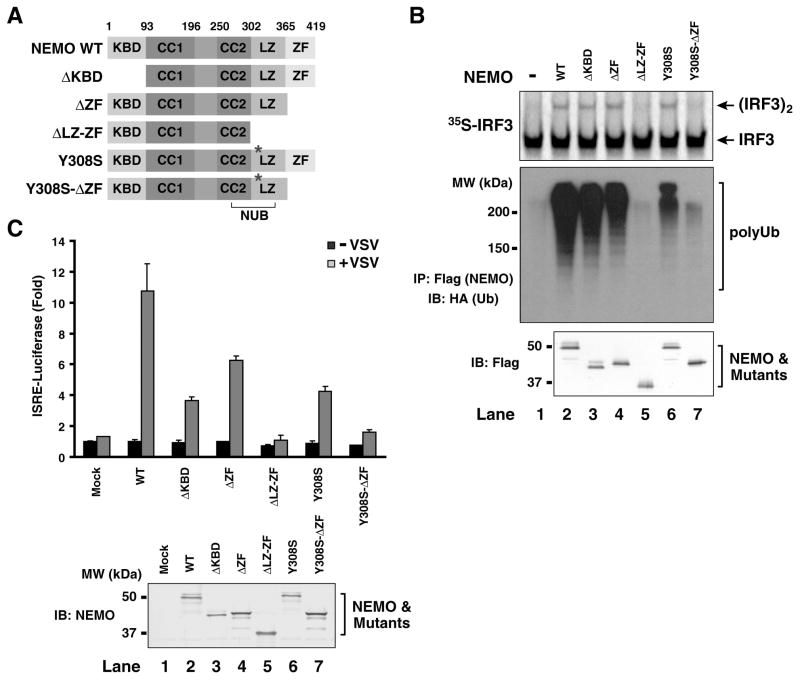
Recent studies have shown that the NEMO-deficient cells fail to activate both NF-κB and IRF3 in response to viral infection (Zhao et al., 2007). Consistent with these results, we found that cytosolic extracts from NEMO−/− MEF cells could not support IRF3 activation in the presence of virus-activated mitochondria, but this activation was restored by adding back Flag-NEMO protein (Fig. 6B; compare lanes 1 & 2). This reconstitution system allowed us to test the domains of NEMO required for IRF3 activation. NEMO contains an N-terminal IKK-binding domain (KBD), two coiled coil domains (CC1 and CC2), a leucine-zipper (LZ) and a C-terminal zinc finger (ZF) domain (Fig. 6A). Part of the CC2-LZ region constitutes a ubiquitin-binding domain called NUB (Ea et al., 2006; Wu et al., 2006). In addition, the C-terminal ZF domain has been shown to bind ubiquitin and this binding is important for IKK activation (Cordier et al., 2008).
Figure 6. Ubiquitin-binding domains of NEMO are required for IRF3 activation.
(A) Diagrams of NEMO domains and its mutants. KBD, IKK-binding domain; CC1 and CC2, coiled-coil domain 1 and 2; LZ, leucine-zipper motif; ZF, zinc-finger domain; NUB: NEMO-ubiquitin binding. (B) Ubiquitin-binding of NEMO is required for IRF3 activation in vitro. Expression vectors for Flag-NEMO and its mutants as shown in (A) were transfected into HEK293T cells, then the proteins were purified using anti-Flag (M2) agarose. Equal amounts of the NEMO proteins (bottom panel) were incubated with cytosolic extracts from NEMO-deficient MEF cells together with the virus-activated mitochondrial fraction (P5). Dimerization of 35S-IRF3 was analyzed by native gel electrophoresis followed by autoradiography (upper panel). Aliquots of NEMO proteins were incubated with K63-linked HA-Ub chains and immunoprecipitated with anti-Flag agarose beads. Co-immunoprecipitated polyubiquitin chains were analyzed by immunoblotting (middle panel). (C) Ubiquitin-binding of NEMO is required for viral activation of IRF3. NEMO−/− MEF cells were transiently transfected with NEMO and its mutants together with the ISRE- luciferase reporter and a control plasmid (pRL-CMV). The cells were infected with VSV (ΔM51)-GFP for 14 hours before cell lysates were prepared for luciferase assays. The error bars represent variation ranges of duplicate experiments.
A series of Flag-tagged NEMO mutants were expressed and purified from HEK293T cells, and tested for their ability to bind K63 polyUb chains and activate IRF3 (Fig. 6A & B). To facilitate detection, the K63 polyUb chains were synthesized using HA-Ub, and the K63 linkage was verified using an antibody specific for this linkage (Supplementary Fig. S5). Wild type NEMO bound K63 polyUb and activated IRF3, and deletion of the N-terminal KBD or C-terminal ZF did not affect polyUb binding or IRF3 activation (Fig. 6B, lanes 2–4). In contrast, deletion of both LZ and ZF (ΔLZ-ZF) abolished polyUb binding by NEMO as well as its ability to activate IRF3 (lane 5). A point mutation of a conserved tyrosine (Y308S) of NEMO, which is known to reduce ubiquitin binding (Ea et al., 2006), did not significantly impair its ability to activate IRF3 (lane 6), presumably because the residual ubiquitin-binding by the ZF domain was sufficient to mediate IRF3 activation. Indeed, further deletion of ZF from the Y308S mutant abrogated IRF3 activation as well as polyUb binding (lane 7). These results indicate that both LZ and ZF domains of NEMO contribute to its polyUb binding and IRF3 activation in vitro. Control experiments showed that mutations of the ubiquitin-binding domains of NEMO (ΔLZ-ZF and Y308S-ΔZF) did not impair its ability to associate with TANK and TBK1 (Supplementary Fig. S6A). Similarly, RNAi of Ubc5 did not interfere with the binding of NEMO with TANK and TBK1 (Fig. S6B), suggesting that sensing of polyUb chains by NEMO does not affect its association with TANK and TBK1, but is required for TBK1 activation possibly by affecting the conformation or oligomerization of TBK1.
To determine if ubiquitin-binding of NEMO is required for IRF3 activation in vivo, we transfected NEMO-deficient MEF cells with the same series of NEMO mutants together with the ISRE-luciferase reporter plasmid (Fig. 6C). These cells were then infected with a vesicular stomatitis virus strain [VSV(ΔM51)-GFP], which is known to strongly induce type-I interferons (Stojdl et al., 2003). While wild type NEMO restored viral induction of the ISRE-luciferase reporter, the NEMO mutants lacking both polyUb-binding domains (ΔLZ-ZF and Y308S-ΔZF) failed to rescue ISRE-luciferase induction (Fig. 6C). The mutants with a defect in only one ubiquitin-binding domain (ΔZF or Y308S) and that lacking the IKK-binding domain (ΔKBD) also showed reduced activation of ISRE. Taken together, these in vitro and cell-based assays show that the ubiquitin-binding domains of NEMO are required for IRF3 activation.
DISCUSSION
Studies of biochemical mechanisms of signal transduction often require a cell-free system that faithfully recapitulates cell signaling in vivo. By using purified recombinant TBK1 and IRF3 proteins in phosphorylation reactions in vitro, Panne et al have convincingly demonstrated that TBK1 is sufficient to phosphorylate IRF3 at specific serine residues, resulting in its dimerization (Panne et al., 2007). However, since TBK1 expressed in insect cells is already active, it is necessary to use endogenous TBK1 in a cell-free system in order to understand how this kinase is activated by upstream signals. Herein we report the development of a cell-free system in which mitochondria from virus-infected cells can activate IRF3 in the cytosol. Using this system, we identify Ubc5 as an activator of IRF3 dimerization in a manner that depends on its ubiquitin-conjugating activity. We also show that the ubiquitin-binding domains of NEMO are required for IRF3 activation by MAVS in vitro and by viral infection in cells. Furthermore, by using a new strategy to replace endogenous ubiquitin with its K63R mutant in a human cell line we have demonstrated the essential role of K63 polyubiquitination in IRF3 activation by virus. These results suggest a broader role of K63 polyubiquitination in cell signaling pathways, including those of NF-κB and IRF3.
An important question remaining to be resolved is the identity of the E3 (or E3s) that mediates IRF3 activation by MAVS. A strong candidate for this E3 is TRAF3, which has been shown to be important for IFNα induction by TLRs and RLRs (Hacker et al., 2005; Oganesyan et al., 2006; Saha et al., 2006). Indeed, we found that TRAF3 could catalyze its auto-polyubiquitination in the presence of Ubc5c in vitro (Supplementary Fig. S7A), and that polyubiquitinated TRAF3 exhibited enhanced ability to bind to NEMO (Fig. S7B). However, we found that TRAF3−/− MEF cells could still produce IFNβ and activate IRF3, albeit at a reduced level (Fig. S8A & B). In addition, cytosolic extracts from TRAF3−/− MEF cells could still support IRF3 dimerization (Fig. S8C). It is known that MAVS contains binding sites for TRAF2 and TRAF6 in addition to TRAF3 (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). However, MEF cells lacking TRAF6 or both TRAF2 and TRAF5 were still capable of activating IRF3 dimerization following Sendai virus infection (Fig. S9). Another candidate E3 is TRIM25, which has been shown to catalyze polyubiquitination of RIG-I (Gack et al., 2007). Consistent with previous reports, we found that TRIM25-deficient MEF cells were defective in IFN production in the early phase of Sendai virus infection (Fig. S10A). However, at the later stage, copious amounts of IFNβ were still produced in TRIM25−/− MEF cells. The early defect in IFNβ induction was likely at a step upstream of MAVS activation (i.e, RIG-I ubiquitination) because the cytosol (S5) from TRIM25-deficient cells were still capable of supporting IRF3 activation by virus-activated mitochondrial fraction (P5; Fig. S10B), whereas P5 from virus-infected cells lacking TRIM25 failed to activate IRF3 in the cytosol (Fig. S10C). These results suggest that there is a yet-to-be identified E3 required for IRF3 activation by MAVS. It is also possible that this unknown E3 functions redundantly with TRAF3 to mediate IRF3 activation. Further fractionation of cytosolic extracts may shed light on the identity of the E3 required for IRF3 activation by MAVS.
Our results show that K63 polyubiquitination is involved in at least two steps of the RIG-I pathways. The first step, which is upstream of MAVS, involves K63 polyubiquitination of RIG-I by TRIM25 (Gack et al., 2007). The second step, which is downstream of MAVS, involves K63 polyubiquitination by Ubc5 and an unknown E3, and the recognition of the polyubiquitin chains by NEMO. It is surprising that Ubc5, rather than Ubc13/Uev1A, is involved in K63 polyubiquitination in the MAVS pathway, because Ubc5 is known to be quite promiscuous in promoting synthesis of polyubiquitin chains of different linkages, including K63 and K48. It is possible that the types of polyubiquitin chains synthesized by Ubc5 depend on specific E3s employed in the pathways. Indeed, it has been shown that the HECT domain E3 E6AP functions together with Ubc5 to catalyze the synthesis of homogenous K48-linked polyubiquitin chains, whereas another HECT domain E3, Nedd4, functions with the same E2 (Ubc5) to produce K63-linked polyubiquitin chains (Kim et al., 2007). It is also possible that Ubc5 functions with an E3 to produce polyubiquitin chains of different linkages, but only the K63-linked polyubiquitin chains can lead to activation of IRF3.
The target of polyubiquitination required for IRF3 activation by MAVS remains to be determined. Several proteins have been reported to undergo polyubiquitination in the TLR or RLR pathways, and these include TRAF3, TANK, NEMO, and MAVS (Chau et al., 2008; Gatot et al., 2007; Kayagaki et al., 2007; Paz et al., 2009). We found that MAVS was polyubiquitinated in response to Sendai virus infection (Supplementary Fig. S11A). However, a MAVS mutant lacking all lysines was still capable of rescuing viral-activation of IRF3 and induction of IFNβ in Mavs-deficient MEF cells (Fig. S11B-D). Similarly, Paz et. al. (2009) have reported that ubiquitination of MAVS recruits IKKε to the mitochondria but this event negatively regulates IFN induction. We have also examined potential ubiquitination of several other proteins known to function in the RIG-I pathway, including TRAF3, NEMO, TANK, TBK1, IKKε and IRF3 (Fig. S12). Among these proteins, only TRAF3 displayed MAVS-induced polyubiquitination. However, since even the complete absence of TRAF3 does not abolish IRF3 activation (Fig. S8), it is unlikely that TRAF3 auto-polyubiquitination is solely responsible for IRF3 activation. We also found that a point mutation of the major ubiquitination site of NEMO (K285R) did not impair its ability to support IRF3 activation in vitro (data not shown). However, deletion of both ubiquitin-binding domains (NUB and ZF) of NEMO blocked IRF3 activation (Fig. 6B & C). Therefore, similar to its role in the IKK pathway, NEMO functions as a sensor of K63 polyubiquitin chains to mediate the activation of TBK1. A model of TBK1 activation by K63 polyubiquitination is presented in Supplementary Figure S13. In this model, we propose that viral infection leads to oligomerization of MAVS. MAVS then binds to and activates TRAF3 and/or another E3, which function with Ubc5 to catalyze K63 polyubiquitination of target proteins including TRAF3 itself. The polyubiquitin chains bind to NEMO, which in turn binds to TANK and TBK1, leading to activation of TBK1. TBK1 then phosphorylates IRF3, causing IRF3 to dimerize and translocate into the nucleus to induce type-I interferons. Validation of this model requires identification of additional molecular components required for IRF3 activation. The development of a signal-dependent cell-free system of IRF3 and IKK activation should pave the way for the eventual reconstitution of the RIG-I - MAVS signaling pathway with defined components and contribute to a detailed understanding of the signal transduction mechanism in this antiviral pathway.
EXPERIMENTAL PROCEDURES
Materials and Standard Methods
SDS-PAGE, native gel electrophoresis for IRF3 dimerization, and immunoblot analysis were carried out as previously described (Seth et al., 2005). The antibodies used in this study were purchased from the following vendors: Cell Signaling: TBK1 and phospho-IκBα (Ser32/36); Santa Cruz Biotech: ubiquitin, rabbit anti-NEMO, TRAF3, IκBα and human IRF3; Zymed: mouse IRF3; Covance: mouse anti-HA; Sigma: rabbit anti-HA and mouse anti-Flag (M2); Qiagen: mouse anti-His5; Upstate Biotech: phospho-IRF3 (Ser-396); BIOMOL: K63-polyUb; BD Pharmingen: mouse anti-NEMO. The antibody against human Ubc5 was kindly provided by Dr. Allan Weissman (NIH). Antibodies against MAVS and RIG-I were generated as previously described (Seth et al., 2005). Rat anti-HA agarose was purchased from Roche. Tetracycline (Sigma) was added to culture medium at a final concentration of 1 μg/ml. Sendai virus (Cantell strain, Charles River Laboratories) was used at a final concentration of 100 hemagglutinating-units/ml. VSV(ΔM51)-GFP virus was kindly provided by Dr. John Bell (University of Ottawa) and propagated in Vero cells (Stojdl et al., 2003). All chromatography columns were from GE Healthcare. Methylated ubiquitin (MeUb) was purchased from Boston Biochem, and other chemicals were purchased from Sigma unless otherwise specified.
Recombinant Proteins
His6-E1, His8-TRAF3, His8-IRF3 and His6-TRAF6 were expressed and purified in Sf9 cells, and His6-Ubc13 and His6-Uev1A were expressed and purified in E. coli, as previously described (Deng et al., 2000). Human E2-25K, Ubc3, Ubc5a, Ubc5b, Ubc5c, Ubc5c (C85A) and Ubc7 were expressed in E. coli as GST-fusion proteins and purified with glutathione affinity chromatography. After releasing GST-tag by thrombin cleavage, the proteins were further purified on SP-Sepharose column (Ubc5a, Ubc5b, Ubc5c, Ubc5c-C85A and Ubc7), or Q-Sepharose column (E2-25K and Ubc3). Ubiquitin and its mutant versions were expressed in a modified E.coli strain BL21(DE3)-pJY2 to prevent mis-incorporation of lysine residues (You et al., 1999).
In Vitro Assay for IRF3 Activation
All procedures were carried out at 4°C unless otherwise specified. After homogenization of culture cells in hypotonic buffer (10 mM Tris-Cl at pH 7.5, 10 mM KCl, 0.5 mM EGTA, 1.5 mM MgCl2, and Roche EDTA-free protease inhibitor cocktail), the homogenates were centrifuged at 1,000 g for 5 minutes to pellet nuclei and unbroken cells (P1). The supernatant (S1) was subjected to centrifugation at 5,000 g for 10 minutes to separate crude mitochondrial pellet from cytosolic supernatant (S5). Mitochondrial pellet was washed once with Mitochondria Resuspension Buffer (MRB; 20 mM HEPES-KOH at pH 7.4, 10% glycerol, 0.5 mM EGTA, and Roche EDTA-free protease inhibitor cocktail), and re-suspended in MRB buffer containing 1% CHAPS. After centrifugation at 10,000 g for 15 minutes, the supernatant was designated as P5 and used in all assays. For preparation of S5 and P5 after viral infection, the cells were infected with Sendai virus for 16 hours before the fractionation procedure. Most of the assays were carried out using P5 and S5, but for biochemical fractionation, S5 was further centrifuged at 100,000 g for 60 minutes to obtain cytosolic extract (S100).
Wild type 35S-IRF3 or its mutant (S385A/S386A) was synthesized using TNT Coupled Reticulocyte Lysate Kit (Promega) supplemented with 35S-methionine. For each 10 μl reaction, 4 μg of P5, 30 to 40 μg of cytosolic fraction (S5 or S100) and 0.5 μl of 35S-IRF3 were mixed in a buffer containing 20 mM HEPES-KOH (pH 7.0), 2 mM ATP, and 5 mM MgCl2. After incubation at 30°C for 1 hour, the samples were subjected to native gel electrophoresis, and dimerization of 35S-IRF3 was visualized by autoradiography using a PhosphorImager (GE Healthcare). To measure ubiquitin-dependent IRF3 activation, Q-B fraction (Fig. 2A) was fractionated on Superdex-200 column and fractions with apparent molecular mass lager than 40 kDa were pooled for the assay. 35S-IRF3 was also purified from reticulocyte lysates using anti-Flag M2 beads (Sigma).
In Vitro Assay for IKK Activation
For each 10-μl reaction, 4 μg of P5, 20 to 40 μg of cytosolic fraction (S5) were mixed in a buffer containing 20 mM Tris-Cl (pH 7.5), 2 mM ATP, 5 mM MgCl2 and 0.1 μM okadaic acid. After incubation at 30°C for 1 hour, the samples were subjected to SDS-PAGE, and phosphorylation of I B was determined by immunoblot analysis using an antibody against phospho-IκBα or total IκBα.
Biochemical Fractionation of Cytosolic Extract and Purification of Ubc5
HeLa cytosolic extract (S100) was prepared as described above from 50 liter of cells purchased from National Cell Culture Center. S100 was loaded onto 60 ml Q-Sepharose column equilibrated with buffer Q-A (20 mM Tris-HCl at pH 7.5, 10% glycerol, and 0.02% CHAPS), and flow-through was precipitated with 40% to 80% (w/v) of ammonium sulfate. After centrifugation at 10,000 g for 30 min, the precipitates were re-suspended in buffer SP-A (20 mM HEPES-KOH at pH 7.0, 10% glycerol, and 0.02% CHAPS), and subjected to extensive dialysis against buffer SP-A. The sample was further fractionated on 1 ml Heparin-Sepharose column with a linear gradient of NaCl (0 mM to 300 mM) in buffer SP-A, and active fractions eluted around 150 mM NaCl were pooled. After the salt was reduced by repeated dilution in buffer SP-A and concentration, the sample was fractionated on 1 ml SP-Sepharose column with a linear gradient of NaCl (0 to 300 mM) in buffer SP-A. Active fractions eluted around 150 mM NaCl were concentrated, and loaded onto Superdex-75 column. In vitro assay for IRF3 activation was performed following each step of chromatography. 1% of the gel-filtration fractions were subjected to SDS-PAGE, and the proteins were visualized by silver staining. The protein band correlating with IRF3-stimulatory activity was excised for analysis by tandem mass spectrometry.
Tetracycline-induced Expression of shRNA and Transgenes
The procedures for establishing U2OS cells stably incorporating tetracycline-inducible shRNA and/or transgenes are described in a companion manuscript (Xu, M., Skaug, B., Zeng, W., and Chen, Z., manuscript submitted). These cells were induced with tetracycline (1 μg/ml) for 3 (ubiquitin knockdown cells), 4 (ubiquitin rescue cells) or 7 days (Ubc5 knockdown and rescue cells), then infected with Sendai virus for indicated times before the cell lysates were prepared for native gel electrophoresis as described above.
Ubiquitin-Binding Assays
pcDNA3-Flag-NEMO and its mutants were transiently expressed in HEK293T cells and purified using the FLAG antibody (M2) beads (Sigma). Synthesis of K63-linked polyubiquitin chains was carried out at 30°C for 2 hours in 100-μl reaction mixture containing 0.5 μg His6-E1, 2 μg His6-Ubc13/Uev1A, 3 μg His6-TRAF6 and 100 μg HA-Ub in the presence of 5 mM MgCl2 and 2 mM ATP. The Flag-NEMO proteins (~ 20 pmol) were incubated with K63-linked HA-ubiquitin polymers at room temperature for 30 min in a 200-μl mixture containing 20 mM HEPES-KOH at pH 7.0, 10% glycerol, 50 mM NaCl, and 0.1% NP-40. After immunoprecipitation with the anti-Flag beads, polyubiquitin chains on the beads were detected by immunoblotting with an anti-HA antibody.
Supplementary Material
Acknowledgments
We thank Dr. Inder Verma (Salk Institute) for providing NEMO-deficient MEF cells, Dr. Genhong Cheng (UCLA) for TRAF3-deficient MEF cells, Dr. Hiroyasu Nakano (Juntendo University School of Medicine) for TRAF2/5-deficient MEF cells, Dr. Jae Jung (University of Southern California) for TRIM25-deficient MEF cells and Dr. John Bell (University of Ottawa) for VSV(ΔM51)-GFP virus. This work was supported by grants from NIH (RO1-AI09919 and RO1-GM63692) and the Welch Foundation (I-1389). Z.J.C. is an Investigator of Howard Hughes Medical Institute.
Footnotes
Supplemental Data include Supplemental Experimental Procedures and thirteen figures and can be found with this article online at http://www.cell.com/molecular-cell.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chau TL, Gioia R, Gatot JS, Patrascu F, Carpentier I, Chapelle JP, O’Neill L, Beyaert R, Piette J, Chariot A. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends Biochem Sci. 2008;33:171–180. doi: 10.1016/j.tibs.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Cordier F, Grubisha O, Traincard F, Veron M, Delepierre M, Agou F. The zinc finger of NEMO is a functional ubiquitin-binding domain. J Biol Chem. 2008 doi: 10.1074/jbc.M806655200. [DOI] [PubMed] [Google Scholar]
- Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Friedman CS, O’Donnell MA, Legarda-Addison D, Ng A, Cardenas WB, Yount JS, Moran TM, Basler CF, Komuro A, Horvath CM, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008 doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Gatot JS, Gioia R, Chau TL, Patrascu F, Warnier M, Close P, Chapelle JP, Muraille E, Brown K, Siebenlist U, et al. Lipopolysaccharide-mediated interferon regulatory factor activation involves TBK1-IKKepsilon-dependent Lys(63)-linked polyubiquitination and phosphorylation of TANK/I-TRAF. J Biol Chem. 2007;282:31131–31146. doi: 10.1074/jbc.M701690200. [DOI] [PubMed] [Google Scholar]
- Guo B, Cheng G. Modulation of the interferon antiviral response by the TBK1/IKKi adaptor protein TANK. J Biol Chem. 2007;282:11817–11826. doi: 10.1074/jbc.M700017200. [DOI] [PubMed] [Google Scholar]
- Hacker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG, Kamps MP, Raz E, Wagner H, Hacker G, et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2005 doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Phung Q, Chan S, Chaudhari R, Quan C, O’Rourke KM, Eby M, Pietras E, Cheng G, Bazan JF, et al. DUBA: a deubiquitinase that regulates type I interferon production. Science. 2007;318:1628–1632. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- Lin R, Heylbroeck C, Pitha PM, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter SM, Tenoever BR, Maniatis T. Connecting mitochondria and innate immunity. Cell. 2005;122:645–647. doi: 10.1016/j.cell.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Oganesyan G, Saha SK, Guo B, He JQ, Shahangian A, Zarnegar B, Perry A, Cheng G. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439:208–211. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- Panne D, McWhirter SM, Maniatis T, Harrison SC. Interferon regulatory factor 3 is regulated by a dual phosphorylation-dependent switch. J Biol Chem. 2007;282:22816–22822. doi: 10.1074/jbc.M703019200. [DOI] [PubMed] [Google Scholar]
- Paz S, Vilasco M, Arguello M, Sun Q, Lacoste J, Nguyen TL, Zhao T, Shestakova EA, Zaari S, Bibeau-Poirier A, et al. Ubiquitin-regulated recruitment of IkappaB kinase epsilon to the MAVS interferon signaling adapter. Mol Cell Biol. 2009;29:3401–3412. doi: 10.1128/MCB.00880-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Ryzhakov G, Randow F. SINTBAD, a novel component of innate antiviral immunity, shares a TBK1-binding domain with NAP1 and TANK. Embo J. 2007;26:3180–3190. doi: 10.1038/sj.emboj.7601743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha SK, Pietras EM, He JQ, Kang JR, Liu SY, Oganesyan G, Shahangian A, Zarnegar B, Shiba TL, Wang Y, Cheng G. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. Embo J. 2006;25:3257–3263. doi: 10.1038/sj.emboj.7601220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai M, Shingai M, Funami K, Yoneyama M, Fujita T, Matsumoto M, Seya T. NAK-associated protein 1 participates in both the TLR3 and the cytoplasmic pathways in type I IFN induction. J Immunol. 2006;177:8676–8683. doi: 10.4049/jimmunol.177.12.8676. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Chen ZJ. Antiviral innate immunity pathways. Cell Res. 2006;16:141–147. doi: 10.1038/sj.cr.7310019. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Stojdl DF, Lichty BD, tenOever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003;4:263–275. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA Is an Adapter Protein Required for Virus-Triggered IFN-beta Signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. Embo J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Cohen RE, Pickart CM. Construct for high-level expression and low misincorporation of lysine for arginine during expression of pET-encoded eukaryotic proteins in Escherichia coli. Biotechniques. 1999;27:950–954. doi: 10.2144/99275st01. [DOI] [PubMed] [Google Scholar]
- Zhang M, Wu X, Lee AJ, Jin W, Chang M, Wright A, Imaizumi T, Sun SC. Regulation of IkappaB kinase-related kinases and antiviral responses by tumor suppressor CYLD. J Biol Chem. 2008;283:18621–18626. doi: 10.1074/jbc.M801451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Yang L, Sun Q, Arguello M, Ballard DW, Hiscott J, Lin R. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat Immunol. 2007;8:592–600. doi: 10.1038/ni1465. [DOI] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.