Spermatogonial stem cells have an innate ability to choose, with constant probability, between different fates independently of cues from the microenvironment.
Abstract
Mammalian spermatogenesis is initiated and sustained by spermatogonial stem cells (SSCs) through self-renewal and differentiation. The basic question of whether SSCs have the potential to specify self-renewal and differentiation in a cell-autonomous manner has yet to be addressed. Here, we show that rat SSCs in ex vivo culture conditions consistently give rise to two distinct types of progeny: new SSCs and differentiating germ cells, even when they have been exposed to virtually identical microenvironments. Quantitative experimental measurements and mathematical modeling indicates that fate decision is stochastic, with constant probability. These results reveal an unexpected ability in a mammalian SSC to specify both self-renewal and differentiation through a self-directed mechanism, and further suggest that this mechanism operates according to stochastic principles. These findings provide an experimental basis for autonomous and stochastic fate choice as an alternative strategy for SSC fate bifurcation, which may also be relevant to other stem cell types.
Introduction
In mammalian testes, spermatogonial stem cells (SSCs) sustain continuous spermatogenesis throughout a mammal’s reproductive years. The power of SSCs depends on their ability to produce two types of progeny: one type replicates the mother stem cell (self-renewal); the other acquires specialized function and morphology to become sperm (differentiation). Although no known molecular markers can unambiguously identify SSCs, it is generally accepted that SSCs abut the basement membrane of the seminiferous tubules as single cells (Asingle). The earliest identifiable differentiating progeny are sibling spermatogonia (Apaired) that do not complete cytokinesis but form intercellular bridges, which connect their cytoplasm into a syncytium (Huckins, 1971; Oakberg, 1971; de Rooij and Grootegoed, 1998; Oatley and Brinster, 2006, 2008). Apaired then develop into Aaligned (chains of 4, 8, 16, and 32 cells) through a series of synchronous mitotic divisions with incomplete cytokinesis. Subsequently, Aaligned undergo a lengthy differentiation process that eventually leads to meiosis and formation of haploid spermatids (Russell et al., 1990).
How SSC progeny adopt different cell fates has not been elucidated. Most current models presume a causal correlation between particular environmental cues and a specific cell fate outcome. For instance, in the niche model, it is proposed that a stem cell can only self-renew within a specialized microenvironment (niche) that promotes “stemness” and prevents differentiation, whereas differentiation only occurs outside the niche environment (Schofield, 1978; Spradling et al., 2001). Support for the niche model comes from many studies of diverse tissue stem cells in various organisms (Spradling et al., 2001; Fuchs et al., 2004; Li and Xie, 2005), especially in Drosophila testis, where the niche constituents as well as their detailed role in SSC fate determination have been characterized (Gilboa and Lehmann, 2004; Yamashita and Fuller, 2005; Fuller and Spradling, 2007). However, within mammalian seminiferous tubules, SSCs are intermingled with differentiating germ cells as a monolayer on the basement membrane, where no specialized niche has been found (Ogawa et al., 2005). Recently, a few studies suggest that peritubular blood vessels or interstitial cells might serve as niches (Chiarini-Garcia et al., 2001; Yoshida et al., 2007). However, due to their much larger scale relative to individual germ cells and the lack of direct contact with them, the peritubular structures are unlikely to differentially influence the two intermingled cell types to account for alternative fate specification, and their role in SSC self-renewal and differentiation remains to be examined.
An outstanding question is whether mammalian SSCs are able to specify self-renewal and differentiation cell autonomously and independently of differential extrinsic stimuli. Evaluating this possibility is essential for determining the causes and mechanisms governing SSC alternative fate decisions. Autonomous fate choice can be tested through determining if mammalian SSCs can give rise to both SSCs and differentiating germ cells under identical environmental conditions. We therefore characterized the fate outcomes of rat SSC daughter cells in a well-characterized homogeneous ex vivo culture setting that supports expansion of rodent SSCs and preserves their ability to generate offspring upon transplantation back into testes (Hamra et al., 2005; Kanatsu-Shinohara et al., 2003, 2005a; Kubota et al., 2004b; Ryu et al., 2005; Wu et al., 2009).
The culture method employs fibroblast feeder cells and defined serum-free media supplemented with glial cell line–derived neurotrophic factor (GDNF), a growth factor essential for SSC survival/proliferation in vivo (Meng et al., 2000). The presence, quantity, and physiological properties of SSCs in these cultures have been assessed by the testis cell transplantation assay, a functional and quantitative SSC assay previously developed by Brinster and co-workers (Brinster and Avarbock, 1994; Brinster and Zimmermann, 1994; Brinster, 2002). In this assay, germ cells are transplanted into seminiferous tubules that are depleted of endogenous germ cells, and only donor SSC-derived progeny develop as discrete colonies of spermatogenesis over time. Because each colony typically arises through the self-renewal and differentiation of a single stem cell, the resulting colony number offers a quantitative measure of SSC content (Dobrinski et al., 1999; Nagano et al., 1999; Brinster, 2002; Zhang et al., 2003; Kanatsu-Shinohara et al., 2006; Oatley and Brinster, 2006). Using this assay, the cultured SSCs have been shown to repopulate recipient testes and produce functional sperm, indicating that their potential for both self-renewal and differentiation are fully preserved. Furthermore, SSC number continues to increase as the total germ cell population expands over time; hence, SSC self-renewal is taking place in culture (Kanatsu-Shinohara et al., 2003, 2005a; Kubota et al., 2004b; Hamra et al., 2005; Ryu et al., 2005; Wu et al., 2009). However, it is not clear if self-renewal is the only outcome of SSC divisions or whether differentiation is also taking place in the cultures.
Here, we find that under the established uniform culture conditions, SSCs consistently give rise to both new SSCs and differentiating progeny. The differentiating progeny have lost stemness and unlimited self-renewal capacity but have developed functional intercellular bridges, forming syncytia among sibling cells similar to Apair and Aaligned in testis. Twin daughter cells of single SSCs often undergo self-renewal and differentiation side by side even though they have been exposed to virtually identical microenvironments. Moreover, quantitative experimental measurements and mathematical modeling indicates that fate decision is stochastic, with constant probability. These results demonstrate the capacity of a mammalian SSC to specify both self-renewal and differentiation autonomously, and this capacity operates according to stochastic principles.
Results
Germ cells in culture remain heterogeneous in terms of stemness and proliferation potential
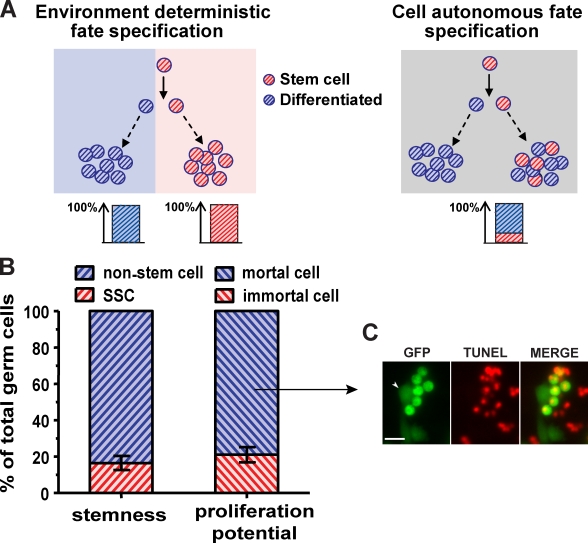
If self-renewal and differentiation are ordained by environmental cues, a uniform environment should result in uniform cell fate specification (Fig. 1 A, left). In contrast, if fate decision is intrinsic to the cell, different fates could arise in a homogeneous culture (Fig. 1 A, right). To distinguish between these two scenarios, we first determined whether all the rat germ cells in culture are SSCs. Previous studies have shown that cultured rat germ cells remain heterogeneous during the first few months outside the testes (Hamra et al., 2005; Ryu et al., 2005). To examine whether this was due to transient proliferation of differentiating germ cells from testes, we derived germ cell lines from rat testes that express EGFP specifically in all germ cells (Cronkhite et al., 2005), and cultured them continuously for much more extended periods. Even after 20 mo of propagation, SSC content in the cultures remained ∼17% of the total germ cells, as measured by the transplantation assay (Fig. 1 B), which is similar to the initial reports (Hamra et al., 2005; Ryu et al., 2005). Thus, besides SSCs, a nonstem germ cell population persists and constitutes a significant fraction (>80%) of the culture, which suggests that they either have unlimited proliferation capacity or are continuously replenished by an immortal cell type, presumably the SSCs.
Figure 1.
Persistent heterogeneity of germ cells in stemness and proliferation potential under uniform culture conditions is compatible with a cell-autonomous fate specification model. (A) Environment deterministic and cell-autonomous fate specification models predict homogeneous and heterogeneous composition of SSC progeny, respectively, in a uniform environment. (A, left) In the environment deterministic model, self-renewal and differentiation are controlled by distinct environmental cues (red and blue), which result in homogeneous yet distinct fate outcomes in the red and the blue area. (A, right) In the cell-autonomous fate specification model, occurrence of self-renewal and differentiation does not depend on differential environmental cues, and both SSCs and differentiating progeny can be generated in a uniform environment (gray). (B) Transplantation of germ cells that have been cultured for 20 mo into sterile testes shows that only 16.5 ± 3.9% (mean ± SEM, n = 6) of the population are SSCs, and that 83.5% do not possess stemness. Tracing of sorted single germ cells in culture for 6 mo shows that 79.0 ± 4.1% (mean ± SEM, n = 84 in three indepenent experiments) of the cells perish within a month after transient proliferation, and only 21.0% of the single cells can continue proliferate for at least 6 mo. (C) Dying germ cells show condensed cell bodies and are positive for TUNEL staining. The arrowhead points to a healthy TUNEL-negative germ cell that remains flattened on the culture substrate. Bar, 20 µm.
We next examined whether the proliferation potential of the germ cells in culture is also heterogeneous. Because the cultures have been shown to expand indefinitely (Kanatsu-Shinohara et al., 2003; Kubota et al., 2004a; Hamra et al., 2005; Kanatsu-Shinohara et al., 2005a, 2008; Ryu et al., 2005), and offspring can be produced from single SCC-derived long-lasting clones (Kanatsu-Shinohara et al., 2005c), it is evident that at least some of the germ cells, especially some SSCs, must be immortal in culture. Nevertheless, we observed widespread and continued cell death in culture through long-term time lapse imaging (Fig. S1); only ∼18% (n = 221) of identified single germ cells gave rise to clones that continued to proliferate beyond 2 wk. To confirm this and to measure the longevity of the mortal cells, single EGFP-positive germ cells were sorted into 96-well plates and cultured. We found that 21.0 ± 4.1% (mean ± SEM) of these single cells continue to proliferate for at least 6 mo and can be considered as immortal, whereas the other 79% proliferate only transiently and form small clones (Fig. 1 B). Of the transient clones, 91.8% perish within the first two weeks. TUNEL staining indicates that the dying cells contain double-strand DNA breaks (Fig. 1 C), which suggests that their death is by apoptosis. Because unlimited proliferation potential is a defining attribute of stem cells, the similar percentages of immortal cells and SSCs implies that all the immortal cells are SSCs and the non–stem cells are mortal.
Immortal germ cells are SSCs, nonstem germ cells are mortal
To investigate whether all the immortal cells are SSCs, we randomly selected 26 of the single cell–derived long-lasting clones and used the transplantation assay to determine whether they contain SSCs (outlined in Fig. 2 A). Given that only a stem cell can give rise to stem cells, a positive result would indicate retrospectively that the founding cell of the clone was an SSC. From each clone, ∼5–25 × 103 cells were transplanted into individual rat recipient testes. 14 testes were analyzed 2 mo later, and the other 12 testes were examined 5 mo after transplantation. We found that 100% of the 26 transplanted clones had colonized the recipient seminiferous tubules, forming between 2 and 50 EGFP-positive colonies per testis. A representative recipient testis is shown in Fig. 2 B. This result suggests that all immortal cells are SSCs. Conversely, the non–stem cells are mortal, for they never form long lasting clones.
Figure 2.
Immortal germ cells are SSCs. (A) Outline of the experimental protocol to yield single cell–derived long-lasting clones and subsequent transplantation of cells from these clones into presterilized recipient rat testes. (B) A wild-type recipient testis transplanted with a long-lasting clone derived from single EGFP-positive germ cell. The donor SSC-derived EGFP-positive spermatogenic colonies (left) indicate the founding cell of the long-lasting clone was an SSC. An enlarged view of individual donor SSC derived colonies (right) showed multiple layers of differentiating germ cells. This testis is representative of the 26 recipient testes, all of which were colonized similarly by donor SSCs from long-lasting clones. White boxes indicate sections that have been enlarged. Bars: (left) 500 µm; (right) 100 µm.
The mortal nonstem germ cells develop functional intercellular bridges, a conserved marker for germ cell differentiation
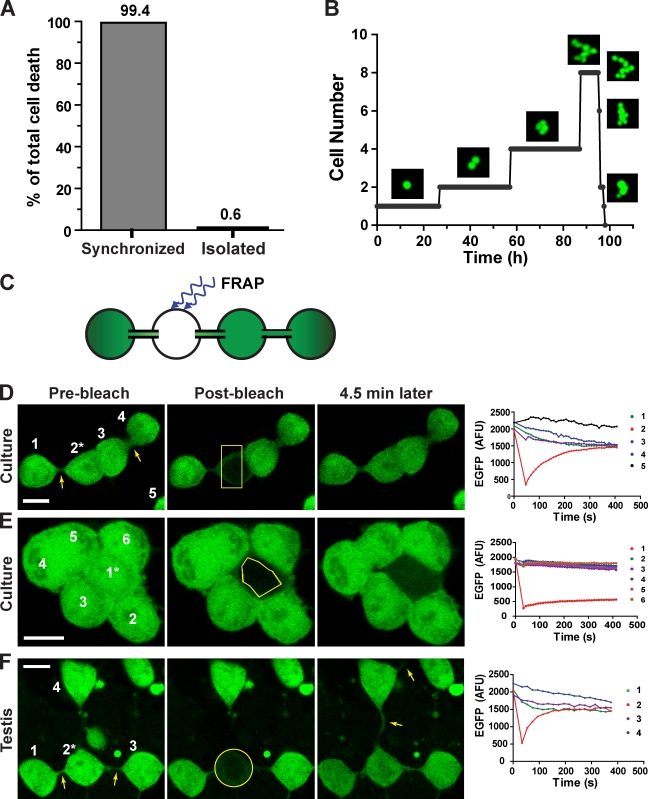
We next investigated whether the mortal non–stem cells have differentiated. Time-lapse imaging revealed that essentially all the mortal non–stem cells (>99%; Table S1) exhibited highly synchronized cell divisions and deaths as sibling groups (Fig. 3, A and B; and Video 1). Coordinated cell cycle and apoptosis have been extensively documented for differentiating germ cells in rodent testes and have been shown to be mediated by intercellular bridges that connect the cytoplasm of sibling cells into a syncytium (Fawcett et al., 1959; Fawcett, 1961; Huckins, 1978; Hamer et al., 2003). Therefore, the synchrony observed in culture suggests that the mortal non–stem cells have formed intercellular bridges, a marker for differentiating germ cells that is conserved from invertebrates to human.
Figure 3.
Mortal nonstem germ cells in culture are differentiating. (A) Analysis of time-lapse image sequences showed that among germ cells that have divided at least once in culture, 99.4% of the cell deaths (n = 314) are synchronized among sibling groups; only 0.6% died as isolated cells. (B) Images from a time-lapse sequence of a typical mortal germ cell–derived clone (Video 1) and a plot of the cell number within the clone over time show the synchrony of cell divisions and cell death of sibling germ cells in the clone. (C) The experimental scheme for a functional assay specific for intercellular bridges. It involves photobleaching of EGFP within one cell of a group and monitoring of EGFP fluorescence intensity of each cell by time-lapse imaging. (D–F) Time-lapse image sequences before and after photobleaching of the cell indicated by an asterisk, and a time trace of EGFP fluorescence intensity in each cell of a germ cell group in culture (D and E, also see Video 2) or in testis (F). Bleached regions are delineated by yellow lines. Arrows point to visible intercellular bridges. The numbers in D–F indicate which cells correspond to the fluorescence intensity traces shown on the right. Bars, 10 µm.
Consistent with this notion, germ cells in culture often appear to be interconnected by thin strands, like beads on a string (Fig. 3 D, arrows). To test for cytoplasmic continuity among these cells, we developed a novel assay specific for intercellular bridges using FRAP. In this assay, as outlined in Fig. 3 C, EGFP is photobleached specifically in one cell of a group, and the fluorescence intensities of all cells in the group are monitored by time-lapse imaging. If the cells are interconnected, diffusion should eventually result in equilibration of EGFP fluorescence intensity throughout the group. Because EGFP is too large to pass through gap junctions, the only other known junction that allows molecule exchange between mammalian cells, this assay is specific for intercellular bridges, which are usually 1–3 µm in diameter (Weber and Russell, 1987; Greenbaum et al., 2007).
When this method was applied in culture, in most cases, we observed EGFP fluorescence recovery in the bleached germ cell accompanied by a corresponding fluorescence decrease in its contiguous neighbors, eventually reaching equilibrium (Fig. 4 D and Video 2). As a control, when EGFP in isolated cells that have no neighbors was photobleached, the fluorescence intensity in these cells never recovered significantly within a similar time frame (unpublished data). Hence, the majority of the germ cells in culture are coupled by intercellular bridges, which suggests that they are differentiating. This is consistent with the results from the transplantation assay showing that most germ cells in culture are not SSCs (Fig. 1 B). We also found a small fraction of the germ cells that do not exchange EGFP with any contiguous cells (Fig. 3 E), which indicates that differentiation has not been initiated or has not progressed as far in these cells. Therefore, some of these uncoupled cells are probably SSCs, in agreement with morphological observations in vivo indicating that SSCs are not interconnected with other cells (Huckins, 1971; Oakberg, 1971; de Rooij and Grootegoed, 1998).
Figure 4.
Individual SSCs consistently give rise to SSCs and differentiating germ cells independently of environmental cues. (A) Cultures of single germ cells from two randomly selected long-lasting clones (Nos. 1 and 6) derived from single SSCs showed that they contained 82.7% and 78.4% mortal differentiating cells and 17.3% and 21.6% immortal SSCs, respectively (n = 84). This was similar to their parent cell line (82.7% differentiating and 17.3% SSC, n = 84), which indicates that individual SSCs consistently give rise to both SSCs and differentiating germ cells. (B) Schematic diagram based on the long-term time-lapse imaging of twin daughter cells of a single SSC that gave rise to two clones with different fates. In one clone, all cells died synchronously by day 10, whereas the other continued to live by day 18, which indicates that one of the twin daughter cells differentiated, whereas the other became an SSC. (C and D) Relative positions of the two daughter cells during the time between their birth and next division (Video 3). (C) The solid line indicates the mean diameter (∼13 µm) of the two cells, and black dots mark the distance between the geometric centers of the two cells at the indicated time points. (D) Centroid positions of each daughter cell during the time window are represented by the centers of the red and blue circles, respectively. The diameter of the circles is equal to the mean diameter of the cells to show the area transiently visited by the two cells.
To compare the intercellular bridges formed in culture with those formed in vivo, seminiferous tubules containing EGFP-positive spermatogonia were freshly dissected, and germ cell chains (Aaligned) on the basement membrane were subjected to the FRAP assay (Fig. 3 F). The kinetics of EGFP exchange in testis were indistinguishable from those observed in culture (Fig. 3, compare D and F), which suggests that the intercellular bridges formed in culture and in testes are physiologically comparable. Therefore, the mortal non–stem cells in culture appear to have initiated the same differentiation process as differentiating germ cells in vivo.
SSCs both self-renew and differentiate in culture
Given that the lifespan of the differentiating germ cells does not exceed 1 mo, their persistence in culture over many months implies that they must be continually replenished by the immortal SSCs. To verify this, we examined whether long-lasting clones derived from single SSCs contain a mortal population. When single germ cells from these clones and from their parent lines were sorted into 96-well plates and cultured in parallel, a similar proportion of mortal cells were observed in each of them (Fig. 4 A).
In addition, by long-term time-lapse imaging, we also directly observed fate bifurcation in clones derived from single SSCs. In these experiments, SSCs were identified retrospectively by their long-lasting progeny (>17 d), and differentiating germ cells were identified by progeny that formed intercellular bridges and died synchronously. We observed examples in which twin daughter cells of single SSCs acquired different cell fates, and others in which they both became SSCs. However, the method cannot unambiguously distinguish between an SSC that gives rise to two differentiating daughter cells and a mother germ cell that is already differentiating.
Within the SSC lineages observed, no obvious pattern of cell fate outcome was identified. However, divergence of SSC daughter cell fates, similar to the example diagrammed in Fig. 4 B, was always observed (100%, n > 50). Based on these observations, we concluded that individual SSCs not only undergo self-renewal, but also give rise to differentiating germ cells in culture.
Because under certain conditions differentiating germ cells can de-differentiate and become SSCs in Drosophila (Brawley and Matunis, 2004; Cheng et al., 2008) and mouse testes (Nakagawa et al., 2007; Barroca et al., 2009; Yoshida, 2009), we therefore examined if such fate conversion occurs in rat germ cell cultures. Even though the majority of rat germ cells in culture are c-kit negative (>98%; Fig. S2), which indicates an early state of differentiation, none of the cells identified as differentiating germ cells by our criteria gave rise to long-lasting clones (n > 200 cells). Consistently, in the SSCs examined (n > 50), none of them were derived from a cell that seemed to be connected to other cells through intercellular bridges. These observations indicate that de-differentiation, if it occurs at all in the culture, is rare.
SSC self-renewal and differentiation are independent of extrinsic heterogeneity
The fact that self-renewal and differentiation occur concomitantly in a uniform culture setting suggests an independence of cell fate choice with respect to the environment. However, it is formally possible that fate bifurcation is dependent on microscale heterogeneity associated with the feeder cells or extracellular matrix. Although direct and thorough evaluation of microscale heterogeneity is impossible, a detailed analysis of germ cell behaviors in culture suggests it is unlikely to be the reason for fate bifurcation. We found that SSC daughter cell pairs, including those that have adopted different fates, typically remained closely associated the whole time from their birth until they divided again to give rise to clones of either identical or distinct fates. This observation was quantified by measuring the distance between the geometric centers of twin daughter cells from time-lapse recordings. In most cases, as for the cell pair indicated by an asterisk in Fig. 4 B, the center-to-center distance was always less than their mean diameter, which indicated that the two cells were never separated into discrete microenvironments (Fig. 4 C and Video 3). Furthermore, twin daughter cells in all cases, instead of statically occupying one spot, constantly move around together, dynamically probing the same microenvironment. Typically, the areas transiently visited by each daughter cell during the fate bifurcation time window are nearly identical, as shown in Fig. 4 D. The intimate association and the dynamic movement of the two cells minimize the chances for their exposure to different extrinsic cues, including microheterogeneity. Nevertheless, as in this case, twin daughter cells derived from a single SSC often adopted different fates. Therefore, fate bifurcation does not seem to depend on extrinsic heterogeneity. Instead, cell-autonomous fate specification is most consistent with our data.
SSC self-renewal and differentiation are stochastic
Because SSC self-renewal results in continued proliferation, whereas differentiation leads to cell death in culture, fate specification will directly influence germ cell population growth, death, and composition. We noted that the proportions of SSC and differentiating cells in the parent cultures and in long-lasting clones derived from single SSCs are essentially invariant (Fig. 4 A), which suggests a constant ratio of SSC self-renewal and differentiation. In addition, we did not observe any consistent pattern of fate outcome along the lineage trees of long-lasting clones. These observations suggest that self-renewal and differentiation might be stochastic events occurring with a consistent probability. Based on this premise, we derived the following mathematical models for the kinetics of total germ cell growth (Eq. 1) and for the decay of single cell–derived clones over time (Eq. 2, detailed in Materials and methods):
| (1) |
And when t < (q + 1)cmax,
| (2) |
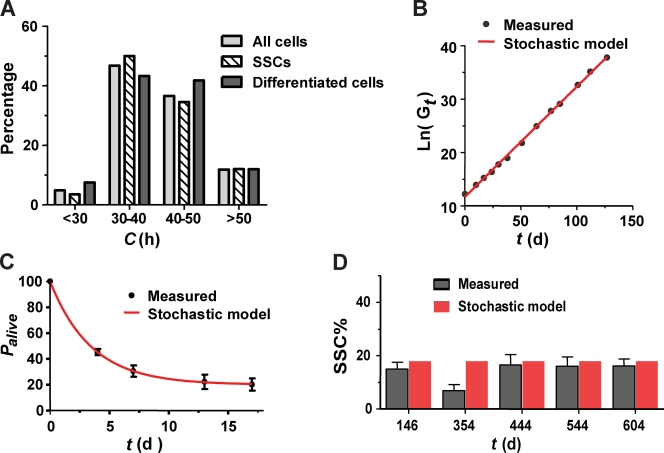
where Gt denotes the number of total live germ cells at time t, S0 denotes the number of SSCs of the given germ cell population at arbitrary time zero during continuous culture, s denotes the chance per cell division that an individual SSC daughter cell will become an SSC. The probability that it will become differentiated is 1 − s, and q denotes the number of cell divisions needed for newly differentiated germ cells to produce the mean number of progeny before their death. c denotes mean cell cycle length (40.0 ± 8.2 h), which is not different for SSC and differentiating germ cells as measured by long-term time-lapse imaging (Fig. 5 A). cmax denotes the observed maximum cell cycle length, which is 86 h. Palive denotes percentage of live clones derived from single germ cells of a given population at time t.
Figure 5.
A mathematical model for stochastic fate specification describes and predicts germ cell population dynamics in culture. (A) Histogram of cell cycle length in groups of retrospectively identified SSCs (n = 110), differentiating mortal cells (n = 76, synchronized sibling divisions are counted as one), and all cells (n = 186). No significant difference was observed between the SSCs and the differentiating cells. Mean cell cycle length for all cells was 40.0 ± 8.2 h. (B and C) The mathematical model (red line) fits the measured germ cell growth (B, black dots), and the measured germ cell survival curve (C, black dots; mean ± SEM, n = 84 in three independent experiments). (D) The mathematical model predicts a constant SSC content of 17.9% over time (red bars). Dark gray bars indicate the measured SSC content at the indicated time points (mean ± SEM, n > 4).
These equations assume that SSCs are immortal and that differentiating germ cells die after transient propagation, as we have shown experimentally.
Eq. 1 fits the measured total germ cell growth well when s = 0.706 (r2 = 0.9987; Fig. 5 B), and Eq. 2 fits the measured survival curve of single cell–derived clones when s = 0.618 and q = 3.3 (r2 = 0.9720; Fig. 5 C). These analyses thus showed that the probabilistic model can coherently describe various uncorrelated aspects of population dynamics in culture. The analyses also revealed that the probability for SSC self-renewal under the particular culture conditions is ∼67% (mean of 0.706 and 0.618); for differentiation, the probability is 33%. Even though fate decisions are biased toward self-renewal, differentiating germ cells actually outnumber the SSCs in culture because 100% of their progeny will be differentiated, whereas the SSC progeny will be a mixture of SSCs and differentiating progeny.
In addition, according to the model, the stem cell fraction in culture is:
| (3) |
which is not a function of time (see Materials and methods). Therefore, the model predicts that the stem cell ratio in culture should remain constant, with a value of 17.9% (assuming s = 0.67 and q = 3.3). To verify this prediction, we used the transplantation assay to measure the number of SSCs in continuous cultures of several independently derived germ cell lines over extended periods of time. One set of such experiments is shown in Fig. 5 D. These measurements confirmed that the SSC fraction remains essentially constant over time. The measured SSC percentages are also comparable to the predicted 17.9%. Hence, our data fully support the model of stochastic fate determination with constant probability.
Discussion
Further discussion of the results and their implications
Using novel assays for SSCs and their differentiating progeny, we demonstrated that both self-renewal and differentiation take place in the same culture. The independence of cell fate choice with respect to the environment reveals a capacity of the rat SSC to autonomously specify its self-renewal and differentiation. Consistent with this conclusion, omitting various components from the current culture system, including feeder cells, does not eliminate the heterogeneous fate outcome (Wu et al., 2009), which suggests that none of the components of the culture system specifically decrees self-renewal or differentiation. In addition, mathematical modeling of dynamic population growth, death, and SSC content of germ cell cultures indicates that autonomous fate decision operates according to probabilistic principles. In our hands, stochastic fate choice does not appear to be specific to rat SSCs, but also characterizes mouse SSC fate specification in culture (unpublished data). However, it remains to be examined whether the capacity for stochastic fate choice is a general attribute of most mammalian SSCs.
SSCs have been shown to display extensive parallel behaviors in culture and in vivo. Both systems support SSC self-renewal, GDNF-dependent perpetual proliferation, and a characteristic gene expression profile (Kanatsu-Shinohara et al., 2003, 2005a,b; Nagano et al., 2003; Kubota et al., 2004a,b; Hamra et al., 2005; Ryu et al., 2005; Lee et al., 2007; Oatley et al., 2007; Wu et al., 2009). Adding to the list of shared attributes, we have now shown that differentiation also occurs alongside self-renewal in both systems. Because the germ cell cultures replicate many important characteristics of SSCs in vivo, it is conceivable that stochastic fate choice may be an authentic feature of SSC behavior in vivo. However, this remains to be tested directly by experimentation in testes.
Stochastic fate choice and environmental regulation are not mutually exclusive
Maintenance of tissue homeostasis usually demands feedback regulation of stem cell activity by extrinsic physiological cues. This raises the question of how an autonomous and stochastic mechanism for SSC self-renewal and differentiation integrates input from its environment. Although our results argue against a deterministic role of extrinsic cues in SSC progeny fate specification, a nondeterministic influence of environment can fit well with the observed stochastic fate choice. For example, rather than ordaining fate of individual cells, extrinsic cues may serve to bias the probability of cell fate choices at the population level. A niche, in this case, can be viewed as an extreme environment in which fate choice is entirely skewed toward self-renewal. Therefore, stochastic fate choice and environmental regulation are not mutually exclusive but could work jointly to achieve SSC self-renewal and differentiation that is minimally dependent on the environment but responsive to its regulation.
Possible mechanisms of SSC stochastic fate choice
One possible mechanism for autonomous cell fate diversification is asymmetric cell division (Knoblich, 2008; Wu et al., 2008). In this scenario, a parent cell segregates fate determinants unequally to its daughter cells, thereby creating intrinsic differences that predestine them for different fates. However, because asymmetric division alone cannot increase stem cell number, symmetric division must also take place to produce the net increase of SSC number we observed in culture. This requires some other mechanism to account for the choice between symmetric and asymmetric divisions, which should be stochastic, for no consistent pattern of cell fate outcome was observed within SSC lineages.
Alternatively, intrinsic variation (noise) in gene expression has been shown in a few simple organisms to cause cell fate divergence within isogenic populations. Examples include entry into the competent state by Bacillus subtilis (Maamar and Dubnau, 2005; Smits et al., 2005; Dubnau and Losick, 2006; Süel et al., 2006, 2007) and generation of photoreceptors with alternative color vision in fly compound eyes (Wernet et al., 2006; Bell et al., 2007). These studies demonstrate that small and transient initial fluctuations in transcription of fate determinant genes can be amplified and stabilized at qualitatively different levels and cause differential cell fate outcome (Losick and Desplan, 2008). A recent study of mouse hematopoietic progenitor cells also indicates that transcription noise may control lineage choice in mammalians (Chang et al., 2008). Thus, it is possible that noisy expression of genes that drive self-renewal or differentiation may play a role in fate decision of SSC daughter cells. The methodology developed in this study can be used in future investigations to identify the molecular machineries responsible for SSC stochastic fate choice.
Materials and methods
Materials and chemicals
Cell culture media, PBS nonessential amino acids, MEM vitamin solution,l-glutamine solution, 0.05% or 0.25% trypsin-EDTA solutions, B27 minus vitamin A supplement, antibiotic-antimycotic solutions, and horse serum were obtained from Invitrogen. BSA and DMSO were obtained from EMD. Mitomycin-C, mouse laminin, mouse EGF, rat GDNF, bovine apo-transferrin, human basic fibroblast growth factor (bFGF), d-(+)-glucose, d-biotin, ascorbic acid, sodium pyruvate, putrescine, progesterone, β-estradiol 17-cypionate, insulin, sodium selenite, 2-mercaptoethanol (ME), dl-lactic acid, and all fatty acids were obtained from Sigma-Aldrich. Mouse leukemia inhibitory factor/ESGRO (LIF) was obtained from Millipore. FBS was obtained from Atlanta Biologicals. Dispase and rat-tail collagen I-coated culture dishes were obtained from Thermo Fisher Scientific, Inc. The In Situ Cell Death Detection kit (TMR red) was obtained from Roche. Busulfan was obtained from MP Biomedicals. Glass-bottom dishes were obtained from MatTek Corporation.
Derivation and maintenance of rat spermatogonial cell cultures
All spermatogonial cell lines used in this study were derived from testes of 20–30-d-old homozygous male SD-Tg(ROSA-EGFP)2-4Reh Sprague Dawley rats, which exhibit germ cell–specific expression of EGFP (GCS-EGFP; Cronkhite et al., 2005). The derivation and maintenance of the rat spermatogonial cell cultures were performed largely as described previously (Hamra et al., 2002, 2005), but without FACS-based cell sorting of EGFP-positive germ cells. In brief, seminiferous tubules were minced and digested with dispase (50 units/testis) at 32°C for up to 30 min. The dissociated testicular cells were cultured at 32°C in DHF12 medium supplemented with 5.5% horse serum, 2.5% FBS, and 1% antibiotic/antimycotic solution at 32°C in humidified incubators with 5% CO2 in order to allow the somatic cells to attach to the plastic or gelatin-coated tissue culture dishes. After 2–3 d of incubation, the germ cells on top of the somatic cells were harvested by washing repeatedly with medium, plated on gelatin-coated dishes, and incubated at 37°C for another 2–3 h to allow remnant somatic cells to attach. At the end of the incubation, the floating germ cells were harvested and plated on laminin-coated dishes in DHF12 medium and supplemented with 10% FBS, 1% antibiotic/antimycotic solution, and 30 µM 2-mercaptoethanol to enrich for stem cells. After 20–40 min of incubation at 32°C, germ cells that did not attach to the laminin were washed away with medium. Germ cells attached to the laminin (LamB cells) were harvested and transferred to gelatin-coated dishes in SA medium to further deplete the remnant somatic cells. After 2 d of incubation at 37°C, the floating LamB germ cells were transferred onto mitomycin-C–treated or γ-irradiated primary mouse embryonic fibroblast (MEF) feeders; then they continued to be cultured in SA medium. Routinely, the culture medium was refreshed every 3–4 d. The cultured germ cells with MEF feeder cells were passaged by trypsinization with 0.05% or 0.25% trypsin-EDTA solution onto new MEF feeders every 2–3 wk.
Single spermatogonial cell culture
To establish single cell cultures, GCS-EGFP spermatogonial cell cultures were trypsinized with 0.25% trypsin-EDTA solution to isolate and harvest the cells. After one wash, the cell pellet was resuspended at 0.5–2 × 106/ml in SA medium and filtered through 40-µm cell strainers. 96-well plates with MEF feeder cells were washed once with PBS and replenished with 100 µl per well of SA medium. FACS using an Aria Cell Sorter (BD) was used to deposit one EGFP-positive germ cell into each well of the 96-well plates. Medium was refreshed every 3–4 d and the growth/death of the clones was monitored using a fluorescence microscope (IX70; Olympus). The long-lasting clones were continually cultured in the original well for 2–3 mo without passaging until transplantation. Then, each clone was passaged by trypsinization with 0.25% trypsin-EDTA solution about every 2 wk.
Germ cell transplantation and analysis
Germ cell transplantations by retrograde injection through the rete of busulfan-treated wild-type rat recipient testes were performed as described previously (Ogawa et al., 1997, 1999; Hamra et al., 2002). In brief, wild-type Sprague Dawley rats at 8 d of age (Harlan Laboratories, Inc.) were injected i.p. with 12.5 mg/kg busulfan (4 mg/ml in 50% DMSO and 50% PBS) to eliminate the endogenous germ cells, then used as recipient males for germ cell transplantation at 24–29 d of age (Ogawa et al., 1999). Donor cells were harvested from long-term in vitro cultures of GCS-EGFP rat spermatogonia by 0.25% trypsin-EDTA and filtered through a 40-µm cell strainer. EGFP-positive germ cells were counted and diluted to 3 × 104–3 × 105 cells/ml of SA or SG medium containing 0.05% trypan blue as a fluid phase marker. About 50 µl of the cell suspension was loaded into an injection needle pulled from a 100-µl glass capillary tube, and 20–50 µl was then transferred into the seminiferous tubules of anesthetized rats by retrograde injection through the rete of each testis (Ogawa et al., 1997, 1999). 2 or 5 mo after transplantation, recipient testes were dissected out and spread out on glass slides. The EGFP-positive colonies in each recipient testis were visualized and scored using a fluorescence microscope (IX70). The SSC number was then calculated based on 8% colonization efficiency (Nagano, 2003; Ogawa et al., 2003). Images of whole recipient testes were taken with a fluorescence stereomicroscope using 0.63× and 1.5× lenses (SteREO Discovery.V12; Carl Zeiss, Inc.).
Long-term time-lapse microscopy and image analysis
Cultured rat germ cells were plated at 0.5–1 × 104 cells/cm2 in 25-T flasks on MEF feeder cells in SA or SG medium immediately before imaging. The flasks were mounted on the computer-controlled stage of a DeltaVision RT microscope (Applied Precision) in a temperature (37°C)- and humidity-controlled chamber. The flasks were perfused continuously with 5% CO2. The medium was manually changed every 2–3 d without disturbing the position of the flask. Multiple fields were selected for automated time-lapse imaging. Images of each field at 4×, 10×, or 20× magnification were collected at regular time intervals of 15–30 min for 2–18 d. Time-lapse image sequences were analyzed using ImageJ (http://rsbweb.nih.gov/ij/). The Manual Tracking plug-in (written by F. Cordelieres, Institut Curie, France; http://rsb.info.nih.gov/ij/plugins/track/track.html) was used to track the centroid positions of selected individual cells.
FRAP assay for intercellular bridges and image analysis
EGFP-expressing germ cells were cultured under normal conditions in glass-bottom dishes for 4–7 d before analysis. Using a confocal microscope (LSM510; Carl Zeiss, Inc.) with a 40×/0.8 NA water immersion objective lens, photobleaching within a region of interest was performed with 488-nm illumination at 100% laser power, scanning at maximum speed 200–500 times (approximately 40 s). Time-lapse images were acquired immediately before and after photobleaching. For FRAP of EGFP germ cells in testis, seminiferous tubules were spread out between a glass slides and a coverslip. Chains of EGFP-positive germ cells near the basement membrane were selected for FRAP as described for cultured cells. The mean EGFP fluorescence intensity within a region of interest of each germ cell was measured as a function of time using ImageJ.
Mathematical models based on probabilistic fate determination
As demonstrated in the results, germ cells in culture are composed of two distinct populations: the SSCs and the differentiating germ cells. SSCs are able to proliferate indefinitely and give rise to both new SSCs and differentiating progeny. The differentiating germ cells eventually die after transient amplification, producing only differentiating cells. In addition, the invariant proportions of SSC and differentiating cells in the parent culture and in long-lasting clones derived from single SSCs (Fig. 3 A) suggest that a constant probability for self-renewal (s) and differentiation (1 − s) can be assumed. Therefore, the growth of the two populations over time (t) can be represented by the following two exponential equations:
where St and S0 are the number of SSCs at time t and time zero, respectively, Dt is the number of differentiating germ cells at time t, and q denotes the number of divisions required to produce the average number of progeny among the differentiating cells before they die. In both equations, c is the mean cell cycle length. Measurements from time-lapse imaging showed that c is the same for SSCs and differentiating cells (Fig. 5 A).
Therefore, at time point t, the total germ cell number (Gt) is:
Taking the natural log of the above equation, we obtained:
When t = 0, then G0 = S0 (s(−q−1)). Therefore, the equation above becomes:
| (1) |
Experimental values for G0 and Gt were measured as the total number of germ cells at the beginning and at subsequent time points. A linear least squares fit to a semilog plot of the measured data (ln Gt vs. t) has a slope of 0.2064 ± 0.002245. Hence, Eq. 1 best fits the measured growth kinetics (Fig. 5 B) when ln (2s)/c = 0.2064. As the measured average cell cycle length c = 1.67 d (40 h), s = 0.706.
As experimentally determined, differentiating cells have limited proliferation capacity, assuming a differentiating cell can divide on an average q number of divisions before it dies, then in a given germ cell group of total number G0 with S0 number of SSCs, the derivative of the percentage of live clone (Palive) derived from this population at a given time point t is
Therefore, Palive is an exponential decay function of t, when t < (q+1) cmax:
| (2) |
Because exponential decay function P = (P0 − Plateau) e(−Kt) + Plateau best fit to the measured data (R2 = 0.9720) when K = 0.2886, P0 = 100%, and Plateau = 20.02%, our model best fit with the measured data (Fig. 5 C) when s = 0.618, S0/G0 = 20.02%, and q = 3.3.
Plugging in the exponential expressions for St and Dt, the stem cell fraction (Rs) is:
| (3) |
Because Rs is not a function of time, this model predicts that the stem cell ratio remains unchanged over time. Assuming s = 0.67 and q = 3.3, it predicts that the stem cell ratio in culture should remain constant, with a value of 17.9%.
Online supplemental material
Fig. S1 shows that a mortal population persists in long-term rat germ cell cultures. Fig. S2 shows flow cytometric analysis of cultured GCS-EGFP rat germ cells for c-kit expression. Table S1 shows that mortal germ cells in culture die synchronously as sibling groups. Video 1 shows synchronized cell division and death in a clone of germ cells. Video 2 shows EGFP exchange between cultured germ cells through intercellular bridges. Video 3 shows that daughter cells of a single SSC dynamically sample the same microenvironment.
Acknowledgments
Z. Wu is especially grateful to Drs. Richard, G. Anderson and David Mangelsdorf for evaluating the work, reading the manuscript, and the mentoring they generously provided following the decease of Dr. David L. Garbers. We thank Dr. F. Kent Hamra for sharing his expertise in rat testis cell transplantation and in culturing rat germ cells; Elizabeth Curry for conducting cell sorting; and Drs. Eric Olson, Luis Parada, Michael Roth, Chengcheng Zhang, and Xiaodong Zhang for critical reading of the manuscript. Time-lapse imaging and FRAP were carried out in the University of Texas Southwestern Live Cell Imaging Facility.
This work received funding from the Cecil H. & Ida Green Center for Reproductive Biology Sciences at the University of Texas Southwestern Medical Center, and from the Howard Hughes Medical Institute to D.L. Garbers, prior to his passing. G.M. Süel acknowledges funding by the Welch Foundation (I-1674) and the James S. McDonnell Foundation (220020141). G.M. Süel is a W.W. Caruth Jr. Scholar of Biomedical Research.
Footnotes
Abbreviations used in this paper:
- GCS
- germ cell–specific
- GDNF
- glial cell line–derived neurotrophic factor
- MEF
- mouse embryonic fibroblast
- SSC
- spermatogonial stem cell
References
- Barroca V., Lassalle B., Coureuil M., Louis J.P., Le Page F., Testart J., Allemand I., Riou L., Fouchet P. 2009. Mouse differentiating spermatogonia can generate germinal stem cells in vivo. Nat. Cell Biol. 11:190–196 10.1038/ncb1826 [DOI] [PubMed] [Google Scholar]
- Bell M.L., Earl J.B., Britt S.G. 2007. Two types of Drosophila R7 photoreceptor cells are arranged randomly: a model for stochastic cell-fate determination. J. Comp. Neurol. 502:75–85 10.1002/cne.21298 [DOI] [PubMed] [Google Scholar]
- Brawley C., Matunis E. 2004. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 304:1331–1334 10.1126/science.1097676 [DOI] [PubMed] [Google Scholar]
- Brinster R.L. 2002. Germline stem cell transplantation and transgenesis. Science. 296:2174–2176 10.1126/science.1071607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster R.L., Avarbock M.R. 1994. Germline transmission of donor haplotype following spermatogonial transplantation. Proc. Natl. Acad. Sci. USA. 91:11303–11307 10.1073/pnas.91.24.11303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster R.L., Zimmermann J.W. 1994. Spermatogenesis following male germ-cell transplantation. Proc. Natl. Acad. Sci. USA. 91:11298–11302 10.1073/pnas.91.24.11298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.H., Hemberg M., Barahona M., Ingber D.E., Huang S. 2008. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 453:544–547 10.1038/nature06965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Türkel N., Hemati N., Fuller M.T., Hunt A.J., Yamashita Y.M. 2008. Centrosome misorientation reduces stem cell division during ageing. Nature. 456:599–604 10.1038/nature07386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarini-Garcia H., Hornick J.R., Griswold M.D., Russell L.D. 2001. Distribution of type A spermatogonia in the mouse is not random. Biol. Reprod. 65:1179–1185 10.1095/biolreprod65.4.1179 [DOI] [PubMed] [Google Scholar]
- Cronkhite J.T., Norlander C., Furth J.K., Levan G., Garbers D.L., Hammer R.E. 2005. Male and female germline specific expression of an EGFP reporter gene in a unique strain of transgenic rats. Dev. Biol. 284:171–183 10.1016/j.ydbio.2005.05.015 [DOI] [PubMed] [Google Scholar]
- de Rooij D.G., Grootegoed J.A. 1998. Spermatogonial stem cells. Curr. Opin. Cell Biol. 10:694–701 10.1016/S0955-0674(98)80109-9 [DOI] [PubMed] [Google Scholar]
- Dobrinski I., Ogawa T., Avarbock M.R., Brinster R.L. 1999. Computer assisted image analysis to assess colonization of recipient seminiferous tubules by spermatogonial stem cells from transgenic donor mice. Mol. Reprod. Dev. 53:142–148 [DOI] [PubMed] [Google Scholar]
- Dubnau D., Losick R. 2006. Bistability in bacteria. Mol. Microbiol. 61:564–572 10.1111/j.1365-2958.2006.05249.x [DOI] [PubMed] [Google Scholar]
- Fawcett D.W. 1961. Intercellular bridges. Exp. Cell Res. Suppl 8:174–187 10.1016/0014-4827(61)90347-0 [DOI] [PubMed] [Google Scholar]
- Fawcett D.W., Ito S., Slautterback D. 1959. The occurrence of intercellular bridges in groups of cells exhibiting synchronous differentiation. J. Biophys. Biochem. Cytol. 5:453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Tumbar T., Guasch G. 2004. Socializing with the neighbors: stem cells and their niche. Cell. 116:769–778 10.1016/S0092-8674(04)00255-7 [DOI] [PubMed] [Google Scholar]
- Fuller M.T., Spradling A.C. 2007. Male and female Drosophila germline stem cells: two versions of immortality. Science. 316:402–404 10.1126/science.1140861 [DOI] [PubMed] [Google Scholar]
- Gilboa L., Lehmann R. 2004. How different is Venus from Mars? The genetics of germ-line stem cells in Drosophila females and males. Development. 131:4895–4905 10.1242/dev.01373 [DOI] [PubMed] [Google Scholar]
- Greenbaum M.P., Ma L., Matzuk M.M. 2007. Conversion of midbodies into germ cell intercellular bridges. Dev. Biol. 305:389–396 10.1016/j.ydbio.2007.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer G., Roepers-Gajadien H.L., Gademan I.S., Kal H.B., De Rooij D.G. 2003. Intercellular bridges and apoptosis in clones of male germ cells. Int. J. Androl. 26:348–353 10.1111/j.1365-2605.2003.00436.x [DOI] [PubMed] [Google Scholar]
- Hamra F.K., Gatlin J., Chapman K.M., Grellhesl D.M., Garcia J.V., Hammer R.E., Garbers D.L. 2002. Production of transgenic rats by lentiviral transduction of male germ-line stem cells. Proc. Natl. Acad. Sci. USA. 99:14931–14936 10.1073/pnas.222561399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamra F.K., Chapman K.M., Nguyen D.M., Williams-Stephens A.A., Hammer R.E., Garbers D.L. 2005. Self renewal, expansion, and transfection of rat spermatogonial stem cells in culture. Proc. Natl. Acad. Sci. USA. 102:17430–17435 10.1073/pnas.0508780102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckins C. 1971. The spermatogonial stem cell population in adult rats. I. Their morphology, proliferation and maturation. Anat. Rec. 169:533–557 10.1002/ar.1091690306 [DOI] [PubMed] [Google Scholar]
- Huckins C. 1978. Spermatogonial intercellular bridges in whole-mounted seminiferous tubules from normal and irradiated rodent testes. Am. J. Anat. 153:97–121 10.1002/aja.1001530107 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Ogonuki N., Inoue K., Miki H., Ogura A., Toyokuni S., Shinohara T. 2003. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol. Reprod. 69:612–616 10.1095/biolreprod.103.017012 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Miki H., Inoue K., Ogonuki N., Toyokuni S., Ogura A., Shinohara T. 2005a. Long-term culture of mouse male germline stem cells under serum-or feeder-free conditions. Biol. Reprod. 72:985–991 10.1095/biolreprod.104.036400 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Ogonuki N., Iwano T., Lee J., Kazuki Y., Inoue K., Miki H., Takehashi M., Toyokuni S., Shinkai Y., et al. 2005b. Genetic and epigenetic properties of mouse male germline stem cells during long-term culture. Development. 132:4155–4163 10.1242/dev.02004 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Toyokuni S., Shinohara T. 2005c. Genetic selection of mouse male germline stem cells in vitro: offspring from single stem cells. Biol. Reprod. 72:236–240 10.1095/biolreprod.104.035659 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Inoue K., Miki H., Ogonuki N., Takehashi M., Morimoto T., Ogura A., Shinohara T. 2006. Clonal origin of germ cell colonies after spermatogonial transplantation in mice. Biol. Reprod. 75:68–74 10.1095/biolreprod.106.051193 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Muneto T., Lee J., Takenaka M., Chuma S., Nakatsuji N., Horiuchi T., Shinohara T. 2008. Long-term culture of male germline stem cells from hamster testes. Biol. Reprod. 78:611–617 10.1095/biolreprod.107.065615 [DOI] [PubMed] [Google Scholar]
- Knoblich J.A. 2008. Mechanisms of asymmetric stem cell division. Cell. 132:583–597 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Kubota H., Avarbock M.R., Brinster R.L. 2004a. Culture conditions and single growth factors affect fate determination of mouse spermatogonial stem cells. Biol. Reprod. 71:722–731 10.1095/biolreprod.104.029207 [DOI] [PubMed] [Google Scholar]
- Kubota H., Avarbock M.R., Brinster R.L. 2004b. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA. 101:16489–16494 10.1073/pnas.0407063101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kanatsu-Shinohara M., Inoue K., Ogonuki N., Miki H., Toyokuni S., Kimura T., Nakano T., Ogura A., Shinohara T. 2007. Akt mediates self-renewal division of mouse spermatogonial stem cells. Development. 134:1853–1859 10.1242/dev.003004 [DOI] [PubMed] [Google Scholar]
- Li L., Xie T. 2005. Stem cell niche: structure and function. Annu. Rev. Cell Dev. Biol. 21:605–631 10.1146/annurev.cellbio.21.012704.131525 [DOI] [PubMed] [Google Scholar]
- Losick R., Desplan C. 2008. Stochasticity and cell fate. Science. 320:65–68 10.1126/science.1147888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H., Dubnau D. 2005. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 56:615–624 10.1111/j.1365-2958.2005.04592.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Lindahl M., Hyvönen M.E., Parvinen M., de Rooij D.G., Hess M.W., Raatikainen-Ahokas A., Sainio K., Rauvala H., Lakso M., et al. 2000. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 287:1489–1493 10.1126/science.287.5457.1489 [DOI] [PubMed] [Google Scholar]
- Nagano M.C. 2003. Homing efficiency and proliferation kinetics of male germ line stem cells following transplantation in mice. Biol. Reprod. 69:701–707 10.1095/biolreprod.103.016352 [DOI] [PubMed] [Google Scholar]
- Nagano M., Avarbock M.R., Brinster R.L. 1999. Pattern and kinetics of mouse donor spermatogonial stem cell colonization in recipient testes. Biol. Reprod. 60:1429–1436 10.1095/biolreprod60.6.1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M., Ryu B.Y., Brinster C.J., Avarbock M.R., Brinster R.L. 2003. Maintenance of mouse male germ line stem cells in vitro. Biol. Reprod. 68:2207–2214 10.1095/biolreprod.102.014050 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Nabeshima Y., Yoshida S. 2007. Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev. Cell. 12:195–206 10.1016/j.devcel.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Oakberg E.F. 1971. Spermatogonial stem-cell renewal in the mouse. Anat. Rec. 169:515–531 10.1002/ar.1091690305 [DOI] [PubMed] [Google Scholar]
- Oatley J.M., Brinster R.L. 2006. Spermatogonial stem cells. Methods Enzymol. 419:259–282 10.1016/S0076-6879(06)19011-4 [DOI] [PubMed] [Google Scholar]
- Oatley J.M., Brinster R.L. 2008. Regulation of spermatogonial stem cell self-renewal in mammals. Annu. Rev. Cell Dev. Biol. 24:263–286 10.1146/annurev.cellbio.24.110707.175355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J.M., Avarbock M.R., Brinster R.L. 2007. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J. Biol. Chem. 282:25842–25851 10.1074/jbc.M703474200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Aréchaga J.M., Avarbock M.R., Brinster R.L. 1997. Transplantation of testis germinal cells into mouse seminiferous tubules. Int. J. Dev. Biol. 41:111–122 [PubMed] [Google Scholar]
- Ogawa T., Dobrinski I., Brinster R.L. 1999. Recipient preparation is critical for spermatogonial transplantation in the rat. Tissue Cell. 31:461–472 10.1054/tice.1999.0060 [DOI] [PubMed] [Google Scholar]
- Ogawa T., Ohmura M., Yumura Y., Sawada H., Kubota Y. 2003. Expansion of murine spermatogonial stem cells through serial transplantation. Biol. Reprod. 68:316–322 10.1095/biolreprod.102.004549 [DOI] [PubMed] [Google Scholar]
- Ogawa T., Ohmura M., Ohbo K. 2005. The niche for spermatogonial stem cells in the mammalian testis. Int. J. Hematol. 82:381–388 10.1532/IJH97.05088 [DOI] [PubMed] [Google Scholar]
- Russell L.D., Ettlin R.A., Hikim A.P., Clegg E.D. 1990. Mammalian spermatogenesis. Histological and Histopathological Evaluation of the Testis. Cache River Press, Clearwater, FL: 1–40 [Google Scholar]
- Ryu B.Y., Kubota H., Avarbock M.R., Brinster R.L. 2005. Conservation of spermatogonial stem cell self-renewal signaling between mouse and rat. Proc. Natl. Acad. Sci. USA. 102:14302–14307 10.1073/pnas.0506970102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield R. 1978. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 4:7–25 [PubMed] [Google Scholar]
- Smits W.K., Eschevins C.C., Susanna K.A., Bron S., Kuipers O.P., Hamoen L.W. 2005. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol. Microbiol. 56:604–614 10.1111/j.1365-2958.2005.04488.x [DOI] [PubMed] [Google Scholar]
- Spradling A., Drummond-Barbosa D., Kai T. 2001. Stem cells find their niche. Nature. 414:98–104 10.1038/35102160 [DOI] [PubMed] [Google Scholar]
- Süel G.M., Garcia-Ojalvo J., Liberman L.M., Elowitz M.B. 2006. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 440:545–550 10.1038/nature04588 [DOI] [PubMed] [Google Scholar]
- Süel G.M., Kulkarni R.P., Dworkin J., Garcia-Ojalvo J., Elowitz M.B. 2007. Tunability and noise dependence in differentiation dynamics. Science. 315:1716–1719 10.1126/science.1137455 [DOI] [PubMed] [Google Scholar]
- Weber J.E., Russell L.D. 1987. A study of intercellular bridges during spermatogenesis in the rat. Am. J. Anat. 180:1–24 10.1002/aja.1001800102 [DOI] [PubMed] [Google Scholar]
- Wernet M.F., Mazzoni E.O., Celik A., Duncan D.M., Duncan I., Desplan C. 2006. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 440:174–180 10.1038/nature04615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P.S., Egger B., Brand A.H. 2008. Asymmetric stem cell division: lessons from Drosophila. Semin. Cell Dev. Biol. 19:283–293 10.1016/j.semcdb.2008.01.007 [DOI] [PubMed] [Google Scholar]
- Wu Z., Falciatori I., Molyneux L.A., Richardson T.E., Chapman K.M., Hamra F.K. 2009. Spermatogonial culture medium: an effective and efficient nutrient mixture for culturing rat spermatogonial stem cells. Biol. Reprod. 81:77–86 10.1095/biolreprod.108.072645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y.M., Fuller M.T. 2005. Asymmetric stem cell division and function of the niche in the Drosophila male germ line. Int. J. Hematol. 82:377–380 10.1532/IJH97.05097 [DOI] [PubMed] [Google Scholar]
- Yoshida S. 2009. Casting back to stem cells. Nat. Cell Biol. 11:118–120 10.1038/ncb0209-118 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Sukeno M., Nabeshima Y.I. 2007. A vasculature-associated niche for undifferentiated spermatogonia in the mouse testis. Science. 317:1722–1726 10.1126/science.1144885 [DOI] [PubMed] [Google Scholar]
- Zhang X., Ebata K.T., Nagano M.C. 2003. Genetic analysis of the clonal origin of regenerating mouse spermatogenesis following transplantation. Biol. Reprod. 69:1872–1878 10.1095/biolreprod.103.019273 [DOI] [PubMed] [Google Scholar]