Reevaluation of Lte1’s involvement in the mitotic exit network reveals that it is involved in localizing Bfa1 to the spindle pole body but does not function as a GEF for Tem1.
Abstract
Lte1 is a mitotic regulator long envisaged as a guanosine nucleotide exchange factor (GEF) for Tem1, the small guanosine triphosphatase governing activity of the Saccharomyces cerevisiae mitotic exit network. We demonstrate that this model requires reevaluation. No GEF activity was detectable in vitro, and mutational analysis of Lte1’s putative GEF domain indicated that Lte1 activity relies on interaction with Ras for localization at the bud cortex rather than providing nucleotide exchange. Instead, we found that Lte1 can determine the subcellular localization of Bfa1 at spindle pole bodies (SPBs). Under conditions in which Lte1 is essential, Lte1 promoted the loss of Bfa1 from the maternal SPB. Moreover, in cells with a misaligned spindle, mislocalization of Lte1 in the mother cell promoted loss of Bfa1 from one SPB and allowed bypass of the spindle position checkpoint. We observed that lte1 mutants display aberrant localization of the polarity cap, which is the organizer of the actin cytoskeleton. We propose that Lte1’s role in cell polarization underlies its contribution to mitotic regulation.
Introduction
In the eukaryotic cell cycle, mitotic exit and cytokinesis must be coordinated with the partition of the duplicated chromosomes to opposite ends of the extended mitotic spindle. In budding yeast, the site of cytokinesis is established well before mitosis, so the mitotic spindle has to be correctly aligned along the longitudinal axis of the dividing cell so that mother and bud compartments each receive a complement of DNA. Similarly, in asymmetric stem cell divisions of more advanced eukaryotes, the axis of the mitotic spindle has to be correctly aligned with the axis of polarized growth and the plane of cell division (Yamashita et al., 2007; for review see Yamashita and Fuller, 2008). In Saccharomyces cerevisiae, a complex spatial-sensing system called the spindle position checkpoint (SPoC) ensures that mitosis is only completed if the extended mitotic spindle is correctly positioned. A similar process for transiently arresting cell cycle progression in metazoan cells with misoriented spindles has been recognized (Cheng et al., 2008). The SPoC regulates the mitotic exit network (MEN), which governs the transition from late M to G1. The key switch element in the MEN is a small, Ras-like GTPase called Tem1. Activated Tem1, presumably Tem1-GTP, triggers a signaling cascade of Cdc15 and Dbf2-Mob1 kinases to activate Cdc14 phosphatase. Cdc14 has multiple targets whose dephosphorylation drives the cell cycle out of mitosis (for review see Stegmeier and Amon, 2004).
Regulation of the MEN is complex, with multiple, partially redundant pathways. The most studied regulators are Lte1 and the Bfa1–Bub2 complex, although Cdc5, Kin4, Kel1/2, Gic1 and -2, Ste20, Cla4, Cdc42, and Ras2 are also implicated (Hu et al., 2001; Höfken and Schiebel, 2002, 2004; Jensen et al., 2002a; Seshan et al., 2002; Stegmeier et al., 2002; Yoshida et al., 2003; Bosl and Li, 2005; D’Aquino et al., 2005; Pereira and Schiebel, 2005; Seshan and Amon, 2005). Lte1 was seen as a positive mitotic regulator because lte1 mutants undergo a telophase arrest at low temperature (Shirayama et al., 1994a). Lte1 shares homology with the guanosine nucleotide exchange domain of the Ras–guanosine nucleotide exchange factor (GEF) Cdc25 (Shirayama et al., 1994a). Thus, when TEM1 was isolated as a high copy number suppressor of the cold sensitivity of an lte1 mutant, it was proposed that Lte1 might be a GEF for this small GTPase (Keng et al., 1994; Shirayama et al., 1994b). Bfa1 and Bub2 are negative regulators of the MEN (Alexandru et al., 1999; Fesquet et al., 1999; Fraschini et al., 1999; Li, 1999) that form a two-component GTPase-activating protein (GAP) for Tem1 in vitro (Geymonat et al., 2002), although a more complex pattern of negative regulatory activity in vivo may also operate (Ro et al., 2002; Fraschini et al., 2006; Kim et al., 2008).
MEN regulation has a spatial dimension, with many components occupying discrete intracellular sites. Lte1 localization to the bud cortex (Bardin et al., 2000; Pereira et al., 2000) is important for Lte1 activity. It requires interaction with Ras-GTP (Yoshida et al., 2003) and Cla4-dependent phosphorylation (Höfken and Schiebel, 2002; Jensen et al., 2002a; Seshan et al., 2002; Seshan and Amon, 2005). Bub2 and Bfa1 as well as Tem1 and many other MEN constituents localize at the spindle pole bodies (SPBs; for review see Pereira and Schiebel, 2001). Initially, there is dynamic, low affinity binding of Bfa1–Bub2 to both SPBs of short metaphase spindles. When the daughter-directed SPB (dSPB) approaches the neck and the spindle elongates, Bfa1–Bub2 disappears from the maternal SPB (mSPB) and becomes more tightly bound to the dSPB. At the same time Kin4, a kinase which regulates Bfa1, appears at the mSPB. Thus, Bfa1–Bub2 and Kin4 become asymmetrically localized at opposite SPBs (Pereira et al., 2000, 2001; Molk et al., 2004; D’Aquino et al., 2005; Pereira and Schiebel, 2005; Fraschini et al., 2006; Maekawa et al., 2007). Importantly, Bfa1–Bub2 and Kin4 remain on both SPBs when the spindle is misaligned in the mother and the SPoC is activated and when there are defects in cytoplasmic microtubules (Pereira et al., 2000, 2001; Molk et al., 2004; Pereira and Schiebel, 2005; Fraschini et al., 2006; Maekawa et al., 2007). Thus, the switch from symmetrical to asymmetrical distributions of Bfa1–Bub2 and its regulator Kin4 precedes MEN activation.
Several elements of the spatially polarized actin cytoskeleton also influence MEN activation either directly or through the asymmetric localization of Bfa1 at dSPBs. The MEN regulators Ste20, Cla4, and Gic1 and -2 are effectors of the polarity coordinator Cdc42; Bni1 is a constituent of the polarity cap required for the formation of actin filaments, and Kel1 is a polarity cap protein that interacts with Lte1 (Höfken and Schiebel, 2002, 2004; Seshan et al., 2002; Monje-Casas and Amon, 2009).
It was initially proposed that a properly aligned dSPB delivered Tem1 to the daughter cortex, where Tem1 was activated by Lte1’s putative GEF activity (Bardin et al., 2000; Pereira et al., 2000). However, MEN activation without Lte1 and kinetic measurements linked activation to either passage of microtubules or the dSPB through the bud neck or contact of microtubules with the daughter cell cortex (Adames et al., 2001; Molk et al., 2004; Nelson and Cooper, 2007). If so, it is somewhat difficult to understand how the putative GEF activity of cortical Lte1 could impinge on Tem1 located elsewhere in the cell.
The purpose of this work was to test directly whether purified Lte1 could provide GEF activity for Tem1 in vitro. A point mutation analysis was also conducted to reveal the effects of precise changes in the GEF domain in full-length Lte1 in vivo. We report that neither approach gave any support for Lte1 being a GEF for Tem1. Instead, we show that Lte1 can contribute to the asymmetric accumulation of Bfa1 on the dSPB, which precedes activation of the MEN. Finally, we show that Lte1 is required for fixing the position of the polarity cap and suggest that it is through this activity that Lte1 influences the MEN and cell cycle progression. Together, our results prompt a reevaluation of the activity of Lte1 in the regulation of the MEN.
Results
Tem1 GDP to GTP turnover is not stimulated by Lte1 in vitro
The putative guanosine nucleotide exchange activity of Lte1 on Tem1 was tested directly in an in vitro assay. For this purpose, 6His-Lte1 was expressed and purified from its native yeast source (Geymonat et al., 2007), whereas Tem1 protein was produced in Escherichia coli as described previously (Geymonat et al., 2002). The purified 6His-Lte1 protein was functional, as it could interact in vitro with GST-Ras2V19 but not with control GST (Fig. 1 D) as previously reported (Yoshida et al., 2003). The functionality of the bacterially produced Tem1 has been confirmed in an earlier study of GTPase activity (Geymonat et al., 2002). Using a filter-based assay, we monitored the loss of tritiated GDP from purified Tem1 with and without additional Lte1 (Fig. 1 A). As reported, Tem1 alone showed a high intrinsic exchange activity (Geymonat et al., 2002). However, the addition of Lte1 protein did not affect these high rates of GDP exchange (Fig. 1 A), suggesting that Lte1 has no GEF activity toward Tem1 in vitro.
Figure 1.
In vitro assays of protein activity. (A and B) Guanosine nucleotide exchange assays. (A) Release of [3H]GDP from 150 nM Tem1 without (open circles) or with 790 nM Lte1 (closed circles). (B) Release of [3H]GDP from a Tem1–Bfa1 complex (150 nM/340 nM) was assayed in the presence of 0 (open circles), 420 (triangles), or 840 nM (closed circles) Lte1. (C) Bfa1 inhibits intrinsic nucleotide exchange of Tem1. Increasing concentrations of Bfa1 were added to 1.8 µM Tem1. The percentage of GDP bound is the amount of nucleotide remaining bound to Tem1 after a 4-min incubation at 25°C. (D) Lte1–Ras2V19 interaction. Binding of 6His-Lte1 to glutathione beads carrying GST or GST-Ras2V19 was assayed in a Western blot (WB) using anti-6His antibody. Input amounts of GST and GST-Ras2V19 are visualized by Coomassie blue staining. Molecular mass is indicated in kilodaltons.
A second assay examined exchange in a Tem1–Bfa1 complex (Fig. 1 B). This mimics the in vivo situation in which Bfa1–Tem1 on the dSPB would be exposed to cortical Lte1 in the bud (Pereira et al., 2000; Lee et al., 2001). In vitro, Bfa1 inhibited Tem1’s intrinsic nucleotide exchange when stoichiometric amounts of proteins were used (Fig. 1 C). This is contrary to results with the Schizosaccharomyces pombe homologue Byr4 (Furge et al., 1998) and differs from our earlier study, which used suboptimal levels of Bfa1 (Geymonat et al., 2002). Again, the addition of Lte1 at different concentrations did not stimulate any release of GDP from Tem1 (Fig. 1 B). Collectively, our results provide no direct in vitro evidence that Lte1 stimulates nucleotide exchange activity of Tem1 or of the Tem1–Bfa1 complex, suggesting that Lte1 may not function as a GEF for Tem1 in vivo.
Site-specific mutagenesis of the GEF domain of Lte1
As an alternative test of the putative GEF activity of Lte1, we determined the in vivo effects of site-specific mutations in the GEF-like domain of Lte1. Lte1 interacts with Ras via its GEF domain (Yoshida et al., 2003; Seshan and Amon, 2005), which shows homology with hSos and scCdc25, two known GEFs for Ras proteins (for reviews see Cherfils and Chardin, 1999; Quilliam et al., 2002). Therefore, previous mutational analyses of scCdc25 and hSos (Park et al., 1994; Hall et al., 2001) served as models for site-specific mutagenesis of Lte1 (Fig. S1). The idea that Lte1 is a GEF for Tem1 is complicated by the additional requirement of the Lte1 GEF domain for interaction with Ras (Yoshida et al., 2003). The putative interaction with Tem1 would facilitate nucleotide exchange, whereas the established interaction with Ras might activate Lte1 exchange activity and, coupled with Lte1 phosphorylation, allow localization of Lte1 to the bud cortex (Yoshida et al., 2003; Seshan and Amon, 2005).
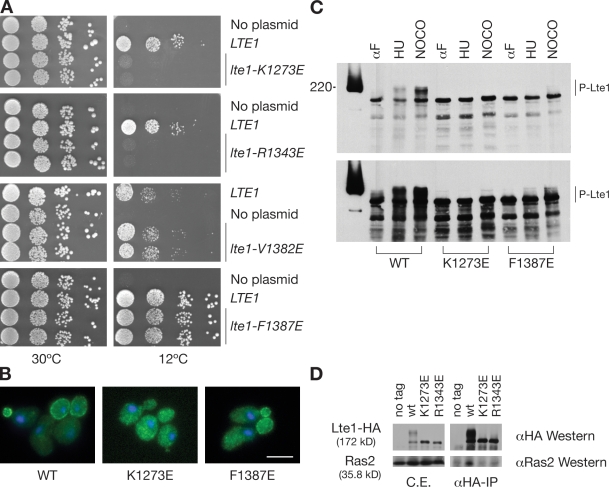
Mutations that are predicted to block interaction of Lte1 with small G proteins were modeled on scCdc25 glutamic acid substitutions at conserved R1374 and R1444 residues. These mutations block interaction with Ras, thereby preventing complementation of cdc25ts mutants (Park et al., 1994). The equivalent K1273E and R1343E mutations in the GEF region of LTE1 genes were expressed from native promoters on integrative plasmids. Neither mutated form of LTE1 rescued the cold sensitivity of a deletion mutant lacking the entire LTE1 ORF, indicating that the GEF region of Lte1 is required for activity of the full-length protein in vivo (Fig. 2 A).
Figure 2.
Analysis of point mutations in the putative GEF domain of Lte1. (A) Complementation assays for rescue of cold sensitivity of the lte1Δ strain SY144 with integrative YIplac128-based plasmids expressing the mutant forms of LTE1 indicated. (B) Subcellular localization of wild-type (WT), K1273E, and F1387E forms of Lte1-GFP expressed in the lte1Δ strain SY144. (C) Cell cycle–dependent phosphorylation of Lte1. The lte1Δ strain SY144 carried the integrative plasmid expressing the 3HA-tagged forms of Lte1 indicated. After cell arrest with α factor (αF), hydroxyurea (HU), or nocodazole (NOCO), Lte1 status was revealed by Western blotting with anti-HA antibody. Short and long exposures of the same blot are presented. Molecular mass is indicated in kilodaltons. (D) In vivo interaction between wild type (wt) and mutant Lte1-3HA with Ras2. After cell arrest with hydroxyurea, proteins were extracted from the strains indicated. Lte1 was immunoprecipitated with anti-HA antibody. Cell extracts (C.E.) and immunoprecipitates (αHA-IP) were analyzed by Western blotting with anti-HA or anti-Ras2 antibodies. Bar, 5 µm.
Lte1-F1387E is a second type of mutational change that was modeled on Cdc25-R1489E, which prevents complementation of a cdc25 mutant but, in contrast to R1374E and R1444E, retains its interaction with Ras (Park et al., 1994). Similarly, Lte1-V1382E was modeled on hSos-I956E, which has reduced exchange activity in vitro but again retains almost full interaction with Ras (Hall et al., 2001). Importantly, both Lte1-F1387E and -V1382E complemented the cold sensitivity of lte1Δ mutants at 12°C (Fig. 2 A). Thus, two independently selected forms of Lte1 that are expected to lose exchange activity but retain interaction with Ras complemented the cold sensitivity of lte1Δ cells.
Cortical localization is important for Lte1 activity and requires coordinated interaction with Ras and phosphorylation (Yoshida et al., 2003; Seshan and Amon, 2005). Thus, Lte1-F1387E, which is predicted to retain some Ras interaction and rescued lte1Δ cold sensitivity, did show some cortical localization (clear in all small to medium buds) and some phosphorylation, albeit less than wild type (Fig. 2, B and C). In contrast, Lte1-K1273E and -R1343E showed no cortical localization (Fig. 2 B and not depicted), no phosphorylation (Fig. 2 C and not depicted), and no complementation activity, which is consistent with its predicted and experimental lack of interaction with Ras2 in vivo (Fig. 2 D). Therefore, the putative GEF domain of Lte1 is required for cortical localization through interactions with Ras rather than providing GEF activity per se.
Multiple domains of Lte1 are required for its complete activity in vivo
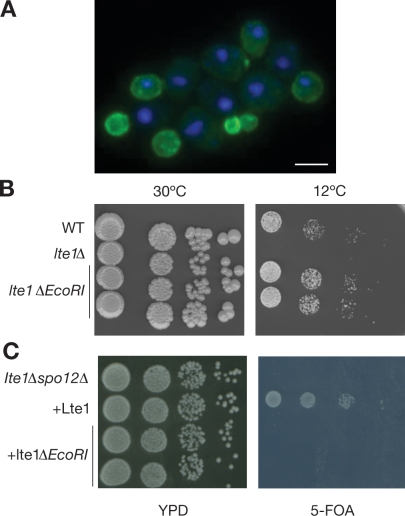
As well as causing cold sensitivity, lte1 mutations are synthetically lethal with deletions of the Cdc14 early anaphase release (FEAR) genes SPO12 and SLK19 and of several genes involved in microtubule dynamics (Heil-Chapdelaine et al., 2000). To test whether Lte1’s GEF-like domains alone are required for complementation of synthetic lethality, we examined complementation by lte1-ΔEcoRI. Lte1-ΔEcoRI has an internal deletion of residues 327–892 but retains its N- and C-terminal GEF domains. Lte1-ΔEcoRI localizes at the bud cortex–like full-length Lte1 (Fig. 3 A) and complements the cold sensitivity of lte1Δ mutants as wild-type Lte1 (Fig. 3 B). However Lte1-ΔEcoRI was unable to complement the inviability of lte1Δ spo12Δ or lte1Δ slk19Δ, even if overexpressed (Fig. 3 C and not depicted), nor was it able to suppress the SPoC in dyn1Δ cells when overexpressed. Equally, overexpression of Lte1-K1273E and -F1387E was also unable to bypass the SPoC in dyn1Δ cells with mispositioned spindles (Fig. S2). Therefore, differing Lte1 activities may complement the different physiological challenges posed by low temperature, synthetic lethality, and mispositioned spindles. Thus, activity of the GEF-like domain of Lte1 alone is insufficient to explain all of Lte1’s in vivo roles. Rather, the complex pattern of complementation by different alleles of Lte1 points to multiple activities of the molecule.
Figure 3.
Lte1 activity in vivo requires multiple domains. (A) Subcellular localization of Lte1-ΔEcoRI–GFP expressed from pMG158 in lte1Δ SY144. (B) Complementation of cold sensitivity of the lte1Δ strain SY144 with wild-type (WT) or ΔEcoRI forms of Lte1 expressed from the integrative plasmid pMG157. (C) Complementation of synthetic lethality of lte1Δ spo12Δ strain SY169 by Lte1 and Lte1-ΔEcoRI. The lte1Δ spo12Δ strain carried a URA3-based plasmid expressing wild-type LTE1. After transformation with integrative plasmids expressing Lte1 (pMG52) or Lte1-ΔEcoR1 (pMG157), cells were tested for growth on fluoroorotic acid (FOA) plates. YPD, YEP dextrose. Bar, 5 µm.
Lte1 contributes to the correct localization of Bfa1 at the SPBs
If Lte1 is not a GEF for Tem1, how does it promote mitotic progression? Recently, a new aspect of MEN regulation has been described. Before MEN activation, Bfa1–Bub2 shifts from having a similar, low affinity localization at both SPBs in early anaphase to displaying an asymmetric, higher affinity localization at the dSPBs of correctly elongated anaphase spindles (Caydasi and Pereira, 2009; Monje-Casas and Amon, 2009). As Lte1 has such a striking spatial distribution, we considered whether Lte1 affects the MEN through influencing the distribution of Bfa1 on the SPBs.
Surprisingly, we were unable to construct an lte1Δ BFA1-GFP strain to test whether loss of Lte1 activity affected Bfa1 localization because this genetic combination proved lethal. However, Bfa1-GFP with wild-type activity in vivo has been previously described (Pereira et al., 2001), and BFA1-GFP has been successfully combined with mutant lte1 by others (Kim et al., 2008). Synthetic lethality with our BFA1-GFP construct was specific to lte1Δ and did not occur with the MEN mutations cdc15-1, mob1-77, or cdc14-1 nor with deleted FEAR genes SPO12 and SLK19. Moreover, in wild-type cells, our Bfa1-GFP displayed the expected pattern of SPB association and retained spindle assembly checkpoint activity (Fig. 4 C and not depicted). Thus, the reason for the inviability of lte1Δ BFA1-GFP encountered in this study is unclear but may stem from subtle differences in our construction of the fusion between BFA1 and GFP and/or features of the genetic background of S. cerevisiae 15D. Whatever the cause of lte1Δ BFA1-GFP inviability, it importantly provided an experimental system in which Bfa1 localization at SPBs could be monitored at 30°C in cells with an essential requirement for Lte1.
Figure 4.
Lte1 influences localization of Bfa1-GFP at SPBs. (A) Depletion of Lte1 arrests growth in strains expressing Bfa1-GFP. MGY252 (lte1Δ BFA1-GFP YCplac33 URA3-LTE1) and MGY260 (lte1Δ BFA1-GFP GAL1pr–lte1-F1387E–3HA) were tested for growth at 30°C after spotting on YEP galactose (GAL) and YEP glucose (GLU) agar to induce or repress expression of Lte1-F1387E, respectively. (B) BFA1-GFP is colethal with lte1Δ and can be rescued by activation of the MEN. MGY252 (lte1Δ BFA1-GFP YCplac33 URA3-LTE) was transformed with dbf2 HyA or deleted for KIN4 and spotted onto YEP dextrose (YPD) and 5-fluoroorotic acid (5FOA) agar to test for growth at 30°C in the absence of Lte1. (C) lte1Δ BFA1-GFP cells arrest in anaphase with Bfa1-GFP at both SPBs. MGY260 lte1Δ expresses Bfa1-GFP (red) and Tub1-CFP (green) from their endogenous promoters and expresses Lte1-F1387E from the GAL1 promoter of the integrative plasmid pMG75. +Lte1 indicates Lte1 expressed during growth in galactose-containing medium; −Lte1 indicates Lte1 expression repressed by 4–6-h growth with glucose. Expression of Bfa1-GFP and Tub1-CFP was also monitored in MGY272 (lte1Δ BFA1-GFP TUB1-CFP), which is kept alive by pDbf2-1c expressing Dbf2 HyA, and in MMY41 (lte1Δ kin4Δ BFA1-GFP TUB1-CFP). Both strains were cultivated at 30°C. (D) Bfa1 (green) and DNA (blue) were visualized in MGY274 (mob1-77 BFA1-GFP) cells arrested in anaphase by incubation at 37°C for 2.5 h. (E) The percentage of cells expressing Bfa1-GFP at one or two SPBs in late anaphase cells is presented in the yeast strains indicated (n > 100). Bars, 5 µm.
The terminal phenotype of BFA1-GFP lte1Δ was studied in cells carrying an integrative plasmid expressing Lte1-F1387E from a GAL1 promoter. This strain was viable on galactose-containing medium, in which Lte1-F1387E was expressed. However, it was unable to form colonies on medium containing glucose, in which the partially functional Lte1-F1387E permitted an effective depletion of Lte1 activity (Fig. 4 A). After 4–6 h in glucose-containing medium, there was a uniform cell cycle arrest with large buds, elongated spindles, and separated nuclei symptomatic of a MEN arrest. A MEN arrest was also indicated by the rescue of viability by a hyperactive (HyA) form of Dbf2 (Dbf2 HyA; Fig. 4 B). The constitutively active Dbf2 HyA was independent from upstream elements of the MEN pathway, having been isolated by its ability to complement a deletion of TEM1 (Fig. S3). Similarly, deletion of the Bfa1 regulator KIN4 restored the viability of BFA1-GFP lte1Δ cells (Fig. 4 B) just as it rescued the cold sensitivity of lte1 mutants (Pereira and Schiebel, 2005) or lte1 synthetic lethality with FEAR mutations (D’Aquino et al., 2005).
The conditional expression of Lte1 allowed us to monitor the localization of Bfa1-GFP at anaphase in the presence or absence of Lte1. When Lte1-F1387E was expressed, Bfa1 localized, as expected, on both SPBs on short spindles. With spindle elongation, the dSPB approached the bud neck, and Bfa1 at the mSPB disappeared, whereas the Bfa1-GFP signal intensified at the dSPB until spindle disassembly and cytokinesis occurred (Fig. 4, C and E). In contrast, most cells depleted for Lte1 arrested in late anaphase with elongated spindles and Bfa1-GFP visible on both SPBs. This symmetrical distribution of Bfa1-GFP was not caused by the late anaphase arrest per se because the Bfa1-GFP was only seen at the dSPBs in mob1-77, cdc15-1, and cdc14-1 mutants undergoing similar anaphase arrests (Fig. 4 D and not depicted).
As mentioned in a previous paragraph, the growth arrest of lte1Δ BFA1-GFP cells could be bypassed with Dbf2 HyA, allowing the effects of Lte1 loss to be observed in cycling lte1Δ BFA1-GFP DBF2 HyA cells. In this situation, Bfa1-GFP was found at both SPBs, even when anaphase spindles were correctly orientated. This again contrasts to the preferential localization of Bfa1-GFP to the dSPBs of cells expressing Lte1 (Fig. 4, C and E) and demonstrates that Lte1 can contribute to the distribution of Bfa1 at SPBs in anaphase cells independently of MEN activity.
Finally, the relationship between the synthetic lethality of lte1Δ BFA1-GFP and Bfa1 localization was examined after deletion of KIN4. Loss of KIN4 in cells with a misorientated spindle was found to reduce the turnover of Bfa1 at SPBs so that Bfa1-GFP accumulated asymmetrically at one SPB (Caydasi and Pereira, 2009). Similarly, the localization of Bfa1 at both SPBs in arrested lte1Δ BFA1-GFP cells was switched to the dSPB when viability was restored through deletion of KIN4 (Fig. 4, C and E).
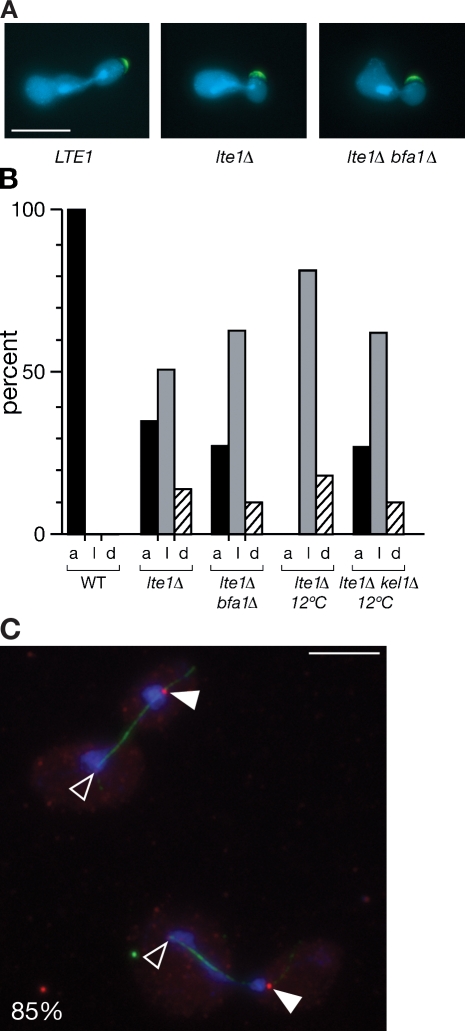
Although these different approaches consistently show a need for Lte1 activity in driving the asymmetric distribution of Bfa1 onto the dSPB, it remained important to exclude any possible artifacts arising from the altered properties of the tagged Bfa1-GFP. Therefore, the localization of native, untagged Bfa1 in wild type and lte1Δ mutants was also monitored by indirect immunofluorescence with anti-Bfa1 antibody. Wild-type and lte1Δ cells were cultured at 30 or 12°C to reveal the lte1Δ mutant defect in mitotic progression. Extended anaphase spindles of wild-type cells at both temperatures and of lte1Δ mutants at 30°C showed, as expected, a predominantly asymmetric distribution of Bfa1 on dSPBs. However, Bfa1 was seen at both SPBs on >90% of the extended mitotic spindles of lte1Δ mutants undergoing anaphase arrest at 12°C (Fig. 5). Thus, a role of Lte1 in generating the asymmetric localization of Bfa1 to the dSPB has been confirmed using untagged Bfa1.
Figure 5.
Lte1 is required for asymmetric localization of Bfa1 at dSPBs at 12°C. (A) Cells were grown overnight at 12°C. Bfa1 and tubulin were monitored by indirect immunofluorescence in wild-type 15D, SY144 lte1Δ, and MGY389 bfa1Δ cells with extended anaphase spindles. Open arrowheads indicate SPBs where Bfa1 is absent. Closed arrowheads indicate SPBs with Bfa1. Cells are outlined in white. (B) The percentage of wild-type or lte1Δ cells cultivated at 30 or 12°C in late anaphase with Bfa1 at 0, 1, or 2 SPBs was determined by the presence of Bfa1 signals at the ends of the elongated spindle (n > 100). Bar, 5 µm.
Mislocalization of Lte1 in mother cells increases asymmetric distribution of Bfa1 on SPBs of misoriented spindles
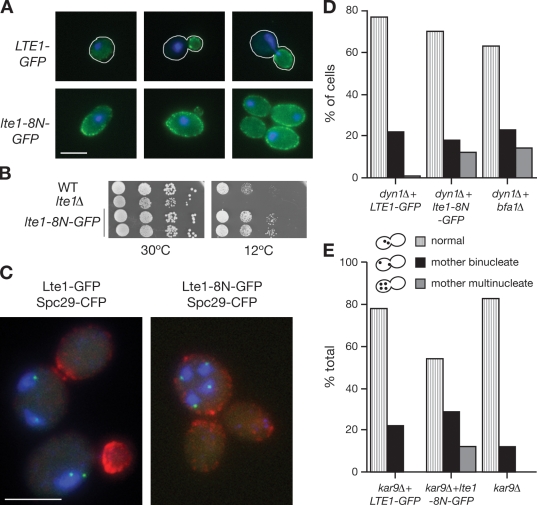
Localization of Lte1 at the mother bud cortex inactivates the SPoC and allows cell cycle progression even if nuclei are not partitioned between mother and daughter cells (Bardin et al., 2000; Castillon et al., 2003). In this study, we have used the novel lte1-8N allele of LTE1 to investigate how mislocalization of Lte1 at the mother cortex affects Bfa1 distribution at the SPBs of misaligned spindles. Lte1-8N was initially isolated as a serendipitous insertion of eight residues (APDLSFAA) at position 212 during a routine point mutagenesis protocol. This position is outside both putative N- and C-terminal GEF domains. The phenotype of lte1-8N was recognized because of its effects on Lte1 localization: Lte1-8N is present at both mother and daughter cortexes throughout the whole cell cycle, whereas wild-type Lte1 is restricted to the bud cortex from bud emergence to late M (Fig. 6 A). Lte1-8N is active, as judged by complementation of the cold sensitivity of an lte1Δ strain (Fig. 6 B). Also, the predicted abrogation of the SPoC by Lte1-8N was confirmed by monitoring the SPB marker Spc29-CFP in dyn1Δ mutants cultivated at low temperature (Yeh et al., 1995). In control LTE1 dyn1Δ mutants, binucleate mother cells containing only two SPBs accumulated, indicating that spindle misalignment had triggered the SPoC (Fig. 6 C). However, in lte1-8N dyn1 mutants, multinucleate mother cells with more than two SPBs appeared, such as the tetranucleate example with multiple empty buds shown in Fig. 6 C. A similar result was seen when the SPoC was impaired by the deletion of BFA1 (Fig. 6 D). lte1-8N also abrogated the SPoC response to spindle misalignment triggered by a kar9 mutation at normal temperatures (Fig. 6 E). Thus, Lte1-8N impairs the activity of the SPoC regardless of culture temperature or the genetic defect causing spindle misorientation.
Figure 6.
Lte1-8N is localized at mother and daughter cell cortexes and abrogates the SPoC. (A) Subcellular localization of wild-type and 8N forms of Lte1 expressed in SY144 lte1Δ in unbudded cells (left), cells with a small bud (middle), and large-budded cells (right). Cells are outlined in white. (B) Complementation of the cold sensitivity of lte1Δ mutants by lte1-8N–GFP. WT, wild type. (C) SPoC activity in MGY407 (LTE1-GFP dyn1) and MGY408 (lte1-8N–GFP dyn1) cells expressing SPC29-CFP grown overnight at 14°C. The left panel shows examples of LTE1 cells with binucleate mother cells, and the right panel shows lte1-8N mutants with multiple nuclei in the mother and multiple anucleate buds. Lte1 is shown in red, Spc29 is shown in green, and DNA is shown in blue. (D) The frequencies of normal and bi- and multinucleate cells in C (n > 100). (E) SPoC activity at 30°C in logarithmically growing MGY459 (kar9Δ), MGY462 (kar9Δ LTE1-GFP), and MGY463 (kar9Δ lte1-8N–GFP) cells expressing Spc29-CFP. The histogram shows frequencies of normal and bi- and multinucleate cells (n > 100). Bars, 5 µm.
The effect of the maternally localized Lte1 on Bfa1 distribution at the SPBs was then investigated by indirect immunofluorescence of Bfa1 and tubulin in LTE1 dyn1Δ and lte1-8N dyn1Δ cells cultivated at 14°C (Fig. 7 A). The intensities of Bfa1 foci were quantified in binucleate mother cells in which the nuclei had entirely separated on extended but intact mitotic spindles. The ratio of Bfa1 signal intensities at the two SPBs indicates the degree of symmetry, with a value of 1 indicating completely symmetrical localization. In LTE1 dyn1 cells, the intensities of Bfa1 at the two SPBs were within a factor of two of each other in >65% of cells, indicating a largely symmetrical distribution of Bfa1 on both SPBs of the cells undergoing a SPoC arrest. In contrast, 64% of dyn1Δ lte1-8N mutants had relative intensities of Bfa1 signal that differed by more than threefold, indicating increased asymmetric localization of Bfa1 on one SPB (Fig. 7 B). Thus, in cells with a misoriented spindle, lte1-8N abrogates the SPoC and does so with an increase in the asymmetrical distribution of Bfa1 at the SPBs.
Figure 7.
Expression of Lte1 in the mother increases the asymmetric SPB localization of Bfa1 on misaligned spindles. (A) Bfa1, tubulin, and nuclei were monitored in binucleate mother cells with elongated spindles after overnight growth at 14°C. MGY407 (LTE1 dyn1) cells are shown in the top panels, and MGY408 (lte1-8N dyn1) cells are shown in the bottom panels. Open arrowheads indicate SPBs where Bfa1 signal is diminished. Closed arrowheads indicate SPBs with Bfa1. Cells are outlined in white. (B) The ratios of Bfa1 signal intensities were recorded in binucleate cells observed in A (n > 50). Bar, 5 µm.
Bfa1 localization at SPBs is independent of Tem1
Fraschini et al. (2006) proposed that the mechanism driving the asymmetrical accumulation of the Bfa1–Bub2 complex at the dSPB depended on Bub2 GAP activity acting on Tem1 at the mSPB. To test this idea, we investigated the distribution of Bfa1-GFP in the absence of Tem1 using a tem1Δ strain kept alive by the expression of HyA Dbf2. We analyzed Bfa1 distribution in cells with short, correctly oriented spindles (early anaphase), in cells with a correctly elongated spindle (late anaphase), and in cells in which the spindle was elongated in the mother (mispositioned). The large majority of cells in early anaphase or with a mispositioned spindle showed a symmetrical Bfa1 distribution with signal ratios within a factor of two (Fig. 8). On the contrary, in late anaphase cells, Bfa1 was clearly asymmetrical with a signal ratio always >4, which is in accordance with a previous study (Pereira et al., 2000). These results indicate that neither the switch from symmetrical to asymmetrical localization of Bfa1 during a normal cell cycle nor the persistent symmetrical localization at both SPBs on misaligned spindles is regulated by Tem1 activity.
Figure 8.
Distribution of Bfa1 at SPBs is Tem1 independent. (A) Bfa1-GFP localization in MGY504 (tem1Δ BFA1-GFP dbf2 HyA) cells in metaphase–early anaphase and late anaphase cultivated at 30°C. (B) Bfa1-GFP localization in anaphase cells with a mispositioned spindle. Strain MGY547 (tem1Δ dyn1Δ BFA1-GFP dbf2 HyA) was cultivated at 14°C to increase the proportion of cells containing a mispositioned spindle. (A and B) Cells are outlined in white. (C) The ratios of Bfa1 signal intensities at SPBs were recorded in cells observed in A and B (n > 100). Bars, 5 µm.
Lte1 affects polarity cap behavior
Whatever the mechanism that alters the affinity of the Bfa1–Bub2 complex to the SPBs after correct alignment of the mitotic spindle, it seems that the actin and microtubule networks are responsible for communicating this spindle position information to the SPBs (Pereira et al., 2001; Maekawa et al., 2007; Caydasi and Pereira, 2009; Monje-Casas and Amon, 2009). Therefore, the effect of Lte1 on Bfa1 localization at SPBs may stem from perturbations in the actin and/or microtubule networks, from elements that control these networks, or from the signal generation process itself. Previously, no alterations were noted in the actin distribution of lte1 mutants (Höfken and Schiebel, 2002). Similarly, we were unable to see any gross changes in microtubules in wild type and lte1 mutants, although submicroscopic events cannot be excluded. However, we did see a marked difference between polarity cap behavior of wild-type and lte1Δ cells. The polarity cap coordinates polarization of the actin cytoskeleton with exocytosis and endocytosis to focus growth at the bud tip (for review see Moseley and Goode, 2006). It also influences microtubule behavior through transport of tubulin along actin filaments (for review see Pruyne et al., 2004). The polarity cap was visualized for live imaging using Spa2- and Kel1-GFP together. Although similar results were obtained using either tagged protein alone (unpublished data), the combined tags were easier to monitor. Spa2 is a core polarizome subunit of the polarity cap (for review see Moseley and Goode, 2006), whereas Kel1 affects cell morphology and cell fusion (Philips and Herskowitz, 1998). Importantly, Kel1 interacts with Lte1, and Kel1 deletion is able to rescue the cold sensitivity of an lte1Δ strain (Höfken and Schiebel, 2002). During the early stages of bud development in wild-type cells, Spa2 and Kel1 occupy a crescent-shaped zone at the bud tip. They then disperse and reappear at the bud neck before cytokinesis (Video 1). In lte1 mutants, the Spa2 and Kel1 signals did not always remain at the tip of the bud as it emerged from the mother but instead veered to one side of the bud (Video 2). This behavior could be observed and quantified in still images in which Spa2 and Kel1 were found at the sides of the buds in more than half of the lte1 mutants observed, whereas in wild-type cells, the polarity cap remained apical (Fig. 9, A and B). This phenotype was complemented by expression of wild-type or 8N forms of Lte1 (unpublished data). If lte1Δ cells were incubated at the restrictive temperature of 12°C, the polarizome accumulated at the side of the bud in >80% of arrested cells, and no apical signals were detected (Fig. 9 B). In addition, a faint signal could sometimes be observed at the bud neck. However, deletion of BFA1, which rescues the MEN defect of lte1 mutants, did not restore apical localization of the polarity cap (Fig. 9, A and B). Thus, Lte1 has a hitherto unknown, MEN-independent role in maintaining the apical axis of the polarity cap.
Figure 9.
Lte1 maintains apical localization of the polarity cap through a MEN-independent mechanism. (A) The polarity caps of MGY308 wild type, MGY309 lte1Δ, and MGY470 lte1Δ bfa1Δ cells with medium-sized buds were visualized using Spa2- and Kel1-GFP. (B) Quantification of wild-type (WT; n = 70), lte1Δ (n = 100), and lte1Δ bfa1Δ (n = 100) medium-sized budding cells cultivated at 30°C and lte1Δ (n = 100) and lte1Δ kel1Δ (n = 100) cells cultivated at 12°C with apical (a), lateral (l), or dispersed (d) Kel1- and/or Spa2-GFP. (C) Bfa1 and tubulin were visualized in SY161 lte1Δ kel1Δ by indirect immunofluorescence after overnight growth at 12°C. Open arrowheads indicate SPBs where Bfa1 is absent. Closed arrowheads indicate SPBs with Bfa1. Cells (n = 100) were scored for asymmetric (85%) or symmetric (15%) localization of Bfa1. Bars, 5 µm.
To test further the proposal that polarity cap behavior influences the distribution of Bfa1 at SPBs, we made a deletion of the polarity cap gene KEL1 in an lte1Δ mutant. As deletion of KEL1 rescues the cold sensitivity of lte1Δ mutants (Höfken and Schiebel, 2002; unpublished data), we asked whether this was accompanied with a change in the distribution of Bfa1 at the SPBs. lte1Δ kel1Δ cells were cultivated overnight at 12°C, and the localization of Bfa1 and tubulin were observed. In contrast to the mainly symmetrical localization of Bfa1 at SPBs seen in lte1Δ cells cultivated at the same temperature (Fig. 5), lte1Δ kel1Δ mutants displayed an asymmetrical distribution of Bfa1 at one SPB in the majority of the late anaphase cells observed (Fig. 9 C).
Although deletion of KEL1 restored the asymmetric localization of Bfa1 at SPBs, only 27% regained apical polarizome localization in anaphase cells cultivated at 12°C. 63% of cells continued to have their polarizome mislocalized at the side of the bud, whereas in the remaining 10% of cells, a dispersed signal was seen. The interpretation of this result is somewhat complicated by the morphological perturbations caused by the deletion of KEL1 (Philips and Herskowitz, 1998). Nevertheless, deletion of KEL1 in an lte1Δ mutant clearly did not fully restore normal localization of Spa2-GFP at 12°C.
Discussion
The idea that Lte1 positively regulates mitotic exit by acting as a GEF for the small GTPase Tem1 has long been part of the consensus view of how the MEN is regulated despite any direct proof that this is actually the case. Moreover, the role of Lte1’s GEF domain in MEN regulation has been controversial, with contradictory results being reported (Jensen et al., 2002a; Yoshida et al., 2003; Seshan and Amon, 2005; Zhao et al., 2007). Given this uncertainty, our aim was to resolve how Lte1 affects MEN activity. Our data indicate that Lte1 does not rely on nucleotide exchange activity for its role in mitotic regulation. Instead, we show that Lte1 acts by influencing the localization of Bfa1 at the SPBs.
Obviously, a negative result showing the absence of in vitro GEF activity is difficult to evaluate, especially as additional cofactors like Cla4-dependent phosphorylation and Ras may have been required. Nevertheless, the requirement for Ras in vivo is not absolute because overexpression of Lte1 can rescue the mitotic arrest defects of rasΔ cells (Morishita et al., 1995; Yoshida et al., 2003). Furthermore, Lte1-Cdk, lacking cell cycle–dependent phosphorylation in vivo, could still complement an lte1Δ mutant (Jensen et al., 2002a), indicating that the bulk of phosphorylation is not essential for Lte1 function in MEN regulation. Thus, we believe that our in vitro assay would not have been absolutely reliant on these additional factors.
The lack of demonstrable GEF activity in vitro is consistent with our mutational analysis of the role of Lte1’s GEF domain in which complementation of the cold sensitivity of an lte1Δ strain relied on the interaction of Lte1 with Ras and localization at the bud cortex rather than GEF activity itself. The idea that Tem1 does not require a GEF for its activity is also supported by observations of the septation initiation network (SIN) pathway in S. pombe, in which most MEN-like homologues are conserved but no GEF for Spg1 has been described (Bardin and Amon, 2001). Furthermore, Tem1 and Spg1 have atypically high intrinsic nucleotide exchange activities (Furge et al., 1998; Geymonat et al., 2002) that question the need for additional specific exchange factors.
Instead of regulation of the MEN by GEF activity, we show by several experimental approaches that Lte1 can contribute to the asymmetric localization of Bfa1 at dSPBs. This pattern of localization has already been linked to mitotic progression (Pereira et al., 2001; Molk et al., 2004; Pereira and Schiebel, 2005; Piatti et al., 2006; Maekawa et al., 2007; for review see Pereira and Schiebel, 2001), and one proposal is that it allows any Tem1 remaining in the maternal cell to activate the MEN (Fraschini et al., 2006; Piatti et al., 2006). Lte1’s influence on the association of Bfa1 at the SPBs was revealed in two different situations under which LTE1 is essential for viability, namely growth at 12°C and in combination with a form of BFA1-GFP. In both instances, depletion of LTE1 caused late anaphase arrest, leading to the appearance of Bfa1 on both SPBs, which is in contrast to the accumulation of Bfa1 at the dSPBs of cells expressing Lte1 (Pereira et al., 2001).
The synthetic lethality of an lte1Δ BFA1-GFP strain could be complemented by the expression of a HyA form of Dbf2 or by deleting KIN4, but the localization pattern of Bfa1 in these two conditions was different. With Dbf2 HyA, Bfa1 remained on both SPBs in late anaphase cells, as might be expected by bypass of the regulatory aspects of the MEN by a constitutively active form of the pathway’s effector kinase. On the contrary, deletion of KIN4 restored the asymmetric distribution of Bfa1 observed in wild-type cells, confirming the role of Kin4 in Bfa1 localization (Caydasi and Pereira, 2009).
The role of Lte1 in Bfa1 localization was further demonstrated by the properties of Lte1-8N, which localizes to both mother and bud cortexes. As expected from earlier work in which Lte1 was present in mother cells, the LTE1-8N allele abrogated the SPoC in cells with a misaligned spindle (Bardin et al., 2000; Castillon et al., 2003), and this coincided with a switch of Bfa1 localization to one SPB. Collectively, these observations confirm the link between the localization pattern of Bfa1 at the SPBs with cell cycle progression and, for the first time, demonstrate the involvement of Lte1 in this process.
How might Lte1 at the bud cortex affect events at the SPBs? First, whatever the mechanism, it does not act through Tem1 because deletion of TEM1 neither alters the accumulation of Bfa1 at the dSPB in normal cells nor does it prevent the continued localization of Bfa1 at both SPBs in cells with misaligned spindles (Fig. 8; Pereira et al., 2000). Nevertheless, this result cannot exclude that, when present, Tem1 can still contribute to Bfa1 distribution at the SPBs. In considering how Lte1 might influence Bfa1 binding at SPBs, it was shown recently that the formin Bni1 is required for the asymmetric association of Bfa1 to one SPB (Monje-Casas and Amon, 2009). Bni1 nucleates the formation of polarized actin filaments that extend along the mother–bud cell axis from the bud tip. Localization of Bni1 at the bud tip occurs through interactions with polarity cap components, including Spa2 (for review see Park and Bi, 2007). In this study, we found that the polarity cap behaved abnormally in lte1Δ mutants and often took up nonapical positions in the developing bud. Moreover, we showed that deletion of the polarity cap component KEL1 rescues the cold sensitivity of an lte1Δ strain with restitution of the asymmetrical distribution of Bfa1 to one SPB in late anaphase. This demonstrates a connection between the polarity cap and Bfa1 localization. Thus, collectively, Lte1 and Bni1 both affect aspects of polarity cap activity: Lte1 affects polarity cap localization, whereas Bni1 is needed for actin filament formation from the polarity cap. Therefore, it is particularly striking that, under certain conditions, loss of either Lte1 or Bni1 activity can prevent accumulation of Bfa1 at the dSPB. Therefore, we suggest that Bni1 and Lte1 exert their effects on Bfa1 localization at SPBs by similar means. In the Bni1 study, the signal linking the polarity cap to the SPBs was not described (Monje-Casas and Amon, 2009). However, it was suggested that cues from the asymmetry of the actin cytoskeleton were transmitted to the SPBs via cytoplasmic microtubules. Indeed, other work also identified microtubules as the signaling conduits between the bud cortex and SPBs (Pereira et al., 2001; Maekawa et al., 2007; Caydasi and Pereira, 2009). We propose that the apparent requirement for a polarized actin cytoskeleton might stem from the role of actin filaments in directing the ends of cytoplasmic microtubules in their exploration of the bud neck and cortex (for reviews see Bretscher, 2003; Pruyne et al., 2004). This process aligns the axis of the mitotic spindle with that of mother and bud and so would prime the cell for activation of the MEN.
What then happens at 12°C that makes Lte1 essential? A well-described consequence of low temperature is a change in the kinetics of microtubule polymerization and depolymerization (Hamel et al., 1984). Therefore, we envisage that the alteration in polarization in the absence of Lte1 combined with low temperature may be sufficient to disturb the signaling cascade that appears to exist between polarity, microtubule behavior, and the dynamics of Bfa1 association with the SPBs. It is especially notable that lte1 mutants are sensitive to drugs that depolymerize microtubules (Shirayama et al., 1994a) and that lte1Δ is synthetically lethal or sick with defects in several genes involved in microtubule metabolism, including BIM1, CIN8, CIK1, DYN1, JNM1, and KAR3 and -9 (summarized at Saccharomyces Genome Database http://www.yeastgenome.org/). Intriguingly, many of these same mutations are also synthetically lethal with deletions of BFA1 or BUB2, and bfa1Δ/bub2Δ strains are also sensitive to microtubule-depolymerizing drugs (Hoyt et al., 1991; Li, 1999). These phenotypic similarities are difficult to reconcile with a model of Tem1 regulation by antagonistic Lte1 GEF and Bfa1–Bub2 GAP activities. Instead, they are consistent with different altered responses to microtubule perturbation. The essential activity of Lte1 is not just limited to growth at 12°C because at higher temperatures, the continued activity of Lte1 is indicated by the multiple examples of synthetic lethality and by the aberrant polarity cap behavior of lte1Δ mutants.
To summarize, Lte1’s role in controlling mitotic exit requires reevaluation. Rather than providing guanosine exchange activity for Tem1, as previously supposed (Shirayama et al., 1994a; Bardin et al., 2000; Pereira et al., 2000), Lte1 can influence the localization of Bfa1 at SPBs, which in turn appears to be an important factor in mitotic progression. The possible linkage between Lte1’s role in cell polarization and SPB behavior clearly offers an exciting new area for study that is being actively pursued.
After submission of this manuscript, the S. pombe ETD1 gene was characterized as a functional homologue of budding yeast LTE1 (García-Cortés and McCollum, 2009). The Etd1 protein influences activity of the SIN pathway, and it displays cortical asymmetry, which may be coordinated with differential SIN activity at the SPBs. Interestingly, the authors also question whether the Etd1 protein acts as a GEF for Spg1, the S. pombe homologue of Tem1.
Materials and methods
Strains and culture
Most strains are derivatives of BF264-15DU: a ura3Δns ade1 his2 leu2-3, 112 trp1-1a (Table I; Richardson et al., 1989). Yeast extract peptone (YEP) complete medium or yeast nitrogen base selective medium was used. For cold sensitivity assays, 10-fold serial dilutions of stationary phase cultures were spotted on YEP agar plates and incubated at 30°C for 2 d or at 12°C for 10–14 d.
Table I.
Yeast strains
| Strain name | Relevant genotype | Source or reference |
| 15D | MATa, ura3Δns ade1 his2 leu2-3,112 trp1-1 | Richardson et al., 1989 |
| SY144 | MATa lte1::KANR | Jensen et al., 2002a |
| SY161 | MATa lte1::KANR kel1::TRP1 | Derived from 15D; S. Jensena |
| SY169 | MATa lte1::KANR spo12::TRP1 + YCplac33-LTE1 | Jensen et al., 2002a |
| MMY40 | MATa lte1::KANR BFA1::BFA1-GFP(TRP1) kin4::LEU2 | Derived from 15D |
| MMY41 | MATa lte1::KANR BFA1::BFA1-GFP(TRP1) kin4::LEU2 ura3::CFP-TUB1(URA3) | Derived from 15D |
| MGY104 | MATa ura3-1 trp1-28 leu2Δ0 lys2Δ0 his7 mob1::kanMX4 pep4::LEU2 + pMH919-LTE1 | Geymonat et al., 2007 |
| MGY170 | MATa ura3-1 trp1-28 leu2Δ0 lys2Δ0 his7 mob1::kanMX4 pep4::LEU2 + pMH903-RAS2V19 | Derived from MGY70 (Geymonat et al., 2007) |
| MGY252 | MATa lte1::KANR BFA1::BFA1-GFP(TRP1) + YCplac33-LTE1 | Derived from 15D |
| MGY260 | MATa lte1::KANR BFA1::BFA1-GFP(TRP1) leu2::GAL–lte1-F1387E:HA3(LEU2) ura3::CFP-TUB1(URA3) | Derived from 15D |
| MGY272 | MATa lte1::KANR BFA1::BFA1-GFP(TRP1) ura3::CFP-TUB1(URA3) + pRS315-dbf2 HyA | Derived from 15D |
| MGY274 | MATa mob1-77 ura3-52 trp1D1 his3D200 leu2-3,112 BFA1::BFA1-GFP(TRP1) | Derived from FLY30 (Luca and Winey, 1998) |
| MGY308 | MATa KEL1::KEL1:GFP(KANR) SPA2::SPA2-GFP(URA3) | Derived from 15D |
| MGY309 | MATa lte1::KANR KEL1::KEL1-GFP(KANR) SPA2::SPA2-GFP(URA3) | Derived from 15D |
| MGY386 | MATa cla4::URA3 | Derived from 15D |
| MGY389 | MATa bfa1::LEU2 | Derived from 15D |
| MGY396 | MATa dyn1::URA3 SPC29::SPC29-CFP(TRP1) | Derived from 15D |
| MGY406 | MATa dyn1::URA3 bfa1::LEU2 SPC29::SPC29-CFP(TRP1) | Derived from 15D |
| MGY407 | MATa dyn1::URA3 leu2::LTE1-GFP(LEU2) SPC29::SPC29-CFP(TRP1) | Derived from 15D |
| MGY408 | MATa dyn1::URA3 leu2::lte1-8N–GFP(LEU2) SPC29::SPC29-CFP(TRP1) | Derived from 15D |
| MGY457 | MATa lte1::KANR kel1::TRP1 SPA2::SPA2-GFP(URA3) | Derived from 15D |
| MGY459 | MATa kar9:: KANR SPC29::SPC29-CFP(TRP1) | Derived from 15D |
| MGY462 | MATa kar9:: KANR leu2::LTE1-GFP(LEU2) SPC29::SPC29-CFP(TRP1) | Derived from 15D |
| MGY463 | MATa kar9:: KANR leu2::lte1-8N–GFP(LEU2) SPC29::SPC29-CFP(TRP1) | Derived from 15D |
| MGY470 | MATa lte1::KANR bfa1::LEU2 KEL1::KEL1-GFP(KANR) SPA2::SPA2-GFP(URA3) | Derived from 15D |
| MGY504 | MATα tem1::KANR BFA1::BFA1-GFP(TRP1) + pRS315-dbf2 HyA | Derived from YPH501 |
| MGY547 | MATα tem1::KANR dyn1::URA3 BFA1::BFA1-GFP(TRP1) + pRS315-dbf2 HyA | Derived from YPH501 |
Evolva A/S, Frederiksberg, Denmark.
Plasmids
Plasmids are listed in Table II. YIplac128-based plasmids expressed Lte1 from its own promoter with C-terminal 3HA or GFP epitope tags and were integrated at the LEU2 locus (Jensen et al., 2002b). lte1-K1273E, -R1343E, -F1387E, and -V1382E mutations were introduced by PCR ligation. lte1-8N encodes Lte1 with an APDLSFAA replacement of S212 and arose serendipitously during point mutagenesis with a QuikChange Site-Directed Mutagenesis kit (Agilent Technologies). lte1-ΔEcoRI was obtained by digestion with EcoRI and relegation of LTE1 plasmids.
Table II.
Plasmids
| Name | Backbone | Content | Source or reference |
| pCFP-Tub1 | pRS306 | CFP-TUB1 | (Jensen et al., 2002a) |
| pKGFP-Kel1 | pKGFP | Last 674 bp of KEL1 in frame with GFP | (Jensen et al., 2002a) |
| pDbf2-1c | pRS315 | HyA of DBF2 | This study |
| pMG52 | YIplac128 | LTE1-HA3 | This study |
| pMG56 | YIplac128 | lte1-R1343E–HA3 | This study |
| pMG58 | YIplac128 | LTE1-GFP | This study |
| pMG59 | YIplac128 | lte1-K1273E–HA3 | This study |
| pMG60 | YIplac128 | lte1-F1387E–HA3 | This study |
| pMG66 | YIplac128 | lte1-K1273E–GFP | This study |
| pMG67 | YIplac128 | lte1-F1387E–GFP | This study |
| pMG75 | YIplac128 | 6His–lte1-F1387E–HA3; under GAL1 promoter control | This study |
| pMG77 | YIplac128 | 6His–lte1-K1273E–HA3; under GAL1 promoter control | This study |
| pMG110 | pRS306 | Last 947 bp of SPA2 in frame with GFP | This study |
| pMG112 | pRS304 | Last 418 bp of SPC29 in frame with CFP | This study |
| pMG118 | pRS304 | Last 875 bp of BFA1 in frame with GFP | This study |
| pMG152 | pRS304 | 6His–lte1-ΔEcoRI–HA3; under GAL1 promoter control | This study |
| pMG157 | YIplac128 | lte1-ΔEcoRI–HA3 | This study |
| pMG158 | YIplac128 | lte1-ΔEcoRI–GFP | This study |
| pMG159 | YIplac128 | 6His–lte1-R1343E–HA3; under GAL1 promoter control | This study |
| pMG164 | YIplac128 | lte1-V1382E–HA3 | This study |
| pMG234 | YIplac128 | 6His-lte1-HA3; under GAL1 promoter control | This study |
| pMG183 | YIplac128 | lte1-8N–HA3 | This study |
| pMG180 | YIplac128 | lte1-8N–GFP | This study |
For conditional expression of Lte1, a 5′ 1,058-bp fragment of LTE1 in frame with a 6His tag under the control of the GAL1 promoter was amplified by PCR using pMH919-Lte1 (Geymonat et al., 2007) as template and was cloned into YIplac128-based plasmids. Endogenous Bfa1 was C-terminally tagged with GFP using a fusion between BFA1’s final 875 bp and GFP in pRS304 containing the ADH1 terminator. Integrants at the BFA1 locus were verified by PCR and visual screening. Endogenous Spc29 was C-terminally tagged using a fusion between SPC29’s final 418 bp and CFP in a pRS304 vector containing the ADH1 terminator. Integrated transformants were verified by visual screening. Endogenous Spa2 and Kel1 were C-terminally tagged with GFP using a fusion between the last 947 bp of SPA2 or the last 674 bp of KEL1 and GFP cloned, respectively, into pRS306 or pKGFP (Jensen et al., 2002b). Integrants were verified by visual screening.
Protein purification
6His-Lte1 and GST-Ras2V19 were expressed and purified from yeast (Geymonat et al., 2007). Maltose-binding protein (MBP)–Bfa1 and MBP-Tem1 were expressed in E. coli (Geymonat et al., 2002). GST-NT-Bfa1 comprises the N-terminal 230 residues of Bfa1 and was expressed from pGEX-KG in E. coli BL21 (Geymonat et al., 2002).
Lte1 and RasV19 binding assay
Glutathione agarose beads (GE Healthcare) carrying GST and GST-Ras2V19 were mixed at 4°C for 2 h with approximately equal amounts of 6His-Lte1, as described previously (Yoshida et al., 2003). Bound proteins were eluted with 20 mM of reduced glutathione, and Lte1 was detected by Western blotting with anti-6His antibody (EMD).
Lte1-3HA–Ras2 coimmunoprecipitation
Logarithmically growing cultures were arrested with 100 mM hydroxyurea for 2 h. Anti-HA antibody (12CA5; S. Ley, National Institute for Medical Research, London, England, UK) was added to equal amounts of protein extracts (Geymonat et al., 2007) and incubated for 2 h at 4°C. Immunocomplexes were purified with protein G–coupled Sepharose beads (GE Healthcare). Beads were washed extensively with lysis buffer without phosphatase and protease inhibitors. The proteins were separated by 6–10% step SDS-PAGE for Western blot analysis with anti-HA and anti-Ras2 (Santa Cruz Biotechnology, Inc.) antibodies. The relative amounts of Ras2 bound to Lte1 were estimated using ImageJ (version 1.41; National Institutes of Health) on scanned images on x-ray film.
Guanosine nucleotide exchange assay
Nucleotide exchange was assayed by loss of [3H]GDP from Tem1 as described previously (Geymonat et al., 2002). In brief, for [3H]GDP loading, purified Tem1 or the Tem1–Bfa1 complex was incubated with 0.1 Mbq [3H]GDP at 25°C for 10 min in loading buffer (20 mM Tris-HCl, pH 7.5, 25 mM NaCl, 5 mM MgCl2, and 0.1 mM DTT). Then, for GDP release, 10 µl was added to 50 µl of reaction buffer (20 mM Tris, pH 7.5, 2 mM GTP, and 0.6 µg/µl BSA) in the presence or absence of Lte1. The tube was transferred to 25°C, and, at the indicated times, 10 µl was diluted in 990 µl of cold washing buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, and 5 mM MgCl2). The mixture was passed through nitrocellulose filters, and filter-bound radioactive nucleotide was determined by scintillation counting. When the effect of Bfa1 on Tem1 was examined, the indicated amount of Bfa1 was added to Tem1 in loading buffer, and activity was assessed after 4 min of incubation in reaction buffer at 25°C.
Antibody purification
Rabbit antibodies were raised to GST-NT-Bfa1 (Harlan Sera Lab Ltd). IgG from 10 ml of serum was purified with a HiTrap protein A column (GE Healthcare). Anti-Bfa1 antibodies were obtained by affinity purification with MBP-Bfa1 immobilized on Protran nitrocellulose transfer membrane (Whatman Ltd; Pringle et al., 1991). Antibody specificity was confirmed in Western blots with cell extracts of wild-type and bfa1Δ cells (Fig. S4) and by indirect immunofluorescence microscopy of wild-type and bfa1Δ cells (Fig. 5).
Microscopy and imaging
Indirect immunofluorescence (Geymonat et al., 2004) used anti-Bfa1 (previous section) and monoclonal anti–α-tubulin (clone B-5-1-2; Sigma-Aldrich). Secondary antibodies were Alexa Fluor 594 (anti–rabbit) and 488 (anti–mouse). GFP- and CFP-labeled proteins were analyzed by fluorescence microscopy after fixation with 3.7% formaldehyde added directly to the medium for 10 min at room temperature. DNA was stained with 2 µg/ml DAPI contained in the mounting medium, Vectashield (Vector Laboratories). Fluorescence microscopy used a liquid-cooled charge-coupled device camera (CH350L; Photometrics) on an inverted microscope (IX70; Olympus) with a 100× F1.4 objective. Cell images were captured and manipulated using SoftWoRx software (Applied Precision) and Photoshop CS (Adobe). Relative fluorescence intensities at m- and dSPBs were measured using the SoftWoRx software as the difference between the maximal fluorescence at the SPB and the background fluorescence in the immediate proximity of the SPB. For time lapse experiments, images were taken at 5-min intervals of cells growing at 30°C on agar plugs in a temperature-controlled chamber.
Online supplemental material
Fig. S1 shows the alignment of the GEF-like domain of Lte1 with other known GEFs. Fig. S2 is an assay for the bypass of the SPoC by overexpression of different alleles of LTE1. Fig. S3 describes the isolation and characterization of the dbf2 HyA allele. Fig. S4 demonstrates the specificity of the anti-Bfa1 antibody. Video 1 shows the polarity cap behavior in wild-type cells. Video 2 shows how polarity cap behavior is affected in an lte1Δ mutant. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200905114/DC1.
Acknowledgments
We thank Rita Cha, Sanne Jensen, Lee Johnston, Stephen Ley, Margaret Migocki, Katrin Rittinger, Marisa Segal, Stephen Smerdon, Kate Sullivan, and Wai Han Yau for advice, materials, and help.
This research was financed by the UK Medical Research Council.
Footnotes
Abbreviations used in this paper:
- dSPB
- daughter-directed SPB
- FEAR
- Cdc14 early anaphase release
- GAP
- GTPase-activating protein
- GEF
- guanosine nucleotide exchange factor
- HyA
- hyperactive
- MBP
- maltose-binding protein
- MEN
- mitotic exit network
- mSPB
- maternal SPB
- SIN
- septation initiation network
- SPB
- spindle pole body
- SPoC
- spindle position checkpoint
- YEP
- yeast extract peptone
References
- Adames N.R., Oberle J.R., Cooper J.A. 2001. The surveillance mechanism of the spindle position checkpoint in yeast. J. Cell Biol. 153:159–168 10.1083/jcb.153.1.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandru G., Zachariae W., Schleiffer A., Nasmyth K. 1999. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 18:2707–2721 10.1093/emboj/18.10.2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A.J., Amon A. 2001. Men and sin: what’s the difference? Nat. Rev. Mol. Cell Biol. 2:815–826 10.1038/35099020 [DOI] [PubMed] [Google Scholar]
- Bardin A.J., Visintin R., Amon A. 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 102:21–31 10.1016/S0092-8674(00)00007-6 [DOI] [PubMed] [Google Scholar]
- Bosl W.J., Li R. 2005. Mitotic-exit control as an evolved complex system. Cell. 121:325–333 10.1016/j.cell.2005.04.006 [DOI] [PubMed] [Google Scholar]
- Bretscher A. 2003. Polarized growth and organelle segregation in yeast: the tracks, motors, and receptors. J. Cell Biol. 160:811–816 10.1083/jcb.200301035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon G.A., Adames N.R., Rosello C.H., Seidel H.S., Longtine M.S., Cooper J.A., Heil-Chapdelaine R.A. 2003. Septins have a dual role in controlling mitotic exit in budding yeast. Curr. Biol. 13:654–658 10.1016/S0960-9822(03)00247-1 [DOI] [PubMed] [Google Scholar]
- Caydasi A.K., Pereira G. 2009. Spindle alignment regulates the dynamic association of checkpoint proteins with yeast spindle pole bodies. Dev. Cell. 16:146–156 10.1016/j.devcel.2008.10.013 [DOI] [PubMed] [Google Scholar]
- Cheng J., Türkel N., Hemati N., Fuller M.T., Hunt A.J., Yamashita Y.M. 2008. Centrosome misorientation reduces stem cell division during ageing. Nature. 456:599–604 10.1038/nature07386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J., Chardin P. 1999. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci. 24:306–311 10.1016/S0968-0004(99)01429-2 [DOI] [PubMed] [Google Scholar]
- D’Aquino K.E., Monje-Casas F., Paulson J., Reiser V., Charles G.M., Lai L., Shokat K.M., Amon A. 2005. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol. Cell. 19:223–234 10.1016/j.molcel.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Fesquet D., Fitzpatrick P.J., Johnson A.L., Kramer K.M., Toyn J.H., Johnston L.H. 1999. A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. EMBO J. 18:2424–2434 10.1093/emboj/18.9.2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., Formenti E., Lucchini G., Piatti S. 1999. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J. Cell Biol. 145:979–991 10.1083/jcb.145.5.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., D’Ambrosio C., Venturetti M., Lucchini G., Piatti S. 2006. Disappearance of the budding yeast Bub2–Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J. Cell Biol. 172:335–346 10.1083/jcb.200507162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furge K.A., Wong K., Armstrong J., Balasubramanian M., Albright C.F. 1998. Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr. Biol. 8:947–954 10.1016/S0960-9822(98)70394-X [DOI] [PubMed] [Google Scholar]
- García-Cortés J.C., McCollum D. 2009. Proper timing of cytokinesis is regulated by Schizosaccharomyces pombe Etd1. J. Cell Biol. 186:739–753 10.1083/jcb.200902116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Smith S.J., Wheatley E., Rittinger K., Johnston L.H., Sedgwick S.G. 2002. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J. Biol. Chem. 277:28439–28445 10.1074/jbc.M202540200 [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Wells G.P., Smerdon S.J., Sedgwick S.G. 2004. Clb6/Cdc28 and Cdc14 regulate phosphorylation status and cellular localization of Swi6. Mol. Cell. Biol. 24:2277–2285 10.1128/MCB.24.6.2277-2285.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Sedgwick S.G. 2007. A Saccharomyces cerevisiae autoselection system for optimised recombinant protein expression. Gene. 399:120–128 10.1016/j.gene.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Hall B.E., Yang S.S., Boriack-Sjodin P.A., Kuriyan J., Bar-Sagi D. 2001. Structure-based mutagenesis reveals distinct functions for Ras switch 1 and switch 2 in Sos-catalyzed guanine nucleotide exchange. J. Biol. Chem. 276:27629–27637 10.1074/jbc.M101727200 [DOI] [PubMed] [Google Scholar]
- Hamel E., del Campo A.A., Lin C.M. 1984. Stability of tubulin polymers formed with dideoxyguanosine nucleotides in the presence and absence of microtubule-associated proteins. J. Biol. Chem. 259:2501–2508 [PubMed] [Google Scholar]
- Heil-Chapdelaine R.A., Oberle J.R., Cooper J.A. 2000. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J. Cell Biol. 151:1337–1344 10.1083/jcb.151.6.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfken T., Schiebel E. 2002. A role for cell polarity proteins in mitotic exit. EMBO J. 21:4851–4862 10.1093/emboj/cdf481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfken T., Schiebel E. 2004. Novel regulation of mitotic exit by the Cdc42 effectors Gic1 and Gic2. J. Cell Biol. 164:219–231 10.1083/jcb.200309080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Totis L., Roberts B.T. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 66:507–517 10.1016/0092-8674(81)90014-3 [DOI] [PubMed] [Google Scholar]
- Hu F., Wang Y., Liu D., Li Y., Qin J., Elledge S.J. 2001. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 107:655–665 10.1016/S0092-8674(01)00580-3 [DOI] [PubMed] [Google Scholar]
- Jensen S., Geymonat M., Johnson A.L., Segal M., Johnston L.H. 2002a. Spatial regulation of the guanine nucleotide exchange factor Lte1 in Saccharomyces cerevisiae. J. Cell Sci. 115:4977–4991 10.1242/jcs.00189 [DOI] [PubMed] [Google Scholar]
- Jensen S., Geymonat M., Johnston L.H. 2002b. Mitotic exit: delaying the end without FEAR. Curr. Biol. 12:R221–R223 10.1016/S0960-9822(02)00756-X [DOI] [PubMed] [Google Scholar]
- Keng T., Clark M.W., Storms R.K., Fortin N., Zhong W., Ouellette B.F., Barton A.B., Kaback D.B., Bussey H. 1994. LTE1 of Saccharomyces cerevisiae is a 1435 codon open reading frame that has sequence similarities to guanine nucleotide releasing factors. Yeast. 10:953–958 10.1002/yea.320100710 [DOI] [PubMed] [Google Scholar]
- Kim J., Jang S.S., Song K. 2008. Different levels of Bfa1/Bub2 GAP activity are required to prevent mitotic exit of budding yeast depending on the type of perturbations. Mol. Biol. Cell. 19:4328–4340 10.1091/mbc.E08-02-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Jensen S., Frenz L.M., Johnson A.L., Fesquet D., Johnston L.H. 2001. The Bub2-dependent mitotic pathway in yeast acts every cell cycle and regulates cytokinesis. J. Cell Sci. 114:2345–2354 [DOI] [PubMed] [Google Scholar]
- Li R. 1999. Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc. Natl. Acad. Sci. USA. 96:4989–4994 10.1073/pnas.96.9.4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F.C., Winey M. 1998. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell. 9:29–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H., Priest C., Lechner J., Pereira G., Schiebel E. 2007. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J. Cell Biol. 179:423–436 10.1083/jcb.200705197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molk J.N., Schuyler S.C., Liu J.Y., Evans J.G., Salmon E.D., Pellman D., Bloom K. 2004. The differential roles of budding yeast Tem1p, Cdc15p, and Bub2p protein dynamics in mitotic exit. Mol. Biol. Cell. 15:1519–1532 10.1091/mbc.E03-09-0708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje-Casas F., Amon A. 2009. Cell polarity determinants establish asymmetry in MEN signaling. Dev. Cell. 16:132–145 10.1016/j.devcel.2008.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T., Mitsuzawa H., Nakafuku M., Nakamura S., Hattori S., Anraku Y. 1995. Requirement of Saccharomyces cerevisiae Ras for completion of mitosis. Science. 270:1213–1215 10.1126/science.270.5239.1213 [DOI] [PubMed] [Google Scholar]
- Moseley J.B., Goode B.L. 2006. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol. Mol. Biol. Rev. 70:605–645 10.1128/MMBR.00013-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson S.A., Cooper J.A. 2007. A novel pathway that coordinates mitotic exit with spindle position. Mol. Biol. Cell. 18:3440–3450 10.1091/mbc.E07-03-0242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.O., Bi E. 2007. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 71:48–96 10.1128/MMBR.00028-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Mosteller R.D., Broek D. 1994. Amino acid residues in the CDC25 guanine nucleotide exchange factor critical for interaction with Ras. Mol. Cell. Biol. 14:8117–8122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. 2001. The role of the yeast spindle pole body and the mammalian centrosome in regulating late mitotic events. Curr. Opin. Cell Biol. 13:762–769 10.1016/S0955-0674(00)00281-7 [DOI] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. 2005. Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol. Cell. 19:209–221 10.1016/j.molcel.2005.05.030 [DOI] [PubMed] [Google Scholar]
- Pereira G., Höfken T., Grindlay J., Manson C., Schiebel E. 2000. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell. 6:1–10 10.1016/S1097-2765(00)00002-2 [DOI] [PubMed] [Google Scholar]
- Pereira G., Tanaka T.U., Nasmyth K., Schiebel E. 2001. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 20:6359–6370 10.1093/emboj/20.22.6359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips J., Herskowitz I. 1998. Identification of Kel1p, a kelch domain-containing protein involved in cell fusion and morphology in Saccharomyces cerevisiae. J. Cell Biol. 143:375–389 10.1083/jcb.143.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Venturetti M., Chiroli E., Fraschini R. 2006. The spindle position checkpoint in budding yeast: the motherly care of MEN. Cell Div. 1:2 10.1186/1747-1028-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J.R., Adams A.E., Drubin D.G., Haarer B.K. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565–602 10.1016/0076-6879(91)94043-C [DOI] [PubMed] [Google Scholar]
- Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. 2004. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 20:559–591 10.1146/annurev.cellbio.20.010403.103108 [DOI] [PubMed] [Google Scholar]
- Quilliam L.A., Rebhun J.F., Castro A.F. 2002. A growing family of guanine nucleotide exchange factors is responsible for activation of Ras-family GTPases. Prog. Nucleic Acid Res. Mol. Biol. 71:391–444 10.1016/S0079-6603(02)71047-7 [DOI] [PubMed] [Google Scholar]
- Richardson H.E., Wittenberg C., Cross F., Reed S.I. 1989. An essential G1 function for cyclin-like proteins in yeast. Cell. 59:1127–1133 10.1016/0092-8674(89)90768-X [DOI] [PubMed] [Google Scholar]
- Ro H.S., Song S., Lee K.S. 2002. Bfa1 can regulate Tem1 function independently of Bub2 in the mitotic exit network of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 99:5436–5441 10.1073/pnas.062059999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshan A., Amon A. 2005. Ras and the Rho effector Cla4 collaborate to target and anchor Lte1 at the bud cortex. Cell Cycle. 4:940–946 [DOI] [PubMed] [Google Scholar]
- Seshan A., Bardin A.J., Amon A. 2002. Control of Lte1 localization by cell polarity determinants and Cdc14. Curr. Biol. 12:2098–2110 10.1016/S0960-9822(02)01388-X [DOI] [PubMed] [Google Scholar]
- Shirayama M., Matsui Y., Tanaka K., Toh-e A. 1994a. Isolation of a CDC25 family gene, MSI2/LTE1, as a multicopy suppressor of ira1. Yeast. 10:451–461 10.1002/yea.320100404 [DOI] [PubMed] [Google Scholar]
- Shirayama M., Matsui Y., Toh-E A. 1994b. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol. Cell. Biol. 14:7476–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38:203–232 10.1146/annurev.genet.38.072902.093051 [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Visintin R., Amon A. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 108:207–220 10.1016/S0092-8674(02)00618-9 [DOI] [PubMed] [Google Scholar]
- Yamashita Y.M., Fuller M.T. 2008. Asymmetric centrosome behavior and the mechanisms of stem cell division. J. Cell Biol. 180:261–266 10.1083/jcb.200707083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y.M., Mahowald A.P., Perlin J.R., Fuller M.T. 2007. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 315:518–521 10.1126/science.1134910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Skibbens R.V., Cheng J.W., Salmon E.D., Bloom K. 1995. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130:687–700 10.1083/jcb.130.3.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Ichihashi R., Toh-e A. 2003. Ras recruits mitotic exit regulator Lte1 to the bud cortex in budding yeast. J. Cell Biol. 161:889–897 10.1083/jcb.200301128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Chang A.Y., Toh-E A., Arvan P. 2007. A role for Lte1p (a low temperature essential protein involved in mitosis) in proprotein processing in the yeast secretory pathway. J. Biol. Chem. 282:1670–1678 10.1074/jbc.M610500200 [DOI] [PMC free article] [PubMed] [Google Scholar]