Viral subversion of cholesterol homeostasis provides insights into sterol trafficking, autophagy, and lysosomal storage diseases.
Abstract
Host–pathogen interactions are important model systems for understanding fundamental cell biological processes. In this study, we describe a cholesterol-trafficking pathway induced by the adenovirus membrane protein RID-α that also subverts the cellular autophagy pathway during early stages of an acute infection. A palmitoylation-defective RID-α mutant deregulates cholesterol homeostasis and elicits lysosomal storage abnormalities similar to mutations associated with Niemann-Pick type C (NPC) disease. Wild-type RID-α rescues lipid-sorting defects in cells from patients with this disease by a mechanism involving a class III phosphatidylinositol-3-kinase. In contrast to NPC disease gene products that are localized to late endosomes/lysosomes, RID-α induces the accumulation of autophagy-like vesicles with a unique molecular composition. Ectopic RID-α regulates intracellular cholesterol trafficking at two distinct levels: the egress from endosomes and transport to the endoplasmic reticulum necessary for homeostatic gene regulation. However, RID-α also induces a novel cellular phenotype, suggesting that it activates an autonomous cholesterol regulatory mechanism distinct from NPC disease gene products.
Introduction
Lysosomal storage diseases (LSDs) comprise >40 human genetic disorders (Neufeld, 1991). Although a majority of LSDs involve mutations in lysosomal acid hydrolases, others such as Niemann-Pick type C (NPC) disease have underlying defects in intracellular trafficking (Patterson et al., 2001). NPC is a fatal autosomal recessive disorder caused by mutations in the polytopic membrane protein NPC1 located in late endosomes (LEs) and lysosomes in 95% of cases or more rarely in the soluble protein NPC2, which is concentrated in lysosomes (Chang et al., 2005). NPC1 and -2 coordinate egress of unesterified cholesterol from LEs/lysosomes, and mutations in either protein cause cholesterol overload in these organelles. As a result, elevated cholesterol levels are not counterbalanced by sterol homeostatic mechanisms in the ER, and cholesterol and other lipids continue to accumulate, causing the formation of abnormal lysosomal storage organelles (LSOs; Goldstein et al., 2006). NPC cholesterol dysfunction also increases basal levels of autophagy (Ko et al., 2005; Pacheco et al., 2007), indicating a possible role for sterol trafficking in this pathway as well. Perturbed autophagy has been implicated in cell death associated with NPC and other neuropathies, including Alzheimer’s and Huntington’s diseases, suggesting a common molecular basis for disorders with extensive endocytic–autophagic–lysosomal neuropathology (Shacka et al., 2008).
In contrast to the endocytic–lysosomal pathway, which degrades extracellular and plasma membrane (PM) proteins, autophagy mediates turnover of cytosolic constituents (Klionsky and Emr, 2000; Mizushima, 2007). Although autophagy occurs at low basal levels in virtually all cells, multiple stimuli, including nutrient depletion, accumulation of protein aggregates, and organelle obsolescence, up-regulate this pathway. Autophagy is controlled by a unique set of autophagy-related (Atg) proteins that sequester cytosolic components in double-membrane vesicles known as autophagosomes (Klionsky and Emr, 2000; Mizushima, 2007). One of these proteins, LC3 (the mammalian homologue of yeast Atg8), is lipidated by an Atg8 ubiquitin-like conjugation system, facilitating its insertion into nascent autophagic membranes (Tanida et al., 2004). Although the functional significance of this modification is unknown, LC3 translocation provides a convenient means of identifying autophagy-derived membranes (Tanida et al., 2004). Despite differences in substrates and compartmental structure, cellular homeostasis requires coordinated activity of endocytic–autophagic–lysosomal pathways. Some of the key molecules linking these pathways include the class III phosphatidylinositol-3-kinase (PI3K) Vps34, which regulates early endosome (EE) biogenesis as well as autophagosome membrane expansion (Backer, 2008), and the small GTPase Rab7 (Bucci et al., 2000; Gutierrez et al., 2004). In mammalian cells, autophagosomes have also been shown to fuse with endosomes en route to lysosomes, resulting in intermediate structures known as amphisomes (Eskelinen, 2005). Recently, mutations in components of the ESCRT (endosomal sorting complex required for transport) machinery responsible for sorting ubiquitinated endocytic protein cargo in multivesicular bodies (MVBs) have been shown to block autophagy by inhibiting autophagosome–endosome fusion (Nara et al., 2002; Lee et al., 2007; Rusten et al., 2007). Furthermore, autophagy is impaired by loss of COPI coatomer necessary for normal EE function (Razi et al., 2009). However, despite recent progress, there are relatively few mechanistic insights as to how endocytosis and autophagy are coordinated. Continued examination of the molecular basis for connectivity between these two degradative pathways is crucial to identify common therapeutic targets for LSDs and other disorders in which accumulation of undegraded substrates is a prominent feature.
Adenovirus (Ad) is a nonenveloped DNA virus internalized by receptor-mediated endocytosis that escapes to cytosol by lysing endosomal membranes (Fig. 1 a; Meier and Greber, 2003; Wiethoff et al., 2005). Although the majority of Ad infections are subclinical, approximately half of Ad serotypes are associated with human diseases (Horwitz, 1996). The molecular basis for Ad-induced disease involves signaling pathways elicited during viral uptake and early Ad gene products that modulate host innate immune responses (Ginsberg and Prince, 1994; Stewart et al., 2007). RID-α (formerly E3-13.7), which is encoded by an early 3 region (E3) Ad transcript, was first identified by its ability to redirect constitutively recycling EGF receptors (EGFRs) to lysosomes independent of ligand and EGFR Tyr kinase activity or ubiquitination (Fig. 1 b; Hoffman et al., 1992a; Hoffman and Carlin, 1994; Tsacoumangos et al., 2005). Subsequent studies demonstrated that RID-α cooperates with a second E3 protein, RID-β, to down-regulate proapoptotic receptors TNFR1, TRAIL-R1, and Fas (for review see Horwitz, 2004). Recently, we discovered that RID-α regulates protein sorting by mimicking GTP-Rab7 through the binding of two Rab7 effectors, Rab7-interacting lysosomal protein (RILP) and oxysterol-binding protein–related protein 1L (ORP1L; Shah et al., 2007). However, RID-α shares no sequence homology with Rab7 and has no intrinsic catalytic activity, providing a unique example of how a viral protein mimics the function of an endogenous protein to hijack an intracellular trafficking process. In this study, we report that RID-α coordinates trafficking in endocytic and autophagic pathways by activating a cholesterol homeostasis mechanism that operates independent of NPC1/NPC2 by a class III PI3K–dependent mechanism.
Figure 1.
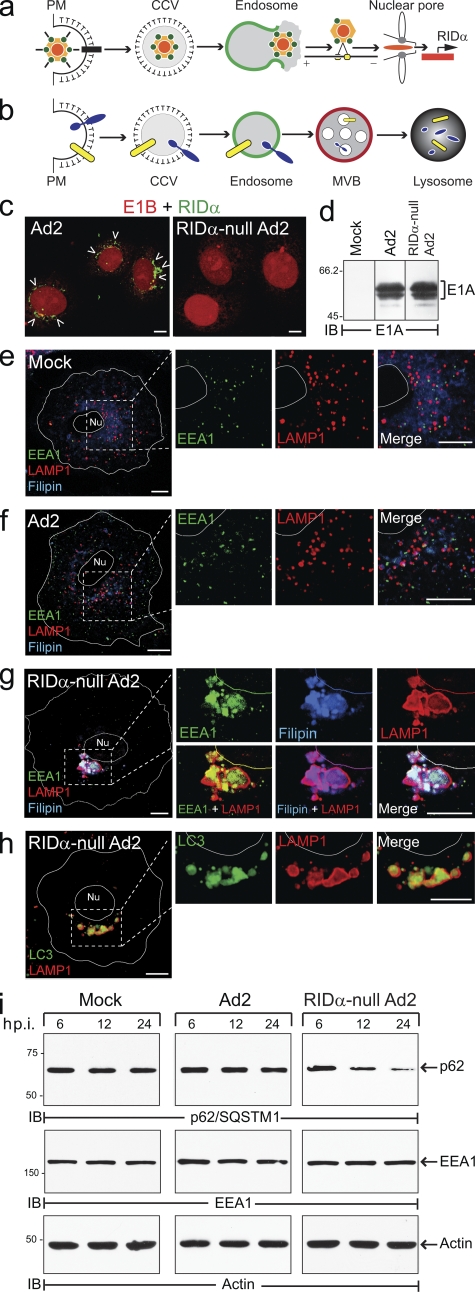
RID-α counterbalances Ad-induced autophagy. (a) Ad2 internalized by clathrin-mediated endocytosis escapes from EEs by membrane lysis followed by MT-dependent transport to the nucleus and viral gene expression. (b) RID-α, which is expressed within the first few hours of an acute infection, facilitates targeted degradation of EGFRs (blue) and proapoptotic receptors (yellow). (c) Confocal images of A549 cells infected with wild-type or RID-α–null Ad2 viruses stained with E1B and RID-α antibodies. Arrowheads indicate RID-α–positive compartments. (d) Equal aliquots of total cellular protein from mock- and Ad-infected A549 cells resolved by SDS-PAGE and immunoblotted with E1A antibody. (e–g) Confocal images of mock (e), Ad2 (f), and RID-α–null Ad2-infected (g) A549 cells stained with EEA1 and LAMP1 antibodies and filipin; magnified images of single and merged channels are shown on the right. (h) Confocal images of RID-α–null Ad2-infected A549 cells stained for LC3 and LAMP1. (e–h) Cell and nucleus (Nu) boundaries were drawn using MetaMorph software. Boxed areas show regions of the image that were magnified. (i) Equal aliquots of total cellular protein from mock- and Ad-infected cells immunoblotted with antibodies to p62/SQSTM1, EEA1, and actin as a function of time postinfection (p.i.). CCV, clathrin-coated vesicle; IB, immunoblot. (d and i) Molecular mass is indicated in kilodaltons. Bars, 10 µm.
Results
RID-α counteracts Ad-induced autophagy
To gain further insight into RID-α function, Ad-infected cells were examined with antibodies to early (EEA1) and late (LAMP1) endocytic markers. Cells were also stained with filipin, an intrinsically fluorescent antibiotic which binds free cholesterol. Intracellular cholesterol levels vary greatly among different organelles, and specific sorting mechanisms are required to establish and maintain these distinct compositions (Maxfield and Tabas, 2005). Because Ad triggers endosome membrane lysis, we hypothesized that this might lead to altered sterol balance in infected cells. Human A549 cells were infected with wild-type group C Ad2 and a mutant Ad2 virus with an internal deletion in the RID-α ORF that both produced equivalent levels of major early region (E1) proteins (Fig. 1, c and d). Similar to mock-infected cells (Fig. 1 e), EEA1 and LAMP1 vesicles were dispersed throughout the cytosol in cells infected with wild-type Ad2 (Fig. 1 f). In addition, intracellular distributions of free cholesterol were essentially identical in mock-treated and Ad2-infected cells. In contrast, cells infected with the RID-α–null Ad2 virus displayed relatively few enlarged LAMP1 compartments that were concentrated in a perinuclear cluster. These cells were essentially devoid of typical cytosolic EEA1 vesicles, and EEA1 was sequestered in the enlarged LAMP1 compartments (Fig. 1 g). The LAMP1 structures were also enriched for free cholesterol (Fig. 1 g) and the autophagosomal membrane protein LC3 (Fig. 1 h). Altogether, these data suggest that Ad infection by itself induces an autophagic response characterized by a pronounced redistribution of intracellular cholesterol and EEA1 that is offset by RID-α expression. This hypothesis was further investigated by monitoring levels of the LC3-binding protein p62/SQSTM1, which regulates the formation of protein aggregates and is selectively degraded by autophagy (Ichimura et al., 2008). In contrast to mock- and Ad2-infected cells in which levels were essentially unchanged, cells infected with RID-α–null Ad2 virus exhibited significant p62/SQSTM1 turnover by 24 h postinfection compared with a control protein (actin), which was unaffected by autophagy (Fig. 1 i). Interestingly, although EEA1 was sequestered in enlarged LAMP1/LC3-positive autophagosomes in RID-α–null Ad2-infected cells, EEA1 protein levels were essentially unchanged (Fig. 1 i), which is consistent with a recent study showing that fusion with EEs is required for efficient autophagy (Razi et al., 2009). These data suggest that RID-α regulates homeostatic balance between endocytosis and autophagy to facilitate endosome–lysosome degradation of select membrane cargo in Ad-infected cells.
RID-α is palmitoylated at Cys67
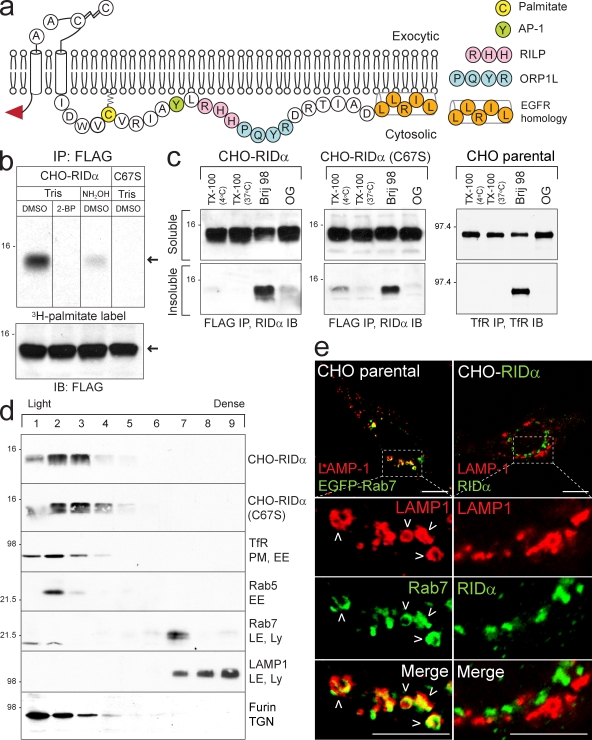
Despite a functional relationship to Rab7 (Shah et al., 2007), RID-α lacks intrinsic GTPase activity, raising questions as to how this viral protein is regulated in cells. The RID-α carboxyl tail has a Cys residue located 6 aa from its second transmembrane domain that could modulate RID-α function by undergoing reversible palmitoylation (Fig. 2 a). Because RID-α down-regulates EGFRs independent of other Ad proteins (Hoffman et al., 1990; Cianciola et al., 2007), the following experiments were performed in CHO cells stably expressing amino-terminal Flag-tagged RID-α to facilitate biochemical analysis. We found that RID-α was labeled with [3H]palmitate and that radioactive incorporation was blocked in cells incubated with the palmitoylation inhibitor 2-bromopalmitate (2-BP; Fig. 2 b). Furthermore, as expected for thioester linkages, the 3H radiolabel was significantly reduced when RID-α immunocomplexes were incubated with hydroxylamine after SDS-PAGE (Fig. 2 b). [3H]palmitate incorporation was also completely blocked by introducing a Ser mutation at position 67 (C67S; Fig. 2 b). Although palmitoylation can have an important role in sorting proteins lacking transmembrane peptide sequences to detergent-resistant membranes (DRMs), this modification is less likely to affect DRM partitioning of integral membrane proteins (Greaves and Chamberlain, 2007; Linder and Deschenes, 2007). This notion was tested for RID-α by extracting cell membranes with Triton X-100 at different temperatures to determine whether the viral protein partitions into classical Triton X-100–insoluble DRMs (Brown and Rose, 1992). Results in Fig. 2 c indicate that wild-type RID-α and mutant RID-α (C67S) proteins were completely solubilized by Triton X-100 at low temperature, which is in contrast to the glycosylphosphatidylinositol-anchored protein CD73 found exclusively in the Triton X-100–insoluble fraction (Fig. S1 a). Similar to transferrin receptor (TfR), which is a nonraft protein, both of the viral proteins also exhibited modest insolubility with Brij 98 and essentially complete solubility with octyl glucoside (Fig. 2 c and Fig. S1 b; Pike et al., 2005). Altogether, these data suggest that the viral protein is found in bulk membrane independent of palmitoylation status.
Figure 2.
RID-α is palmitoylated at Cys67. (a) Schematic of RID-α membrane topology showing the amino-terminal Flag epitope (red arrowhead), luminal signal peptidase cleavage (which generates 13.7- or 11.3-kD RID-α species) and disulfide bond formation sites, carboxyl tail sequence with palmitoylation and protein interaction sites, and the EGFR homology domain (Hoffman et al., 1992b; Vinogradova et al., 1998; Tsacoumangos et al., 2005; Cianciola et al., 2007; Shah et al., 2007). (b) CHO–RID-α and CHO–RID-α (C67S) cells treated with 2-BP or DMSO and radiolabeled with [3H]palmitate. Flag immunocomplexes were separated by SDS-PAGE, and gels were incubated with Tris or hydroxylamine solutions before fluorography. The bottom panel shows Flag immunocomplexes from duplicate samples analyzed by RID-α immunoblotting for loading control. Arrows denote 13.7-kD and RID-α species. (c) CHO cell lines extracted with the indicated detergents and RID-α (left and middle) or TfR (right) immunocomplexes from detergent-soluble and -insoluble fractions immunoblotted with antibodies to the same protein. (d) Membranes from stable CHO cells expressing wild-type RID-α or RID-α (C67S) fractionated on 27% Percoll gradients and equal aliquots of total membrane protein immunoblotted with antibodies to RID-α (top two panels) are shown. Equal aliquots of total membrane protein from parental CHO cells immunoblotted with antibodies to marker proteins for intracellular membrane compartments listed in the figure (bottom five panels) are shown. (b–d) Molecular mass is indicated in kilodaltons. (e) Confocal images of CHO cells transfected with EGFP-Rab7 plasmid and stained for LAMP1 or CHO–RID-α cells stained for LAMP1 and RID-α. Boxed areas show regions of the image that were magnified. Arrowheads denote compartments with colocalized markers. IB, immunoblot; IP, immunoprecipitation; Ly, lysosome; OG, octyl glucoside; TX-100, Triton X-100. Bars, 10 µm.
RID-α induces accumulation of an autophagy-like compartment
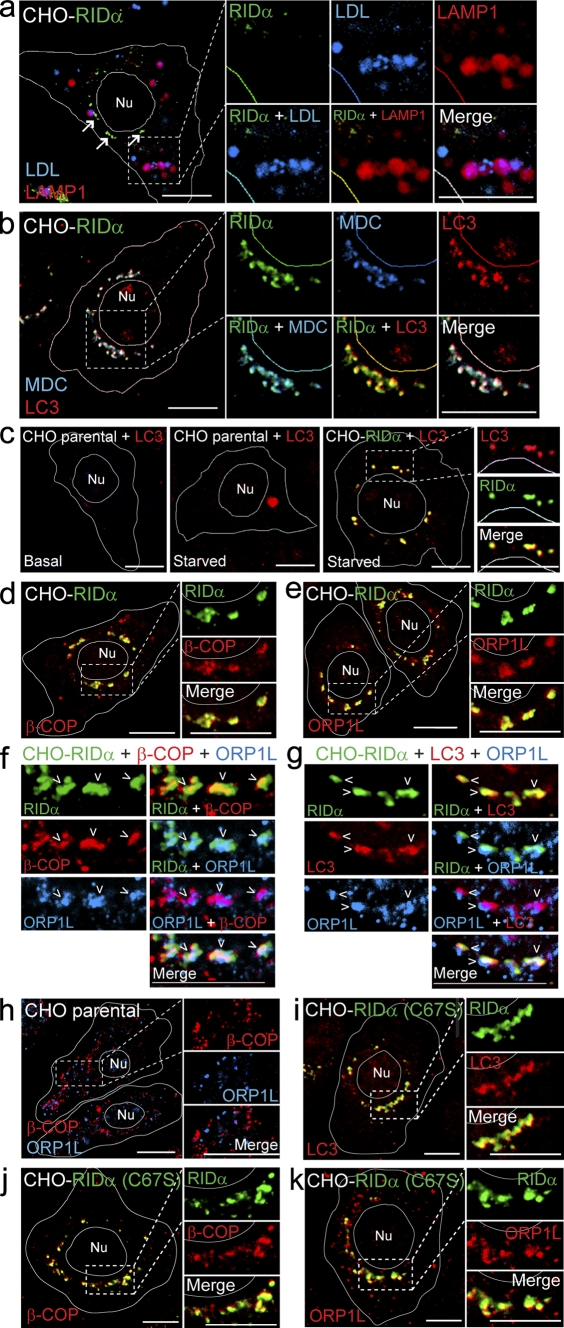
A previous study showed that RID-α binds GTP-Rab7 effectors and that at least one of these interactions is necessary for RID-α biological activity in infected cells (Shah et al., 2007). To determine whether Rab7 and RID-α are in the same or different membrane compartments, cell membranes were fractionated on isoosmotic Percoll gradients. Wild-type and RID-α (C67S) mutant proteins were enriched in relatively light fractions overlapping EE markers as well as the TGN marker furin (Fig. 2 d). RID-α sedimentation profiles were in sharp contrast to Rab7 in parental CHO cells, which cosedimented with LAMP1 in a relatively dense fraction (Fig. 2 d). Similarly, confocal microscopy revealed that EGFP-Rab7 but not wild-type RID-α colocalized with LAMP1-positive LEs/lysosomes (Fig. 2 e). Instead, wild-type RID-α was associated primarily with a discrete population of perinuclear vesicles (Fig. 2 e). To determine whether RID-α compartments were accessible to endocytic probes, cells with ectopic RID-α expression were incubated with low density lipoprotein (LDL) conjugated to a highly fluorescent lipophilic dye (DiI-LDL) and then fixed and stained for the viral protein and LAMP1. LDL colocalized with LAMP1 but not RID-α (Fig. 3 a), indicating that this ligand is largely excluded from RID-α compartments. However, in contrast to the endocytic probe, the RID-α compartment was readily accessible to the autofluorescent compound monodansylcadaverine (MDC), which is a specific in vivo marker for autophagic vesicles (Fig. 3 b; Biederbick et al., 1995). Furthermore, RID-α was extensively colocalized with the autophagic membrane marker LC3 (Fig. 3 b and Fig. S2, a and b). The LC3 staining pattern in RID-α–expressing cells is striking for two reasons. First, RID-α/LC3-positive compartments were devoid of LAMP1, suggesting that they do not fuse with lysosomes. Second, relatively little LC3 was membrane associated in parental CHO cells unless the cells were cultured under autophagy-inducing conditions to trigger the formation of LC3-positive autophagosomes (Fig. 3 c, left and middle). Interestingly, LC3-positive vesicles retained the same perinuclear distribution in autophagy-induced cells expressing ectopic RID-α (Fig. 3 c, right) that we had originally observed under basal conditions (Fig. 3 b), suggesting that RID-α modulates starvation-induced autophagic machinery.
Figure 3.
RID-α induces formation of a hybrid organelle with characteristics of both endocytic and autophagic vesicles. (a) CHO–RID-α cells stained with LAMP1 and RID-α antibodies after incubation with DiI-LDL. Arrows denote the RID-α compartment. (b) CHO–RID-α cells stained for LC3 and RID-α after incubation with MDC. (c) Parental CHO or CHO–RID-α cells stained for LC3 or LC3 and RID-α under basal conditions (left) or after 6-h incubation in Earl’s balanced salt solution to induce autophagy (middle and right). (d and e) CHO–RID-α cells stained for RID-α and β-COP (d) or ORP1L (e). (f and g) Magnified images of single and merged channels of CHO–RID-α cells triple stained for RID-α and ORP1L and either β-COP (f) or LC3 (g). Arrowheads denote compartments with colocalized markers. (h) Parental CHO cells stained for β-COP and ORP1L. (i–k) CHO–RID-α (C67S) cells stained for RID-α and LC3 (i), β-COP (j), or ORP1L (k). (a–e and h–k) Cell and nucleus (Nu) boundaries were drawn using MetaMorph software. Boxed areas show regions of the image that were magnified. Bars, 10 µm.
Several other cellular proteins were enriched in RID-α compartments, including the COPI coatomer subunit β-COP (Fig. 3 d and Fig. S2 c), which is involved in retrograde Golgi to ER transport, endocytosis, and autophagy (McMahon and Mills, 2004), and the Rab7 effector protein ORP1L (Fig. 3 e and Fig. S2 d), which is normally present on LEs but which also binds directly to the RID-α carboxyl tail (Johansson et al., 2005; Shah et al., 2007). In addition, we demonstrated that some RID-α compartments contained β-COP and ORP1L (Fig. 3 f) or LC3 and ORP1L (Fig. 3 g), suggesting the presence of all three marker proteins in a common set of vesicles. Contrary to the pattern of staining in RID-α–expressing cells, β-COP and ORP1L were found primarily on nonoverlapping small vesicles dispersed throughout the cytosol in parental CHO cells (Fig. 3 h). Introduction of the C67S mutation had no apparent effect on the accumulation of RID-α–positive compartments enriched for LC3, β-COP, and ORP1L (Fig. 3, i–k; and Fig. S2, e–g). Previous studies showed that RID-α exhibits only minimal overlap with other EE markers (EEA1 and Rab5) and none with the Golgi (Crooks et al., 2000; Cianciola et al., 2007). Together with the new data presented in this study, RID-α compartments do not correspond to any well-defined intracellular organelle. In addition, assembly of the organelle is unaffected by RID-α palmitoylation status.
Palmitoylation regulates RID-α function
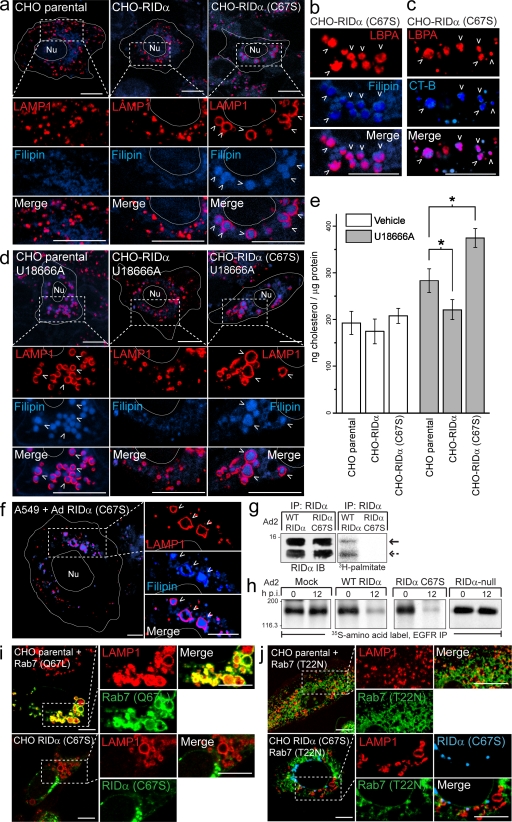
In contrast to membrane partitioning and intracellular localization, our data indicate that palmitoylation does influence RID-α function. This conclusion is based on confocal images showing that RID-α (C67S) alters morphology of LAMP1-positive LEs/lysosomes in the absence of other Ad proteins compared with parental cells or cells expressing wild-type RID-α (Fig. 4 a). LAMP1-postive structures are enlarged and also accumulate high levels of several molecules, including cholesterol (Fig. 4, a and b), the atypical LE/lysosome lipid lysobisphosphatidic acid (LBPA; Fig. 4, b and c), and cholera toxin subunit B (CT-B), which binds GM1 ganglioside (Fig. 4 c). These data suggest that RID-α controls a cholesterol egress mechanism unmasked in cells with normal NPC1/NPC2 machinery by a mutation affecting its ability to undergo palmitoylation. The RID-α (C67S)–induced phenotype is similar to the phenotype produced by U18666A, which impairs intracellular cholesterol biosynthesis and perturbs cholesterol egress from endosomes by an unknown mechanism (Fig. 4 d; Liscum, 1990). Interestingly, ectopic expression of wild-type RID-α substantially reduced the U18666A-induced phenotype compared with parental CHO cells, in contrast to RID-α (C67S), which appeared to exacerbate U18666A-induced accumulation of free cholesterol in enlarged LAMP1-positive LEs/lysosomes (Fig. 4 d). Furthermore, ectopic RID-α was associated with a statistically significant reduction in intracellular levels of free cholesterol compared with parental CHO cells after U18666A treatment (Fig. 4 e). Similarly RID-α (C67S) produced a statistically significant increase in intracellular free cholesterol after U18666A treatment compared with parental CHO cells (Fig. 4 e).
Figure 4.
RID-α (C67S) induces the formation of enlarged lipid-filled LAMP1 structures. (a) Confocal images of CHO cell lines stained with LAMP1 antibody and filipin. (b and c) Magnified images of single and merged channels from CHO–RID-α (C67S) cells stained with LBPA antibody and filipin (b) or LBPA antibody after incubation with Alexa Fluor 647 CT-B (c). (d) CHO cell lines treated with U18666A for 8 h and stained with LAMP1 antibody and filipin. (e) Cholesterol quantification in CHO cell lines treated with DMSO (vehicle) or U18666A for 8 h using the Amplex red cholesterol assay kit. Values were normalized to total cellular protein and are displayed as mean ± SEM (*, P < 0.01). (f) Confocal images of A549 cells infected with a mutant RID-α (C67S) Ad2 virus and stained with LAMP1 antibody and filipin 24 h postinfection. (g) A549 cells infected with wild-type (WT) or RID-α (C67S) Ad2 viruses radiolabeled with [3H]palmitate and RID-α immunocomplexes separated by SDS-PAGE for fluorography. Arrows denote 13.7 (solid)- or 11.3 (dashed)-kD RID-α species. (h) A549 cells radiolabeled with 35S-Express Protein Labeling mix and mock infected or infected with wild-type or mutant Ad viruses and EGFR immunocomplexes analyzed by SDS-PAGE and fluorography at the times indicated. (g and h) Molecular mass is indicated in kilodaltons. (i) Parental CHO cells transfected with constitutively active (Q67L) EGFP-Rab7 and stained for LAMP1 or CHO–RID-α (C67S) cells stained for LAMP1 and RID-α. (j) Parental CHO or CHO–RID-α (C67S) cells transfected with dominant-negative (T22N) EGFP-Rab7 and stained for LAMP1 (parental) or LAMP1 and RID-α (CHO–RID-α (C67S)). (a–d and f) Arrowheads indicate examples of costained vesicles. (a, d, f, i, and j) Boxed areas show regions of the image that were magnified. (a, d, and f) Cell and nucleus (Nu) boundaries were drawn using MetaMorph software. IB, immunoblot; IP, immunoprecipitation; p.i., postinfection. Bars, 10 µm.
Aberrant LAMP1-positive, cholesterol-loaded vesicles were also found in A549 cells infected with a RID-α (C67S) Ad2 mutant virus (Fig. 4 f). Similar to results obtained in stable RID-α–expressing CHO cell lines, RID-α underwent palmitoylation at Cys67 in Ad2-infected cells (Fig. 4 g). Cells infected with RID-α (C67S) mutant Ad2 nevertheless degraded EGFRs similar to cells infected with wild-type Ad2 and in contrast to cells infected with a RID-α–null virus (Fig. 4 h). This result is not unexpected because LEs contain lysosomal enzymes that are active at the low pH of the compartment (Bright et al., 2005). Altogether, these data indicate that RID-α (C67S) affects lipid trafficking but not EGFR degradation, suggesting that these two RID-α functions are dissociable.
Analysis of GTP-Rab7 function has been greatly aided by the design of mutant proteins that are either constitutively active or dominant inhibitory because of defective GTP-GDP cycling (Press et al., 1998). To determine whether RID-α (C67S) has constitutively active or dominant-inhibitory properties, its effects on LE morphology were compared with those elicited by mutant Rab7 proteins. Although constitutively active Rab7 and RID-α (C67S) both induced the formation of enlarged LAMP1-positive vesicles, constitutively active Rab7 colocalized with the enlarged vesicles, whereas RID-α (C67S) did not (Fig. 4 i). Dominant-inhibitory Rab7 dispersed LAMP1-positive vesicles throughout the cytosol (Fig. 4 j) by interfering with microtubule (MT)-dependent MVB/LE motility (Bucci et al., 2000). RID-α (C67S) coexpression counteracted the effect of dominant-inhibitory Rab7 and altered the morphology of LAMP1 compartments (Fig. 4 j), which is similar to results in cells with normal levels of wild-type Rab7 (Fig. 4 a). These results suggest that RID-α (C67S) alters LE morphology similar to constitutively active Rab7 and that wild-type RID-α regulates a pathway that parallels Rab7-dependent trafficking or acts upstream of GTP-Rab7 in a common pathway.
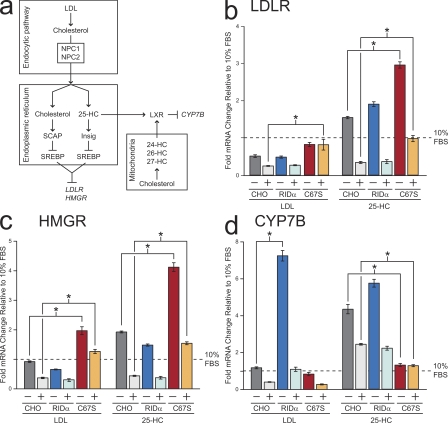
RID-α regulates cholesterol homeostatic gene expression
Cholesterol levels are tightly regulated by classical end-product feedback mechanisms that control sterol regulatory element–binding protein (SREBP)–dependent gene expression at the level of the ER (Fig. 5 a; Chang et al., 2006). These mechanisms are set by rapid cholesterol flux between the ER and endosomes, and they become exaggerated or erratic if this dynamic flux is perturbed. If RID-α regulates cholesterol trafficking to the ER, it should also modulate SREBP-dependent gene transcription. In addition, results in the previous section showing LSO-like structures in CHO cells expressing RID-α (C67S) predict that this mechanism should be highly dependent on RID-α palmitoylation. These hypotheses were tested using real-time PCR to examine the expression of two SREBP-dependent genes: LDL receptor (LDLR), which is responsible for endosomal transport of LDL-cholesterol, and 3-hydroxy-3-methyglutaryl-CoA reductase (HMGR), the rate-limiting enzyme in de novo cholesterol synthesis (Fig. 5 a). Homeostatic responses were assayed by comparing mRNA levels in sterol-depleted CHO cell lines with and without supplemental sterol (see Materials and methods). In addition to LDL-cholesterol, levels of 25-hydroxycholesterol (HC), an ER cholesterol metabolite and another potent regulator of the SREBP pathway (Fig. 5 a; Goldstein et al., 2006) were manipulated. Similar to parental CHO cells, LDLR and HMGR mRNA levels declined after LDL-cholesterol loading in CHO–RID-α cells (Fig. 5, b and c). CHO–RID-α (C67S) cells exhibited exaggerated LDL-cholesterol homeostatic responses compared with the other two cell lines, which were statistically significant for both genes. In contrast to HMGR (Fig. 5 c), LDLR mRNA was not reduced by LDL-cholesterol loading (Fig. 5 b), suggesting that these two genes respond differently to fluctuations in ER cholesterol pool size in CHO–RID-α (C67S) cells. Parental CHO and CHO–RID-α cells both exhibited elevated LDLR and HMGR gene expression after sterol depletion, which sharply declined with the addition of 25-HC (Fig. 5, b and c). Although CHO–RID-α (C67S) cells displayed a similar pattern, the homeostatic response for both genes was significantly exaggerated, suggesting that RID-α (C67S) also perturbs the 25-HC ER pool. These results support the hypothesis that RID-α regulates the SREBP pathway by a cholesterol-trafficking mechanism that is phenotypically silent unless it is blocked by the C67S RID-α mutation. They also suggest that the RID-α–dependent cholesterol-trafficking pathway is the prevalent mechanism in CHO cells with functional NPC proteins.
Figure 5.
RID-α modulates sterol-regulated gene expression. (a) Transcriptional mechanisms controlling expression of the target genes LDLR, HMGR, and CYP7B described in Results. (b–d) LDLR (b), HMGR (c), and CYP7B (d) mRNA levels quantified by real-time PCR. Values are expressed as relative units after internal normalization to glyceraldehyde 3-phosphate dehydrogenase mRNA levels and compared with control samples from the same cell lines cultured in 10% FBS from three independent experiments. Data are presented as mean ± SEM (*, P < 0.001).
In addition to the SREBP pathway, oxysterols are potent activators of nuclear liver X receptors (LXRs), which regulate genes involved in sterol storage, transport, and catabolism (Fig. 5 a; Ory, 2004). Thus, cells were also examined for expression of the gene encoding CYP7B, a P450 enzyme which is transcriptionally suppressed by activated LXRs (Ory, 2004). Whereas LDL-cholesterol loading provoked a modest homeostatic response in CHO parental and CHO–RID-α (C67S) cells, CHO–RID-α cells displayed an exaggerated sevenfold increase in CYP7B mRNA expression in sterol-depleted cells, which declined with LDL-cholesterol loading (Fig. 5 d). In contrast, CHO–RID-α cells exhibited a CYP7B homeostatic response comparable with parental cells after 25-HC loading (Fig. 5 d). Because LXRs are activated by multiple oxysterols, these data suggest that RID-α regulates LXR gene transcription by perturbing a different oxysterol pool (Fig. 5 a). For example, NPC fibroblasts have reduced levels of the cholesterol metabolite 27-HC produced in mitochondria, indicating that the NPC1/NPC2 machinery also regulates cholesterol egress to this compartment (Zhang et al., 2008). In contrast to LDL-cholesterol, 25-HC loading did not elicit a CYP7B homeostatic response in CHO–RID-α (C67S) cells (Fig. 5 d). These results are consistent with data obtained for the SREBP pathway, indicating that RID-α regulates 25-HC–dependent gene transcription by a palmitoylation-dependent mechanism.
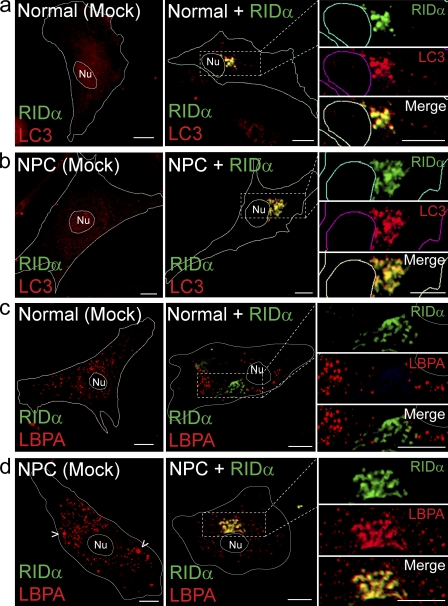
RID-α reduces LSOs and intracellular cholesterol levels in NPC cells
Production of LSO-like structures in CHO–RID-α (C67S) cells with functional NPC1/NPC2 proteins suggests that RID-α (C67S) either disables the NPC1/NPC2 machinery or deregulates a novel NPC1/NPC2-independent pathway. We determined whether or not the viral protein complements genetic defects in NPC cells to distinguish between these two possibilities. Despite some cell type–specific variability in compartment morphology, our data indicate that RID-α induced the accumulation of a compartment that contains the autophagic membrane protein LC3 in normal fibroblasts (Fig. 6 a) and fibroblasts from an NPC patient (Fig. 6 b), which is similar to results obtained in stable CHO cell lines (Fig. 3). By comparison, membrane-associated LC3 was virtually undetectable in either cell type after mock transfection under basal conditions (Fig. 6, a and b). Similarly, RID-α colocalized with β-COP and ORP1L in both normal and NPC fibroblasts (unpublished data). However, in contrast to normal fibroblasts in which LBPA was present in dispersed cytosolic vesicles with or without ectopic RID-α expression (Fig. 6 c), LBPA was highly concentrated in RID-α compartments in NPC cells expressing the viral protein (Fig. 6 d). By comparison, LBPA was present in a few enlarged vesicles that presumably correspond to LSOs (Fig. 6 d, arrowheads) in addition to smaller dispersed vesicles in mock-transfected NPC cells. These data imply that RID-α vesicles are dynamic compartments that may be remodeled in response to intracellular cholesterol levels or other abnormalities associated with NPC proteins.
Figure 6.
RID-α induces formation of dynamic hybrid organelles with characteristics of both endocytic and autophagic vesicles in NPC fibroblasts. (a and b) Confocal images of normal (a) or NPC cells (b) stained for RID-α and LC3 after mock transfection or transfection with a RID-α expression plasmid. (c and d) Confocal images of normal (c) or NPC cells (d) stained for RID-α and LBPA after mock transfection or transfection with a RID-α expression plasmid. (d) Arrowheads show presumptive LBPA-positive LSOs. (a–d) Cell and nucleus (Nu) boundaries were drawn using MetaMorph software. Boxed areas show regions of the image that were magnified. Bars, 10 µm.
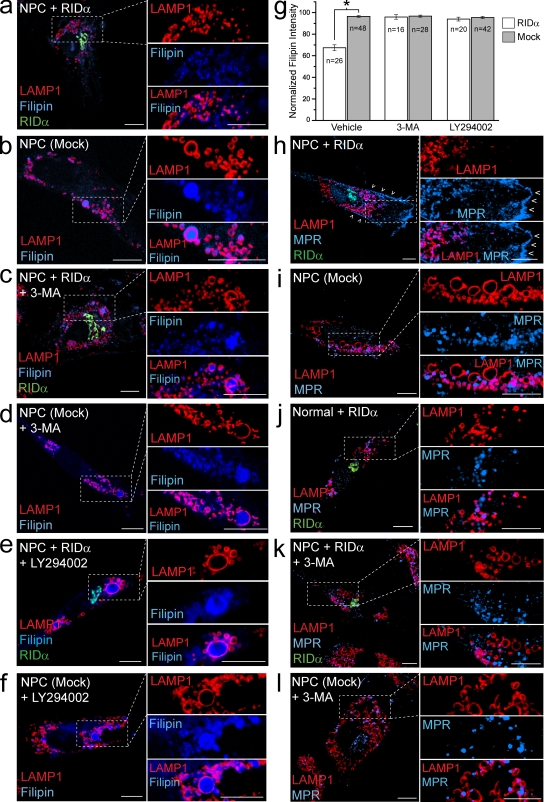
We next determined whether RID-α offsets NPC cholesterol dysfunction. Cholesterol-enriched LSOs were virtually undetectable in NPC cells expressing RID-α (Fig. 7 a) compared with mock-transfected NPC cells (Fig. 7 b). The finding that RID-α and LC3 colocalize suggests that RID-α–dependent lipid trafficking subverts the autophagy machinery. This hypothesis was tested by treating cells with the class III PI3K inhibitor 3-methyladenine (3-MA), which prevents autophagic sequestration after LC3 membrane insertion (Eskelinen, 2005). 3-MA substantially reduced the ability of the viral protein to clear cholesterol-filled LSOs in NPC cells (Fig. 7 c) compared with mock-transfected NPC cells, in which it had no apparent effect (Fig. 7 d). Similar results were obtained using a second PI3K inhibitor, LY294002 (Fig. 7, e and f). Morphological data were corroborated by quantification of filipin fluorescence, which revealed that cholesterol levels were reduced by ∼33% in NPC cells expressing RID-α versus mock-transfected cells in the same microscopic field (Fig. 7 g). Furthermore, cholesterol levels were essentially identical in NPC cells treated with 3-MA or LY294002 with and without ectopic RID-α expression (Fig. 7 g).
Figure 7.
RID-α expression rescues cholesterol trafficking and associated defects in NPC cells by a class III PI3K–dependent mechanism. (a–f) NPC cells transfected in the absence or presence of PI3K inhibitors, as indicated, and stained for LAMP1, filipin, and RID-α (a, c, and e) or LAMP1 and filipin (b, d, and f). (g) Quantification of normalized filipin fluorescence intensity in cells treated similarly as in a–f and as described in Materials and methods. Data are presented as mean ± SEM (*, P < 0.001). (h–l) NPC and normal cells transfected in the absence or presence of PI3K inhibitors, as indicated, and stained for LAMP1, MPR, and RID-α (h, j, and k) or LAMP1 and MPR (i and l). Cells in h–l were cultured in 10% LPDS media for 3 d followed by 50 µg/ml LDL for 24 h before staining. (h) Arrowheads denote PM localization of MPR. (a–f and h–l) Boxed areas show regions of the image that were magnified. Bars, 10 µm.
NPC mutations also adversely affect trafficking of protein cargo such as mannose 6-phosphate receptors (MPRs; Kobayashi et al., 1998). MPRs deliver newly synthesized acid hydrolases to the endocytic pathway, where hydrolases are subsequently transferred to lysosomes and MPRs recycle back to the TGN (Ghosh et al., 2003). MPRs may also reach the cell surface followed by rapid uptake from clathrin-coated pits. RID-α expression caused MPRs to accumulate at the PM (Fig. 7 h), in contrast to mock-transfected NPC cells in which MPRs were trapped in LSOs because of a lipid imbalance imposed by cholesterol accumulation (Fig. 7 i). Expression of RID-α in normal fibroblasts had no discernable effect on intracellular MPR distribution (Fig. 7 j). These data suggest that RID-α does not reconstitute MPR EE to LE transport enabling increased cell surface expression and/or that it prevents MPR reinternalization from the PM. Similar to results obtained for cholesterol, 3-MA blocked MPR rerouting to the PM in RID-α–expressing NPC cells (Fig. 7 k), in contrast to mock-transfected cells in which 3-MA had no apparent effect on intracellular MPR distribution (Fig. 7 l).
Discussion
In this study, we demonstrate that RID-α is a potent regulator of intracellular cholesterol in gene transfer experiments as well as during acute Ad infections. Our data indicate that RID-α facilitates two distinct steps in cholesterol trafficking: egress from endosomes and transport to the ER necessary for homeostatic gene regulation (Fig. 8). RID-α rescues the cholesterol storage phenotype in NPC fibroblasts, suggesting that the viral protein regulates a molecular mechanism that operates independent of NPC1/NPC2. This supposition is further supported by evidence obtained with a palmitoylation-defective RID-α (C67S) mutant, which alters LE morphology to produce NPC-like LSOs and deregulates ER homeostatic mechanisms in CHO cells with functional NPC proteins (Cruz et al., 2000; Naureckiene et al., 2000). Even though the viral protein binds GTP-Rab7 effectors (Shah et al., 2007), RID-α is not present in LAMP1-positive LEs but influences endosome function at a distance from a membrane compartment with characteristics of autophagic vesicles. Furthermore, RID-α is excluded from cholesterol-rich DRMs, suggesting that its function is not restricted by NPC mutations, in contrast to Rab7 which is inhibited by high levels of cholesterol (Kobayashi et al., 1998). The RID-α (C67S) mutant also abrogates the cellular phenotype associated with a dominant-negative form of Rab7, providing further evidence that RID-α and GTP-Rab7 are not necessarily interchangeable. A previous study indicated that newly synthesized RID-α is sorted at the level of TGN via AP-1 clathrin adapters (Cianciola et al., 2007), and the working model in Fig. 8 suggests that RID-α regulates cholesterol trafficking in endosomes after TGN exit. A similar mechanism has been reported in T cells of the adaptive immune system, where antigen-loading compartments for major histocompatibility complex class II molecules receive continuous input from autophagosomes (Schmid et al., 2007). However, we cannot exclude the possibility that RID-α facilitates retrograde cholesterol transport to the ER via a TGN intermediate (Urano et al., 2008). The presence of β-COP in RID-α compartments is consistent with either role because COP1 coatomer regulates endocytosis and autophagy as well as Golgi to ER retrograde transport (McMahon and Mills, 2004). It is also conceivable that RID-α regulates transport in both directions depending on sterol load and cell physiology.
Figure 8.
Model of the role of RID-α as a coordinator of endosome trafficking. LDL-cholesterol (Chol) esters are internalized via the PM LDLR (1a), unesterified, and trafficked to MVBs/LEs (1b). Cholesterol is egressed out of these organelles by the coordinated action of NPC1/NPC2 and transported to the ER, where it is sensed by cholesterol and 25-HC homeostatic machinery (2a), and to mitochondria, where it is converted to other oxysterols (2b). NPC1/NPC2 mutations block cholesterol egress to both of these compartments, deregulate cholesterol homeostasis, reduce oxysterol production, and induce LSO formation (3). Although RID-α vesicles resemble autophagic vesicles, they also have distinct molecular properties that distinguish them from bona fide autophagosomes (4a). Specialized RID-α compartments that sequester the MDC indicator dye from bulk cytosol may supply an unknown rate-limiting factor important for endocytic maintenance and/or cholesterol homeostasis. RID-α activates an autonomous class III PI3K–dependent cholesterol egress mechanism that restores ER cholesterol trafficking in NPC1-defective cells (4b). RID-α also suppresses Ad-induced autophagy (5a) and facilitates endosome to lysosome targeting of select membrane protein cargo (5b). Positive and negative RID-α–mediated actions are highlighted in red.
Our results indicate the RID-α–containing membranes do not correspond to any well-defined intracellular organelle. Although the origin of these compartments remains unclear, it is possible that RID-α arrests maturation of a physiologically short-lived early autophagic compartment. Alternatively RID-α could create a virus-specific compartment by remodeling intracellular membranes. Our data favor the first possibility because RID-α vesicles are enriched in LC3 recruited to nascent autophagic membranes but devoid of LAMP1 found in mature autophagosomes. This is not the first example of a pathogen-hijacking host autophagic machinery to carry out a novel function, as poliovirus triggers formation of unique LC3-positive compartments (Taylor and Kirkegaard, 2007). However, in contrast to poliovirus, which utilizes virally induced autophagosomal membranes to facilitate viral RNA replication, RID-α regulates innate immune responses (Ginsberg and Prince, 1994; Stewart et al., 2007). We have also demonstrated that RID-α promotes endosome to lysosome transport of select membrane cargo at the expense of enhanced autophagic flux in acutely infected cells. It is conceivable that RID-α compartments sequester key proteins required for autophagosome–lysosome fusion or alternatively provide a surplus of rate-limiting molecules required for efficient endocytosis. Evidence that RID-α regulates endosome sterol balance independent of NPC1/NPC2 favors the latter hypothesis. However, it is conceivable that RID-α integrates endocytosis and autophagy by a combination of these two mechanisms. The finding that RID-α function requires class III PI3K activity necessary for trafficking in both pathways (Backer, 2008) also supports a coordinating role for RID-α.
Another molecule enriched in RID-α compartments is ORP1L, a member of the family of oxysterol-binding proteins implicated in a variety of cellular functions (Olkkonen et al., 2006). ORP1L is a GTP-Rab7 effector linked to LE MT-dependent motility and lipid trafficking that also binds RID-α (Johansson et al., 2005, 2007; Shah et al., 2007). ORP1L is comprised of multiple amino-terminal ankyrin repeats, a pleckstrin homology domain, and a carboxyl-terminal oxysterol regulatory domain capable of binding cholesterol and 25-HC in vitro (Suchanek et al., 2007). Contrary to GTP-Rab7, which recognizes ORP1L ankyrin repeats, RID-α interacts with oxysterol regulatory domain sequences (Shah et al., 2007), suggesting that it may directly regulate sterol binding. Although palmitoylation does not alter RID-α membrane partitioning or intracellular compartmentalization, this modification does influence RID-α function presumably by regulating conformation of the RID-α carboxyl tail and its ability to interact with protein-binding partners (Charollais and Van Der Goot, 2009). Thus, failure of RID-α to undergo palmitoylation may sequester ORP1L and/or inhibit its sterol-binding properties, leading to deregulated cholesterol homeostasis. RID-α also binds a second GTP-Rab7 effector, RILP, and this interaction is required for RID-α biological activity in Ad2-infected cells (Shah et al., 2007). It will be interesting in the future to determine whether RILP is also recruited to RID-α–induced compartments because RILP binds ESCRT components involved in the dual regulation of endocytosis and autophagy independent of GTP-Rab7 (Progida et al., 2006; Wang and Hong, 2006; Fader et al., 2008).
In contrast to CHO cells and normal fibroblasts, RID-α compartments in NPC fibroblasts are also enriched for LBPA, which is known to regulate cholesterol clearance from endosomes (Chevallier et al., 2008). The NPC phenotype is partially reversed by the addition of exogenous LBPA, suggesting that LBPA is a limiting factor contributing to disease pathology (Chevallier et al., 2008). Therefore, it is possible that RID-α restores NPC cholesterol trafficking by up-regulating a basal mechanism involved in LBPA transport to the endocytic system. The finding that LBPA is only detectable in RID-α compartments in cells with inherent cholesterol imbalance reveals the plasticity of RID-α–based mechanisms.
Although its role in controlling innate immune responses is well known, our experiments have identified at least three new RID-α–associated functions during acute Ad infections. First, RID-α subverts virus-induced autophagy. Although the molecular basis for Ad2-induced autophagy is currently unknown, it is conceivable that this pathway is activated to remove endosomes damaged by membrane lysis immediately after Ad internalization (Fig. 1 a). Ad-induced endosome membrane lysis may also deplete a critical rate-limiting factor sufficient to disrupt trafficking throughout the endocytic pathway that is reintroduced to the system via a RID-α–dependent mechanism (Fig. 8). Second, in contrast to receptors involved in innate immune responses, RID-α may divert MPRs to the PM, where they fulfill distinct functions compared with their normal role in shuttling acid hydrolases to lysosomes (Ghosh et al., 2003). Third, RID-α regulates endosome cholesterol egress in acutely infected cells. Cholesterol is required for Ad2 internalization from the PM and endosome escape, suggesting that Ads may be taken up to specialized cholesterol-enriched domains that execute membrane lysis (Imelli et al., 2004). Therefore, it is conceivable that RID-α restores the cholesterol balance that is perturbed during the early stages of an acute Ad infection. RID-α–induced cholesterol trafficking may also be important in latent infections in which E3 proteins are thought to have a prominent role (McNees et al., 2002). However, in contrast to NPC1/NPC2, RID-α selectively regulates cholesterol trafficking to the ER and consequently could trigger cholesterol imbalance in other intracellular compartments during an acute infection.
This study highlights several new areas for future investigation. First, given its capacity to reconstitute cholesterol trafficking in cells with defective NPC proteins, it will now be of interest to fully characterize the newly identified RID-α–induced pathway. Second, this study raises the possibility that the NPC1/NPC2 machinery is impaired during acute Ad infections. Third, RID-α–containing vesicles may shed new light on the intracellular membrane origin of autophagic vesicles.
Materials and methods
Antibodies and reagents
The following antibodies were used: actin and Flag-BioM2 mouse mAb (Sigma-Aldrich), β-COP mouse mAb (Novus Biologicals), E1A mouse mAb (BD), E1B rat mAb (EMD), furin rabbit pAb (Thermo Fisher Scientific), LBPA mouse mAb (provided by J. Gruenberg, University of Geneva, Geneva, Switzerland), LC3 mouse mAb (MBL), LC3 rabbit pAb (Abgent), cation-dependent MPR chicken pAb (Millipore), human-specific IL2R-α mAb (American Type Culture Collection), ORP1L goat pAb (Imgenex), Rab5 mouse mAb (BD), Rab7 goat pAb and CD73 rabbit pAb (Santa Cruz Biotechnology, Inc.), RID-α rabbit pAb produced with a synthetic peptide corresponding to residues 76–91 (Hoffman et al., 1992b), p62/SQSTM1 mouse mAb (Abnova), TfR mouse mAb (Invitrogen), and hamster- and human-specific LAMP1 mouse mAbs (Developmental Studies Hybridoma Bank). Fluorescent- and HRP-tagged secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc.; filipin, MDC, U18666A, and 3-MA were purchased from Sigma-Aldrich; LY294002 was purchased from EMD; and Alexa Fluor 674 CT-B and DiI-LDL were purchased from Invitrogen. EGFP-Rab7 mammalian expression plasmids were a gift from C. Bucci (University of Helsinki, Helsinki, Finland).
Mutagenesis and cloning
Wild-type RID-α with an amino-terminal Flag epitope (Flag–RID-α) was amplified by PCR from a pExchange2/RID-α template (Cianciola et al., 2007) using forward, 5′-GCGGAGCTAGCATGGATTACAAGGATG-3′; and reverse, 5′-GATCAGCTAGCCTTAAAGAATTCTGAGAAGATCA-3′ primers incorporating flanking NheI restriction sites (underlined) and subcloned in pCR2.1 supplied with the TOPO TA cloning kit (Invitrogen). Flag–RID-α was excised from pCR2.1 and ligated to the NheI restriction site in multiple cloning site A of the bicistronic mammalian expression vector pIRES (BD). A cDNA encoding the human IL2R-α subunit excised from pBK–IL2R-α (Tsacoumangos et al., 2005) was ligated to BamHI and NotI restriction sites in pIRES/RID-α multiple cloning site B. A RID-α C67S point mutation was incorporated into pIRES/Flag–RID-α/IL2R-α using the QuikChange XL site-directed mutagenesis kit (Agilent Technologies) and forward, 5′-CAGTTCATTGACTGGGTTAGTGTCCGCATTGCGTAC-3′; and reverse, 5′-GTACGCAATGCGCACACTAACCCAGTCAATGAACTG-3′ primers (C67S substitution in bold). Flag–RID-α (C67S) was amplified using forward, 5′-TTACAGCTCTTGCGGCCGCAGTACTTAATACGACTCAC-3′; and reverse, 5′-ATGCTCGACGCGCGTACGCTCGAGGCTAG-3′ primers incorporating 5′ NotI and 3′ BsiWI restriction sites (underlined) and subcloned into pCR2.1. Flag–RID-α (C67S) excised from pCR2.1 was ligated to the retroviral bicistronic expression vector pQCXIN (BD) digested with NotI and BsiWI. PCR primers were designed using the DNASTAR software package (DNASTAR, Inc.) and purchased from Operon. PCR-amplified sequences were verified by automated DNA sequencing.
Cell lines and Ad stocks
To create a permanent CHO cell line expressing Flag–RID-α (referred to as CHO–RID-α), parental CHO cells were transfected with pIRES/Flag–RID-α/IL2R-α using Trans-IT CHO transfection reagent (Mirus Bio) and selected for drug resistance in media supplemented with 200 µg/ml G418 (EMD). Drug-selected cells were further enriched for IL2R-α surface expression by sterile sorting on a flow cytometer (Aria; BD) after surface staining with an IL2R-α mAb and FITC-conjugated secondary antibody. To create a permanent CHO cell line expressing Flag–RID-α (C67S) (referred to as CHO–RID-α (C67S)), GP2-293 retrovirus packaging cells were transfected with pQCXIN/Flag–RID-α (C67S) using Trans-IT 293 transfection reagent (Mirus Bio). Pantropic retrovirus was generated upon subsequent transfection of drug-selected packaging cells with pVSV-G plasmid. Retrovirus-containing media was collected 48 h later and added to CHO cells, followed by G418 selection. Stable RID-α expression was verified by immunoblotting and immunostaining with Flag and RID-α antibodies.
GP2-293 cells were grown in DME, and CHO cells were grown in MEM-α. A549 cells were grown in Ham’s F12 media, and normal (GM05659) and NPC (GM03123) fibroblasts (Coriell Institute for Medical Research) were grown in MEM. All media were supplemented with 10% FBS (unless otherwise noted) and 2 mM Glu. For cholesterol loading, cells were cultured in media with 10% lipoprotein-deficient serum (LPDS) obtained commercially (Sigma-Aldrich), followed by LPDS-containing media supplemented with freshly prepared 50 µg/ml LDL-cholesterol (provided by R. Morton and D. Greene, Cleveland Clinic Foundation, Cleveland, OH). For 25-HC loading, cells were cultured in media with 10% delipidated serum that was prepared by incubating FBS with 20 mg/ml Cab-O-Sil (Sigma-Aldrich), which is known to deplete lipoproteins (Vance et al., 1984), for 16 h at 4°C with rotation, clarified by centrifugation (15,000 rpm for 1 h at 4°C), and sterilized sequentially with 0.45- and 0.22-µm filters (Lala et al., 1997; Haas et al., 2007). The cells were incubated for 24 h in the delipidated serum–containing media with or without 5 µg/ml of supplemented 25-HC (diluted from a 2.5-mg/ml ethanol stock; Sigma-Aldrich) for the final 24 h.
Wild-type Ad2 was obtained from the American Type Culture Collection. Mutant Ads with an internal RID-α deletion or a RID-α (C67S) substitution were described previously (Carlin et al., 1989; Hoffman et al., 1992b). Ad stocks were grown in HEK293 cells, and titers were determined by plaque assay using standard techniques. Cells were acutely infected with ∼200 plaque-forming units per cell (Hoffman and Carlin, 1994).
Cell harvesting
Cells were lysed under denaturing conditions using 1% SDS in 10 mM Tris, pH 7.4, preheated to 100°C to analyze total cellular protein. Cells were lysed using radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 2 mM EGTA, 5 mM EDTA, 1% NP-40, 0.5% Na deoxycholate, and 0.1% SDS) to recover immunocomplexes on protein A–Sepharose beads (Sigma-Aldrich; Kostenko et al., 2006). Differential detergent extractability experiments were performed according to previously established methods (Skibbens et al., 1989). In brief, cells were washed twice with PBS supplemented with 1 mM MgCl2 and 1 mM CaCl2, scraped in the same buffer, and centrifuged for 3 min at 3,200 rpm. Cells were lysed with extraction buffer (25 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 0.2 mM PMSF, and 1 µM leupeptin) supplemented with 1% Triton X-100, 1% Brij 98, or 60 mM octyl glucoside. Cells were passed through a 22-gauge needle 10 times and incubated at 4 or 37°C for 30 min, followed by five passages through a 22-gauge needle. Lysates were centrifuged at high speed for 10 min at 4°C, and supernatants (detergent-soluble fraction) were adjusted to 0.1% SDS. Membrane pellets were washed once with appropriate extraction buffer, resuspended in 100 µl of solubilization buffer (50 mM Tris, pH 8.8, 1% SDS, and 5 mM EDTA), passed through a 22-gauge needle 12 times, brought up to 1 ml with appropriate extraction buffer, and centrifuged at high speed for 10 min at 4°C to generate a detergent-insoluble fraction. Detergent-soluble and -insoluble fractions were subjected to immunoprecipitation, and immunocomplexes were washed extensively, solubilized with Laemmli buffer, resolved by SDS-PAGE, and transferred to nitrocellulose filters for immunoblotting using standard methods.
Cell fractionation
Cells were homogenized and fractionated on Percoll (GE Healthcare) gradients as previously described (Cianciola et al., 2007). In brief, cells were rinsed twice with PBS supplemented with 2 mM EDTA and 5 mM EGTA and then scraped in ice-cold homogenization buffer (HB) consisting of 10 mM Hepes, pH 7.5, 0.25 M sucrose, 1 mM EDTA, and protease inhibitors. Cells were collected by centrifugation, resuspended in HB buffer, and homogenized with 22 strokes of a Dounce homogenizer. The homogenate was diluted with an equal volume of fresh HB and centrifuged at 400 g for 10 min at 4°C to pellet unbroken cells and nuclei. Postnuclear supernatants were adjusted to a final concentration of 27% Percoll in 0.25 M sucrose using a 90% Percoll stock solution and then layered over a 1-ml sucrose cushion consisting of 10× HB. Gradients were centrifuged for 90 min at 25,000 g in an SS34 fixed angle rotor (Thermo Fisher Scientific) without braking. A total of nine 1.2-ml fractions were collected manually starting from the top of the gradient. Fractions were analyzed by comparing equal aliquots of total cellular protein determined by the Bradford assay (Bio-Rad Laboratories) or by immunoprecipitation after membranes were solubilized with RIPA buffer and centrifuged at 100,000 g for 30 min at 4°C in a TL 100.3 fixed angle rotor (Beckman Coulter) to precipitate Percoll.
Metabolic labeling
For metabolic labeling with [3H]palmitate, cells were pretreated with 100 µM 2-BP or DMSO vehicle for 24 h and labeled with 1 mCi [3H]palmitate (American Radiolabeled Chemicals, Inc.) in 3 ml of complete media supplemented with 2-BP or DMSO for 3 h. Cells were lysed with RIPA buffer, and lysates were immunoprecipitated with anti-Flag BioM2 antibody. After SDS-PAGE, gels were incubated for 16 h in 1 M hydroxylamine, pH 7, or a control solution of 1 M Tris, pH 7, and prepared for fluorography. For metabolic labeling with 35S-labeled amino acids, cells were preincubated in Cys- and Met-free medium for 1 h. Amino acid–starved cells were pulse labeled for 30 min with 35S-Express Protein Labeling mix (PerkinElmer) diluted in amino acid–deficient medium supplemented with 10% dialyzed FBS. Radiolabeled cells were incubated with medium supplemented with 10% dialyzed FBS and a 10-fold excess of nonradioactive Cys and Met (chase medium) for 2 h before Ad infection, followed by additional incubation with chase medium for up to 12 h.
Confocal microscopy
Normal and NPC fibroblasts were mock treated or transfected with 3 µg of plasmid DNA using the Normal Human Dermal Fibroblast Nucleofector kit and the Nucleofector device (Lonza). CHO cells were transfected using Trans-It CHO transfection reagent. All cells were seeded on poly-l-Lys–treated coverslips, perforated with 0.5% β-escin, fixed with 3% paraformaldehyde, incubated with appropriate primary antibodies for 1 h at room temperature and fluorochrome-conjugated secondary antibodies for 16 h at 4°C, and mounted on glass slides using SlowFade (Invitrogen). For PI3K inhibitor experiments, cells were incubated with 10 mM 3-MA (diluted from a 200-mM stock made in water) or 100 µM LY294002 (diluted from a 10-mM stock made in DMSO) for 3 h before staining. Some cells were incubated with 10 µg/ml DiI-LDL for 15 min at 4°C or 5 µg/ml Alexa Fluor 674 CT-B for 30 min at 4°C, followed by a 2-h chase in serum-free media at 37°C before staining. Other cells were incubated with 50 µM MDC diluted in PBS for 10 min at 37°C before staining. Fixed cells were incubated with 50 µg/ml filipin for 16 h at 4°C to detect free cholesterol. Confocal images were acquired with a laser-scanning microscope (LSM 510 Meta; Carl Zeiss, Inc.) and accompanying software using diode (excitation 405 nm), argon (excitation 488 nm), and HeNe (excitation 543 and 633 nm) lasers and 63× or 100× Plan-Apochromat NA 1.4 objectives. Filipin was excited with the diode laser, and emissions were collected between 411 and 486 nm. When necessary, phase contrast images were collected, and cell and nucleus boundaries were drawn using MetaMorph software (MDS Analytical Technologies) and overlaid onto respective confocal images. All images were processed with Photoshop CS3 and Illustrator CS3 software (Adobe).
Quantification of total cellular cholesterol
CHO cell lines were treated with 500 nM U18666A for 8 h, washed twice with PBS, scraped in 1 ml PBS, and homogenized by 10 passages through a 22-gauge needle. Aliquots were analyzed for cholesterol content using the Amplex red cholesterol assay (Invitrogen) according to the manufacturer’s instructions, and absorbance was read at 568 nm using a SpectraMax M2 plate reader and SoftMax Pro v5 software (MDS Analytical Technologies). Cholesterol content was calculated using cholesterol standards supplied with the kit and normalized to total denatured cellular protein, as determined by the Bradford assay.
Quantification of filipin fluorescence intensity
Confocal images were collected using a 40× C-Apochromat NA 1.2 objective and a 6-µm optical slice. Data were collected for untransfected and transfected cells that were discriminated based on RID-α staining. Free cholesterol content was determined using MetaMorph software to quantify the mean filipin intensity of individual cells. The highest value obtained for all of the untransfected cells in a single microscopic field was set to 100, and the data for cells expressing RID-α in the same field were normalized to this value.
Autophagy induction
Before staining, cells were washed twice with PBS and incubated with Earl’s balanced salt solution for 6 h, with the addition of 10 µg/ml pepstatin A and 10 µg/ml E64d (Sigma-Aldrich) for the final 3 h.
Gene expression experiments
RNA extracted using the ToTALLY RNA kit (Applied Biosystems) was reverse transcribed using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). Target gene mRNA levels were analyzed by real-time PCR using standard hydrolysis probe (TaqMan) techniques and a glyceraldehyde 3-phosphate dehydrogenase internal control. The mRNA expression levels for cells receiving various sterol treatments were plotted as fold change relative to mRNA levels for control cells cultured in standard media containing 10% FBS for each individual cell line. PCR was performed on a real-time PCR system (model 7500; Applied Biosystems), and products were analyzed using 7500 System SDS software version 1.3 (Applied Biosystems). Probe and primer pairs listed in Table S1 were designed using the Universal Probe Library Assay Design Center (Roche). Probes and PCR master mix were purchased from Roche, and primers were purchased from Operon.
Statistical analysis
Statistical analyses for Amplex red cholesterol quantification were performed using the Student’s t test (SigmaStat). Data are expressed as mean ± SEM from three independent experiments. A p-value of <0.01 was considered statistically significant. Statistical analyses for real-time PCR were performed using a one-way repeated measures analysis of variance and Bonferroni post-hoc testing (SigmaStat). Values obtained from three independent experiments are expressed as mean ± SEM. A p-value of <0.001 was considered statistically significant. Statistical analyses for filipin fluorescence quantification were performed using the Student’s t test. Data are expressed as mean ± SEM from three independent experiments. A p-value of <0.001 was considered statistically significant.
Online supplemental material
Fig. S1 shows the temperature-sensitive Triton X-100 extraction of lipid raft–associated CD73 as a positive control and quantification of wild-type and mutant RID-α detergent extraction profiles for Fig. 2 c. Fig. S2 is a gallery of colocalization data for wild type and mutant RID-α with various marker proteins, as described in Fig. 3. Table S1 lists the target genes (with accession number), probes, and primer sequences used for real-time PCR. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200903039/DC1.
Acknowledgments
We would like to thank Maryanne Pendergast and the Neurosciences Imaging Center for assistance with the confocal microscopy, Richard Morton and Diane Greene for the purified LDL, Cecilia Bucci for the Rab7 constructs, Jean Gruenberg for the anti-LBPA antibody, Ankur Shah and Gary Landreth for critically reviewing the manuscript, and all laboratory members for their support and discussions.
This work was supported by Public Health Service grant RO1GM081498 to C.R. Carlin. N.L. Cianciola was supported in part by National Institutes of Health grant T32 HL-007653.
Footnotes
Abbreviations used in this paper:
- 2-BP
- 2-bromopalmitate
- 3-MA
- 3-methyladenine
- Ad
- adenovirus
- CT-B
- cholera toxin subunit B
- DRM
- detergent-resistant membrane
- EE
- early endosome
- EGFR
- EGF receptor
- HB
- homogenization buffer
- HC
- hydroxycholesterol
- HMGR
- 3-hydroxy-3-methyglutaryl-CoA reductase
- LBPA
- lysobisphosphatidic acid
- LDL
- low density lipoprotein
- LDLR
- LDL receptor
- LE
- late endosome
- LPDS
- lipoprotein-deficient serum
- LSD
- lysosomal storage disease
- LSO
- lysosomal storage organelle
- LXR
- liver X receptor
- MDC
- monodansylcadaverine
- MPR
- mannose 6-phosphate receptor
- MT
- microtubule
- MVB
- multivesicular body
- NPC
- Niemann-Pick type C
- ORP1L
- oxysterol-binding protein–related protein 1L
- PI3K
- phosphatidylinositol-3-kinase
- PM
- plasma membrane
- RILP
- Rab7-interacting lysosomal protein
- RIPA
- radioimmunoprecipitation assay
- SREBP
- sterol regulatory element–binding protein
- TfR
- transferrin receptor
References
- Backer J.M. 2008. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem. J. 410:1–17 10.1042/BJ20071427 [DOI] [PubMed] [Google Scholar]
- Biederbick A., Kern H.F., Elsässer H.P. 1995. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur. J. Cell Biol. 66:3–14 [PubMed] [Google Scholar]
- Bright N.A., Gratian M.J., Luzio J.P. 2005. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr. Biol. 15:360–365 10.1016/j.cub.2005.01.049 [DOI] [PubMed] [Google Scholar]
- Brown D.A., Rose J.K. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 68:533–544 10.1016/0092-8674(92)90189-J [DOI] [PubMed] [Google Scholar]
- Bucci C., Thomsen P., Nicoziani P., McCarthy J., van Deurs B. 2000. Rab7: a key to lysosome biogenesis. Mol. Biol. Cell. 11:467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin C.R., Tollefson A.E., Brady H.A., Hoffman B.L., Wold W.S. 1989. Epidermal growth factor receptor is down-regulated by a 10,400 MW protein encoded by the E3 region of adenovirus. Cell. 57:135–144 10.1016/0092-8674(89)90179-7 [DOI] [PubMed] [Google Scholar]
- Chang T.Y., Reid P.C., Sugii S., Ohgami N., Cruz J.C., Chang C.C. 2005. Niemann-Pick type C disease and intracellular cholesterol trafficking. J. Biol. Chem. 280:20917–20920 10.1074/jbc.R400040200 [DOI] [PubMed] [Google Scholar]
- Chang T.-Y., Chang C.C.Y., Ohgami N., Yamauchi Y. 2006. Cholesterol sensing, trafficking, and esterification. Annu. Rev. Cell Dev. Biol. 22:129–157 10.1146/annurev.cellbio.22.010305.104656 [DOI] [PubMed] [Google Scholar]
- Charollais J., Van Der Goot F.G. 2009. Palmitoylation of membrane proteins (Review). Mol. Membr. Biol. 26:55–66 10.1080/09687680802620369 [DOI] [PubMed] [Google Scholar]
- Chevallier J., Chamoun Z., Jiang G., Prestwich G., Sakai N., Matile S., Parton R.G., Gruenberg J. 2008. Lysobisphosphatidic acid controls endosomal cholesterol levels. J. Biol. Chem. 283:27871–27880 10.1074/jbc.M801463200 [DOI] [PubMed] [Google Scholar]
- Cianciola N.L., Crooks D., Shah A.H., Carlin C. 2007. A tyrosine-based signal plays a critical role in the targeting and function of adenovirus RIDalpha protein. J. Virol. 81:10437–10450 10.1128/JVI.00399-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks D., Kil S.J., McCaffery J.M., Carlin C. 2000. E3-13.7 integral membrane proteins encoded by human adenoviruses alter epidermal growth factor receptor trafficking by interacting directly with receptors in early endosomes. Mol. Biol. Cell. 11:3559–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz J.C., Sugii S., Yu C., Chang T.Y. 2000. Role of Niemann-Pick type C1 protein in intracellular trafficking of low density lipoprotein-derived cholesterol. J. Biol. Chem. 275:4013–4021 10.1074/jbc.275.6.4013 [DOI] [PubMed] [Google Scholar]
- Eskelinen E.L. 2005. Maturation of autophagic vacuoles in Mammalian cells. Autophagy. 1:1–10 10.4161/auto.1.1.1270 [DOI] [PubMed] [Google Scholar]
- Fader C.M., Sánchez D., Furlán M., Colombo M.I. 2008. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 9:230–250 [DOI] [PubMed] [Google Scholar]
- Ghosh P., Dahms N.M., Kornfeld S. 2003. Mannose 6-phosphate receptors: new twists in the tale. Nat. Rev. Mol. Cell Biol. 4:202–212 10.1038/nrm1050 [DOI] [PubMed] [Google Scholar]
- Ginsberg H.S., Prince G.A. 1994. The molecular basis of adenovirus pathogenesis. Infect. Agents Dis. 3:1–8 [PubMed] [Google Scholar]
- Goldstein J.L., DeBose-Boyd R.A., Brown M.S. 2006. Protein sensors for membrane sterols. Cell. 124:35–46 10.1016/j.cell.2005.12.022 [DOI] [PubMed] [Google Scholar]
- Greaves J., Chamberlain L.H. 2007. Palmitoylation-dependent protein sorting. J. Cell Biol. 176:249–254 10.1083/jcb.200610151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M.G., Munafó D.B., Berón W., Colombo M.I. 2004. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 117:2687–2697 10.1242/jcs.01114 [DOI] [PubMed] [Google Scholar]
- Haas D., Morgenthaler J., Lacbawan F., Long B., Runz H., Garbade S.F., Zschocke J., Kelley R.I., Okun J.G., Hoffmann G.F., Muenke M. 2007. Abnormal sterol metabolism in holoprosencephaly: studies in cultured lymphoblasts. J. Med. Genet. 44:298–305 10.1136/jmg.2006.047258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B.L., Ullrich A., Wold W.S., Carlin C.R. 1990. Retrovirus-mediated transfer of an adenovirus gene encoding an integral membrane protein is sufficient to down regulate the receptor for epidermal growth factor. Mol. Cell. Biol. 10:5521–5524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman P., Carlin C. 1994. Adenovirus E3 protein causes constitutively internalized epidermal growth factor receptors to accumulate in a prelysosomal compartment, resulting in enhanced degradation. Mol. Cell. Biol. 14:3695–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman P.H., Rajakumar P., Hoffman B., Heuertz R., Wold W.S.M., Carlin C.R. 1992a. Evidence for intracellular down-regulation of the epidermal growth factor (EGF) receptor during adenovirus infection by an EGF-independent mechanism. J. Virol. 66:197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman P., Yaffe M.B., Hoffman B.L., Yei S., Wold W.S.M., Carlin C. 1992b. Characterization of the adenovirus E3 protein that down-regulates the epidermal growth factor receptor. Evidence for intermolecular disulfide bonding and plasma membrane localization. J. Biol. Chem. 267:13480–13487 [PubMed] [Google Scholar]
- Horwitz M.S. 1996. Adenoviruses. Fields Virology. Third edition Fields B.N., Knipe D.M., Howley P.M., Lippincott-Raven, Philadelphia: 2149–2171 [Google Scholar]
- Horwitz M.S. 2004. Function of adenovirus E3 proteins and their interactions with immunoregulatory cell proteins. J. Gene Med. 6:S172–S183 10.1002/jgm.495 [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Kominami E., Tanaka K., Komatsu M. 2008. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy. 4:1063–1066 [DOI] [PubMed] [Google Scholar]
- Imelli N., Meier O., Boucke K., Hemmi S., Greber U.F. 2004. Cholesterol is required for endocytosis and endosomal escape of adenovirus type 2. J. Virol. 78:3089–3098 10.1128/JVI.78.6.3089-3098.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M., Lehto M., Tanhuanpää K., Cover T.L., Olkkonen V.M. 2005. The oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol. Biol. Cell. 16:5480–5492 10.1091/mbc.E05-03-0189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M., Rocha N., Zwart W., Jordens I., Janssen L., Kuijl C., Olkkonen V.M., Neefjes J. 2007. Activation of endosomal dynein motors by stepwise assembly of Rab7–RILP–p150Glued, ORP1L, and the receptor βlll spectrin. J. Cell Biol. 176:459–471 10.1083/jcb.200606077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Emr S.D. 2000. Autophagy as a regulated pathway of cellular degradation. Science. 290:1717–1721 10.1126/science.290.5497.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko D.C., Milenkovic L., Beier S.M., Manuel H., Buchanan J., Scott M.P. 2005. Cell-autonomous death of cerebellar purkinje neurons with autophagy in Niemann-Pick type C disease. PLoS Genet. 1:81–95 10.1371/journal.pgen.0010081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Stang E., Fang K.S., de Moerloose P., Parton R.G., Gruenberg J. 1998. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 392:193–197 10.1038/32440 [DOI] [PubMed] [Google Scholar]
- Kostenko O., Tsacoumangos A., Crooks D.M., Kil S.J., Carlin C.R. 2006. Gab1 signaling is regulated by EGF receptor sorting in early endosomes. Oncogene. 25:6604–6617 10.1038/sj.onc.1209675 [DOI] [PubMed] [Google Scholar]
- Lala D.S., Syka P.M., Lazarchik S.B., Mangelsdorf D.J., Parker K.L., Heyman R.A. 1997. Activation of the orphan nuclear receptor steroidogenic factor 1 by oxysterols. Proc. Natl. Acad. Sci. USA. 94:4895–4900 10.1073/pnas.94.10.4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.A., Beigneux A., Ahmad S.T., Young S.G., Gao F.B. 2007. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 17:1561–1567 10.1016/j.cub.2007.07.029 [DOI] [PubMed] [Google Scholar]
- Linder M.E., Deschenes R.J. 2007. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8:74–84 10.1038/nrm2084 [DOI] [PubMed] [Google Scholar]
- Liscum L. 1990. Pharmacological inhibition of the intracellular transport of low-density lipoprotein-derived cholesterol in Chinese hamster ovary cells. Biochim. Biophys. Acta. 1045:40–48 [DOI] [PubMed] [Google Scholar]
- Maxfield F.R., Tabas I. 2005. Role of cholesterol and lipid organization in disease. Nature. 438:612–621 10.1038/nature04399 [DOI] [PubMed] [Google Scholar]
- McMahon H.T., Mills I.G. 2004. COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr. Opin. Cell Biol. 16:379–391 10.1016/j.ceb.2004.06.009 [DOI] [PubMed] [Google Scholar]
- McNees A.L., Garnett C.T., Gooding L.R. 2002. The adenovirus E3 RID complex protects some cultured human T and B lymphocytes from Fas-induced apoptosis. J. Virol. 76:9716–9723 10.1128/JVI.76.19.9716-9723.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier O., Greber U.F. 2003. Adenovirus endocytosis. J. Gene Med. 5:451–462 10.1002/jgm.409 [DOI] [PubMed] [Google Scholar]
- Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21:2861–2873 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- Nara A., Mizushima N., Yamamoto A., Kabeya Y., Ohsumi Y., Yoshimori T. 2002. SKD1 AAA ATPase-dependent endosomal transport is involved in autolysosome formation. Cell Struct. Funct. 27:29–37 10.1247/csf.27.29 [DOI] [PubMed] [Google Scholar]
- Naureckiene S., Sleat D.E., Lackland H., Fensom A., Vanier M.T., Wattiaux R., Jadot M., Lobel P. 2000. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 290:2298–2301 10.1126/science.290.5500.2298 [DOI] [PubMed] [Google Scholar]
- Neufeld E.F. 1991. Lysosomal storage diseases. Annu. Rev. Biochem. 60:257–280 10.1146/annurev.bi.60.070191.001353 [DOI] [PubMed] [Google Scholar]
- Olkkonen V.M., Johansson M., Suchanek M., Yan D., Hynynen R., Ehnholm C., Jauhiainen M., Thiele C., Lehto M. 2006. The OSBP-related proteins (ORPs): global sterol sensors for co-ordination of cellular lipid metabolism, membrane trafficking and signalling processes? Biochem. Soc. Trans. 34:389–391 10.1042/BST0340389 [DOI] [PubMed] [Google Scholar]
- Ory D.S. 2004. Nuclear receptor signaling in the control of cholesterol homeostasis: have the orphans found a home? Circ. Res. 95:660–670 10.1161/01.RES.0000143422.83209.be [DOI] [PubMed] [Google Scholar]
- Pacheco C.D., Kunkel R., Lieberman A.P. 2007. Autophagy in Niemann-Pick C disease is dependent upon Beclin-1 and responsive to lipid trafficking defects. Hum. Mol. Genet. 16:1495–1503 10.1093/hmg/ddm100 [DOI] [PubMed] [Google Scholar]
- Patterson M.C., Vanier M.T., Suzuki K., Morris J.A., Carstea E., Neufeld E.B., Blanchette-Mackie J.E., Pentchev P.G. 2001. Niemann-Pick disease type C: a lipid trafficking disorder. The Online Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill. Available at: http://www.ommbid.com/(accessed March 1, 2008)
- Pike L.J., Han X., Gross R.W. 2005. Epidermal growth factor receptors are localized to lipid rafts that contain a balance of inner and outer leaflet lipids: a shotgun lipidomics study. J. Biol. Chem. 280:26796–26804 10.1074/jbc.M503805200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press B., Feng Y., Hoflack B., Wandinger-Ness A. 1998. Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J. Cell Biol. 140:1075–1089 10.1083/jcb.140.5.1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Progida C., Spinosa M.R., De Luca A., Bucci C. 2006. RILP interacts with the VPS22 component of the ESCRT-II complex. Biochem. Biophys. Res. Commun. 347:1074–1079 10.1016/j.bbrc.2006.07.007 [DOI] [PubMed] [Google Scholar]
- Razi M., Chan E.Y., Tooze S.A. 2009. Early endosomes and endosomal coatomer are required for autophagy. J. Cell Biol. 185:305–321 10.1083/jcb.200810098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten T.E., Vaccari T., Lindmo K., Rodahl L.M., Nezis I.P., Sem-Jacobsen C., Wendler F., Vincent J.P., Brech A., Bilder D., Stenmark H. 2007. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 17:1817–1825 10.1016/j.cub.2007.09.032 [DOI] [PubMed] [Google Scholar]
- Schmid D., Pypaert M., Münz C. 2007. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 26:79–92 10.1016/j.immuni.2006.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shacka J.J., Roth K.A., Zhang J. 2008. The autophagy-lysosomal degradation pathway: role in neurodegenerative disease and therapy. Front. Biosci. 13:718–736 10.2741/2714 [DOI] [PubMed] [Google Scholar]
- Shah A.H., Cianciola N.L., Mills J.L., Sönnichsen F.D., Carlin C. 2007. Adenovirus RIDα regulates endosome maturation by mimicking GTP-Rab7. J. Cell Biol. 179:965–980 10.1083/jcb.200702187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens J.E., Roth M.G., Matlin K.S. 1989. Differential extractability of influenza virus hemagglutinin during intracellular transport in polarized epithelial cells and nonpolar fibroblasts. J. Cell Biol. 108:821–832 10.1083/jcb.108.3.821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V.A., McGrath S.M., Dubois P.M., Pau M.G., Mettens P., Shott J., Cobb M., Burge J.R., Larson D., Ware L.A., et al. 2007. Priming with an adenovirus 35-circumsporozoite protein (CS) vaccine followed by RTS,S/AS01B boosting significantly improves immunogenicity to Plasmodium falciparum CS compared to that with either malaria vaccine alone. Infect. Immun. 75:2283–2290 10.1128/IAI.01879-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchanek M., Hynynen R., Wohlfahrt G., Lehto M., Johansson M., Saarinen H., Radzikowska A., Thiele C., Olkkonen V.M. 2007. The mammalian oxysterol-binding protein-related proteins (ORPs) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochem. J. 405:473–480 10.1042/BJ20070176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Ueno T., Kominami E. 2004. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 36:2503–2518 10.1016/j.biocel.2004.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.P., Kirkegaard K. 2007. Modification of cellular autophagy protein LC3 by poliovirus. J. Virol. 81:12543–12553 10.1128/JVI.00755-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsacoumangos A., Kil S.J., Ma L., Sönnichsen F.D., Carlin C. 2005. A novel dileucine lysosomal-sorting-signal mediates intracellular EGF-receptor retention independently of protein ubiquitylation. J. Cell Sci. 118:3959–3971 10.1242/jcs.02527 [DOI] [PubMed] [Google Scholar]
- Urano Y., Watanabe H., Murphy S.R., Shibuya Y., Geng Y., Peden A.A., Chang C.C., Chang T.Y. 2008. Transport of LDL-derived cholesterol from the NPC1 compartment to the ER involves the trans-Golgi network and the SNARE protein complex. Proc. Natl. Acad. Sci. USA. 105:16513–16518 10.1073/pnas.0807450105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance D.E., Weinstein D.B., Steinberg D. 1984. Isolation and analysis of lipoproteins secreted by rat liver hepatocytes. Biochim. Biophys. Acta. 792:39–47 [DOI] [PubMed] [Google Scholar]
- Vinogradova O., Carlin C.R., Sonnichsen F.D., Sanders C.R., II 1998. A membrane setting for the sorting motifs present in the adenovirus E3-13.7 protein which down-regulates the epidermal growth factor receptor. J. Biol. Chem. 273:17343–17350 10.1074/jbc.273.28.17343 [DOI] [PubMed] [Google Scholar]
- Wang T., Hong W. 2006. RILP interacts with VPS22 and VPS36 of ESCRT-II and regulates their membrane recruitment. Biochem. Biophys. Res. Commun. 350:413–423 10.1016/j.bbrc.2006.09.064 [DOI] [PubMed] [Google Scholar]
- Wiethoff C.M., Wodrich H., Gerace L., Nemerow G.R. 2005. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 79:1992–2000 10.1128/JVI.79.4.1992-2000.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.R., Coleman T., Langmade S.J., Scherrer D.E., Lane L., Lanier M.H., Feng C., Sands M.S., Schaffer J.E., Semenkovich C.F., Ory D.S. 2008. Niemann-Pick C1 protects against atherosclerosis in mice via regulation of macrophage intracellular cholesterol trafficking. J. Clin. Invest. 118:2281–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]