Abstract
We performed a cross-sectional study to estimate the prevalence of 2 gamma-2-herpesviruses, rhesus rhadinovirus (RRV) and retroperitoneal fibromatosis herpesvirus (RFHV), in breeding colonies of rhesus macaques. Of 90 animals selected for sampling, 73 (81%) were positive for RRV, which was detected only in blood in 22 (24%), only in saliva in 15 (16%), and in both blood and saliva in 36 (40%). Detection of RRV DNA in blood and saliva was significantly higher in animals younger than 2 y. In comparison, RFHV was detected in 40 (44%) of the 90 animals: only in blood in 5 (6%), only in saliva in 26 (29%), and in both blood and saliva in 9 (10%). Dual infection was detected in 38 (42%) animals; RFHV was only detected in coinfections. The mean RRV genome copy number in blood was significantly higher than that for RFHV. Age was a significant predictor of RRV copy number in blood and RFHV copy number in saliva. Of the 90 animals, 88 (98%) were positive for rhadinoviral antibodies on an immunofluorescent assay. Both RRV and RFHV are highly endemic in socially housed breeding colonies of rhesus macaques, and their patterns of infection are similar to that for the betaherpesvirus rhesus cytomegalovirus.
Abbreviations: CNPRC, California National Primate Research Center; GE, genome equivalents (copy number); KSHV, Kaposi sarcoma-associated herpesvirus; RFHV, retroperitoneal fibromatosis herpesvirus; RRV, rhesus rhadinovirus; OSM, oncostatin M gene
The Rhadinovirus genus of gamma-2-herpesviruses is divided into 2 subgroups, RV1 and RV2, based on genomic sequence comparisons.36,44 Rhadinovirus infections are generally subclinical in immunocompetent natural hosts, and overt disease is thought to arise only when hosts are immunocompromised.28 In addition, the ability to establish both lytic and latent infections, a hallmark of the Herpesviridae family, occurs during rhadinovirus infections.1,43 The RV1 subgroup includes Kaposi sarcoma-associated herpesvirus (KSHV; also referred to as human herpesvirus 8)12,32 the causative agent of Kaposi sarcoma, an angioproliferative lesion composed of a mixed population of endothelial, inflammatory and spindle cells.19,24 Furthermore, KSHV has been linked etiologically to 2 different B-cell lymphomas: primary effusion lymphoma and multicentric Castleman disease.17 Retroperitoneal fibromatosis herpesvirus (RFHV) is also a member of the RV1 subgroup and is thought to be the macaque homolog of KSHV.4,8,14,36,37,40 DNA sequences specific for RFHV have been detected in retroperitoneal fibromatosis in macaques coinfected with the potentially immunosuppressive simian betaretrovirus type 2.7 Histologic similarities between retroperitoneal fibromatosis and KS lesions seen in humans coinfected with KSHV and HIV have been previously described.7,9,21,37 During outbreaks of simian betaretrovirus type 2 disease at 2 national primate research centers in the 1980s, the incidence of retroperitoneal fibromatosis was reported to be 5% to 7% for animals younger than 2 y and 1% across all age groups.7,37,45 Since the end of these outbreaks in the late 1980s, retroperitoneal fibromatosis has occurred only rarely in primate colonies. The majority of published RFHV studies have focused on animals with recognized retroperitoneal fibromatosis lesions.9-11 However, RFHV has proven extremely difficult to isolate and, to date, has not been propagated successfully in vitro, and only a small portion of the RFHV genome has been sequenced.36,37,40,44 In this study we determined the prevalence of RFHV infection in nondiseased animals and address aspects of the natural history of this virus infection in captive macaque populations.
Rhesus rhadinovirus (RRV) is a member of the RV2 subgroup, which naturally infects rhesus macaques.15,38,44 RRV was isolated independently at 2 national primate research centers in the late 1990s from rhesus macaques.15,42 Both RRV isolates were shown to have noteworthy sequence similarity to KSHV and RFHV.2,8,15,42 Unlike RFHV, RRV can be propagated readily in vitro, thus facilitating studies of the lytic replication cycle.5,6,16 Experimental coinfection of rhesus macaques with SIV and RRV resulted in a lymphoproliferative disease resembling multicentric Castleman disease, but variations in disease outcome between the 2 RRV isolates were noted.30,49 More recently, RRV has been shown to be associated with nonHodgkin lymphoma and retroperitoneal fibromatosis in SIV-infected rhesus macaques.34 Therefore, RRV infection in macaques is a highly useful animal model for the study of KSHV infection in humans, including studies of viral pathogenesis, factors affecting prevalence of infection, viral shedding, and transmission.2,25,31,42 In addition, RRV is a persistent virus targeted for elimination in some specific pathogen free (SPF) macaque breeding populations. A better understanding of the natural history of RRV and RFHV infections will lead to improved characterization of host–virus interactions, contribute to the refinement of these nonhuman primate models, and allow more efficient management of SPF colonies.
Here we report estimates of the prevalence of viremia and oral shedding of RRV and RFHV in large age-structured breeding colonies of rhesus macaques. Both viruses were highly endemic in the breeding populations we tested, and coinfection with both viruses was common.
Materials and Methods
Animals.
The California National Primate Research Center (CNPRC) currently maintains 14 half-acre conventional breeding corrals each housing 80 to 120 nonSPF rhesus macaques in large social groups. Other similar corrals housing SPF rhesus macaque breeding groups were not included in this study. All monkeys housed in breeding corrals are examined every 3 mo, at which time animals are given a physical exam, weighed, and skin-tested for tuberculosis, and new offspring are tattooed with a unique animal identification number. An extensive database, accessible by animal identification number, is maintained for all animals at the CNPRC and contains diverse physical and biologic information, including date of birth, gender, current and previous housing locations, pedigree and genetic profile, clinical data, and immunization records. Animals included in the present study were selected by applying a systematic sampling method to sampling frames generated from the primate center database and containing a list of all animals in 3 different corrals. Collection of heparinized blood and saliva samples was incorporated into the quarterly routine examination process for the completion of this study. The CNPRC is fully AAALAC-accredited. All animals are maintained in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals and the Animal Welfare Act. Sample collection procedures were performed according to a protocol approved by our institutional animal care and use committee.23
Sampling scheme.
Corrals 8, 13, and 15 were chosen at random from among the 14 available for inclusion in this study. These breeding corrals are established as closed populations. Adjacent corrals (for example, 13 and 15) are separated by 75 ft. Corral 8 is located in a separate row of enclosures more than 100 yards from the other sampled corrals. By using the CNPRC database, a list of animal identification numbers, ordered by date of birth, was generated for each corral. Systematic sampling methods were used to calculate the number of animals to be sampled from each of the 3 corrals to estimate the true prevalence of RRV and RFHV with an error bound of 10%.39 Because no previous statistical studies of RRV and RFHV prevalence in captive macaque populations have been published, we used a default estimate of 50% prevalence, which yields the largest calculated sample size for each corral. In addition, a correction for small sample populations (fewer than 120 subjects) was performed as part of the sample size calculation.39 At the time of sampling, the total populations of corrals 8, 13, and 15 were 154, 100, and 37 animals respectively. By using the small-population correction, the appropriate sampling intervals (that is, every kth animal on the list) were calculated to be 4 (corral 8), 3 (corral 13), and 2 (corral 15). According to this sampling design, a total of 39 animals were sampled from corral 8, 33 animals were sampled from corral 13, and 18 animals were sampled from corral 15. Each of the 3 corrals were sampled independently. All conventional breeding corrals are maintained by using identical management and husbandry practices, thereby allowing the findings of this study to be applied to all conventional breeding corrals at CNPRC.
Samples.
Samples used in this study consisted of 3 mL heparinized blood and saliva swabs collected from each animal. Blood samples were collected into vacuum phlebotomy tubes containing heparin (catalog no. 367961, BD, Franklin Lakes, NJ). Saliva was collected by using swabs (catalog no. GD-1000, Salivary Diagnostic Systems, Brooklyn, NY). Blood samples were centrifuged in a standard tabletop centrifuge at approximately 1000 × g for 15 min; plasma was removed, aliquoted, and stored at −20 °C for future antibody testing. DNA was extracted from 200 µL of each saliva and buffy-coat sample by using spin columns (catalog no. 159914, Generation Capture Kit, Qiagen, Valencia, CA).
Determination of cell copy number.
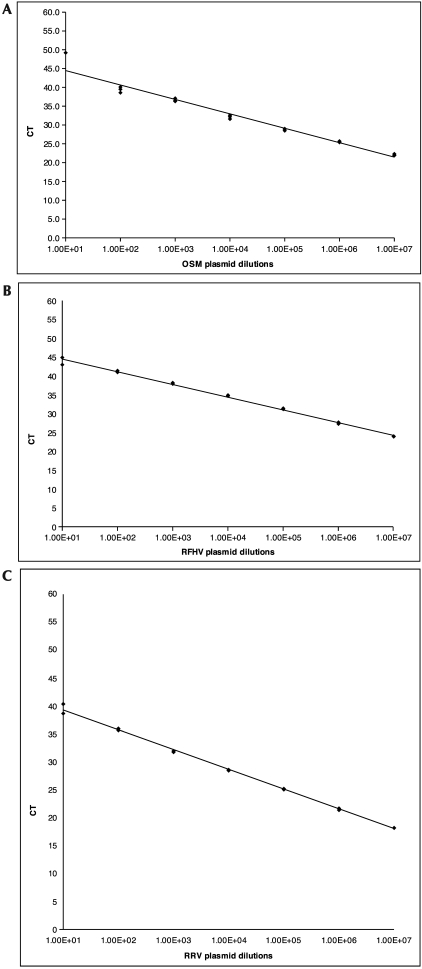
Cell copy number was determined by using the housekeeping gene oncostatin M (OSM).10,29 Because OSM is a diploid gene, real-time PCR could be used for determination of cell number, and a previously published method10 was adapted for use on the TaqMan platform (Applied Biosystems, Foster City, CA). Briefly, DNA from both saliva and buffy-coat samples were tested by using the previously published forward (5′ CCT CGG GCT CAG GAA CAA C 3′) and reverse (5′ GGC CTT CGT GGG CTC AG 3′) primers and the TaqMan probe (5′ VIC–TAC TGC ATG GCC CAG CTG CTG GAC AA–MGB–NFQ 3′) labeled with the fluorescent reporter dye VIC and the nonfluorescent quencher MGB (Applied Biosystems, Foster City, CA).10 Each reaction (volume, 50 µL) contained 0.25 µL of each primer (10 µM), 1 µL of 5µM probe, 25 µL TaqMan Universal master mix(catalog no. 4304437, Applied Biosystems), 7 µL 1× TE buffer, and 15 µL template DNA (approximately 30 ng/µL). Thermocycling conditions consisted of 55 cycles at 50 °C for 2 min, 95 °C for 10 min, 95 °C for 15 s, and 62 °C for 1 min. A plasmid containing the OSM amplicon was diluted in 1× TE and was used to generate a standard curve with an R2 value of 0.9664 (Figure 1A). A relative quantification was performed in which the number of viral (RRV or RFHV) genome copies was divided by the number of cellular genomes (that is, copies of OSM) detected to obtain the number of genome equivalents (GE). By using serial dilutions of plasmid, the detection limit of the real-time PCR assay was determined to be between 10 and 100 copies, with a dynamic range of 106 to 10 copies in a 50-µL reaction. Samples were tested in duplicate, and the cycle threshold values obtained were averaged to determine the cell number tested. Once determined, the number of cells was compared with the calculated virus copy number by using the specific viral standard curve to allow for the calculation of the GE or viral genome copies per number of cells tested in all of the blood samples. Because much of the virus detected in the saliva samples is cell-free, GE values for saliva samples were calculated as number of viral genome copies per volume tested. Given the constraints of the assay system used, we cannot determine the number of infectious virions in each sample.
Figure 1.
Standard curves of real-time PCR assays for the detection of (A) OSM, (B) RFHV, and (C) RRV. CT, cycle threshold. The number of plasmids tested is shown on the x axis.
RFHV PCR.
We used the Applied Biosystems 7900 TaqMan platform to amplify RFHV DNA sequences through real-time PCR. DNA samples from buffy coat and saliva were tested for the presence of RFHV orf7–8 junction sequences as previously described.9 Briefly, 50-µL reactions contained 1 µL (50 µM) each forward (5′ TTA AAG ACA TCT ACG CCC TCC 3′) and reverse (5′ GCC ACC AGG ACG CAG AGC AG 3′) primers and 10 µM probe (5′ FAM– TCA CCT TCA GCT CGG CGA CCA CAA T–TAMARA 3′) labeled with the fluorescent reporter dye FAM and the quencher TAMARA; 25 µL TaqMan Gene Expression master mix (catalog no. 4370048, Applied Biosystems); 12 µL 1× TE buffer; and 10 µL template DNA (approximately 30 ng/µL) . Thermocylcing conditions consisted of 55 cycles of 50 °C for 2 min, 95 °C for 1 min, 95 °C for 15 s, 62 °C for 30 s and 72 °C for 1 min. A plasmid containing the specific RFHV amplicon for this assay was diluted in RFHV-negative monkey DNA to generate a standard curve with an R2 value of 0.9944 (Figure 1 B); the lower limit of detection of the real time PCR assay thus was determined to be between 10 and 100 copies in a 50-µL reaction. All samples were initially tested in duplicate for the detection of RFHV DNA. Samples with discrepant results were repeated in duplicate. Samples were considered positive for RFHV DNA if they tested positive on the initial 2 tests; samples that amplified once, twice, or 3 times in 4 tests were interpreted as indeterminate. Samples that showed no amplification in the initial 2 tests were interpreted as negative for RFHV DNA.
RRV PCR.
Like RFHV, RRV DNA sequences were evaluated by using the Applied Biosystems 7900 real-time PCR TaqMan platform. DNA samples from buffy coat and saliva were tested for the presence of the RRV DNA polymerase gene: forward primer, 5′ TTT AAC CGG CTA TAA CAT CTC AAA CTT 3′; reverse primer, 5′ CCG GTT TTT ATT TTT GTG TAT TCG T 3′; and probe, 5′ FAM– CGA TCT CCC GTA CCT AAT –MGB-NFQ 3′ labeled with the fluorescent reporter dye FAM and the nonfluorescent quencher MGB.5,42 Each 50-µL reaction contained 1 µL each of 20 µM forward primer, reverse primer, and probe; 25 µL TaqMan universal master mix (catalog no. 4304437, Applied Biosystems); 2 µL 1× TE buffer; and 20 µL template DNA (approximately 30 ng/µL). Thermocycling conditions consisted of 55 cycles of 50 °C for 2 min, 95 °C for 10 min, 95 °C for 15 s, and 60 °C for 1 min. This real-time PCR assay was standardized by using a plasmid containing the RRV polymerase gene diluted in a background of RRV-negative monkey DNA to generate a standard curve with an R2 value of 0.9972 (Figure 1 C). By using serial dilutions of the plasmid, the detection limit of the real-time PCR assay was determined to be between 10 and 100 copies in a 50-µL reaction. All DNA samples initially were run in duplicate for the detection of RRV polymerase gene. Samples with discrepant results between the initial 2 duplicate tests were repeated in duplicate. Samples were considered positive for RRV DNA if they tested positive on the initial 2 tests, samples that amplified once, twice, or 3 times in 4 tests were considered indeterminate, and samples that showed no amplification in duplicate were considered negative for RRV DNA.
Amplified products from both RRV and RFHV real-time PCR assays sent to Qiagen Genomic Services (Hilden, Germany) for single-read sequencing confirmed the specificity of both PCR reactions (data not shown). Samples from each corral were sequenced to control for any variation among corrals.
Immunofluorescent assay.
Plasma samples were tested for the presence of rhadinovirus antibodies by using an immunofluorescent assay as previously described.6,20,22,27 Briefly, telomerized rhesus fibroblasts were grown and infected on 16-well chamber slides. Cells were plated on slides at a concentration of 5.4 × 104cells/well in 200 µL media; 100 µL infectious RRV supernatant was added to 8 of the wells, whereas the other 8 wells received 100 µL uninfectious media to serve as uninfected cell controls. Cells were allowed to reach confluence (approximately 3 d) and fixed in acetone. RRV-positive and negative-plasma were run as controls during each assay. All immunofluorescent assays were evaluated independently by 2 readers blinded to sample identification and PCR status. Any samples that gave discrepant results were repeated in duplicate and rescored by the same 2 readers. Both antibody negative and antibody positive plasma samples were tested on infected and uninfected cells as controls. Plasma samples were considered antibody-positive when they yielded at least 1 fluorescing cell per field (magnification, 400×) and fluorescence was greater than that in the uninfected-cell and negative-plasma controls. For plasma to be considered immunofluorescent-assay–negative, fluorescence had to be no greater than that in the uninfected-cell and negative-plasma controls. Plasma samples yielding results that were weakly positive or not repeatable were interpreted as indeterminate.
Western blot.
Rhesus rhadinovirus strain 17577 was purified essentially as described.49 Purified virus was solubilized in RIPA buffer with protease inhibitors and the protein concentration was determined by Bradford analysis. Approximately 25 µg protein was resolved on a denaturing 10% polyacrylamide gel and transferred to a nylon membrane. The membrane was rinsed in TBST (8% NaCl, 3% Tris, 0.2% KCl, 0.1% Tween 20, pH 7.6), soaked in TBST supplemented with 5% nonfat milk for 1 h at room temperature to block nonspecific binding sites and then incubated with serum samples (diluted 1:100 in fresh TBST with 5% nonfat milk) overnight at 4 °C. The samples were removed the following day, and the membrane was washed extensively in TBST buffer. The membrane was incubated with secondary antibody, goat antirhesus IgG conjugated with horseradish peroxidase (Southern Biotech, Birmingham, AL) diluted 1:8000 in TBST with 5% nonfat milk, for 1 h at room temperature and proteins detected by chemiluminescence. A set of 6 randomly selected immunofluorescence-positive samples (2 from each corral) as well as 6 SPF samples were run on an RRV-specific Western blot to confirm reaction to specific viral proteins.
Statistical analysis.
The proportions of RRV- and RFHV-positive animals in each corral were compared to determine whether housing location was a significant determinant of prevalence for either virus. The 95% confidence intervals for the proportions of RRV and RFHV evaluated overlapped between cages, indicating that location was not a significant factor and that all animals could be evaluated as a single group. Results regarding distribution of animals by age and gender were statistically analyzed to determine which covariates are most influential in RRV and RFHV prevalence. The Cochran Q test was used to assess equality of proportions in matched samples to compare RRV, RFHV, and coinfection outcomes by using STATA 9.2.26 The t test for populations with unpaired and unequal variances was used to compare the copy number values between RRV and RFHV in both blood and saliva in STATA 9.2. The nonparametric one-way analysis of variance was performed using SAS 9.1 to determine the significance of age group as a predictor for viral copy number.
Results
The distribution by age and gender of all rhesus macaques in the current study is summarized in Table 1.
Table 1.
Distribution by age and gender of 90 rhesus macaques randomly selected for sampling from 3 outdoor breeding corrals
| Corral |
|||||
| Age group (y) | Gender | NC8 | NC13 | NC15 | Total |
| 0 to <1.6 | Female | 4 | 2 | 1 | 16 |
| Male | 1 | 3 | 5 | ||
| 1.6 to <2 | Female | 3 | 12 | 0 | 23 |
| Male | 0 | 8 | 0 | ||
| 2 to <3 | Female | 5 | 3 | 0 | 13 |
| Male | 1 | 2 | 2 | ||
| 3 to <5 | Female | 3 | 1 | 1 | 11 |
| Male | 5 | 1 | 0 | ||
| 5 to <9 | Female | 7 | 0 | 4 | 13 |
| Male | 2 | 0 | 0 | ||
| 9 to <25 | Female | 8 | 0 | 5 | 14 |
| Male | 0 | 1 | 0 | ||
| Total | 39 | 33 | 18 | 90 | |
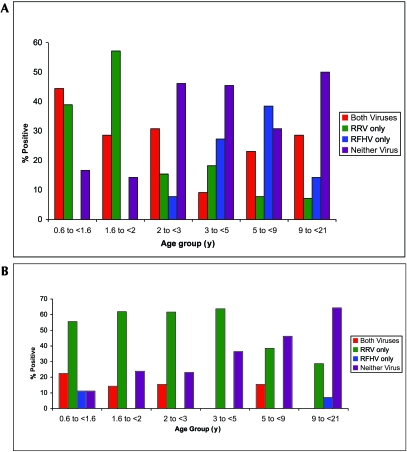
All blood and saliva samples were tested by all 3 real-time PCR assays for the detection of DNA sequences corresponding to OSM, RFHV, and RRV. All samples were positive for OSM sequences, allowing for determination of the number of cells tested in each assay. Of the 90 animals sampled, 73 (81%) were PCR-positive for RRV; RRV-specific DNA was detected only in blood in 22 (24%), only in saliva in 15 (16%), and in both blood and saliva in 36 (40%) macaques. The prevalence of RRV-specific DNA detection was significantly (P = 0.001; Fisher exact probability test) higher in animals younger than 2 y compared with animals 3 y or older (age groups 1 and 2 compared to age groups 4 through 6). Of the 90 animals tested, 40 (44%) carried RFHV-specific DNA; RFHV-specific DNA was detected only in blood in 5 (6%), only in saliva in 26 (29%), and in both blood and saliva in 9 (10%) of the tested macaques. Combining all age groups, the proportion of animals PCR-positive for RRV (81%) is significantly greater than that of animals PCR-positive for RFHV (40%; P < 0.0001; Cochran Q). Among macaques younger than 3 y, none were PCR-positive for RFHV alone, whereas positivity for RRV alone was detected in 25 (48%) of the macaques tested (P < 0.0001; Cochran Q). For all animals younger than 3 y, the proportion of macaques PCR-positive for both RRV and RFHV was significantly lower than that of animals PCR-positive for RRV (regardless of RFHV status) (P < 0.0001; Cochran Q). No significant differences in these proportions were found for animals 3 y or older (age groups 4 through 6). Coinfection with both RRV and RFHV was detected in 38 (42%) of macaques tested (Figure 2).
Figure 2.
Real-time PCR results for RRV and RFHV by age group in (A) saliva and (B) blood.
Of 90 animals sampled for this cross-sectional survey, 88 (98%) had detectable antibody reactivity in the rhadinovirus immunofluorescent assay (Table 2). Samples from 2 animals from 1 corral (corral 8) showed equivocal or weak reactivity that was not reproducible on repeat testing; these results were considered indeterminate. The high prevalence of rhadinovirus antibody reactivity in the immunofluorescent assay was unexpected and led us to test additional animals for confirmation of testing accuracy.
Table 2.
Rhadinovirus immunofluorescent assay results
| No. tested | No. (%) positive | No. (%) indeterminate | No. (%) negative | |
| No. of study samples | 90 | 88 (98) | 2 (2) | 0 (0) |
| No. of negative controls | 2 | 0 (0) | 0 (0) | 2 (100) |
| No. of positive controls | 1 | 1 (100) | 0 (0) | 0 (0) |
| No. samples from SPF level 1 animals | 43 | 17 (40) | 12 (28) | 14 (33) |
| No. of samples from SPF level 2 animals | 31 | 0 (0) | 0 (0) | 31 (100) |
| Total | 167 | 106 (63) | 14 (8) | 47 (28) |
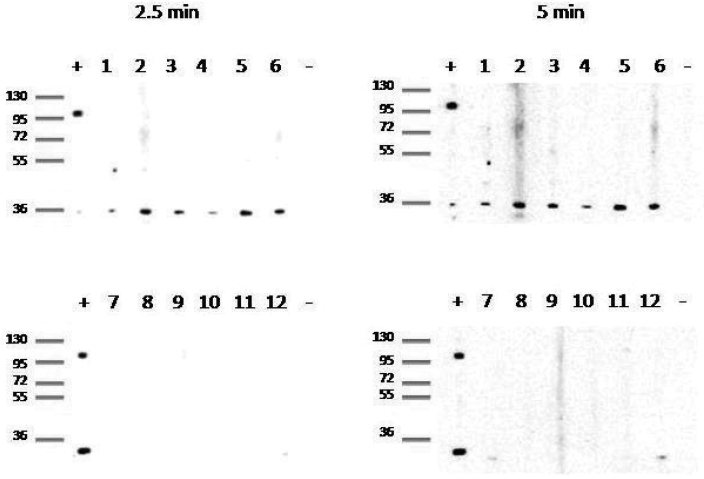
Two additional corrals of different SPF levels (1 and 2) were sampled to confirm the immunofluorescent assay results. Once each quarter, SPF level 1 animals are screened for 4 specific pathogens (simian betaretrovirus, SIV, simian T-cell leukemia virus, and B virus), and SPF level 2 animals are screened for 7 specific pathogens (simian betaretrovirus, SIV, simian T-cell leukemia virus, simian foamy virus, rhesus cytomegalovirus, RRV, and B virus). To obtain comparable samples from these 2 additional corrals, we applied the same systematic sampling method as described earlier. All 31 animals screened by the immunofluorescent assay from SPF level 2 were negative for rhadinovirus antibodies, as expected. Of the 43 animals from SPF level 1 tested, 17 (40%) were positive, 12 (28%) were indeterminate, and 14 (33%) were negative for rhadinovirus antibodies (Table 2). We randomly selected 6 samples (2 from each corral) that were positive by immunofluorescent assay for Western blot testing, which confirmed that the reactivity detected in the rhadinovirus immunofluorescent assay was against specific rhadinovirus proteins and was not nonspecific reactivity. In addition, 6 SPF level 2 samples were tested by Western blot and showed no reactivity to rhadinovirus-specific proteins (Figure 3).
Figure 3.
Western blot results to confirm specificity of immunofluorescent assay. Random samples positive by immunofluorescent assay (numbers 1 and 2 from corral 8, 3 and 4 from corral 13, and 5 and 6 from corral 15) and samples from SPF level 2 animals (numbers 7 through 12) were included. The positive-control sample was from an animal experimentally inoculated with RRV17577, and the negative control was from an SPF animal. Exposure times of 2.5 min (left panels) and 5 min (right panels) were used; and molecular weight standards (kDa) are shown at the far left of each panel. The presence of a band at 36 kDa indicates a positive result.
The mean RRV GE in blood was significantly higher than that for RFHV (P = 0.0003; t test). The mean GE for RRV in saliva did not differ significantly from the RFHV salivary GE (P = 0.0572; t test). No significant differences in GE were present between animals PCR-positive for only 1 virus (RRV or RFHV) compared with animals PCR-positive for both viruses in either blood or saliva. Age was a significant predictor of RRV blood GE (P = 0.0276; nonparametric 1-way ANOVA) and RFHV saliva GE (P = 0.0343; nonparametric 1-way ANOVA), with younger animals having higher GE. However, because of the small numbers of animals in each of these age groups, the statistical power was too low for pairwise testing.
Discussion
Our results indicate that both RRV and RFHV are highly endemic in socially housed rhesus macaques, with RRV approximately twice as prevalent as RFHV. Shedding of both viruses in oral secretions is common, and infection occurs at an early age in a pattern similar to that observed for the betaherpesvirus rhesus cytomegalovirus.46 Younger animals had a higher proportion of PCR positives for both viruses and RRV alone than for RFHV alone. Significant differences in the prevalence of RRV and RFHV were not noted in animals older than 3 y. The increased number of infections and younger age at which animals became infected suggests a higher transmission efficiency for RRV compared with RFHV. The increased GE of RRV in saliva may contribute to the increased transmission efficiency.
Our results confirm that infection with RFHV is common in breeding colonies of rhesus macaques, in which RF disease is rare or absent. Although not sufficient for disease, KSHV is a necessary factor for development of KS,41 and the finding of RFHV without RF in the absence of immunosuppressive cofactors indicates a similar pathogenesis for these 2 closely related viruses. Similar findings were obtained in serologic studies on 178 healthy baboons, which exhibited serologic reactivity to both RV1 (41.5%) and RV2 (68.5%) antigens.48 Despite high levels of antibody reactivity and detectable viral load in peripheral mononuclear cells, the baboons lacked overt clinical signs of disease.47 As with our study in macaques, antibody cross-reactivity between the RV1 and RV2 antigens in baboons confounded type-specific estimates of prevalence.
The GE (viral copy numbers) calculated in our macaque study highlight important areas for further investigation of both RRV and RFHV. Because the current study used samples from only a single time point, we could not assess changes in shedding frequency and GE over time. Extended prospective follow-up of animals over time are underway and will allow analysis of temporally associated variables.
Our immunofluorescent assay was useful for detecting rhadinovirus-specific antibody, but antigenic crossreactivity precluded differentiation of RRV- and RFHV-specific antibodies. No animals were negative for rhadinovirus antibody. The 2 animals with indeterminate antibody results (ages, 15 y and 7 mo; both female) were both PCR-negative for viral sequences in blood and saliva. The unexpectedly high number of positive results (that is, no negative animals) from the rhadinovirus immunofluorescent assay prompted us to test specific-pathogen–free animals to validate the immunofluorescent assay. Level 2 SPF animals, which are repeatedly test-negative for RRV and RFHV in blood, were universally negative by immunofluorescent assay. Level 1 SPF animals, which are not specifically tested for RRV and RFHV but which are derived in a similar fashion to Level 2 animals, showed intermediate rhadinovirus seroprevalence in the immunofluorescent assay. All 6 random samples that were positive by immunofluorescent assay showed reactivity to specific viral proteins in the Western blot assay. Discordant serologic results for the detection of KSHV antibodies limits the use of current serologic testing for diagnostic purposes and highlights the need for improvement of rhadinovirus antibody testing.33,47 Development of improved antibody tests for RRV and RFHV is underway in our laboratory.
Our results indicate that both RRV and RFHV are shed in oral secretions, consistent with findings for KSHV,3,18 which is often found at higher copy numbers in saliva than blood. In addition, oral shedding of virus is thought to be the main route of transmission of KSHV in endemic regions (for example, equatorial Africa), where KSHV infection is more common in women and children than in men.3,35 The high prevalence of RFHV and RRV in young animals in our study is comparable to levels of KSHV infection in endemic regions (36% to 100%).13,18 Close physical contact resulting from normal nonhuman primate group behavior including grooming and aggressive interactions may be significant determinants of transmission for both rhadinoviruses. Future prospective cohort studies will be required to determine whether viral shedding in oral secretions is intermittent and to identify environmental or seasonal influences on shedding frequency.
Coinfection with 2 closely related gamma-2-herpesviruses is common in socially housed rhesus macaques and had not been reported previously. The significance of dual infection with RRV and RFHV in the development of macaque models of human KSHV infection requires further investigation.
Acknowledgments
We would like to thank Ann Rosenthal for her technical assistance, the CNPRC animal care staff, and Research Services technicians. This study was made possible by grant RR00169 from the National Center for Research Resources (NCRR) of the National Institutes of Health (NIH). Portions of this work were supported by NIH grants CA75922 (SWW) and RR00163 (SWW).
References
- 1.Ackermann M, Engels M, Fraefel C, Metzler A, Schwyzer M, Suter M, Tobler K. 2002. Herpesviruses: balance in power and powers imbalanced. Vet Microbiol 86:175–181 [DOI] [PubMed] [Google Scholar]
- 2.Alexander L, Denekamp L, Knapp A, Auerbach MR, Damania B, Desrosiers RC. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J Virol 74:3388–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkharsah KR, Dedicoat M, Blasczyk R, Newton R, Schulz TF. 2007. Influence of HLA alleles on shedding of Kaposi sarcoma-associated herpesvirus in saliva in an African population. J Infect Dis 195:809–816 [DOI] [PubMed] [Google Scholar]
- 4.Auerbach MR, Czajak SC, Johnson WE, Desrosiers RC, Alexander L. 2000. Species specificity of macaque rhadinovirus glycoprotein B sequences. J Virol 74:584–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergquam EP, Avery N, Shiigi SM, Axthelm MK, Wong SW. 1999. Rhesus rhadinovirus establishes a latent infection in B lymphocytes in vivo. J Virol 73:7874–7876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bilello JP, Lang SM, Wang F, Aster JC, Desrosiers RC. 2006. Infection and persistence of rhesus monkey rhadinovirus in immortalized B-cell lines. J Virol 80:3644–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosch ML, Harper E, Schmidt A, Strand KB, Thormahlen S, Thouless ME, Wang Y. 1999. Activation in vivo of retroperitoneal fibromatosis-associated herpesvirus, a simian homologue of human herpesvirus 8. J Gen Virol 80:467–475 [DOI] [PubMed] [Google Scholar]
- 8.Bosch ML, Strand KB, Rose TM. 1998. Gammaherpesvirus sequence comparisons. J Virol 72:845468–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruce AG, Bakke AM, Bielefeldt-Ohmann H, Ryan JT, Thouless ME, Tsai CC, Rose TM. 2006. High levels of retroperitoneal fibromatosis (RF)-associated herpesvirus in RF lesions in macaques are associated with ORF73 LANA expression in spindleoid tumour cells. J Gen Virol 87:3529–3538 [DOI] [PubMed] [Google Scholar]
- 10.Bruce AG, Bakke AM, Thouless ME, Rose TM. 2005. Development of a real-time qPCR assay for the detection of RV2 lineage-specific rhadinoviruses in macaques and baboons. Virol J 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burnside KL, Ryan JT, Bielefeldt-Ohmann H, Gregory Bruce A, Thouless ME, Tsai CC, Rose TM. 2006. RFHVMn ORF73 is structurally related to the KSHV ORF73 latency-associated nuclear antigen (LANA) and is expressed in retroperitoneal fibromatosis (RF) tumor cells. Virology 354:103–115 [DOI] [PubMed] [Google Scholar]
- 12.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869 [DOI] [PubMed] [Google Scholar]
- 13.Chatlynne LG, Ablashi DV. 1999. Seroepidemiology of Kaposi's sarcoma-associated herpesvirus (KSHV). Semin Cancer Biol 9:175–185 [DOI] [PubMed] [Google Scholar]
- 14.Damania B, Desrosiers RC. 2001. Simian homologues of human herpesvirus 8. Philos Trans R Soc Lond B Biol Sci 356:535–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desrosiers RC, Sasseville VG, Czajak SC, Zhang X, Mansfield KG, Kaur A, Johnson RP, Lackner AA, Jung JU. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J Virol 71:9764–9769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeWire SM, Money ES, Krall SP, Damania B. 2003. Rhesus monkey rhadinovirus (RRV): construction of a RRV–GFP recombinant virus and development of assays to assess viral replication. Virology 312:122–134 [DOI] [PubMed] [Google Scholar]
- 17.Dittmer DP, Gonzalez CM, Vahrson W, DeWire SM, Hines-Boykin R, Damania B. 2005. Whole-genome transcription profiling of rhesus monkey rhadinovirus. J Virol 79:8637–8650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dourmishev LA, Dourmishev AL, Palmeri D, Schwartz RA, Lukac DM. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) epidemiology and pathogenesis. Microbiol Mol Biol Rev 67:175–212 [table of contents.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edelman DC. 2005. Human herpesvirus 8—a novel human pathogen. Virol J 2:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao SJ, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo CR, Saah A, Phair J, Detels R, Chang Y, Moore PS. 1996. KSHV antibodies among Americans, Italians, and Ugandans with and without Kaposi's sarcoma. Nat Med 2:925–928 [DOI] [PubMed] [Google Scholar]
- 21.Giddens WE, Jr, Tsai CC, Morton WR, Ochs HD, Knitter GH, Blakley GA. 1985. Retroperitoneal fibromatosis and acquired immunodeficiency syndrome in macaques. Pathologic observations and transmission studies. Am J Pathol 119:253–263 [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue N, Mar EC, Dollard SC, Pau CP, Zheng Q, Pellett PE. 2000. New immunofluorescence assays for detection of human herpesvirus-8-specific antibodies. Clin Diagn Lab Immunol 7:427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Institute of Laboratory Animal Resources 1996. Guide for the care and use of laboratory animals. Washington (DC): National Academy Press [Google Scholar]
- 24.Kaleeba JA, Berger EA. 2006. Broad target cell selectivity of Kaposi's sarcoma-associated herpesvirus glycoprotein-mediated cell fusion and virion entry. Virology 354:7–14 [DOI] [PubMed] [Google Scholar]
- 25.Kedes DH, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med 2:918–924 [DOI] [PubMed] [Google Scholar]
- 26.Leavens DA, Hostetter AB, Wesley MJ, Hopkins WD. 2004. Tactical use of unimodal and bimodal communication by chimpanzees, Pan troglodytes. Anim Behav 67:467–476 [Google Scholar]
- 27.Lennette ET, Blackbourn DJ, Levy JA. 1996. Antibodies to human herpesvirus type 8 in the general population and in Kaposi's sarcoma patients. Lancet 348:858–861 [DOI] [PubMed] [Google Scholar]
- 28.Lerche NW. 2005. Common viral infections of laboratory primates. Wolfe-Coote S. The laboratory primate. London (UK): Elsevier Academic Press; p 75–89 [Google Scholar]
- 29.Malik N, Kallestad JC, Gunderson NL, Austin SD, Neubauer MG, Ochs V, Marquardt H, Zarling JM, Shoyab M, Wei CM, Linsley PS, Rose TM. 1989. Molecular cloning, sequence analysis, and functional expression of a novel growth regulator, oncostatin M. Mol Cell Biol 9:2847–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mansfield KG, Westmoreland SV, DeBakker CD, Czajak S, Lackner AA, Desrosiers RC. 1999. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J Virol 73:10320–10328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayama S, Cuevas LE, Sheldon J, Omar OH, Smith DH, Okong P, Silvel B, Hart CA, Schulz TF. 1998. Prevalence and transmission of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in Ugandan children and adolescents. Int J Cancer 77:817–820 [DOI] [PubMed] [Google Scholar]
- 32.Moore PS, Gao SJ, Dominguez G, Cesarman E, Lungu O, Knowles DM, Garber R, Pellett PE, McGeoch DJ, Chang Y. 1996. Primary characterization of a herpesvirus agent associated with Kaposi's sarcomae. J Virol 70:549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nascimento MC, de Souza VA, Sumita LM, Freire W, Munoz F, Kim J, Pannuti CS, Mayaud P. 2007. Comparative study of Kaposi's sarcoma-associated herpesvirus serological assays using clinically and serologically defined reference standards and latent class analysis. J Clin Microbiol 45:715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orzechowska BU, Powers MF, Sprague J, Li H, Yen B, Searles RP, Axthelm MK, Wong SW. 2008. Rhesus macaque rhadinovirus-associated nonHodgkin's lymphoma: animal model for KSHV-associated malignancies. Blood 112:4227–4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pica F, Volpi A. 2007. Transmission of human herpesvirus 8: an update. Curr Opin Infect Dis 20:152–156 [DOI] [PubMed] [Google Scholar]
- 36.Rose TM, Ryan JT, Schultz ER, Raden BW, Tsai CC. 2003. Analysis of 4.3 kb of divergent locus B of macaque retroperitoneal fibromatosis-associated herpesvirus reveals a close similarity in gene sequence and genome organization to Kaposi's sarcoma-associated herpesvirus. J Virol 77:5084–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose TM, Strand KB, Schultz ER, Schaefer G, Rankin GW, Jr, Thouless ME, Tsai CC, Bosch ML. 1997. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol 71:4138–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruff K, Baskin GB, Simpson L, Murphey-Corb M, Levy LS. 2003. Rhesus rhadinovirus infection in healthy and SIV-infected macaques at Tulane National Primate Research Center. J Med Primatol 32:1–6 [DOI] [PubMed] [Google Scholar]
- 39.Scheaffer RL, Mendenhall W, Ott L. 1995. Elementary survey sampling, 5th ed Belmont (CA): Duxbury Press [Google Scholar]
- 40.Schultz ER, Rankin GW, Jr, Blanc MP, Raden BW, Tsai CC, Rose TM. 2000. Characterization of 2 divergent lineages of macaque rhadinoviruses related to Kaposi's sarcoma-associated herpesvirus. J Virol 74:4919–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz RA, Micali G, Nasca MR, Scuderi L. 2008. Kaposi sarcoma: a continuing conundrum. J Am Acad Dermatol 59:179–206 [quiz 207-8] [DOI] [PubMed] [Google Scholar]
- 42.Searles RP, Bergquam EP, Axthelm MK, Wong SW. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8). J Virol 73:3040–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevenson PG. 2004. Immune evasion by gammaherpesviruses. Curr Opin Immunol 16:456–462 [DOI] [PubMed] [Google Scholar]
- 44.Strand K, Harper E, Thormahlen S, Thouless ME, Tsai C, Rose T, Bosch ML. 2000. Two distinct lineages of macaque gammaherpesviruses related to the Kaposi's sarcoma-associated herpesvirus. J Clin Virol 16:253–269 [DOI] [PubMed] [Google Scholar]
- 45.Tsai CC, Giddens WE, Jr, Morton WR, Rosenkranz SL, Ochs HD, Benveniste RE. 1985. Retroperitoneal fibromatosis and acquired immunodeficiency syndrome in macaques: epidemiologic studies. Lab Anim Sci 35:460–464 [PubMed] [Google Scholar]
- 46.Vogel P, Weigler BJ, Kerr H, Hendrickx AG, Barry PA. 1994. Seroepidemiologic studies of cytomegalovirus infection in a breeding population of rhesus macaques. Lab Anim Sci 44:25–30 [PubMed] [Google Scholar]
- 47.Westmoreland SV, Mansfield KG. 2008. Comparative pathobiology of Kaposi sarcoma-associated herpesvirus and related primate rhadinoviruses. Comp Med 58:31–42 [PMC free article] [PubMed] [Google Scholar]
- 48.Whitby D, Stossel A, Gamache C, Papin J, Bosch M, Smith A, Kedes DH, White G, Kennedy R, Dittmer DP. 2003. Novel Kaposi's sarcoma-associated herpesvirus homolog in baboons. J Virol 77:8159–8165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong SW, Bergquam EP, Swanson RM, Lee FW, Shiigi SM, Avery NA, Fanton JW, Axthelm MK. 1999. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi's sarcoma-associated herpesvirus. J Exp Med 190:827–840 [DOI] [PMC free article] [PubMed] [Google Scholar]