Abstract
We examined how remote enhancers establish physical communication with target promoters to activate gene transcription in response to environmental signals. Although the natural IFN-β enhancer is located immediately upstream of the core promoter, it also can function as a classical enhancer element conferring virus infection-dependent activation of heterologous promoters, even when it is placed several kilobases away from these promoters. We demonstrated that the remote IFN-β enhancer “loops out” the intervening DNA to reach the target promoter. These chromatin loops depend on sequence-specific transcription factors bound to the enhancer and the promoter and thus can explain the specificity observed in enhancer–promoter interactions, especially in complex genetic loci. Transcription factor binding sites scattered between an enhancer and a promoter can work as decoys trapping the enhancer in nonproductive loops, thus resembling insulator elements. Finally, replacement of the transcription factor binding sites involved in DNA looping with those of a heterologous prokaryotic protein, the λ repressor, which is capable of loop formation, rescues enhancer function from a distance by re-establishing enhancer–promoter loop formation.
Keywords: DNA looping, transcription, transcription factors
The accurate execution of gene expression programs during development and differentiation and in response to environmental cues requires 3 types of regulatory DNA elements in higher eukaryotes: core promoters, upstream promoter elements, and enhancers (1). Core promoters function by providing the blueprints for the assembly of functional pre-initiation complexes executing both the mechanics and the accurate initiation of mRNA synthesis (2). Upstream promoter elements are localized within the first 100–200 bp upstream of the core promoter and contain transcription factor binding sites, functioning to increase the rate of transcription by promoting assembly of pre-initiation complexes (3). Enhancers, on the other hand, can be located either upstream or downstream of the promoter, on the same or on different chromosomes (4). Enhancer elements do not simply fine-tune promoter activity but rather are critical for defining the expression patterns of genes (5, 6).
Recent studies have shown that gene activation by remote enhancers is associated with long-range interactions between regulatory elements (chromatin loop formation) (7–10). It is hypothesized that proteins bound to remote enhancers interact directly with proteins bound to promoters, with the intervening DNA being “looped out” (11–14). Furthermore, it also has been proposed that enhancer complexes migrate along the chromatin fiber until they encounter a functional promoter (facilitated tracking model) (15, 16). The intervening chromatin between the enhancer and the promoter loops out as the enhancer complex moves progressively along the chromatin fiber toward the promoter. The physical proximity between the enhancer and the target promoter stimulates assembly of a functional pre-initiation complex on the promoter, thus causing activation of transcription. Interestingly, some studies suggest that RNA polymerase II (PolII) is recruited to the enhancers and then via DNA looping and/or facilitated tracking appears on the promoter (17).
The molecular basis for DNA looping is not yet clear, although interactions between structural proteins, transcription factors, or general transcription factors within transcription factories have been implicated in enhancer–promoter interactions (18). We do not know, in molecular terms, how these interactions are established and maintained. This paper reports our investigations of the mechanisms by which a remote enhancer reaches a target promoter; that is, whether the transcription factors bound to enhancers and promoters are the ones that provide the contact surfaces for these interactions.
We addressed these issues by examining how the IFN-β enhancer activates transcription when positioned away from its promoter. Although, the natural IFN-β enhancer is located immediately upstream of the core promoter, it also can function as a classical enhancer element conferring virus inducibility to heterologous promoters, even when it is placed several kilobases away from these promoters (19, 20). The nearly complete molecular picture of the mechanisms by which the IFN-β enhancer activates transcription in its natural context (21–24) provided a useful tool for investigating the nature of the interactions between enhancers and promoters. Our experiments suggest that sequence-specific transcription factors bound to enhancers and promoters mediate loop formation and can thus explain the specificity observed in enhancer–promoter interactions, especially in complex genetic loci.
Results
Enhancer Action from a Distance Requires Upstream Promoter Elements.
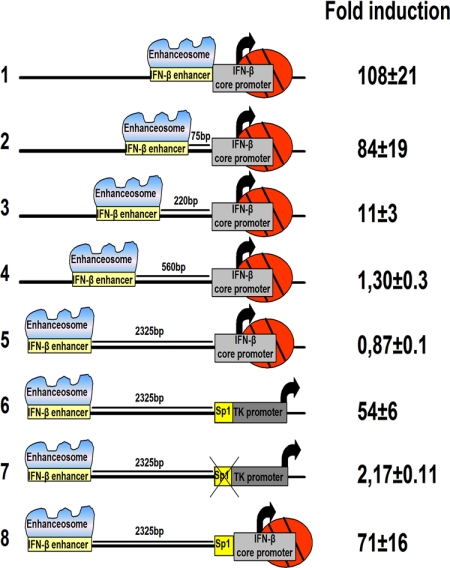
To investigate whether the IFN-β enhancer can activate transcription at a distance from the core promoter, we transfected HeLa cells with the constructs shown in Fig. 1. In the natural cis arrangement, the IFN-β enhancer/core promoter responds to virus infection by stimulating transcription ≈100-fold (line 1). Roughly equivalent levels of transcriptional activation were obtained when the IFN-β enhancer was positioned 75 bp away from the core promoter (line 2). However, when the IFN-β enhancer was placed 220 bp upstream of the core promoter, the levels of activated transcription were significantly reduced (line 3). The IFN-β enhancer was practically inactive when placed 560 bp away (line 4) or 2.3 kbp away (line 5). These results indicate that the IFN-β enhancer requires proximal promoter elements to operate at a distance. A similar prerequisite was observed for the Ig enhancer, which depends on a proximal (promoter) bound octamer factor to mediate enhancer function from a distance (25). Therefore, we replaced the IFN-β core promoter with the thymidine kinase (TK) promoter, which, in addition to the core promoter elements, bears upstream regulatory binding sites for the Sp1 and CCAAT enhancer-binding protein (C/EBP) transcription factors (3). As seen in Fig. 1 (line 6), the IFN-β enhancer strongly activated transcription from the TK promoter from a distance. The rescue of the IFN-β enhancer action from a distance requires intact upstream promoter transcription factor binding sites, because deletion of the Sp1 and C/EBP sites eliminated enhancer-dependent transcriptional activation (line 7). The experiment shown in line 8 indicates that the IFN-β enhancer can activate transcription from the IFN-β core promoter when an Sp1 site is placed upstream of the TATA box. Taken together, these experiments support the notion that the IFN-β enhancer can communicate with either the native or a heterologous core promoter from a distance; however, the information decoded by the enhancer can be conveyed only when transcription factor binding sites are present upstream of the target core promoters.
Fig. 1.
Enhancer action from a distance requires upstream promoter elements. HeLa cells were transfected with the indicated chloramphenicol acetyl transferase (CAT) reporter plasmids. The cells were mock or virus infected for 24 h before being harvested. Then CAT activity was determined.
DNA Looping Mediates the Interaction Between a Remote Enhancer and a Promoter.
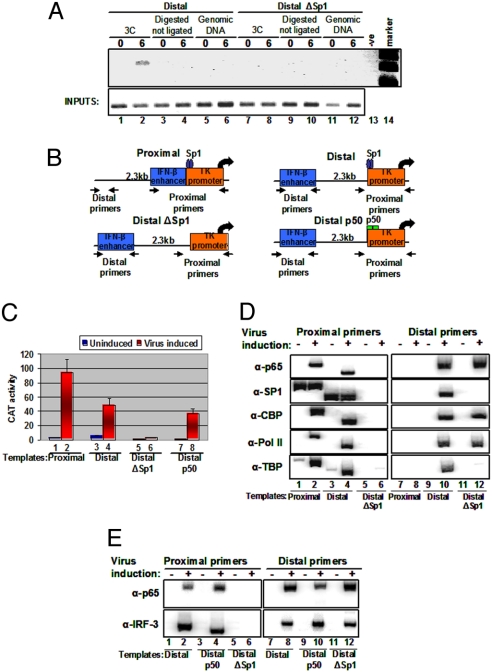
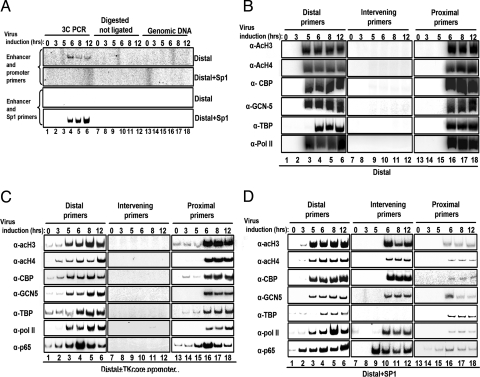
To test whether enhancer–promoter communication involves contacts between the distant regulatory elements, we carried out chromosome conformation capture (3C) assays (26) in which enhancer DNA sequences are ligated to promoter DNA sequences only if they are in close physical proximity. HeLa cells were transfected with the Distal template bearing the IFN-β enhancer 2.3 kbp upstream of the TK promoter (Fig. 1, line 6, and Fig. 2B) or with the Distal Sp1-deleted template (DistalΔSp1 template) (Fig. 1, line 7, and Fig. 2B), followed by virus infection and formaldehyde cross-linking. The cross-linked chromatin was digested with NlaIII (flanking the IFN-β enhancer and the TK promoter) (Fig. S1). The digested chromatin was diluted, and DNA ligase was added to link covalently DNA sequences that are in physical proximity. After the proteins were removed, PCR primers specific for the enhancer and the promoter were used to detect the interacting sequences. Fig. 2A (lane 1) shows that there is no detectable PCR product in uninfected cells. A PCR product was generated when the chromatin DNA used was prepared from virus-infected cells (lane 2) but was not generated in any of the controls, including genomic DNA (lanes 5 and 6) and NlaIII-cleaved but not ligated chromatin DNA (lanes 3 and 4). The size of the PCR product was that predicted for the ligation of the IFN-β enhancer with the TK promoter, and the identity of the PCR product was confirmed by DNA sequencing. Interestingly, no PCR product was detected when the chromatin was prepared from the DistalΔSp1 template (Fig. 2A, lanes 7–12). Taken together, these results suggest that the basis of communication is the physical proximity of the distant IFN-β enhancer and the TK promoter, consistent with a looping transcription mechanism. Furthermore, the inability of the DistalΔSp1 template to be activated by virus infection results from the inability of the IFN-β enhancer to interact with the promoter.
Fig. 2.
DNA looping mediates the interaction between a remote enhancer and a promoter. (A) Shown is a 3C experiment depicting the PCR products using primers specific for the enhancer and the promoter as seen in Fig. S1. PCR was performed on NlaIII-digested chromatin derived from HeLa cells mock or virus infected for 6 h harboring the Distal (lanes 1 and 2) or the DistalΔSp1 (lanes 7 and 8) plasmids. Genomic DNA (lanes 5, 6, 11, and 12) and cross-linked digested but not ligated chromatin (lanes 3, 4, 9, and 10) derived from mock- or virus-infected (6 h) cells were used as controls. Lane 13 is a negative PCR control, and lane 14 is the size marker. (B) Schematic representation of the CAT constructs used to determine mechanisms of enhancer function. The arrows indicate the position of the primers used in the PCR reactions with immunoprecipitated DNA. The wild-type TK promoter contains an Sp1 site (Proximal and Distal constructs), whereas in the DistalΔSp1 construct the Sp1 site has been mutated. In the Distal p50 construct, the Sp1 site was replaced by a consensus p50 homodimer site. (C) Stable HeLa cells bearing the indicated CAT reporter plasmids were mock or virus infected for 12 h before being harvested; then CAT activity was determined. The error bars indicate SD. (D) Cross-linked chromatin prepared from mock- or virus-infected (6 h) HeLa cells stably transfected with the indicated CAT constructs was immunoprecipitated with the indicated antibodies. The precipitated DNA was subjected to PCR analysis using 32P-dCTP and plasmid-specific primers. (E) The process is as described in (D), except that p65 and IRF-3 antibodies were used, and the Distal p50 construct instead of the proximal construct was included in the experiment.
To provide additional support for the 3C data, we performed ChIP experiments. ChIP allows us to distinguish whether DNA looping interactions occur, because the cross-linking of an enhancer-bound factor to a remote promoter, in the absence of the respective binding sites on the promoter, would indicate direct contact between the physically distant sites. We used 3 templates (Proximal, Distal, and DistalΔSp1) in which the IFN-β enhancer was placed immediately upstream of the TK promoter (Proximal), 2.3 kbp away from the TK promoter (Distal), or 2.3 kbp away from a TK promoter lacking the Sp1 and C/EBP sites (DistalΔSp1) (Fig. 2B). These templates were transfected into HeLa cells along with pCDNA3, and stable clones were obtained in the presence of G418. Consistent with the data in Fig. 1 obtained from transient transfections, the Proximal and Distal templates responded to virus infection, but deletion of the Sp1 sites of the TK promoter abolished the ability of the IFN-β enhancer to activate transcription from this promoter from a distance (Fig. 2C). ChIP experiments were performed on chromatin prepared from either mock- or virus-infected cells for 6 h, followed by PCR amplification using enhancer-specific (Distal) and/or promoter-specific (Proximal) primers. Fig. 2D (lanes 1 and 2) shows that p65 associates with the IFN-β enhancer on the Proximal template in a virus-inducible manner, whereas Sp1 is constitutively bound to the nearby TK promoter. Consistent with previous data (21), the IFN-β enhancer assembled an enhanceosome upon virus infection, recruiting CREB binding protein (CBP), PolII, and TATA binding protein (TBP) to the promoter region of the Proximal template (lanes 1 and 2). As a control, we showed that none of these proteins was recruited 2.3 kb away from the promoter on this construct (lanes 7 and 8). As in the Proximal template, we found that the enhancer at the remote position recruited both p65 and CBP (lanes 9 and 10). Remarkably, immunoprecipitation of the constitutively bound promoter factor Sp1 (lanes 3 and 4) and the general transcription factor TBP also pulled down the remote enhancer in a virus-inducible manner (lanes 9 and 10). Likewise, we found the enhanceosome-recruited CBP and PolII associated with the remote promoter in a virus-inducible manner (lanes 9, 10). Notably, DNA intervening between the remote IFN-β enhancer and the TK promoter in the Distal template were not enriched under any conditions (see later discussion). Taken together, these experiments strongly suggest that enhancer- and promoter-bound factors are in physical proximity upon virus infection via DNA looping. Importantly, deletion of the proximal Sp1 DNA binding sites in the TK promoter eliminated both the association of this factor with the TK promoter and the interaction of the enhancer-bound factors with the promoter and vice versa (lanes 5, 6, 11, and 12). Furthermore, these experiments indicated that enhanceosome assembly and enhanceosome-dependent recruitment of CBP, PolII, and General Control Non-derepressible-5 on the remote enhancer occurs independently of factor interaction with the promoter region (see later discussion).
The requirement for an activator (Sp1) DNA binding site upstream of the core promoter for a functional interaction between the enhancer and the promoter suggests either that enhancers interact with promoters when the latter have been “primed” for transcription by local activators or that local DNA binding proteins mark promoters on DNA. To investigate these possibilities, we replaced the Sp1 site with a DNA binding site specifically recognized by the transcriptionally inert p50 homodimer and not by the activating p50/p65 heterodimer of NF-κΒ (27). As expected, Fig. 2C shows that this promoter displays slightly lower levels of basal transcription, but it can be activated efficiently by the remote IFN-β enhancer upon virus infection. ChIP experiments revealed that the enhancer and the promoter are in physical proximity because the enhancer-bound factors p65 and interferon regulatory factor 3 (IRF-3) can be cross-linked to this promoter in a virus-inducible manner (Fig. 2E, lanes 1–4). Thus, the enhancer–promoter communication is independent of the transcriptional strength of the promoter element (also see later discussion).
Enhancer–Promoter Interactions Are Facilitated by Heterologous Proteins Capable of DNA Looping.
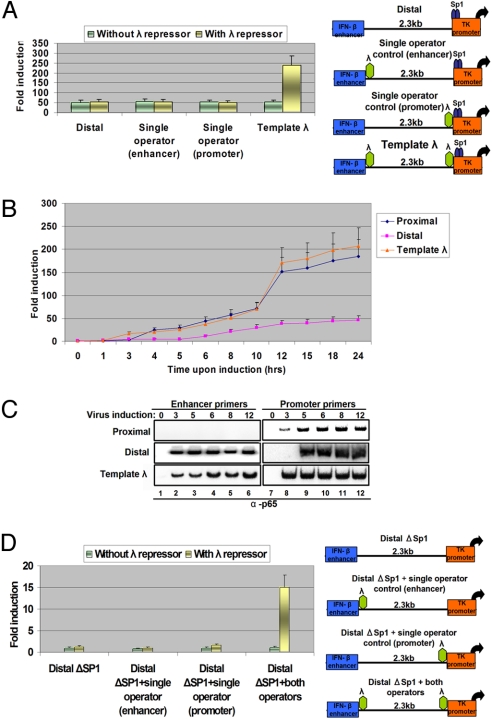
An important prediction derived from our experiments is that the functional interaction between enhancers and promoters via DNA looping should be facilitated by DNA-binding proteins capable of promoting DNA loop formation but incapable of coactivator recruitment. Therefore, we inserted a pair of λ-repressor DNA binding sites upstream of the TK promoter and downstream of the IFN-β enhancer (Fig. 3A Right). The λ repressor is a prokaryotic protein that has been shown to generate DNA loops to regulate the lysogenic phage of bacteriophage λ (28). Fig. 3A shows that expression of the λ repressor stimulated the levels of enhancer-dependent virus-inducible transcription by ≈5-fold only on the template bearing the λ-repressor binding sites at both enhancer and promoter locations (template λ). Furthermore, time-course virus-infection experiments revealed that expression of the λ repressor caused an earlier activation of transcription of the λ template, thus coinciding with the timing of the transcriptional activation of the template bearing the enhancer immediately upstream of the promoter (Fig. 3B). The earlier activation of transcription probably is caused by the existence of preformed DNA loops between the enhancer and the promoter, as revealed by the ChIP experiment of Fig. 3C. To investigate whether enhancer–promoter DNA looping established solely by the λ repressor can lead to activation of transcription, we carried out the experiment shown in Fig. 3D. We inserted the pair of λ-repressor binding sites in the DistalΔSp1 template and tested whether it can be activated by virus infection in the presence of exogenously supplied λ repressor. As shown in Fig. 3D, the remote enhancer can activate the TK promoter in the absence of upstream Sp1 binding sites. Taken together, these experiments demonstrate that a heterologous prokaryotic protein capable of DNA looping but incapable of coactivator recruitment (i.e., the λ repressor) can facilitate the ability of an enhancer to interact with a remote promoter, a result consistent with the DNA looping mechanism of enhancer action from a distance.
Fig. 3.
Enhancer–promoter interactions are facilitated by heterologous proteins capable of DNA looping. (A) HeLa cells were cotransfected with the indicated CAT reporter plasmids in the presence or absence of a λ expression vector; 24 h later, the cells were virus infected for 12 h before being harvested; then CAT activity was determined. The error bars indicate SD. (B) HeLa cells were cotransfected with the indicated CAT reporter plasmids and the λ expression vector; 24 h later, the cells were mock or virus infected for different amounts of time before being harvested. Then CAT activity was determined. The error bars indicate SD. (C) HeLa cells were transfected with the indicated plasmids as described in (A), except that the cells were mock or virus infected for increasing amounts of time. Cross-linked chromatin was immunoprecipitated with the p65 antibody. The precipitated DNA was subjected to PCR analysis using 32P-dCTP and plasmid-specific primers as indicated. (D) HeLa cells were cotransfected with the indicated CAT reporter plasmids in the presence or in the absence of a λ expression vector; 24 h later, the cells were virus infected for 12 h before being harvested. Then CAT activity was determined. The error bars indicate SD.
Trapping Enhancer Function by Interspersed Transcription Factor Binding Sites.
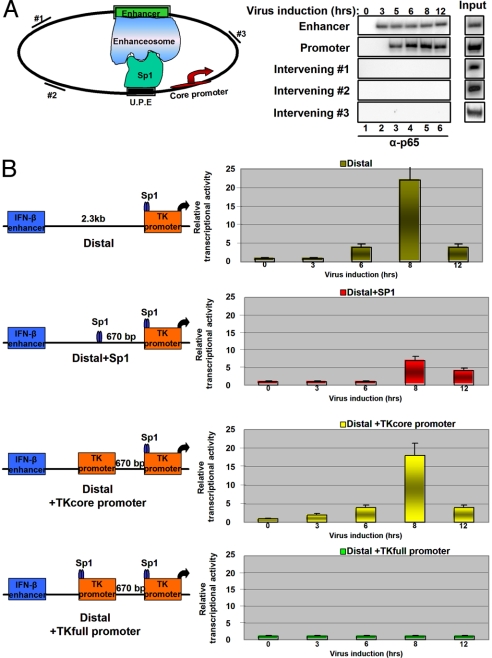
To investigate whether the IFN-β remote enhancer could reach the target promoter via intermediate DNA sites, we carried out ChIP experiments using primers amplifying 3 intervening sequences located between the enhancer and the target promoter. Fig. 4A shows that we were unable to identify stable interactions of the IFN-β enhancer with these intermediate sites. However, the intervening DNA is of a prokaryotic origin, and therefore it is poor in mammalian transcription factor binding sites (29). It is possible that under physiological conditions enhancers could reach the target promoters by hopping between transcription factor binding sites interspersed between the enhancer and promoters until they reach a functional promoter. To test this idea, we inserted 3 different DNA elements between the enhancer and the promoter: a single DNA site for the transcription factor Sp1 (Distal+Sp1), or a core promoter (Distal+TKcore), and/or a fully functional promoter (Distal+TKfull promoter) including both upstream transcription factor binding sites and core promoter elements. Fig. 4B shows that insertion of the Sp1 site decreased the ability of the enhancer to activate the TK promoter located further downstream. By contrast, insertion of a core promoter at the same site did not affect enhancer action significantly. However, the insertion of the same core promoter bearing an upstream Sp1 site dramatically decreased the enhancer action on the downstream target promoter.
Fig. 4.
The effect of inserting transcription factor binding sites between an enhancer and a promoter. (A) HeLa cells were transfected with the Distal reporter plasmid, and 24 h later the cells were mock or virus infected for increasing amounts of time. Cross-linked chromatin was immunoprecipitated with the p65 antibody. The precipitated DNA was subjected to PCR analysis using 32P-dCTP and primers specific for plasmid sequences located between the enhancer and the promoter (intervening #1, #2, and #3), and enhancer- or promote-specific primers as shown at the diagram at the left of the gel. (B) HeLa cells were transfected with the constructs indicated (Left). The cells were infected with virus for different amounts of time, and the isolated RNA was used as a template for RT-PCR analysis using CAT-specific primers. The radioactive bands were quantitated using PhosphorImager (Typhoon), and the data from 3 independent experiments were plotted (Right). Shown are mean values ± SD.
These experiments suggest that transcription factor binding sites (in this case, Sp1) can trap the remote enhancer, interfering with its interaction with promoters and thus resembling insulators. The 3C experiment of Fig. 5A (lanes 1–6) shows that a specific PCR product is generated in the Distal+Sp1 template when primers specific for the enhancer and the sequence next to the inserted Sp1 site were used in the PCR. These interactions appear at 6 h after infection, that is, at the same time as the interaction of the enhancer with the promoter in the case of the Distal template (Fig. 5A, lanes 1–6). The specificity of these interactions is underscored by the inability to generate PCR products using the same primers but the Distal template and/or using digested but not ligated DNA or genomic DNA (Fig. 5A, lanes 7–18).
Fig. 5.
Transcription factor binding sites trap enhancer complexes. (A) A 3C experiment depicts the PCR products using pairs of primers specific for enhancer and promoter (Top) or enhancer and Sp1 (Bottom). PCR was performed on NlaIII-digested chromatin (Fig. S1) derived from HeLa cells mock- or virus-infected for the indicated times transfected with the Distal or Distal+Sp1 (lanes 1–6). Genomic DNA (lanes 13–18) and cross-linked digested but not ligated chromatin (lanes 7–12) derived from mock- or virus-infected cells were used as controls. (B) HeLa cells were transfected with the Distal reporter plasmid; 24 h later the cells were mock or virus infected for increasing amounts of time. Cross-linked chromatin was immunoprecipitated with the indicated antibodies. The precipitated DNA was subjected to PCR analysis using 32P-dCTP and primers specific for the enhancer (lanes 1–6), the intervening #2 (lanes 7–12), and the promoter (lanes 13–18). (C) The process was as described in (B), except the cells were transfected with the Distal+TKcore promoter plasmid. In this case, the intervening primers amplify the inserted TK core promoter. (D) The process was as described in (B), except that the cells were transfected with the Distal+Sp1 plasmid. In this case, the intervening primers amplify the inserted Sp1 DNA binding site.
The ChIP experiments of Fig. 5B show that both CBP and GCN5 are recruited to the enhancer 5 h postinfection, and this recruitment correlates with histone acetylation at the enhancer area (lane 3). Remarkably, although the enhancer and the promoter already have established an interaction at this time point (Fig. 3C), we found that these coactivators and histone acetylation appear at the promoter 1 h later (at the 6-h time point, lane 16), that is, at the time of transcriptional initiation (Fig. 3B). Similarly, PolII is recruited to the enhancer first and then appears at the promoter. By contrast, TBP is found simultaneously in the enhancer and promoter (lanes 4 and 16).
Insertion of the Sp1 site between the enhancer and the promoter led to a strong recruitment of factors and cofactors at this site with a kinetics similar to that observed for the interaction of the same enhancer with the target promoter in the Distal template (Fig. 5D). The only exception is the absence of TBP recruitment at the Sp1 site, a result consistent with the lack of a TATA box. The stable interaction of the enhancer with the Sp1 site antagonizes the interaction of the enhancer with the downstream promoter, thus leading to decreased PolII recruitment (Fig. 5D, lanes 13–18) and lower activation of transcription (Fig. 4B). Notably, the amount of TBP recruitment remains similar between the Distal and Distal+Sp1 templates. This observation indicates that the strength of TBP recruitment does not always correlate with the amount of activated transcription. Interestingly, the ChIP experiments using the template bearing a core promoter inserted between the enhancer and the target promoter revealed a pattern of recruitments and acetylations similar to the Distal template (Fig. 5C). This result is in agreement with our previous data showing that enhancers do not interact stably with core promoters. Taken together, these data suggest that transcription factors bound to proximal promoter elements serve as tethers for distant enhancers. We find that the addition of proximal promoter elements at positions intermediate to the remote enhancer and the site of transcription initiation do not facilitate enhancer–promoter communication but, in fact, compete physically for interactions with the enhancer, thus resulting in decreased transcriptional output.
Discussion
Although previous studies have suggested that transcription factors mediate the interactions between enhancers and promoters (30), the answers to a significant number of important questions have remained elusive. Synthetic enhancer–promoter configurations bearing the IFN-β enhancer at increasing distances from the core promoter were inactive when the enhancer was placed >560 bp away from the promoter because the enhancer could not loop out to reach the promoter. DNA looping and transcriptional activity were restored when a heterologous transcription factor binding site (Sp1) was inserted at the promoter. The interactions between transcription factors could drive DNA loop formation and are independent of ongoing transcription. The λ repressor, a prokaryotic protein incapable of activating transcription in mammalian cells on its own, can establish enhancer–promoter interactions when its binding sites are placed in both elements. Most likely, the λ repressor binds to both sites (at the enhancer and the promoter) and generates a DNA loop by the formation of large protein complexes in which individual repressors bound to the different sites interact with each other. Thus, the forces driving loop formation appear to be protein–protein interactions between widely separated transcription factors and not coactivator proteins.
A prediction derived from this idea is that enhancers could be trapped away from target promoters by interacting with transcription factors scattered throughout the genome. Indeed, we showed that insertion of a single transcription factor binding site between the IFN-β enhancer and the target promoter captured the enhancer at this site and not at the target promoter, thus mimicking the function of insulators. An implication derived from this observation is that in a physiological context enhancers could work from a distance by establishing sequential interactions (loops) with transcription factors until they reach a fully functional promoter. Such a model can explain why these loops are dynamic (12) and how preferences in enhancer–promoter interactions are established. We can imagine that the specificity in enhancer–promoter interactions depends on the strength of the interactions between transcription factors bound to these elements. This idea also can explain the function of promoter-tethering elements that can interact selectively with enhancers during Drosophila development, thus regulating enhancer–promoter interactions at complex genetic loci (31). This model also can explain the functional interaction between enhancers and promoters located on nonhomologous chromosomes, such as the olfactory receptor genes (32).
Ordinarily, the probability of enhancer–promoter interactions and loop formation should depend on the occupancy of the sites by the cognate transcription factors, the distance between the interacting elements, and the flexibility of the looped chromatin. Therefore, we imagine that enhancers and promoters bound by high-affinity and/or abundant transcription factors are more likely to establish functional chromatin loops than are regulatory elements bound by low-abundance and/or low-affinity transcription factors. Furthermore, the shorter the distance between an enhancer and a promoter, the greater is the probability of a productive interaction (18), an observation confirmed by our data. Finally, the recruitment of chromatin modifiers and remodelers to the enhancers and promoters by transcription factors can alter the biophysical properties of the surrounding chromatin, thus facilitating DNA looping (33).
Materials and Methods
Chromosome Conformation Capture (3C).
Cell-culture methods, plasmid constructions, and ChIP approaches are described in the SI Text. The sequences of all primers and oligos used in this study are listed in the SI Text.
The 3C was performed as described in ref. 34 with the following modifications. The transfection mix contained 3 μg of reporter plasmid and 12 μg of empty vector. Approximately 7 × 106 HeLa cells were fixed in 1% formaldehyde at room temperature for 10 min; 1/3 of the fixed cells was digested with the NlaIII restriction enzyme (R0125L; NEB). For intramolecular ligation, the cleaved chromatin was diluted to 2.7 ng/μL and was incubated for 16 h at 16 °C with 100 Weiss units of T4 DNA ligase (M0202M; NEB).
Supplementary Material
Acknowledgments.
We thank A. Hochschild for providing a λ-repressor plasmid. We also thank George Thireos, Ethan Ford, George Panayotou, Effie Apostolou, Giannis Vatsellas, and Mat Lavigne for critical reading of the manuscript. This work was supported by grants (to D.T.) from the Greek Secretariat for Research and Technology (PENED 01–225), the March of Dimes, the Association for International Cancer Research, and the European Union (European Union Marie Currie Action RTN –TAF CHROMATIN).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902454106/DCSupplemental.
References
- 1.Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 2.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 3.McKnight SL, Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982;217:316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- 4.Atchison ML. Enhancers: Mechanisms of action and cell specificity. Annu Rev Cell Biol. 1988;4:127–153. doi: 10.1146/annurev.cb.04.110188.001015. [DOI] [PubMed] [Google Scholar]
- 5.Maniatis T, Goodbourn S, Fischer JA. Regulation of inducible and tissue-specific gene expression. Science. 1987;236:1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- 6.Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- 7.Cook PR. Nongenic transcription, gene regulation and action at a distance. J Cell Sci. 2003;116:4483–4491. doi: 10.1242/jcs.00819. [DOI] [PubMed] [Google Scholar]
- 8.Fraser P. Transcriptional control thrown for a loop. Curr Opin Genet Dev. 2006;16:490–495. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Kadauke S, Blobel GA. Chromatin loops in gene regulation. Biochim Biophys Acta. 2009;1789:17–25. doi: 10.1016/j.bbagrm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vilar JM, Saiz L. DNA looping in gene regulation: From the assembly of macromolecular complexes to the control of transcriptional noise. Curr Opin Genet Devt. 2005;15:136–144. doi: 10.1016/j.gde.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Drissen R, et al. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jing H, et al. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Garrard WT. Long-range interactions between three transcriptional enhancers, active Vκ gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol Cell Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 15.Hatzis P, Talianidis I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol Cell. 2002;10:1467–1477. doi: 10.1016/s1097-2765(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 17.Szutorisz H, Dillon N, Tora L. The role of enhancers as centres for general transcription factor recruitment. Trends Biochem Sci. 2005;30:593–599. doi: 10.1016/j.tibs.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Marenduzzo D, Faro-Trindade I, Cook PR. What are the molecular ties that maintain genomic loops? Trends Genet. 2007;23:126–133. doi: 10.1016/j.tig.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Fan CM, Maniatis T. Two different virus-inducible elements are required for human beta-interferon gene regulation. EMBO J. 1989;8:101–110. doi: 10.1002/j.1460-2075.1989.tb03353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodbourn S, Zinn K, Maniatis T. Human beta-interferon gene expression is regulated by an inducible enhancer element. Cell. 1985;41:509–520. doi: 10.1016/s0092-8674(85)80024-6. [DOI] [PubMed] [Google Scholar]
- 21.Agalioti T, et al. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 22.Lomvardas S, Thanos D. Nucleosome sliding via TBP DNA binding in vivo. Cell. 2001;106:685–696. doi: 10.1016/s0092-8674(01)00490-1. [DOI] [PubMed] [Google Scholar]
- 23.Parekh BS, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 24.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 25.Bertolino E, Singh H. POU/TBP cooperativity: A mechanism for enhancer action from a distance. Mol Cell. 2002;10:397–407. doi: 10.1016/s1097-2765(02)00597-x. [DOI] [PubMed] [Google Scholar]
- 26.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 27.Kunsch C, Ruben SM, Rosen CA. Selection of optimal κ B/Rel DNA-binding motifs: Interaction of both subunits of NF-κ B with DNA is required for transcriptional activation. Mol Cell Biol. 1992;12:4412–4421. doi: 10.1128/mcb.12.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dodd IB, et al. Cooperativity in long-range gene regulation by the lambda CI repressor. Genes Dev. 2004;18:344–354. doi: 10.1101/gad.1167904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minezaki Y, Homma K, Nishikawa K. Genome-wide survey of transcription factors in prokaryotes reveals many bacteria-specific families not found in archaea. DNA Research. 2005;12:269–280. doi: 10.1093/dnares/dsi016. [DOI] [PubMed] [Google Scholar]
- 30.Bartkuhn M, Renkawitz R. Long range chromatin interactions involved in gene regulation. Biochim Biophys Acta. 2008;1783:2161–2166. doi: 10.1016/j.bbamcr.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 31.Calhoun VC, Stathopoulos A, Levine M. Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc Natl Acad Sci USA. 2002;99:9243–9247. doi: 10.1073/pnas.142291299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lomvardas S, et al. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 33.Li Q, Barkess G, Qian H. Chromatin looping and the probability of transcription. Trends Genet. 2006;22:197–202. doi: 10.1016/j.tig.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Apostolou E, Thanos D. Virus infection induces NF-κB-dependent interchromosomal associations mediating monoallelic IFN-β gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.