Abstract
Mitogenic growth factors play an important role in cellular development and differentiation. The purpose of this study was to assess the extent to which epidermal growth factor (EGF) and transforming growth factor α (TGFα) and their cognate receptor (EGFR) are crucial for normal preimplantation embryo development. We used RNA interference to decrease expression of growth factors in preimplantation mouse embryos. We microinjected 1-cell mouse embryos individually with short interfering RNA (siRNA) specific to EGF, TGFα, and EGFR and then analyzed temporal and spatial gene expression patterns at different stages of development before implantation. Transfection with siRNA significantly reduced growth factor expression in 1-cell, 2-cell, morula, early-blastocyst, and late-blastocyst embryos to levels similar to those in untreated ‘cloned’ embryos derived through intracytoplasmic nuclear injection. In addition, siRNA effectively decreased expression of maternally inherited genes between 24 and 72 h after transfection, with recovery of gene expression during late-blastocyst stage at 96 h after transfection. Furthermore, siRNA significantly decreased blastocyst formation, increased the number of apoptotic cells, and reduced the total number of differentiated cells in blastocysts; these changes were greatest after decreasing EGFR and of both EGF and TGFα simultaneously. These results support our hypothesis that EGF and TGFα are crucial for embryo survival and development. Further, dysregulated expression of growth factors is associated with poor development of cloned mouse embryos.
Abbreviations: EGF, epidermal growth factor; EGFR, EGF receptor; BLAST, basic local alignment search tool; FHM, flushing–holding medium; ICM, inner cell mass; ICNI, intracytoplasmic nuclear injection; KSOM, potassium simplex optimized medium; RNAi, RNA interference; siRNA, short interfering RNA; TE, trophoectoderm; TGFα, transforming growth factor α; CZB, Chatot Ziomek Bavister
Mammalian embryonic development is a complex process that involves cell–cell signaling, cell migration, differentiation, apoptosis, and a host of other biological processes. Regulation of embryo development is mediated by specific temporal and spatial expression of numerous growth factors, including epidermal growth factor (EGF) and transforming growth factor α (TGFα).4,6,27 By activating specific intracellular signaling cascades, growth factors regulate cell proliferation and differentiation and prevent apoptosis.9 Genes encoding these growth factors and their cognate receptor (EGFR) are expressed sequentially by cells of the maternal reproductive tract and embryo, optimizing cell cleavage and differentiation during early embryogenesis.31,32
In our previous studies comparing the timing, pattern, and level of expression of EGF, TGFα, and EGFR in mouse embryos,10 we found that gene expression levels are maternally derived in fertilized oocytes and then decline after fertilization until the 2-cell stage, when embryo-derived gene expression is activated and steadily increases until the blastocyst stage. In contrast, growth factor gene expression in cloned mouse embryos fails to increase significantly above baseline. These results are consistent with our earlier work using growth factor immunoneutralization in naturally derived mouse embryos11 and growth factor replacement in cloned mouse embryos,12 which focused on blocking and replacing, respectively, growth factor function.
In contrast, posttranscriptional gene silencing using double-stranded RNA targeted to specific growth factors to induce RNA interference (RNAi) has proven to be a powerful tool to study the function of many endogenous genes in various organisms, including mammals.16,17,39,40 Therefore, in the present study, we sought to determine whether RNAi technology could be used to determine the extent to which growth factor gene expression contributes to the poor survival and development of cloned embryos. After treating embryos derived through natural mating with gene-specific siRNA, we analyzed and correlated gene expression with objective measures of preimplantation development, and we compared these findings to those in cloned embryos derived by intracytoplasmic nuclear injection (ICNI). These results support our previous findings10-12 and indicate that uninterrupted expression of mitogenic growth factors are crucial for embryo survival and development and that deficient expression of these genes leads to poor developmental competence in preimplantation cloned embryos.
Materials and Methods
Experimental design.
Four siRNA experimental treatment groups (EGF, TGFα, EGFR, and combined EGF and TGFα), 2 control groups (no siRNA treatment as negative control and noninhibitory siRNA as positive control), and an untreated ‘cloned’ embryo group were used. The siRNA and noninhibitory siRNA were purchased from Invitrogen (Carlsbad, CA). Embryos for both siRNA and control treatments were derived by natural mating, and cloned embryos were obtained through ICNI. B6D2F1 (C57BL/6J × DBA/2J) male and female mice (age, 6 to 8 wk) were obtained from Jackson Laboratory (Bar Harbor, ME) and were kept in pathogen-free barrier facility according to standard colony management procedures (14:10-h light:dark cycle, 1 mouse per cage; food and water ad libitum). For each treatment and control group, 15 female mice mated to male mice were used to harvest approximately 150 embryos (1-cell stage), which were divided into 5 replicate trials of 30 embryos per treatment. In addition, 150 mature oocytes were harvested from other B6D2F1 female mice for use in cloning by ICNI. All experiments were conducted in accordance with the Institutional Animal Care and Use Committee of University of California, Davis.
siRNA synthesis.
Specific nucleotides sequences for targeted genes (EGF, TGFα, and EGFR) were selected for siRNA constructs from mouse (Mus musculus) cDNA sequences and according to published guidelines.15 siRNA constructs were designed online by using Block-iT RNAi Designer (Invitrogen). Selected sequences were sorted and screened in a basic local alignment search tool BLAST search1 to verify that the mRNAs specific for genes of interest were targeted, and siRNA duplexes corresponding to the genes of interest were custom-synthesized. The siRNA sequences for EGF (locus 13645; GenBank accession number, NM_010113) were: sense RNA, 5′ GCA GAC AUG GAU AUU CAU U 3′; antisense RNA, 5′ AAU GAA UAU CCA UGU CUG C 3′; for TGFα (locus 21802; accession number, NM_031199): sense, 5′ CCC ACA CUC AGU ACU GCU U 3′; antisense, 5′ AAG CAG UAC UGA GUG UGG G 3′; and EGFR (locus 13649; accession number, NM_207655): sense, 5′ GGA AAU UAC CUA UGU GCA A 3′; antisense, 5′ UUG CAC AUA GGU AAU UUC C 3′. Synthetic oligonucleotides were HPLC-purified, and siRNA were generated by mixing 20 µM of each sense and antisense RNA 19mer in annealing buffer as previously described.15 Embryos from natural mating and either transfected with noninhibitory siRNA or nontransfected were used as controls.
siRNA microinjection.
Purified siRNAs were microinjected as described previously.18,20 Briefly, 1-cell stage embryos were collected into warm flushing–holding medium (FHM) from the oviducts of naturally mated B6D2F1 female mice that were superovulated by intraperitoneal injection of 5 IU pregnant mare serum gonadotrophin, followed 48 h later by injection of 5 IU human chorionic gonadotrophin. Mice with vaginal plugs at 0.5 d post coitus were euthanized by CO2 asphyxiation and cervical dislocation.
Cumulus cells attached to zygotes were removed by brief incubation (2 to 3 min) in FHM containing 300 U/mL bovine testis hyaluronidase at room temperature. Zygotes then were washed and cultured in potassium simplex optimized medium (KSOM, Specialty Media, Phillipsburg, NJ), covered with mineral oil, and placed in 5% CO2 in humidified air at 37.5 °C before microinjection of siRNA. Microinjection was performed on an inverted microscope (TE300, Nikon Instruments, Melville, NY) with micromanipulators (Narishige International, East Meadow, NY) at room temperature in FHM containing 0.01% (v/w) polyvinyl alcohol. The siRNA was diluted in PBS to a final concentration of 1 µg/µL, and 4 to 5 pL siRNA solution was injected into the cytoplasm of each 1-cell embryo (zygote). In control groups, 4 to 5 pL PBS or nonfunctional siRNA (Invitrogen) at 1 µg/µL in PBS was microinjected into each pronuclear-stage embryo.
ICNI.
We previously have described10 the ICNI procedure to derive cloned mouse embryos. Briefly, 6- to 8-wk-old B6D2F1 female mice were superovulated by intraperitoneal injection of 5 IU pregnant mare serum gonadotrophin, followed 48 h later by injection of 5 IU human chorionic gonadotrophin. Oocyte–cumulus complexes were collected and incubated briefly in warm FHM containing 300 U/mL bovine testis hyaluronidase to remove cumulus cells. Oocytes then were cultured in microdrops (50 µL each) of KSOM, covered with mineral oil, and placed under 5% CO2 in humidified air at 37.5 °C prior to enucleation. Enucleation was performed in FHM containing 5 µg/mL cytochalasin B. Cumulus cells were used as nuclear donor cells, which were stored in FHM at 4 °C before use. Nuclei of cumulus cells were isolated in BSA-free FHM containing 0.01% (v/w) polyvinyl alcohol by repeated aspiration into and ejection from a micropipette (diameter, 5 to 6 µm). After collection, a single nucleus was injected into the cytoplasm of an enucleated oocyte that had been cultured in KSOM for as long as 2 h after enucleation. Reconstructed oocytes were activated by culture in Ca2+-free CZB medium (EmbryoMax, Millipore, Burlington, MA) containing 10 mM SrCl2 and 5 µg/mL cytochalasin B for 6 h at 37 °C and then collected into KSOM.
Embryo culture.
After injection of siRNA or cloning, embryos were incubated and cultured in 5% CO2 and humidified air at 37.5 °C in KSOM until late-blastocyst stage (4.5 d post coitus). Embryo development to 2-cell, 4-cell, morula, and blastocyst stages was recorded every 12 h. For evaluation of gene expression and development, samples of 25 embryos per treatment were harvested at random from each group (described later). Five replicate trials were conducted for each treatment.
Immunofluorescent labeling.
By using monoclonal mouse antibodies to EGF, TGFα and EGFR (UniTech ABC kit, EMD Biosciences, San Diego, CA), these growth factors underwent immunofluorescent labeling to assess their expression levels as previously described.10 Briefly, embryos were selected randomly and fixed with 4% paraformaldehyde, permeabilized with 0.05% Triton 100, and incubated with the appropriate primary monoclonal antibody conjugated with fluorescein isothiocyanate. In the combined treatment (EGF and TGFα) group, the primary antibody (5µg/mL, Calbiochem, San Diego, CA) to EGF was conjugated with fluorescein isothiocyanate, whereas that to TGFα was conjugated with phycoerythrin. Negative controls were established concurrently by using PBS containing 2% BSA instead of the primary antibody. All images (zygotes and embryos) were captured under a 40× objective (magnification, ×630; resolution, 1300 × 1030 pixels at 150 pixels per inch) by using the same exposure for all images. Fluorescence intensity was measured (Image-Pro Plus 4.5.1, Media Cybernetics, Bethesda, MD) by using integrated optical density. Green fluorescence of the cytoplasm or membrane was considered to indicate positive expression of the individual growth factors, whereas yellowish fluorescence indicated colocalization of EGF (green) and TGFα (red) in the combined treatment group. The mean value of fluorescence intensity for each embryo was corrected by subtracting the mean fluorescence of the negative control.
Embryo cell counting.
Inner cell mass (ICM) and trophectoderm (TE) cells were differentially labeled by using fluorescent propidium iodide (red) and Hoechst 33258 (bisbenzimide, blue) according to published protocols.41 The total numbers of ICM (blue) and TE (pinkish red) cells were counted, and the ICM:TE ratio was calculated.
Terminal dUTP nick-end labeling assay.
To determine apoptosis, 20 blastocysts (4.5 d post coitus) were selected randomly from each experimental group and fixed, permeabilized, and stained as previously described.5 The embryos were observed under a fluorescence microscope by using a blue filter (excitation wave length, 450 to 490 nm) for dUTP-labeled nuclei and a green filter (excitation wave length, 512 to 560 nm) for cells stained with propidium iodide (red). To determine DNA fragmentation, fluorescein-isothiocyanate–labeled (yellowish) nuclei were counted as apoptotic cells, and those stained with propidium iodide were counted as intact cells. The apoptotic index was calculated by dividing the number of apoptotic cells by the total number of live cells.
Western blotting.
For each group, 20 blastocysts were processed for immunoblotting according to the methods described previously.45 Briefly, blastocysts from BSA-free KSOM were transferred into an equal volume of double-strength sodium dodecyl sulfate buffer. Proteins from each sample were resolved on 12% SDS polyacrylamide gels, transferred onto polyvinylidene fluoride membrane (Millipore), blocked with 5% BSA in TBS and 0.1% Tween 20, and incubated with the appropriate monoclonal antibody. AntiEGF and antiTGFα were produced by immunizing BALB/c mice with recombinant human EGF or TGFα, and hybridomas were derived by fusion of mouse spleen cells with P3 × 63 Ag8.653 mouse myeloma cells. AntiEGFR (partially purified from human A431 carcinoma cells) was produced commercially (EMD Biosciences–Calbiochem). After incubation with goat antimouse secondary antibody (conjugated with horseradish peroxidase, EMD Biosciences–Calbiochem), immunoblots were visualized by using X-ray film.
Statistics.
All data were analyzed by using the SPSS 11.0 statistical program (SPSS, Chicago, IL). Univariate ANOVA, Tukey honest significant difference, and Bonferroni multiple-comparison procedures for marginal means (post hoc tests), F tests, and homogeneous subsets were performed for all treatment groups. Data were expressed as mean ± SE, and a P level of less than 0.05 was considered statistically significant.
Results
Protein expression.
Immunofluorescent labeling was performed to assess expression levels of EGF, TGFα, and EGFR. After treatment of embryos derived through natural mating with gene-specific siRNA, expression of EGF, TGFα, and EGFR were significantly decreased at all stages of development in each treatment group compared with controls (P < 0.01) (Table 1, Figures 1, 2 A, C). Protein expression levels after combined treatment with siRNA duplexes to EGF and TGFα together were not significantly different from those after treatment of EGF and TGFα individually (Table 1; Figure 1). In addition, expression levels after treatment with siRNAs were similar to expression levels in cloned embryos (Figures 1 and 2 A, B). Furthermore, levels of expression of EGF, TGFα, and EGFR were greater in late-stage than early-stage blastocysts (P < 0.01), although they were still significantly lower than those of control embryos at the same stage (P < 0.05) (Table 1, Figures 1 and 2 A, C). The effects observed were gene-specific, because expression levels of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase did not differ among embryos in the treatment, control, and cloned embryo groups (Figure 2 D).
Table 1.
Fluorescence intensity (lux; mean ± SE) and percentage of expression of EGF, TGFα, and EGFR.
| Embryos derived through natural mating |
Cloned embryos | ||||||
| Control treatments |
siRNA treatments |
||||||
| Embryonic stage | Positive | Negative | EGF | TGFα | EGFR | EGF + TGFα | Untreated |
| 1-cell | 59.3 ± 0.4a (100%) | 58.9 ± 0.6a (100%) | 42.7 ± 0.5b (72%) | 41.8 ± 1.1b (71%) | 42.6 ± 0.6b (73%) | 41.2 ± 0.7b (70%) | 57.9 ± 0.9a (98%) |
| 2-cell | 47.1 ± 0.6b (100%) | 47.2 ± 0.8b (100%) | 17.2 ± 0.7c (36%) | 16.6 ± 0.9c (35%) | 17.4 ± 0.7c (37%) | 16.3 ± 0.7c (35%) | 17.7 ± 0.6c (38%) |
| Morula | 67.5 ± 0.6d (100%) | 67.6 ± 0.7d (100%) | 32.1 ± 0.9e (48%) | 31.4 ± 0.6e (46%) | 31.7 ± 0.5e (47%) | 30.7 ± 0.6e (45%) | 34.5 ± 0.8e (51%) |
| Early blastocyst | 81.0 ± 1.0f (100%) | 80.9 ± 1.0f (100%) | 41.1 ± 0.7b (51%) | 40.5 ± 1.0b (50%) | 41.6 ± 0.7b (51%) | 39.8 ± 1.2b (49%) | 43.3 ± 1.0b (53%) |
| Late blastocyst | 84.4 ± 0.4g (100%) | 84.6 ± 0.5g (100%) | 67.5 ± 0.5d (80%) | 66.2 ± 0.6d (78%) | 66.8 ± 1.1d (79%) | 65.5 ± 0.6d (77%) | 64.8 ± 0.9d (77%) |
Means with different superscripts in the columns and rows are significantly different (P < 0.01) from values for control treatments. Percentages are relative to values for control treatments (positive, transfection with non-inhibitory siRNA; negative, no transfection or injection of PBS).
Figure 1.
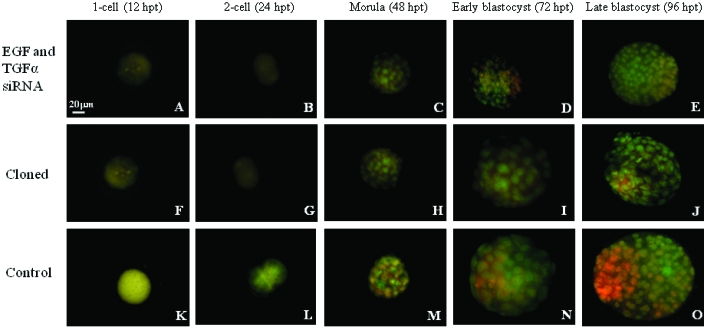
Representative immunofluorescent assay after combined (EGF, green; TGFα, red; colocalization, yellow) transfection of sequence-specific siRNA in 1-cell, 2-cell, morula, early blastocyst, and late blastocyst embryos harvested at 12, 24, 48, 72, and 96 h after transfection (hpt), respectively. (A through E), Transfected embryos derived from natural mating. (F through J) Untransfected, cloned embryos. (K through O) Mock-transfected embryos derived through natural mating. Magnification, ×630.
Figure 2.

Immunoblotting. EGF, TGFα, and EGFR protein expression after treatment with siRNA. (A) Embryos derived from natural mating. (B) Untransfected, cloned embryos. (C) Mock-transfected embryos derived through natural mating. Lanes: 1, 1-cell; 2, 2-cell; 3, morula; 4, early blastocyst; 5, late blastocyst. (D) Expression of glyceraldehyde-3-phosphate dehydrogenase after treatment of embryos derived through natural mating or cloned embryos with siRNA to EGF (lane a), TGFα (lane b), and EGFR (lane c) and mock-transfected embryos derived through natural mating. All embryos harvested at late blastocyst stage. Magnification, ×630.
Embryo development.
We assessed the developmental competence of siRNA-treated embryos. Significantly fewer 1-cell embryos developed to blastocysts after treatment with specific siRNAs to either EGF (16.0 ± 1.0 blastocysts) or TGFα (16.2 ± 0.8 blastocysts), compared with positive (25.4 ± 1.7 blastocysts) and negative (25.6 ± 2.3 blastocysts) controls (P < 0.01 for both comparisons). These results were enhanced by treatment with siRNA to EGFR (11.8 ± 1.5 blastocysts) and with siRNA to both EGF and TGFα simultaneously (11.4 ± 1.3 blastocysts). Cloned embryos showed similar development to that of embryos derived through natural mating and treated with siRNA to either ERGR or EGF and TGFα together (Table 2). Embryos that failed to develop beyond the 1-cell, 2-cell, and compact morula stages were shrunken, and individual cellular nuclei were disintegrated with fragmentation of chromatin (data not shown). In contrast, surviving embryos that developed to blastocysts were not morphologically different from control embryos (Figures 1 and 3).
Table 2.
Development of 1-cell embryos to blastocysts
| No. of embryos (mean ± SE) |
|||||||
| Controls |
siRNA treatments |
||||||
| Embryos | Positive | Negative | EGF | TGF-α | EGF-R | EGF + TGF-α | |
| Natural mating | |||||||
| 1-cell | 29.4 ± 2.0 | 29.8 ± 1.9 | 29.2 ± 1.6 | 30.0 ± 1.5 | 28.6 ± 1.6 | 30.0 ± 2.0 | |
| Blastocyst | 25.4 ± 1.7a (86%) | 25.6 ± 2.3a (86%) | 16.0 ± 1.0b (55%)* | 16.2 ± 0.8b (54%)* | 11.8 ± 1.5c (38%)† | 11.4 ± 1.3c (38%)† | |
| Cloned | |||||||
| 1-cell | not done | 28.3 ± 1.3 | not done | not done | not done | not done | |
| Blastocyst | not done | 10.0 ± 0.3 (35%) | not done | not done | not done | not done | |
Means with different superscripts in the columns and rows are significantly different (P < 0.01) from control treatments. Percentages are relative to values for control treatments (positive, transfection with non-inhibitory siRNA; negative, no transfection or injection of PBS). * (P < 0.05) and †(P < 0.01) indicate significant differences from control treatments.
Figure 3.
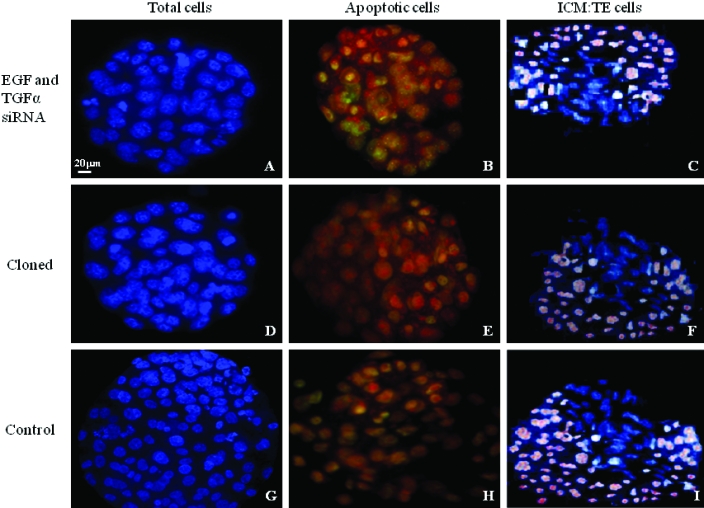
Representative immunofluorescent assay of blastocysts after combined transfection of embryos with siRNA to EGF and TGFα. All cells, blue; apoptotic cells, yellow; ratio of ICM (blue):TE cells (pink to lavender), respectively. (A through C) Transfected embryos derived through natural mating. (D through F), Untreated cloned embryos. (G through I) Mock-transfected embryos derived through natural mating. Magnification, ×630.
Apoptosis.
To assess the influence of siRNA treatment on the apoptotic index, we determined the number of apoptotic cells in treated embryos by using terminal dUTP nick-end labeling. Apoptotic nuclei were detected in all embryos after treatment with siRNA to EGF, TGFα, and EGFR (Table 3, Figure 3). Compared with the mean number of apoptotic cells in control embryos (4.9 ± 0.8 cells), the mean number of apoptotic cells was significantly greater after treatment with siRNA to EGF (9.0 ± 0.7 cells), TGFα (9.2 ± 0.8 cells), EGFR (11.0 ± 0.9 cells), and both EGF and TGFα together (11.2 ± 0.8 cells) (P < 0.01). In comparison, the mean number and percentage of apoptotic cells were similar between untreated cloned embryos and embryos derived through natural mating and treated with siRNA to either EGFR or EGF and TGFα together (Table 3, Figure 3).
Table 3.
Assessment of embryos (mean ± SE)
| No. of embryos (mean ± SE) |
|||||||
| Assessment |
Controls |
siRNA treatments |
|||||
| Embryos | Positive | Negative | EGF | TGF-α | EGF-R | EGF + TGF-α | |
| Natural mating | |||||||
| Apoptosis | 4.8 ± 1.0 (6%) | 5.0 ± 0.8 (6%) | 9.0 ± 0.7b (14%)* | 9.2 ± 0.8b (15%)* | 11.0 ± 0.9c (19%)† | 11.2 ± 0.8c (19%)† | |
| Cell counting | 74.2 ± 1.4 (100%) | 74.5 ± 2.3 (100%) | 63.0 ± 2.0b (84%)* | 62.8 ± 2.3b (84%)* | 58.8 ± 2.1b (79%)* | 58.4 ± 2.0b (78%)* | |
| ICM:TE ratio | 1:2.67 ± 0.05 | 1:2.66 ± 0.03 | 1:2.84 ± 0.09a | 1:2.83 ± 0.08a | 1:3.10 ± 0.07b,* | 1:3.16 ± 0.04b,* | |
| Cloned | |||||||
| Apoptosis | not done | 11.2 ± 0.4 (18%) | not done | not done | not done | not done | |
| Cell counting | not done | 62.8 ± 2.0 (84%) | not done | not done | not done | not done | |
| ICM:TE ratio | not done | 1:3.18 ± 0.05 | not done | not done | not done | not done | |
Means with different superscripts in the columns and rows are significantly different (P < 0.01) from values for control treatments. Percentages are relative to values for control treatments (positive, transfection with non-inhibitory siRNA; negative, no transfection or injection of PBS). *(P < 0.05) and †(P < 0.01) indicate significant differences from values from control treatments.
Embryo cell counting.
Differential labeling techniques were used to assess the total number of cells and ratio of differentiated cells in blastocysts after siRNA treatment. Compared with those in positive (4.8 ± 1.0 cells) and negative (5.0 ± 0.8 cells) controls, the mean total number of cells was decreased significantly after treatment with siRNA to EGF (63.0 ± 2.0 cells), TGFα (62.8 ± 2.3 cells), EGFR (58.8 ± 2.1 cells), and both EGF and TGFα together (58.4 ± 2.0 cells; P < 0.05 for all comparisons; Table 3, Figure 3). These results were similar to the number of cells measured in cloned embryos (63.0 ± 1.6 cells). In addition, the mean ICM:TE ratio after siRNA treatment differed significantly for EGFR (1:3.10 ± 0.07) and combined EGF and TGFα (1:3.16 ± 0.04) as compared with control treatment (1:2.66 ± 0.03) (P < 0.05). However treatment with EGF (1:2.84 ± 0.09) or TGFα (1:2.83 ± 0.08) had no significant effect. The mean ICM:TE ratio in cloned embryos (1:3.18 ± 0.05) was similar to that after downregulation of EGFR and both EGF and TGFα expression (Table 3, Figure 3).
Discussion
In the present study, we demonstrated that sequence-specific siRNAs for EGF, TGFα, and EGFR decreased the expression of maternally inherited and embryonically expressed genes in preimplantation mouse embryos. siRNA microinjected into the cytoplasm of 1-cell embryos, markedly inhibited the expression of growth factor genes and that of the cognate receptor immediately after transfection. Peak suppression of gene expression occurred at the 2-cell stage and decreased gradually yet progressively until returning to control levels by the late-blastocyst stage. These results indicate that siRNA-mediated RNAi effectively ‘knocks down’ both maternally inherited and embryonic gene transcripts. Previous studies have shown successful gene silencing by RNAi by using lipofection,13 a vector-based approach,7,31 electroporation,8,18 and by microinjection.38 Further, our results are consistent with previous studies that RNAi transiently yet effectively inhibited mRNAs recruited during oocyte maturation as well as embryonically expressed genes in early mouse embryos.21,39
In the present study, the observed differences in downregulation of gene expression possibly were due to a switch from maternal to embryonic expression and to the instability of mRNA, particularly at the transition between the 1- and 2-cell stages. The degradation of specific transcripts of maternal mRNA was already underway in 1-cell embryos and maximized by the 2-cell stage. These findings support previous results suggesting that early cleavage-stage embryos rely on maternally synthesized proteins from mRNA during oogenesis23 before embryonic expression initiates. Toward the end of the 2-cell stage, the first wave of gene activation (zygote genome activation)26,33,34 occurs and maternal mRNA continues to degrade as embryonic mRNA transcription and protein synthesis take over.43 The second wave of embryonic gene activation (midpreimplantation genome activation) precedes the morula stage.19,20,29 The slow recovery of gene expression in late blastocyst that followed midpreimplantation genome activation perhaps indicates increases in the numbers of endogenous embryonic transcripts and transient downregulation of growth factors gene expression due to degradation of siRNA. These effects of siRNA treatment were highly gene-specific, in that expression of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase never varied among groups.
In comparison, gene expression of EGF, TGFα, and EGFR after treatment with siRNAs specific to those genes was similar between ICNI-derived, cloned mouse embryos and those due to natural mating. In our previous study,10 we compared the pattern, timing, and level of expression of these same growth factors in ICNI-derived cloned embryos and those of embryos derived in vivo and embryos fertilized in vitro. We found that expression of growth factors occurred first in 1-cell stage embryos and then steadily decreased in 2-cell stage embryos before progressively increasing in compact morula and blastocysts, similar to findings from in-vivo–derived and in-vitro–fertilized embryos. However, expression levels of EGF, TGFα, and EGFR in cloned mouse embryos were less than in embryos generated either by natural mating or in vitro fertilization. Therefore, the accumulated evidence suggests that incomplete or inappropriate epigenetic reprogramming of donor nuclei likely is the primary cause of failures in cloning36 and alters the expression of various genes and transcription factors, such as H19, IGF2R, and Oct4 in cloned embryos.3
Although allele-specific imprints are resistant to genomic-wide demethylation during preimplantation development and imprinted genes of somatic nuclei remain largely unaffected,42 some instances do exist in which imprinted genes are affected and lead to fetal and placenta growth anomalies.37 The change in expression is associated with a loss of DNA methylation involved in the regulation of gene imprinting.46 Similarly, recent studies support our previous studies10 that cloned preimplantation mouse embryos show correct timing but altered levels of gene expression and suggest that genome reprogramming is incomplete.35 In contrast, successful cloning can be expected to occur through appropriate recapitulation of a normal embryonic pattern of gene expression.29 Further, failure to express any of the imprinted genes indicates that epigenetic information associated with imprinted genes is not faithfully retained in cloned embryos.
In the present study, we demonstrated that downregulation of EGF, TGFα, and EGFR markedly increased apoptosis, decreased the embryo development rate and the numbers and ratios of ICM and TE cells, and negatively affected preimplantation embryo development and survival. These results support our hypothesis that RNAi acts in the mouse by either inducing degradation of targeted mRNA or inhibiting its translation.31 Either way, the consequence of these effects is the disruption of normal control and regulation of the expression of targeted genes. In comparison, the changes in the ICNI-derived cloned mouse embryos were very similar to those in embryos derived by natural mating after ablation of EGF, TGFα, and EGFR expression. These results are consistent with our earlier study,10 in which cloned mouse embryos showed decreases in the rate of blastocyst development, numbers and ratio of ICM and TE cells, and total number of differentiated cells and increases in the frequency of apoptotic nuclei consistent with those of naturally derived embryos after combined treatment, with immunoneutralizing antibodies to EFG and TGFα.
Our earlier study tested whether in vitro supplementation of exogenous EGF and TGFα has beneficial effects on the development of cloned embryos.12 Those results demonstrated that supplementation with mitogenic growth factors significantly increased blastocyst formation, the total number of cells, and the ICM:TE cell ratio in cloned embryos. Therefore, our previous results combined with those from the present study indicate that mitogenic growth factors play key regulatory roles in cell proliferation and differentiation.
Delay or interruption of key genes involved in cell growth and differentiation ultimately activates programmed cell death.34 This observation is supported further by other studies indicating that maternally inherited and embryonically expressed mitogenic growth factors play crucial stimulatory and regulatory roles in oocyte maturation and the development of preimplantation embryo.1,20 In particular, EGF and TGFα are thought to play key regulatory roles in the development of preimplantation embryos by promoting nuclear and cytoplasmic maturation, optimizing cellular cleavage, and synchronizing embryonic and maternal maturation to ensure successful implantation.3,13,22 Thus EGF and TGFα are implicated as important factors for embryo survival.3,11
We conclude that siRNA efficiently downregulates maternally inherited and embryonically expressed endogenous EGF and TGFα and their receptor, EGFR. Gene downregulation through RNAi decreases gene expression, thus impairing survival and delaying preimplantation embryo development. Therefore, by nature of its specificity and the timing of inhibition, posttranscriptional gene silencing is an efficient and appropriate tool not only for functional studies of individual genes but also for genome-wide screening. Dysregulation of the expression of these growth factors may explain, at least in part, the poor developmental potential of cloned mammalian embryos. The present study confirms our earlier work,12 in which in vitro supplementation of growth factors improved the maturation and survival of cloned embryos. Further, our previous findings combined with the present results suggest that mitogenic growth factors can support nuclear reprogramming and enhance cloning efficiency in mice.
Acknowledgments
This work was supported in part by a U42 award (no. RR014905) from the National Center for Research Resources, National Institutes of Health, to establish the Mutant Mouse Regional Resource Center (MMRRC) at UC Davis and by UC Davis Mouse Biology Program funds.
References
- 1.Altschul SF, Gish W, miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2.Baker CH, Kedar D, McCarty MF, Tsan R, Weber KL, Bucana CD, Fidler IJ. 2002. Blockade of epidermal growth factor receptor signaling on tumor cells and tumor-associated endothelial cells for therapy of human carcinomas. Am J Pathol 161:929–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boiani M, Eckardt S, Scholer HR, McLaughlin KJ. 2002. Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev 16:1209–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brison DR. 2000. Apoptosis in mammalian preimplantation embryos: regulation by survival factors. Hum Fertil (Camb) 3:36–47 [DOI] [PubMed] [Google Scholar]
- 5.Brison DR, Schultz RM. 1997. Apoptosis during mouse blastocyst formation: evidence for a role for survival factors including transforming growth factor α. Biol Reprod 56:1088–1096 [DOI] [PubMed] [Google Scholar]
- 6.Brison DR, Schultz RM. 1998. Increased incidence in transforming growth factor-α–deficient mouse blastocysts. Biol Reprod 59:136–144 [DOI] [PubMed] [Google Scholar]
- 7.Brummelkamp TR, Bernards R, Agami R. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2:243–247 [DOI] [PubMed] [Google Scholar]
- 8.Calegari F, Marzesco AM, Kittler R, Buchhalz F, Huttner WB. 2004. Tissue-specific RNA interference in postimplantation mouse embryos using directional electroporation and whole embryo culture. Differentiation 72:92–102 [DOI] [PubMed] [Google Scholar]
- 9.Collins MK, Perkins GR, Rodriguez-Tarduchy G, Nieto MA, Lopez-Rivas A. 1994. Growth factors as survival factors: regulation of apoptosis. Bioessays 16:133–138 [DOI] [PubMed] [Google Scholar]
- 10.Dadi TD, Li MW, Lloyd KC. 2004. Expression levels of EGF, TGFα, and EGFR are significantly reduced in preimplantation cloned mouse embryos. Cloning Stem Cells 6:267–283 [DOI] [PubMed] [Google Scholar]
- 11.Dadi TD, Li MW, Lloyd KC. 2006. Development of mouse embryos after immunoneuralization of mitogenic growth factors mimics that of cloned embryos. Comp Med 56:188–195 [PubMed] [Google Scholar]
- 12.Dadi TD, Li MW, Lloyd KC. 2007. EGF and TGFα supplementation enhances development of cloned mouse embryos. Cloning Stem Cells 9:315–326 [DOI] [PubMed] [Google Scholar]
- 13.Dalby B, Cates S, Harris A, Ohki EC, Tilkins ML, Price PJ, Ciccarone VC. 2004. Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods 33:95–103 [DOI] [PubMed] [Google Scholar]
- 14.Diaz-Cueto L, Gerton GL. 2001. The influence of growth factors on the development of preimplantation mammalian embryos. Arch Med Res 32:619–626 [DOI] [PubMed] [Google Scholar]
- 15.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494–498 [DOI] [PubMed] [Google Scholar]
- 16.Fire A. 1999. RNA-triggered gene silencing. Trends Genet 15:358–363 [DOI] [PubMed] [Google Scholar]
- 17.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811 [DOI] [PubMed] [Google Scholar]
- 18.Grabarek JB, Pulsa B, Glover DM, Zemicka-Goetz M. 2002. Efficient delivery of dsRNA into zona-enclosed mouse oocytes and preimplantation embryos by electroporation. Genesis 32:269–276 [DOI] [PubMed] [Google Scholar]
- 19.Hamatani T, Carter MG, Sharov AA, Ko SH. 2004. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 6:117–131 [DOI] [PubMed] [Google Scholar]
- 20.Hamatani T, Ko SH, Yamada M, Kuji N, Mizusawi Y, Shoji M, Hada T, Asada H, Maruyama T, Yoshimura Y. 2006. Global gene expression profiling of preimplantation embryos. Hum Cell 19:98–117 [DOI] [PubMed] [Google Scholar]
- 21.Haraguchi S, Saga Y, Naito K, Inoue H, Seto A. 2004. Specific gene silencing in the pre-implantation stage mouse embryo by an siRNA expression vector system. Mol Reprod Dev 68:17–24 [DOI] [PubMed] [Google Scholar]
- 22.Kaye PL. 1997. Preimplantation growth factor physiology. Rev Reprod 2:121–127 [DOI] [PubMed] [Google Scholar]
- 23.Kidder GM. 1992. The genetic program for pre-implantation development. Dev Genet 13:319–325 [DOI] [PubMed] [Google Scholar]
- 24.Kim MH, Yuan X, Okumura S, Ishikawa F. 2002. Successful inactivation of endogenous cct 3/4 and c-mos genes in mouse preimplantation embryos and oocytes using short interfering RNAs. Biochem Biophys Res Commun 296:1372–1377 [DOI] [PubMed] [Google Scholar]
- 25.Klonisch T, Wolf P, Hombash-Klonisch S, Vogt S, Kuechenhoff A, Tentens F, Fischer B. 2001. Epidermal growth factor-like ligands and ErbB genes in the periimplantation rabbit uterus and blastocyst. Biol Reprod 64:1835–1844 [DOI] [PubMed] [Google Scholar]
- 26.Latham KE. 1999. Mechanisms and control of embryonic genome activation in mammalian embryos. Int Rev Cytol 193:71–124 [DOI] [PubMed] [Google Scholar]
- 27.Lee DC, Fenton SE, Berkowitz EA, Hissong MA. 1995. Transforming growth factor α: expression, regulation, and biological activities. Pharmacol Rev Mar 47:51–85 [PubMed] [Google Scholar]
- 28.Mann MR, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS. 2003. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod 69:902–914 [DOI] [PubMed] [Google Scholar]
- 29.Minami N, Suzuki T, Tsukamoto S. 2007. Zygotic gene activation and maternal factors in mammals. J Reprod Dev 53:707–715 [DOI] [PubMed] [Google Scholar]
- 30.Miyagishi M, Taira K. 2002. Development and application of siRNA expression vector. Nucleic Acids Res Suppl 2:113–114 [DOI] [PubMed] [Google Scholar]
- 31.Ohgane J, Wakayama T, Kogo Y, Senda S, Hattori N, Tanaka S, Yanagimachi R, Shiota K. 2001. DNA methylation variation in cloned mice. Genesis 30:45–50 [DOI] [PubMed] [Google Scholar]
- 32.Reik W, Dean W, Walter J. 2001. Epigenetic reprogramming in mammalian development. Science 293:1089–1093 [DOI] [PubMed] [Google Scholar]
- 33.Schultz RM. 1993. Regulation of zygotic gene activation in the mouse. Bioessays 15:531–538 [DOI] [PubMed] [Google Scholar]
- 34.Schultz RM, Davis W, Jr, Stein P, Svoboda P. 1999. Reprogramming of gene expression during preimplantation development. J Exp Zool 285:276–282 [DOI] [PubMed] [Google Scholar]
- 35.Sebastiano V, Gentile L, Garagna S, Rdi CA, Zuccotti M. 2005. Cloned preimplantation mouse embryos show correct timing but altered levels of gene expression. Mol Reprod Dev 70:146–154 [DOI] [PubMed] [Google Scholar]
- 36.Shi W, Zakhartchenko V, Wolf E. 2003. Epigenetic reprogramming in mammalian nuclear transfer. Differentiation 71:91–113 [DOI] [PubMed] [Google Scholar]
- 37.Solter D. 2000. Mammalian cloning: advances and limitations. Nat Rev Genet 1:199–207 [DOI] [PubMed] [Google Scholar]
- 38.Stein P, Svoboda P, Anger M, Schultz RM. 2003. RNAi: mammalian oocytes do it without RNA-dependent RNA polymerase. RNA 9:187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svaboda P, Stein P, Hayashi H, Schultz RM. 2000. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development 127:4147–4156 [DOI] [PubMed] [Google Scholar]
- 40.Svoboda P, Stein P, Schultz RM. 2001. RNAi in mouse oocytes and pre-implantation embryos: effectiveness of hairpin dsRNA. Biochem Biophys Res Commun 287:1099–1104 [DOI] [PubMed] [Google Scholar]
- 41.Thouas GA, Korfiatis NA, French AJ, Jones GM, Trounson AO. 2001. Simplified technique for differential staining of inner cell mass and trophectoderm cells of mouse and bovine blastocysts. Reprod Biomed Online 3:25–29 [DOI] [PubMed] [Google Scholar]
- 42.Tremblay KD, Saam JR, Ingrm RS, Tilghman SM, Bartolomei MS. 1995. A parental-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet 9:407–413 [DOI] [PubMed] [Google Scholar]
- 43.Wianny F, Zernicka-Goetz M. 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol 2:70–75 [DOI] [PubMed] [Google Scholar]
- 44.Wiley LM, Wu JX, Harare I, Adamson ED. 1992. Epidermal growth factor receptor mRNA and protein increase after the 4-cell preimplantation stage in murine development. Dev Biol 149:247–260 [DOI] [PubMed] [Google Scholar]
- 45.Wu W, Hodges E, Redelius J, Hoog C. 2004. A novel approach for evaluating the efficiency of siRNAs on protein levels in cultured cells. Nucleic Acids Res 32:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young LE, Fernandez K, McEvoy TG, Butterwith SC, Gutierrez CG, Carolan C, Broadbent PJ, Robinson JJ, Wilmut I, Sinclair KD. 2001. Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet 27:153–154 [DOI] [PubMed] [Google Scholar]
- 47.Young LE, Sinclair KD, Wilmut I. 1998. Large offspring syndrome in cattle and sheep. Rev Reprod 3:155–163 [DOI] [PubMed] [Google Scholar]


