FRET analysis of cell lines expressing fluorescently tagged histones on separate nucleosomes demonstrates that variations in chromosome compaction occur during mitosis.
Abstract
We present a quantitative Förster resonance energy transfer (FRET)–based assay using multiphoton fluorescence lifetime imaging microscopy (FLIM) to measure chromatin compaction at the scale of nucleosomal arrays in live cells. The assay uses a human cell line coexpressing histone H2B tagged to either enhanced green fluorescent protein (FP) or mCherry FPs (HeLaH2B-2FP). FRET occurs between FP-tagged histones on separate nucleosomes and is increased when chromatin compacts. Interphase cells consistently show three populations of chromatin with low, medium, or high FRET efficiency, reflecting spatially distinct regions with different levels of chromatin compaction. Treatment with inhibitors that either increase chromatin compaction (i.e., depletion of adenosine triphosphate) or decrease chromosome compaction (trichostatin A) results in a parallel increase or decrease in the FLIM–FRET signal. In mitosis, the assay showed variation in compaction level, as reflected by different FRET efficiency populations, throughout the length of all chromosomes, increasing to a maximum in late anaphase. These data are consistent with extensive higher order folding of chromatin fibers taking place during anaphase.
Introduction
Chromatin is the physiological carrier of genetic and epigenetic information in eukaryotes. The smallest unit of chromatin is the nucleosome, corresponding to a histone octamer complex, with DNA wound around the surface (Luger, 2003). A chromatin fiber consists of arrays of regularly spaced nucleosomes (nucleosomal arrays) bound to linker histones and/or other nucleosome-binding proteins. Chromatin fibers can condense into multiple higher order secondary and tertiary chromatin structures (Luger and Hansen, 2005). Chromatin structure is highly dynamic and sensitive to environmental conditions and imposes profound and ubiquitous effects on DNA-related metabolic processes, including transcription, recombination, DNA repair, replication, and so forth.
Chromatin has historically been classified in two general structural states, depending on how intensely they are stained with DNA dyes (i.e., hetero- and euchromatin, which is related to the level of gene expression; Wolffe and Kurumizaka, 1998). The more highly condensed heterochromatin is generally also replicated later than euchromatin during S phase. Interestingly, hetero- and euchromatin may be differentially and dynamically established by a process that involves interplay between competing repressor complexes and activators of transcription (Elgin, 1996).
Heterochromatin was first defined as the fraction of chromatin that remains condensed after mitosis and further classified as (a) constitutive heterochromatin, which contains centromeres and telomeres and is essential for chromosome function in mitosis and nuclear architecture in interphase, and (b) facultative heterochromatin, which is important for the global and local regulation of gene expression, for instance during differentiation and dosage compensation. Euchromatin has been described as low density, relatively decompacted chromatin, which includes mostly active regions rich in genes and regulatory sequences (Grewal and Elgin, 2002). A recent study indicated that, instead of two discrete chromatin types, a spectrum of intermediate states probably exists in interphase nuclei (Huisinga et al., 2006). Furthermore, the maintenance of higher order chromatin structure and its function is closely related to reversible, posttranslational histone modifications. This is exemplified by the presence of under-acetylated forms of histone H4 as a stable component of heterochromatin regions, which is crucial for gene silencing in organisms throughout evolution (Grunstein, 1998; Turner, 1998).
The complexity of its composition and multiscale nature of chromatin structure represent a formidable challenge for structural biologists (Belmont et al., 1999; Luger and Hansen, 2005). Much of the previous analysis of higher order packaging of DNA into chromatin has involved in vitro biochemical studies, either using chromatin extracted from cells or chromatin reconstituted in vitro from DNA arrays and purified histones (Rhodes and Laskey, 1989; Huynh et al., 2005). This has shown that chromatin can reversibly fold into a 30-nm fiber, dependent on ionic conditions. It remains to be established how the 30-nm fiber relates to differentially compacted regions of chromatin detected throughout the nuclei of living cells (Gilbert et al., 2004; Tremethick, 2007).
In comparison with the high resolution crystal structure available for the nucleosome and the detailed structural information on short regions of chromatin studied in vitro (Luger et al., 1997), analysis of higher order chromosome structures within intact living cells suffers from limitations in the resolution of noninvasive imaging methods that can be used, mostly involving light microscopy. Nonetheless, quantitative multidimensional studies of mitotic chromosome organization in live cells have been achieved. For example, chromatin was shown to be more compacted in metaphase than in prophase or telophase in live Drosophila melanogaster embryos studied in four dimensions by microinjecting fluorescent histones (Swedlow et al., 1993). The emergence of new tools for fluorescence microscopy such as GFP tagging (Lippincott-Schwartz and Patterson, 2003; Shaner et al., 2005) has been instrumental in advancing our understanding of chromosome dynamics over the past decade. In particular, many studies have used core histones fused with either GFP or other FPs (fluorescent proteins) to uniformly label chromatin and allow its detection in live cells by fluorescence microscopy (Kanda et al., 1998; Perche et al., 2000; Kimura and Cook, 2001; Kruhlak et al., 2006; Mora-Bermúdez et al., 2007). The fact that stable cell lines can be established in which core FP-tagged histones are incorporated into nucleosomes has facilitated a wide range of in vivo analyses of chromosome structure and chromatin localization and dynamics within the context of living cells (Kanda et al., 1998; Misteli et al., 2000; Perche et al., 2000; Kimura and Cook, 2001; Keppler et al., 2006).
For example, geometric dimensions of chromosomes have been studied by light microscopy, and volumetric measurements of chromatin in cells progressing through all stages of mitosis have been performed (Gerlich et al., 2006; Mora-Bermúdez and Ellenberg, 2007; Mora-Bermúdez et al., 2007). However, volumetric assays using light microscopy measure compaction at a resolution of ∼800 nm, which is suitable for studying events at the scale of large chromatin fibers and entire chromosomes (Vagnarelli et al., 2006; Mora-Bermúdez and Ellenberg, 2007). To follow the dynamics of mitotic chromosome structure and subchromosomal regions, chromosomes containing arrays of bacterial lac operators have been engineered, which can be visualized by the expression of a GFP-tagged lac repressor, allowing detection in live cells of ∼100-nm chromatin fibers (Robinett et al., 1996; Strukov and Belmont, 2009). An alternative approach to the visualization of chromosomal subregions is based on photolabeling. This led to the discovery of a chromatin compaction step by axial shortening during anaphase (Mora-Bermúdez et al., 2007). However, the resolution limit of several hundreds of nanometers imposed by using the aforementioned microscopy approaches is still lower than the dimensions of chromatin fiber intermediates (11–250 nm).
Förster resonance energy transfer (FRET) can be used to detect direct protein–protein interactions in live cells. While transferring energy from an excited donor to an acceptor (Lakowicz, 2006), FRET decreases the donor fluorescence and increases the acceptor fluorescence. Because FRET is highly distance dependent, FRET requires the two fluorophores to be within ∼1–10 nm, i.e., the distance typically found for directly interacting proteins (Förster, 1949). FRET is also dependent on the geometry of the fluorophores in a FRET pair. The most quantitative readout of a FRET interaction is provided by measuring the fluorescence lifetime of the donor fluorophore, defined as the mean time between fluorophore excitation and photon emission, which is called fluorescence lifetime imaging microscopy (FLIM; Bastiaens and Squire, 1999; Wallrabe and Periasamy, 2005; Treanor et al., 2006; Llères et al., 2007).
In this study, we have used FLIM to measure FRET between histone molecules tagged with either GFP (donor) or mCherry (acceptor) and thus provide an in vivo assay whereby the level of FLIM–FRET is proportional to the level of chromatin compaction. Using this approach provides structural information on chromatin organization in vivo on the scale of nucleosomal arrays. This assay has been used to study differential regions of chromatin compaction in live cells for both interphase and mitotic chromosomes.
Results
A FLIM–FRET assay for chromatin condensation
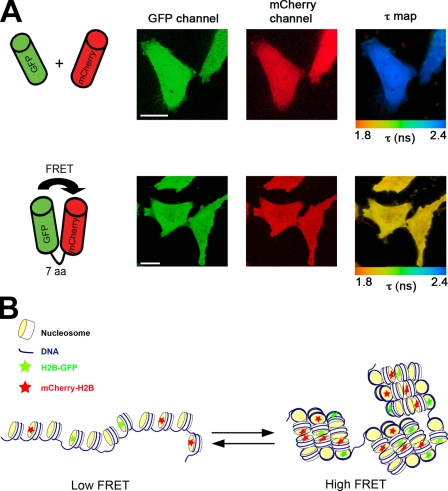
The specificity and sensitivity of using the FLIM technique to detect FRET in vivo was tested using HeLa cells coexpressing GFP and mCherry (Fig. 1 A), either as two separate proteins (top) or fused via a 7-aa linker (bottom). The fluorescence signals in both the GFP and mCherry channels were identical, and therefore, it was not possible to distinguish cells expressing the GFP–mCherry fusion protein from cells expressing GFP and mCherry as two free proteins using standard fluorescence microscopy. However, a difference was evident between these cells in the fluorescence lifetime values recorded for the GFP donor fluorophore, where the reduced mean lifetime for the cells expressing the fusion protein (Fig. 1 A, bottom right) resulted from FRET because of the proximity of the GFP and mCherry fluorophores held together by the linker peptide. Thus, FLIM–FRET analysis can distinguish interactions that are not detectable by imaging the separate red and green fluorescence channels alone.
Figure 1.
In vivo FLIM–FRET assay for chromatin compaction. (A) HeLa cells coexpressing either free EGFP and mCherry (top) or EGFP-fused to mCherry through a 7-aa linker (bottom) were imaged by multiphoton laser-scanning microscopy (MPLSM). The spatial distribution of the mean fluorescence lifetime of the EGFP donor (τ map) is displayed using a continuous pseudocolor scale ranging from 1.8 to 2.4 ns. (B) Representation of the FLIM–FRET chromatin compaction assay. Bars, 10 µm.
We have adapted the FLIM–FRET assay to measure chromatin compaction in live cells, (Fig. 1 B). A double stable HeLa cell line (HeLaH2B-EGFP/mCherry-H2B), hereafter termed HeLaH2B-2FP, was generated using the previously characterized stable cell line expressing histone H2B fused at its carboxy terminus to EGFP (Kanda et al., 1998; Kimura and Cook, 2001). The HeLaH2B-2FP cells coexpress histone H2B fused at its amino terminus to mCherry (see Materials and methods). In this cell line, not all nucleosomes contain an FP-tagged histone, and most will only have a single FP-tagged histone (see next section; Fig. 2 C). In the subset of nucleosomes incorporating both GFP- and mCherry-tagged H2B, molecular modeling based on the published nucleosome structure (Luger et al., 1997) showed that the separation of GFP and mCherry was likely too large to produce FRET within a single nucleosome (Fig. S1 A). Thus, FRET occurs predominantly from interactions between GFP- and mCherry-tagged histones in separate nucleosomes as a result of 3D proximity when chromatin is compacted.
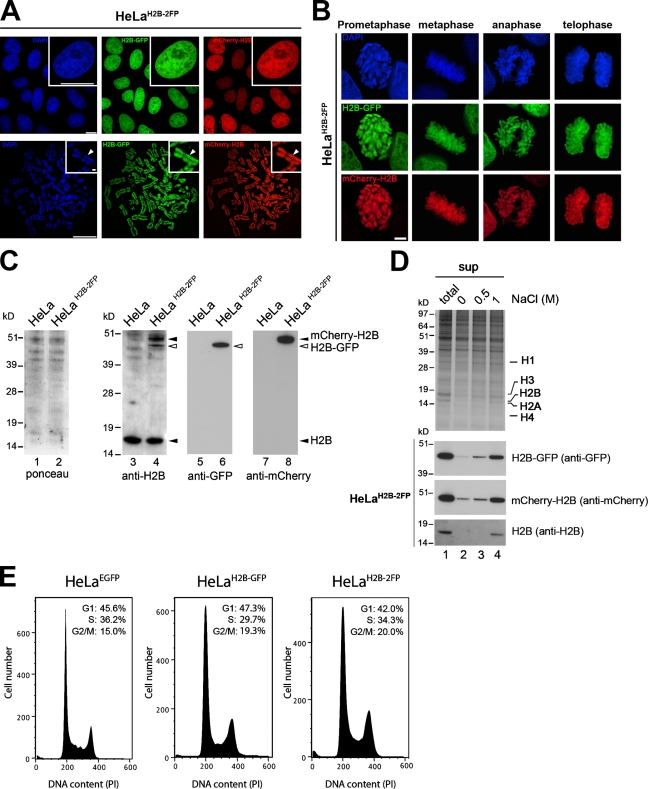
Figure 2.
A stable HeLaH2B-2FP cell line coexpressing H2B-EGFP and mCherry-H2B histones. (A) Fluorescent-tagged H2B histones in HeLaH2B-2FP cells imaged by wide-field fluorescence microscopy in interphase (top) and metaphase spreads (bottom). Insets show high magnification views. Arrowheads indicate concentrated heterochromatin in the chromosome, which was brightly stained with DAPI. (B) High resolution imaging of HeLaH2B-2FP cells at different stages of mitosis. (C) Total cell lysates from HeLa and HeLaH2B-2FP cells analyzed by PAGE and Western blotting with antibodies against H2B (lanes 3 and 4), GFP (lanes 5 and 6), or mCherry (lanes 7 and 8). (D) Salt extraction of histones from HeLaH2B-2FP cells. Proteins either from whole cells (total; lane 1) or the supernatant fractions from 0, 0.5, and 1 M NaCl salt extractions of chromatin (sup; lanes 2–4) were separated on SDS-PAGE gels and either stained with Coomassie blue (top) or blotted and probed with the indicated antibodies. (E) FACS analyses showing cell cycle distribution of HeLaEGFP, HeLaH2B-GFP, and HeLaH2B-2FP stable cell lines. Bars, 10 µm.
Fluorescence imaging of HeLaH2B-2FP cells in both interphase and mitosis showed that both H2B-EGFP and mCherry-H2B colocalized throughout all chromosomes with the same staining pattern as DAPI (Fig. 2, A and B). Immunoblotting showed that endogenous histone H2B, H2B-GFP, and mCherry-H2B were all expressed and migrated at the expected sizes (Fig. 2 C, lanes 3, 4, 6, and 8, arrowheads). H2B-EGFP was expressed at a similar level to that previously reported (Kimura and Cook, 2001), whereas the mCherry-H2B was expressed at a comparable but slightly higher level to H2B-EGFP (Fig. 2 C, lane 4). Furthermore, comparison with HeLa cell lines that did not express any FP-tagged proteins showed that there was a similar level of H2B expression and little or no increase in the pool of endogenous H2B in the HeLaH2B-2FP cells (Fig. 2 C, compare lane 3 with lane 4; and not depicted).
To test the stability of tagged histone incorporation into chromatin, HeLaH2B-2FP cells were lysed in an isotonic buffer supplemented with varying concentrations of NaCl from 0–1 M (Fig. 2 D, top; and Fig. S1 B). Both tagged H2B proteins showed a similar profile of salt extraction to the endogenous, untagged H2B, which is consistent with previous studies reporting that FP tags do not prevent stable incorporation of histones into nucleosomes (Fig. 2 D; Kanda et al., 1998; Misteli et al., 2000; Perche et al., 2000; Kimura and Cook, 2001; Keppler et al., 2006). Comparison of the global nucleosome-banding pattern after digestion with micrococcal nuclease (MNase) showed little or no difference between HeLa, HeLaH2B-GFP, and HeLaH2B-2FP cells (Fig. S1, C and D). Finally, a comparison of the growth profiles of HeLaH2B-2FP with both HeLaH2B-GFP and HeLaEGFP cells showed that their doubling rates were equivalent, and FACS analysis confirmed that the relative populations in G1, S, and G2/M stages were similar (Fig. 2 E).
Together, these data confirm that the HeLaH2B-2FP cells express full-length H2B-EGFP and mCherry-H2B histones, both at a level lower than the endogenous H2B. All three forms of H2B show similar biochemical properties and are uniformly and stably incorporated throughout the full length of all chromosomes. This HeLaH2B-2FP cell line has been propagated in culture for >12 mo and shows similar growth and cell cycle properties to other HeLa cell lines that do not express FP-tagged proteins.
FLIM–FRET measurements spatially discriminate differential chromatin compaction levels in vivo
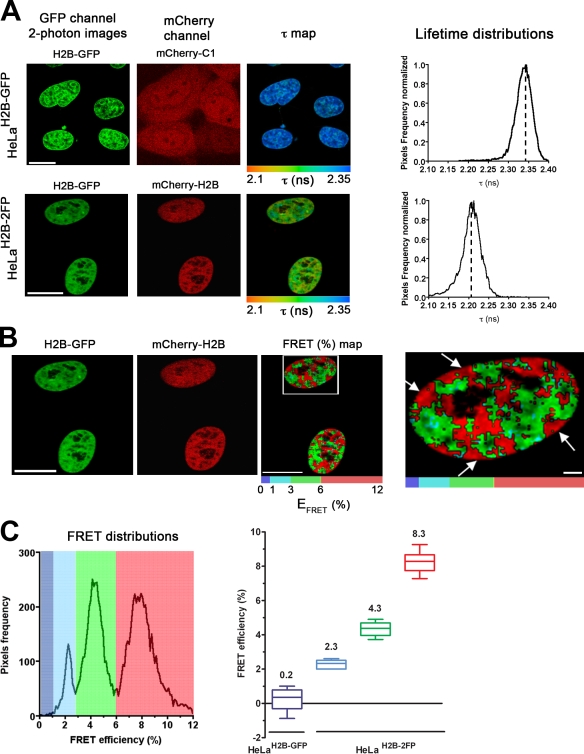
Next, the HeLaH2B-2FP cells were analyzed by FLIM–FRET microscopy to measure chromatin compaction levels in interphase nuclei (Fig. 3 A). As a control, HeLaH2B-GFP cells coexpressing free mCherry were imaged in parallel (Fig. 3 A, top). A decrease in the mean fluorescence lifetime (τ) of H2B-EGFP was detected in HeLaH2B-2FP cells (Fig. 3 A, bottom) but not in the control cells expressing free mCherry (Fig. 3 A, compare the top with the bottom). The decrease of the H2B-EGFP donor lifetime specifically in the presence of the mCherry-H2B fusion protein but not in the presence of free mCherry is also shown by the lifetime distribution curves (Fig. 3 A, right). These data demonstrate specific FRET interactions between the two tagged forms of histone H2B.
Figure 3.
FLIM–FRET measurements spatially discriminate differential chromatin compaction levels in vivo. (A) HeLaH2B-GFP stable cell line transiently transfected with mCherry-C1 empty vector (top) and HeLaH2B-2FP stable cell line (bottom) were imaged using MPLSM. In both cases, the mean fluorescence lifetime τ (nanoseconds) is detected, and its spatial distribution at each pixel of the ROI (τ map) is shown as described in Materials and methods. Fluorescence lifetimes are presented in a continuous pseudocolor scale representing time values ranging from 2.1 to 2.35 ns. The mean lifetime distribution curves of the donor (H2B-GFP) are shown on the right. Dashed lines mark the position of the peaks of the lifetime distribution curves. (B) The FRET efficiency (percentage) and its spatial distribution (FRET (%) map) are depicted in the ROI using discrete colors (see Materials and methods). The ROI comprised the interphase nuclei; the right panel shows a higher magnification of a nucleus (boxed area) revealing high FRET areas at the nuclear periphery (arrows). (C) FRET distributions graph showing three distinct populations distinguished using discrete pseudocolors: blue (low), FRET efficiency up to 3%; green (medium), between 3–6%; and red (high), 6–12%. The statistical analysis of at least 10 cells per condition is presented as box and whisker plots on the right. The three distinct FRET efficiency populations detected in interphase HeLaH2B-2FP cells are represented by colored boxes (i.e., low [blue], medium [green], and high [red] FRET populations, respectively). The mean FRET efficiency value is indicated on top of each box. Bars, 10 µm.
The FRET efficiency was calculated for each pixel in the images (see Materials and methods), and the spatial FRET distribution was depicted using discrete pseudocolors (Fig. 3 B, FRET (%) map). A heterogeneous FRET efficiency map was apparent throughout nuclei, and this is also shown by extracting the FRET efficiency distribution curves related to the EFRET map (Fig. 3 C, left). The FRET efficiency distribution showed three main populations (Fig. 3 C, right). The smaller population had a low FRET efficiency (∼2.3%), and the two larger populations had FRET efficiencies of ∼4.3% (medium) and ∼8.3% (high), respectively. The spatial separation of these three FRET populations is represented using discrete pseudocolors (blue [low], FRET efficiency up to 3%; green [medium], between 3 and 6%; and red [high], 6–12%; Fig. 3, B and C). We interpret these distinct FRET efficiency populations as reflecting chromosome regions with different degrees of compaction. This is consistent with the spatial distribution of the high FRET efficiency regions, which are predicted to include regions of highest chromatin condensation such as heterochromatin clusters around the nucleoli and at the nuclear periphery (Fig. 3 B, right, arrows).
ATP levels reversibly alter the chromatin compaction balance
ATP depletion induces a major change in chromatin organization and the formation of electron-dense barriers of highly condensed chromatin inside the nucleoplasm (Platani et al., 2002; Cushman et al., 2004; Shav-Tal et al., 2004). To test how ATP depletion affects chromatin at the nucleosome array level using the FLIM–FRET assay, we treated HeLaH2B-2FP cells with 2-deoxyglucose (2-DG) and Na azide, which causes energy depletion, and examined any resultant changes in EGFP lifetime and FRET efficiency over time in living cells.
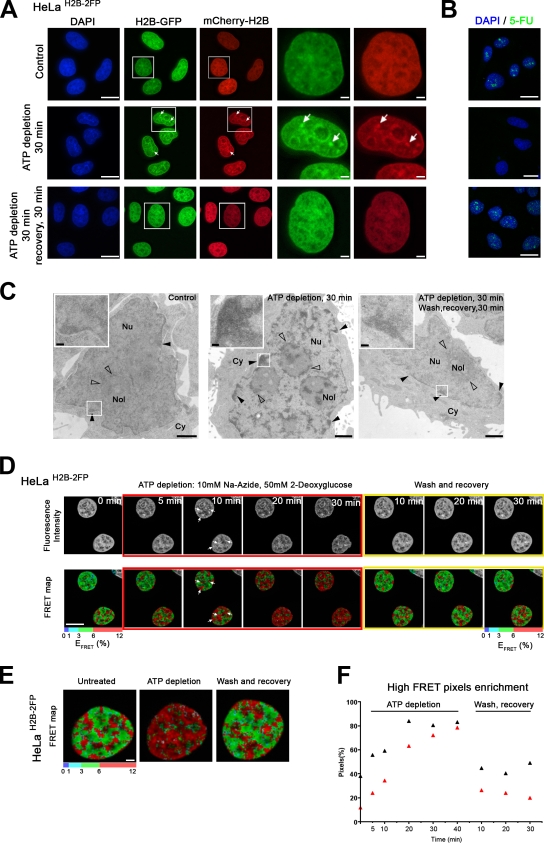
Consistent with previous studies (Platani et al., 2002; Cushman et al., 2004; Shav-Tal et al., 2004), within 30 min of adding 2-DG and Na azide, extensive chromatin compaction was observed. HeLaH2B-2FP cells showed dense chromatin regions throughout the nucleus followed by a reduction in nuclear size (Fig. 4 A, arrows; and Videos 1 and 2). These effects were reversible, and within 30 min of removing the inhibitors, the fluorescence labeling was restored to the same pattern as in untreated cells (Fig. 4 A, bottom; and Videos 1 and 2). Pulse labeling of nascent RNA with 5-fluorouracil (5-FU) confirmed that the ATP depletion treatment inhibited transcription (Fig. 4 B).
Figure 4.
ATP levels reversibly alter the chromatin compaction balance. (A) Effects of ATP depletion on chromatin organization. HeLaH2B-2FP cells were imaged, and the distribution of both H2B-EGFP (green)– and mCherry-H2B (red)–tagged histones before and after 30 min ATP depletion or after subsequent washing away of inhibitors after the 30-min depletion followed by a 30-min recovery was addressed. Arrows indicate dense chromatin regions after ATP depletion. The boxed areas delineate particular ROIs in the field of cells, which are magnified in the panels to the right. (B) Transcription assay using 5-FU incorporation. Images show micrographs of HeLa cells pulse labeled for 30 min with 5-FU and immunolabeled with a mouse anti-BrdU antibody to visualize nascent transcripts either before (top) or after 30 min of ATP depletion (middle) or after subsequent washing and recovery for 30 min (bottom). (C) TEM micrographs of HeLa cells before and after ATP depletion. Untreated control cells (left), HeLa cells ATP depleted for 30 min (middle), and HeLa cells after 30-min ATP depletion followed by washing and recovery for 30 min (right) are shown. Closed arrowheads indicate heterochromatin at the nuclear periphery, and open arrowheads indicate heterochromatin around the nucleolus (Nol). Insets show higher magnification of ROIs in the field of cells (boxed areas). Cy, cytoplasm; Nu, nucleus. (D) Time-lapse FLIM–FRET measurements on HeLaH2B-2FP cells during ATP depletion and recovery. The red boundaries surround micrographs showing HeLaH2B-2FP cells at 5–30 min after ATP depletion, and the yellow boundaries surround micrographs showing HeLaH2B-2FP cells during 10–30 min of recovery. The top panels show MPLSM images of H2B-EGFP fluorescence, and the bottom panels show the calculated FRET efficiency (percentage) over time in the selected ROI. In this case, the ROI includes the two interphase nuclei. Arrows indicate high FRET levels. (E) High magnification views of micrographs showing the spatial distribution of FRET efficiency. (F) Graph showing the time course of enrichment in high FRET pixels (red pixels) of two individual cells during ATP depletion and recovery (see Materials and methods). Bars: (A, B, D, and E) 10 µm; (C) 1 µm; (C, insets) 0.1 µm.
The reversible large-scale condensation of chromatin upon ATP depletion was also observed by transmission EM (TEM; Fig. 4 C, insets). The diffuse distribution of chromatin in untreated cells, together with heterochromatin at the nuclear periphery (Fig. 4 C, left, closed arrowheads) and surrounding nucleoli (Fig. 4 C, left panel, open arrowheads), was replaced with extensive dense regions in the energy-depleted cells, including large electron-dense chromatin barriers throughout the nucleus (Fig. 4 C, middle, arrowheads). After 30 min of recovery, the TEM images showed a similar chromatin distribution to the untreated cells (Fig. 4 C, right). All together, these observations demonstrated that ATP depletion causes a drastic but reversible increase in global chromosome compaction in the HeLaH2B-2FP cells.
Next, we followed the chromatin compaction status upon ATP depletion by time-lapse FLIM–FRET measurements (Fig. 4 D, bottom). The high FRET efficiency population (red pixels) increased after only 5–10 min of depletion and continued increasing throughout the 30-min treatment, particularly surrounding nucleoli and at the nuclear periphery (Fig. 4 D, arrows). This is shown by the increase of red pixels in the FRET map panels during the time course (Fig. 4, D and E). Importantly, the effect of ATP depletion on the FRET signal in HeLaH2B-2FP cells was also reversible, and within 10 min of washing cells to remove the Na azide and 2-DG, the FRET signal had returned to a similar level and pattern as seen in the untreated control cells (Fig. 4, D and E). Quantification of the number of pixels in which high FRET values were recorded over time demonstrated a rapid increase in the fraction of chromatin in the high FRET efficiency population (red pixels) during the first 20 min after ATP depletion and then a slower increase from ∼20–40 min to finally affect >85% of the total chromatin (Fig. 4 F). This major effect on chromatin was almost completely reversed within 10 min of washing the cells to remove 2-DG and Na azide (Fig. 4 F).
As a control, 2-DG and Na azide were also added to HeLaH2B-GFP cells, and the fluorescence lifetime was measured over time under the same conditions (Fig. S2 A). This showed only a minor reduction in the EGFP lifetime, which is not comparable with that seen in the HeLaH2B-2FP cell line, although the chromatin was also highly condensed in these cells after ATP depletion (Fig. S2 A, top). The minor change in EGFP lifetime after ATP depletion in the control cells likely results from an alteration in the pH and/or ion concentration, which is known to influence fluorescence lifetime (van Munster and Gadella, 2005), and is minor in comparison with the large, reversible change in FLIM–FRET seen after ATP depletion in the HeLaH2B-2FP cell line.
Immunodetection of histone H3 lysine trimethylation (H3K9triMe) showed minimal changes in the level of nuclear labeling during ATP depletion or recovery (Fig. S2 B). Instead, there was a partial redistribution of the H3K9triMe labeling, which clustered at the periphery of nucleoli and in nucleoplasmic foci. This was also reversed upon washing away Na azide and 2-DG, which is consistent with the large-scale redistribution of chromatin evident by TEM analysis (compare Fig. 4 C with Fig. S2 B). We conclude that the reversible changes in chromosome compaction caused by ATP depletion are not primarily mediated by alterations in H3K9triMe.
Finally, by performing FRAP experiments on the HeLaH2B-2FP cells before, during, and after ATP depletion (Fig. S3), we excluded that the FRET variations measured by FLIM could be explained primarily by changes in the rate of histone exchange under the different conditions. ATP depletion decreased the mobile fraction of EGFP-H2B, which is consistent with a degree of transcription-dependent exchange (Kimura and Cook, 2001). However, in contrast with the FLIM–FRET measurements, no variation in the mobile histone fraction was observed between ATP depletion and washing/recovery (Fig. S3) and thus does not correlate with the reversible changes in both chromatin compaction and FLIM–FRET values observed.
FLIM–FRET analysis of trichostatin A (TSA)–induced changes in higher order chromatin organization
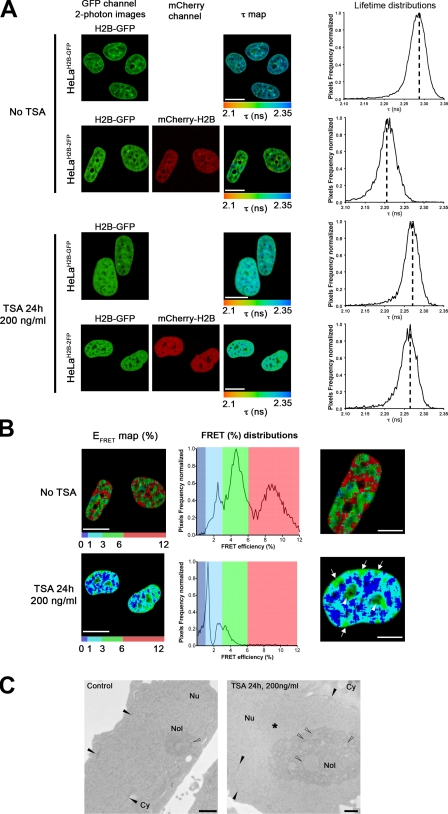
The FLIM–FRET assay was used to assess large-scale chromatin decondensation, which is provoked when histone acetylation levels are affected by the histone deacetylase inhibitor TSA (Tóth et al., 2004). As expected, after 24 h of treatment with TSA, interphase chromatin in HeLaH2B-2FP cells was less condensed, as shown by the more diffuse H2B-EGFP and mCherry-H2B labeling and DAPI staining (Figs. 5 A and S4 B). Next, we measured FLIM–FRET in both HeLaH2B-2FP and HeLaH2B-GFP cells, either in the absence or presence of TSA. First, without any TSA treatment, there was a decrease in the mean fluorescence lifetime of H2B-EGFP specifically in HeLaH2B-2FP cells in comparison with the mean fluorescence lifetime of H2B-EGFP in the control cells expressing H2B-EGFP alone (Fig. 5 A, no TSA panels). After TSA treatment, little or no decrease in the mean lifetime (τ = ∼2.27 ns) of H2B-EGFP is detected in HeLaH2B-2FP cells in comparison with the control cells (Fig. 5 A, TSA panels). This shows that the FLIM assay detects a major reduction in chromatin compaction after TSA treatment.
Figure 5.
FLIM–FRET analysis of TSA-induced changes in higher order chromatin organization. (A) The spatial distribution of the mean fluorescence lifetime (τ map) in the ROI is shown for both HeLaH2B-GFP and HeLaH2B-2FP stable cell lines before and after 24-h treatment with 200 ng/ml TSA. In each panel, the ROI comprised the interphase nuclei. Dashed lines mark the position of the peaks of the lifetime distribution curves. (B) EFRET map (percentage) of an ROI comprising two HeLaH2B-2FP nuclei before and after TSA treatment. The FRET distribution graphs are depicted using discrete colors, with the highest FRET population in red, intermediate population in green, and the lowest population in blue. Highest FRET values after TSA treatment are shown at nucleoli (arrowheads) and at the nuclear periphery (arrows). (C) TEM micrographs of HeLa cells before and after TSA treatment. Control untreated HeLa cells (left) and HeLa cells treated with 200 ng/ml TSA for 24 h (right) are shown. The heterochromatin at the nuclear periphery and associated to nucleoli are indicated by closed and open arrowheads, respectively. The asterisk represents bulk decondensed chromatin. Cy, cytoplasm; Nol, nucleolus; Nu, nucleus. Bars: (A and B) 10 µm; (C) 1 µm.
The effect of TSA treatment on the donor fluorescence lifetime is quantitatively represented by the corresponding lifetime distribution curves (Fig. 5 A, right), which also show that the shift of the mean lifetime values was reduced or abolished after TSA treatment. The FRET efficiency was calculated for both TSA-treated and nontreated cells, and the spatial distribution of the FRET efficiency signal is depicted using discrete pseudocolor to map regions of differential chromatin density (Fig. 5 B). As previously described (Fig. 3), control cells display three main FRET efficiency populations (i.e., low [blue], medium [green], and high [red]), reflecting the presence of both highly compacted heterochromatin as well as less condensed euchromatin throughout interphase nuclei. After inhibition of histone deacetylation with TSA, there is a major reduction in the level of the highest FRET population (red population), although a residual intermediate population remains (green population), predominantly located at nucleoli and at the nuclear periphery (Fig. 5 B, higher magnification, arrowheads and arrows show nucleoli and nuclear periphery, respectively). In contrast, there is a widespread increase throughout most of the nucleoplasm in the level of the low FRET population (Fig. 5 B, blue population). This likely includes chromatin in a predominantly decompacted state that is more accessible to the transcription apparatus.
A statistical analysis of the FRET efficiency distribution for both TSA-treated and control cells indicates that nuclear chromatin is shifted to a lower compaction state by TSA but with a subpopulation of compacted chromatin remaining that is resistant, at least in part, to TSA (Fig. 5 B, FRET (%) distributions; and Fig. S4 A). This is supported by TEM analyses, in which electron-dense regions of condensed chromatin were detected at nucleoli and the nuclear periphery after TSA treatment (Fig. 5 C, open and closed arrowheads, respectively). We also performed immunostaining experiments using antibodies specific for either centromere protein C (CENP-C) or HP1-α (Fig. S4, B and C, respectively), which are markers of pericentric heterochromatin, both before and after TSA treatment (Taddei et al., 2001). After TSA treatment, both antibodies showed a similar relocalization to the perinuclear and nucleolar regions that were detected by FLIM–FRET as retaining condensed chromatin after TSA treatment. Furthermore, for HP1-α, we also noticed nucleolar localization after TSA treatment, as observed by FLIM–FRET.
FLIM–FRET analysis of chromosome compaction during mitosis
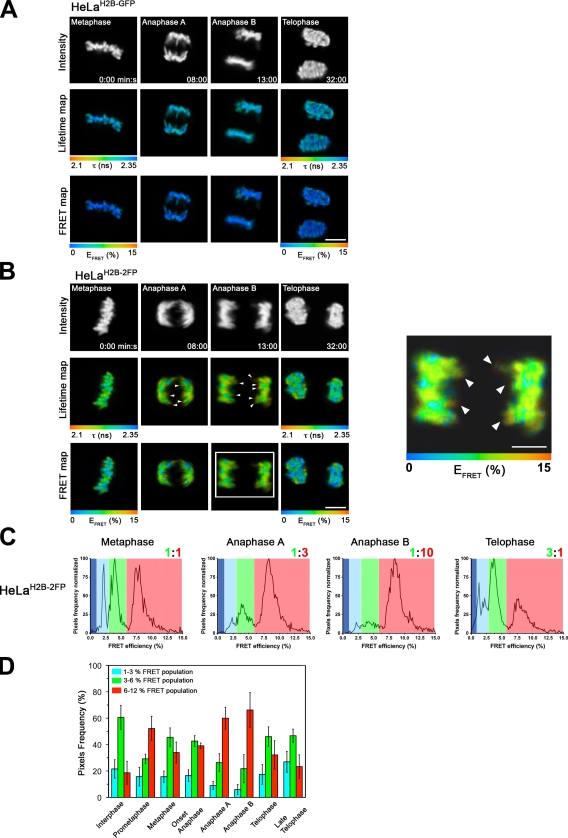
The FLIM–FRET assay was used to assess chromatin compaction within chromosomes of live cells at different stages of mitosis (Fig. 6). First, we checked whether the fluorescence lifetime value of the donor H2B-EGFP alone was affected during the process of image acquisition throughout mitosis (e.g., through photobleaching) using the HeLaH2B-GFP stable cell line (Fig. 6 A). The H2B-EGFP lifetime remained approximately constant (τ = ∼2.3 ns) in sequential images recorded from the same cells from metaphase until telophase. This excludes that photobleaching or other effects alter the lifetime measurements sufficiently to compromise the FRET levels measured during mitosis in HeLaH2B-2FP cells.
Figure 6.
FLIM–FRET analysis of chromosome compaction during mitosis. (A) Time-lapse FLIM–FRET measurements of HeLaH2B-GFP cells at different stages of mitosis. (top) Fluorescence intensity of the donor H2B-EGFP. The spatial map of the mean lifetime (middle) and the FRET percentage (bottom) are shown at the indicated stages of mitosis. At each stage of mitosis, the FRET percentage is depicted for a selected ROI (e.g., metaphasic equatorial plate in metaphase, the two separate paired chromosomes in early and late anaphase, or the two daughter nuclei at telophase). (B) Time-lapse FLIM–FRET of HeLaH2B-2FP cells during mitosis. (top) Fluorescence intensity of the donor H2B-EGFP. (middle) Fluorescence lifetime map. (bottom) FRET percentage map. At each stage of mitosis, the FRET percentage is depicted for a particular ROI (e.g., metaphasic equatorial plate in metaphase, the two separate paired chromosomes in early and late anaphase, or the two daughter nuclei at telophase). A higher magnification view of the FRET efficiency in anaphase B (boxed area) is shown in the right panel, with arrowheads indicating localized high FRET at the extremities of chromosome arms. (C) Quantification of the levels of FRET percentage efficiency populations during mitosis (see Materials and methods). A discrete color is associated to each distinct FRET population (low, blue; middle, green; high, red). The ratio between the medium and high FRET population is indicated. (D) Colored histogram showing the relative fraction of the three distinct FRET efficiency populations in interphase and at different stages of mitosis. Error bars indicate SD. Bars, 10 µm.
Time-lapse FLIM–FRET measurements were performed on individual live HeLaH2B-2FP cells from prometaphase through to telophase (Fig. 6 B and Fig. S5 B). This showed a stepwise decrease of the mean EGFP fluorescence lifetime value between prometaphase and anaphase B, which is reversed in daughter nuclei at telophase (Fig. 6 B and Fig. S5 B, middle rows). Next, the corresponding percentage of FRET efficiency (FRET map) at each pixel of every image was calculated and pseudocolored (Fig. 6 B and Fig. S5 B, bottom rows) to spatially map the degree of compaction for the entire set of chromosomes. A heterogeneous population of different FRET efficiencies was seen along the metaphasic equatorial plate (Fig. 6 B and Fig. S5 B, FRET maps, blue and green regions), which subsequently becomes more homogeneous as the cell progresses into early and late anaphase. This is supported by FRET efficiency distribution curves (Fig. 6 C), showing the presence of at least three distinct FRET populations (i.e., low [blue], medium [green], and high [red]).
The two higher efficiency FRET populations, corresponding to higher condensed chromatin states, were initially at an ∼1:1 ratio in metaphase (Fig. 6 C, metaphase, ratio of green to red). Then, as cells moved into anaphase and during anaphase progression, both the low (blue) and medium (green) FRET populations gradually decreased (ratio of 1:3 and then 1:10 medium [green]/high [red] FRET populations) in favor of a major high FRET population (red in anaphase B). The high FRET population peaked at ∼9% FRET efficiency in anaphase B, corresponding to the maximal level of condensation of the whole set of chromosomes (Fig. 6, B and C, anaphase B). In contrast, when cells reached telophase and chromosome decompaction took place, the three FRET populations reappeared, with a 3:1 ratio of medium (green)/high (red) FRET and a significant chromatin fraction also in the low (blue) FRET population (Fig. 6 C, telophase). The FRET efficiency distribution curves for the control HeLaH2B-GFP stable cell line showed no significant FRET variations throughout mitosis (Fig. S5 A).
The kinetics of mitotic chromosome compaction were also analyzed by quantifying the frequency of pixels in each image that showed either the high FRET population (Fig. 6 D, red bars) or the medium and low FRET populations (Fig. 6 D, green and blue bars, respectively). Together, these data demonstrate that the entire set of mitotic chromosomes increase in condensation as mitosis progresses to a maximum level in anaphase B. Furthermore, high resolution FLIM–FRET measurements revealed some punctate areas of higher FRET, possibly reflecting a particularly high degree of chromosome compaction, located at the extremities of chromosome arms during sister telomere separation in anaphase B (Fig. 6 B, right, red pixels indicated by arrowheads). These data are consistent with a degree of heterogeneity in compaction levels within individual chromosomes remaining even at the stage of maximum overall condensation.
Discussion
We have presented an in vivo assay that allows chromatin compaction to be measured and spatially resolved in live cells. The assay uses FLIM to provide a quantitative measurement of FRET between nucleosomes in a stable cell line, termed HeLaH2B-2FP. As well as endogenous untagged histone H2B, these cells coexpress H2B tagged with either EGFP or mCherry, which act as a FRET pair. Only a subset of nucleosomes contain both EGFP and mCherry, and molecular modeling indicates that, even in this subset of nucleosomes, steric constraints imposed by the spatial separation of the two histone H2B molecules make it unlikely that FRET can occur (Fig. S1 A). Furthermore, when chromatin is predominantly decondensed after treatment of cells with the histone deacetylase inhibitor TSA, the main FRET signal is lost. Therefore, although nucleosomes contain two copies of histone H2B, it is likely that little FRET occurs within a single nucleosome. Instead, FRET detected increases in all of the experimental situations investigated in which the chromatin compaction and the density of nucleosomes increases, thereby bringing the donor and acceptor EGFP and mCherry on separate nucleosomes into close enough proximity for FRET to occur (Fig. 1 B). The HeLaH2B-2FP cells display similar growth and cell cycle properties to HeLa cells and thus provide a convenient in vivo assay system for studying chromatin compaction and its spatial relationship to genome function.
Analysis of higher order chromatin structure in vitro using either chromatin extracted from cells or chromatin reconstituted in vitro from DNA and purified histones (Huynh et al., 2005) has shown that chromatin can reversibly fold into a 30-nm fiber, dependent on ionic conditions (Rhodes and Laskey, 1989). Because of technical limitations, it has been difficult to resolve and quantify the level of chromatin condensation directly in live cells or to investigate higher order chromatin organization in vivo at the nucleosomal array level. The FLIM–FRET assay offers several advantages in this regard. First, higher order chromatin compaction at the level of nucleosome array coiling is in the ∼1–10-nm resolution range of the FRET reporter. FLIM analysis is noninvasive and provides a quantitative method for measuring FRET efficiency at every pixel in an image, providing a mean value for all of the nucleosomes within a pixel (Wouters and Bastiaens, 2001; Wallrabe and Periasamy, 2005). Therefore, the FLIM–FRET assay not only measures the level of chromatin compaction but also provides spatial information showing how regions of differentially compacted chromatin are distributed. Second, the FLIM–FRET map can be obtained using live cells and spatially related to specific subnuclear regions or bodies such as nucleoli. Third, the FLIM–FRET assay can be repeated on the same cell, allowing a time-lapse evaluation of how the compaction and spatial organization of chromatin changes in response to specific perturbations. For example, imaging of individual cells before and after ATP depletion showed a rapid increase in the FLIM–FRET signal that paralleled the increase in condensed chromatin. Furthermore, the FRET signal in the same live cell returned to the same level and pattern seen before ATP depletion within 10 min of washing away the Na azide and 2-DG. It is striking that energy depletion causes a global compaction of chromatin that is reversible upon restoration of ATP levels. This implies that one or more factors are constantly required during interphase to maintain chromatin in a decondensed state accessible to the gene expression machinery and that ATP is either directly or indirectly required for this activity. This is likely not an indirect consequence of preventing transcription because transcription inhibitors do not cause an equivalent effect (unpublished data). The FLIM–FRET assay will provide a useful tool for addressing the mechanism of ATP-dependent chromatin compaction.
Interphase chromatin can exist in at least three separable FRET efficiency populations with low, medium, or high FRET efficiency values (Fig. 3 C). However each FRET efficiency population has a range of values and may include different structures of chromatin rather than a unique compacted structure. Although the absolute values of FRET efficiency measured for each of these populations can vary between experiments and are affected by several parameters (e.g., instrumental fluctuations or variation in the biological sample; Treanor et al., 2006; Llères et al., 2007), the appearance of three separable FRET efficiency populations within chromatin is consistent and likely to reflect a hierarchy of differential chromatin compaction states or distinct classes of chromatin structures existing in vivo.
There is a spatial correlation between the high FRET population and regions defined as heterochromatin. However, the higher compaction regions defined by the FLIM–FRET assay may not correspond precisely with heterochromatin regions defined using other imaging or labeling techniques, which may be heterogeneous. For example, TSA treatment identified a subpopulation of condensed chromatin resistant to decondensation when histone deacetylases are inhibited. Reversible acetylation of amino-terminal histone tails by specific histone acetylases and deacetylases is involved in the regulation of gene expression (Choi and Howe, 2009). Inhibiting histone deacetylation with TSA resulted in a large-scale decompaction of most nuclear chromatin and a corresponding loss of FRET throughout most of the nucleus (Fig. 5). Nonetheless, specific regions of FRET remained clustered at nucleoli and in clumps at the nuclear periphery, which is coincident with densely stained regions of constitutive heterochromatin seen by immunostaining and TEM. Thus, a subset of regions defined as heterochromatin may differ in higher order structure and/or protein composition, preventing the regulation of their compaction through reversible histone acetylation.
The FLIM–FRET assay was also applied to cells at different stages of mitosis, showing an increase in the compaction for all chromosomes as cells progressed from prometaphase to late anaphase and then a subsequent decrease in compaction during telophase. Interestingly, the FLIM–FRET assay reveals that there is heterogeneity in the compaction within individual chromosomes evident at each stage, even during late anaphase when the highest overall level of compaction is detected. For example, the highest regions of compaction appeared at the trailing tips of anaphase chromosomes (Fig. 6 B, arrowheads; and Fig. S5 B). There was a major change in the fraction of each chromosome that displayed the higher FRET efficiency population, with the ratio of high to lower FRET populations changing from ∼1:1 in metaphase to a peak of ∼10:1 in late anaphase (Fig. 6, B–D). These data are consistent with a major structural transition in chromosomes, affecting the compaction of chromatin and increasing the density of nucleosomes, taking place during anaphase. The magnitude of the peak FRET efficiency within the highest FRET population also increases as cells progress through mitosis, from ∼8% in metaphase to ∼9% in late anaphase.
A previous study examined chromosome compaction during mitosis using either a volumetric assay in cells expressing histone GFP-H2B or photoactivatable GFP-H2B (Mora-Bermúdez et al., 2007). The maximal compaction of chromosomes was also detected in late anaphase and reported to result from axial shortening of the chromatid arms from telomere to centromere. Our FLIM–FRET data support these results and extend them by indicating that the major change as chromosomes move toward their maximum compaction state in late anaphase is the result of a structural transition increasing the fraction of chromatin in the high FRET population. Our data suggest that a major change in chromatin structure between metaphase and late anaphase results from additional folding of a higher order condensed chromatin fiber that reduces the volume, shortens the chromosome arms (as seen in the volumetric assay), and thus increases the fraction of chromatin within the higher FRET efficiency population. Finally, the fraction of chromatin in the highest FRET population decreases from a ratio of ∼10:1 high to lower FRET populations in late anaphase to an ∼1:3 ratio in telophase.
The FLIM–FRET assay provides a comparison between interphase and mitotic chromosome structures. Although there is clearly a large-scale reorganization of chromosome structure and an increase in condensation between interphase and prophase/metaphase, the FLIM–FRET assay discriminates two main structural transitions within chromosomes during mitosis, i.e., in prometaphase and later in anaphase, both involving an increase in compaction at the nucleosomal array level (Fig. 6 D and Fig. S5 B). The data show a change in the fraction of each chromosome distributed between three distinct FRET populations when cells enter in mitosis.
In conclusion, we have shown that as cells progress through mitosis, there is a reversible increase in chromosome compaction affecting chromatin organization at the nucleosome array level that can be resolved by the FLIM–FRET assay. It will be interesting in the future to correlate the in vivo FRET signals with specific higher order chromatin structures to determine which factors are responsible for regulating transitions between different higher order chromatin structures during mitosis and to determine whether they have any functional relationship with the factors controlling the energy-dependent transitions in chromosome compaction during interphase.
Materials and methods
Antibodies and plasmids
The antibodies used were as follows: anti-H2B (1:1,000; Millipore), anti-GFP (1:1,000; Roche), and anti-DsRed (1:5,000; Takara Bio Inc.) for Western blotting experiments; and anti–HP1-α (1:500; Euromedex), anti–CENP-C (1:100; gift from J. Swedlow, Wellcome Trust Centre for Gene Regulation and Expression, Dundee, Scotland, UK), and anti-H3K9triMe (1:250; Abcam) for immunostaining experiments.
The H2B-EGFP expression vector (pBOS–H2B-GFP-N1) containing the blasticidin resistance gene as a selection marker was a gift from T. Kanda (Hokkaido University, Kita-ku, Sapporo, Japan) and G. Wahl (Salk Institute for Biological Studies, La Jolla, CA; Kanda et al., 1998). The mCherry-C1 expression vector containing the neomycin resistance gene as a selection marker was a gift from R.Y. Tsien (Howard Hughes Medical Institute, Chevy Chase, MD; Shaner et al., 2004). The mCherry-H2B expression vector was made by cutting the H2B-coding sequence from the pBOS–H2B-GFP vector with KpnI and BamHI restriction enzymes and subcloned into the cloning site of the mCherry-C1 expression vector.
Expression vectors coding for EGFP coupled directly to mCherry through a 7-aa linker were generated as described previously (Llères et al., 2007). In brief, EGFP cDNA was amplified by PCR using primers containing XhoI and BamHI sites added to both the 5′ and 3′ ends of the primers, respectively. PCR product was purified on a PCR purification column (QIAGEN) according to the manufacturer’s instructions. Purified PCR product was digested with the appropriate enzymes and cloned using standard methods into the compatible sites of the mCherry-C1 expression vector. The choice of restriction sites results in 7 aa separating the two proteins.
Establishment of the HeLaH2B-GFP/mCherry-H2B stable cell line
To generate HeLaH2B-GFP/mCherry-H2B, termed HeLaH2B-2FP in the manuscript, which coexpresses histone H2B fused at its carboxy terminus to EGFP (Kanda et al., 1998) and histone H2B fused at its amino terminus to mCherry FP (Tables S1 and S2, and see Results section A FLIM–FRET assay for chromatin condensation), a previously described cell line stably expressing H2B-EGFP alone (HeLaH2B-GFP cells; provided by H. Kimura, Osaka University, Osaka, Japan; Kimura and Cook, 2001) was grown in DME (Invitrogen) supplemented with 10% FBS, 100 U/ml penicillin/streptomycin (Life Technologies), and 2 µg/ml blasticidin-S (Invitrogen). Exponentially growing cells (in 10-cm dishes) were then transfected with 1 µg of mCherry-H2B plasmid using the Effectene reagent protocol (QIAGEN). 48 h after transfection, the medium was changed with fresh DME containing 2 µg/ml blasticidin-S and 400 µg/ml G418 (Roche). After 15 d of drug selection, surviving colonies were checked under fluorescence microscopy, and those colonies expressing both EGFP and mCherry were isolated. Several clones were selected and expanded into cells lines for further analyses. FACS analyses were performed using a FACScan (BD).
Cell culture, treatments, and transfections
HeLa cells were cultured in DME supplemented with 10% FBS and 100 U/ml penicillin/streptomycin in a humidified incubator at 37°C with 5% CO2. The stable HeLa cell line expressing EGFP-C1 alone (HeLaEGFP) was grown in DME supplemented with 10% FBS and 100 U/ml penicillin/streptomycin and containing 200 µg/ml G418 (Trinkle-Mulcahy et al., 2003). The stable HeLa cell line expressing histone H2B-EGFP (HeLaH2B-GFP; Kimura and Cook, 2001) was a gift from P.R. Cook (University of Oxford, Oxford, England, UK) and was cultured in DME supplemented with 10% FBS, 100 U/ml penicillin/streptomycin, and 2 µg/ml blasticidin-S (Kanda et al., 1998; Kimura and Cook, 2001). HeLaH2B-2FP cells were cultured in DME supplemented with 10% FBS and 100 U/ml penicillin/streptomycin and containing 2 µg/ml blasticidin-S and 200 µg/ml G418.
Metaphase spreads were prepared from HeLaH2B-2FP cells after mitotic shake off, swollen in prewarmed 0.56% KCl for 10 min at 37°C, air dried on slides by using a cytocentrifuge (Shandon Cytospin; Thermo Fisher Scientific), and then stained with 20 ng/ml Hoechst 33342. ATP depletion was performed by using 10 mM Na azide (Sigma-Aldrich) in combination with 50 mM 2-DG (Sigma-Aldrich). TSA (Sigma-Aldrich) was added to the cells at a final concentration of 200 ng/ml for 24 h. When indicated, cells were transfected with 1 µg/90-mm dish of the appropriate plasmid DNA using Effectene transfection reagent according to the manufacturer’s instructions.
Cell fixation, immunostaining, and microscopy
Cells were grown on glass coverslips and fixed for 5 min in 3.7% paraformaldehyde in 37°C PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 2 mM MgCl2, pH 6.9). After a 10-min permeabilization with 1% Triton X-100 in PBS, cells were blocked with 1% goat serum for 30 min and then incubated with primary antibodies for 1 h, washed, and incubated with secondary antibodies for 45 min. If required, cells were stained with 0.3 µg/ml DAPI (Sigma-Aldrich). After a final set of washes, cells were mounted in Vectashield media (Vector Laboratories).
All images were acquired with a wide-field fluorescence microscope (DeltaVision Spectris; Applied Precision) and a CoolMax charge-coupled device camera (Roper Industries). Imaging was performed at room temperature using a 60× oil immersion NA 1.4 Plan-Apochromat objective from Olympus. SoftWoRx software (Applied Precision) was used for image acquisition and data deconvolution.
5-FU incorporation assay
HeLa cells, either mock treated or ATP depleted for 30 min or after washing and recovery for 30 min, were incubated with 2 mM 5-FU (Sigma-Aldrich) for 30 min at 37°C. Subsequently, cells were fixed, permeabilized, and incubated with primary anti-BrdU antibody (1:500; Sigma-Aldrich). Immunofluorescence microscopy was performed as indicated in the previous section.
Gel electrophoresis and immunoblotting
In brief, cells were scrapped, pelleted, resuspended in 2× reducing loading buffer (Invitrogen), and denatured at 90°C for 5 min. For Western blot analysis, proteins resolved by 10% Bis-Tris SDS-PAGE were transferred to nitrocellulose filters and probed with specific primary antibodies and the corresponding horseradish peroxidase–conjugated secondary antibodies (GE Healthcare). An ECL reagent (GE Healthcare) was used to visualize protein bands according to the manufacturer’s instructions. Red Ponceau staining of the transferred proteins was used to assess equal loading of the samples.
In Fig. 2 D, HeLaH2B-2FP cells in physiological buffer (PB; Kimura et al., 1999) were divided into five 100-µl aliquots. One (Fig. 2 D, lane 1) was mixed with 100 µl PB. Others were mixed with 100 µl PB plus 0.2% Triton X-100 and NaCl to give a final concentration of 0–1 M (Fig. 2 D, lanes 2–4). After incubation (10 min at 0°C) and pelleting (20,000 g for 1 h at 4°C), 180 µl of the supernatant was collected and mixed with an equal volume of loading buffer (Fig. 2 D). The pellet was resuspended in 180 µl PB, and 200 µl of 2× loading buffer was added (Fig. S1 B). The nucleosomal proteins were also resolved in a 10% polyacrylamide gel stained with Coomassie (Fig. 2 D, top).
MNase digestion of chromatin
HeLa cells and stable cells expressing H2B-EGFP (HeLaH2B-GFP) or both H2B-EGFP and mCherry-H2B (HeLaH2B-2FP) were trypsinized, harvested, and washed once with 1× RSB buffer (10 mM Tris, pH 7.6, 15 mM NaCl, and 1.5 mM MgCl2). After centrifugation, the cell pellet was resuspended in 1× RSB buffer with 1% Triton-X 100 and homogenized by five strokes with a loose-fitting pestle to release nuclei. Nuclei were collected by centrifugation and washed twice with 1 ml of buffer A (15 mM Tris, pH 7.5, 15 mM NaCl, 60 mM KCl, 0.34 M sucrose, 0.5 mM spermidine, 0.15 mM spermine, 0.25 mM PMSF, and 0.1% β-mercaptoethanol). Nuclei were finally resuspended in 1.5 ml of buffer A, and 15 µl of 0.1 M CaCl2 was added.
For making nucleosomal ladders, 0.5 ml of suspended nuclei was digested by adding 1 µl of 200 U/ml MNase (Sigma-Aldrich) at 37°C. 60-µl aliquots were taken at each time point (5, 15, 30, and 60 min), and 1.5 µl EDTA was added to stop the reaction (Telford and Stewart, 1989). To each tube, 18 µl H2O, 12 µl of 10% SDS, and 24 µl of 5 M NaCl were added. The mixtures were extracted with phenol-chloroform, and 5 µl of supernatant was analyzed by 1.5% agarose gel electrophoresis.
Fluorescence lifetime measurements by time-correlated single-photon counting (TCSPC) for FRET measurements
FRET can be used to image direct protein–protein interactions in cells. While transferring energy from an excited donor to an acceptor (Lakowicz, 2006), FRET decreases the donor fluorescence and increases the acceptor fluorescence. Because the energy transfer is highly distance dependent, detection of FRET requires that the two fluorophores must be within ∼1–10 nm, i.e., the distance typically found for directly interacting proteins (Förster, 1949). In this study, as a quantitative readout of FRET, we measured the fluorescence lifetime of the donor, which was defined as the mean time between fluorophore excitation and photon emission by using a FLIM approach.
FLIM was performed using an inverted multiphoton laser-scanning microscope (Radiance 2100MP; Bio-Rad Laboratories) with a 60× oil immersion NA 1.4 Plan-Apochromat objective from Nikon. The software used to run the Radiance 2100MP multiphoton microscope was Lasersharp 2000 (Bio-Rad Laboratories). The microscope was equipped with a Solent Scientific’s incubation chamber suitable to maintain the live cells and optics at constant temperature (usually 37°C). The chamber was constructed with black walls to exclude external sources of light during the sensitive period of FLIM measurement. Two-photon excitation was achieved using a Chameleon Verdi–pumped ultrafast tunable (720–930 nm) laser (Coherent) to pump a mode-locked frequency-doubled Ti:Sapphire laser that provided sub–200-femtosecond pulses at a 90-Mhz repetition rate with an output power of 1.4 W at the peak of the tuning curve (800 nm). Enhanced detection of the scattered component of the emitted (fluorescence) photons was afforded by the use of fast single-photon response direct detectors (model 5783P; Hamamatsu Photonics).
The fluorescence lifetime imaging capability was provided by TCSPC electronics (SPC-830; Becker & Hickl GmbH). TCSPC measures the time elapsed between laser pulses and the fluorescence photons. Indeed, when fluorophore molecules absorb a quantum of light, a valence electron is boosted up into a higher energy orbit, creating an excited state. When this electron returns to its original lower energy orbit (the ground state level), a quantum of light may be emitted. By consequence, the fluorescence lifetime occurs on the nanosecond time scale, and FLIM measurements reflect events occurring in an extremely short period of time. The TCSPC method used in this study is based on the detection of single photons, the measurement of the detection times of the individual photons, and the reconstruction of the waveform from the individual time measurements. Over this integration time, the waveform of the optical pulse builds up and corresponds to a histogram presenting the number of photons recorded for each nanosecond’s detection time interval. Therefore, the FLIM technique provides us with the spatial map distribution of the variations of the fluorescence lifetimes and, indirectly, of the FRET efficiencies at each pixel throughout the nucleus (Becker, 2005).
EGFP and mCherry fluorophores were used as a FRET pair for all of the FLIM–FRET measurements. The optimal two-photon excitation wavelength to excite the donor (EGFP) was determined to be 890 nm. Fluorescence emission of EGFP fusion proteins was collected using a bandpass filter (528 ± 25 nm) to limit detection to only the donor fluorophore (EGFP) and prevent contamination from the acceptor (mCherry) emission (Llères et al., 2007). Laser power was adjusted to give a mean photon count rate of the order 104–105 photons/s. For imaging live cells by FLIM, the standard growth medium was replaced with CO2-independent phenol red–free DME (Invitrogen) supplemented with 10% FBS and 100 U/ml penicillin/streptomycin. Fluorescence lifetime measurements during mitosis were acquired over 90 s. In interphase cells, FLIM measurements were collected over 120 s. Fluorescence lifetimes were calculated for all pixels in the field of view (256 × 256 pixels) or for a particular selected region of interest (ROI; e.g., nucleus) using SPCImage software (Becker & Hickl GmbH).
Analysis of the fluorescence lifetime measurements for FRET experiments
The analysis of the FLIM measurements was performed by using SPCImage software. Because FRET interactions cause a decrease in the fluorescence lifetime of the donor molecules (EGFP), the FRET efficiency can be calculated by comparing the FLIM values obtained for the EGFP donor fluorophores in the presence and absence of the mCherry acceptor fluorophores (Fig. 3 A, right; and Fig. 5 A, right). Mean FRET efficiency images were calculated such as the FRET efficiency, EFRET = 1 − (τDA/τD), where τDA is the mean fluorescence lifetime of the donor (H2B-EGFP) in the presence of the acceptor (mCherry-H2B) expressed in the HeLaH2B-2FP and τD is the mean fluorescence lifetime of the donor (H2B-EGFP) expressed in HeLaH2B-GFP in the absence of acceptor for all of the cells imaged. In the non-FRET conditions, the mean fluorescence lifetime value of the donor in the absence of the acceptor was calculated from a mean of the τD by applying a monoexponential decay model to fit the fluorescence lifetime decays. It was necessary to apply the best-fitting model to extract an accurate fluorescence lifetime value from the decays. For this purpose, several parameters could be adjusted to optimize the χ2 of the fit close to one. For example, we defined a threshold value representing the background to establish a minimum number of photons in the peak of a fluorescence curve. Pixels with lower photon numbers were not analyzed by the fitting procedure. This improved the quality of the lifetime parameter histogram. We adjusted the shift parameter to optimize the χ2 value to as close to one as possible. The scatter parameter (amount of scattered excitation light detected) should be adjusted to a value of either zero or close to zero.
Then, in the FRET conditions, we applied a biexponential fluorescence decay model to fit the experimental decay curves f(t) = a e−t/τDA + b e−t/τD. We obtained information about the lifetime of two populations of molecules, i.e., the noninteracting donor population τD and the donor population that was interacting with the acceptor (τDA) as well as the intensity factors a and b of the two decay components. By fixing the noninteracting proteins lifetime τD using data from control experiments (in the absence of FRET), the value of τDA was estimated. Then, by knowing both values τDA and τD at each single pixel, the FRET efficiency (EFRET) was derived by applying the following equation: EFRET = 1 − (τDA/τD) at each pixel in a selected ROI using SPCImage software. For FLIM–FRET measurements during interphase, the selected ROI generally comprised several interphase nuclei (see the legends for Figs. 3–5 and Fig. S2 for more details). During mitosis acquisitions, the particular ROI comprised the metaphasic equatorial plate in metaphase, the two separate paired chromosomes in early and late anaphase, or the two daughter nuclei during telophase (see the legends for Fig. 6 and Fig. S5). Furthermore, the FRET distribution curves (Fig. 3 A, right) from these ROIs were displayed from the extracted associated matrix using SPCImage software and then normalized. In each experiment, FLIM was performed on multiple cells, as indicated in figure legends, in three independent experiments.
Enrichment in short lifetime pixels (Fig. 4 F, high FRET pixels) over time was quantified from the lifetime distribution histograms. The lifetime histogram for each time point was normalized by the total number of pixels Fi = fi/Ni, where fi and Ni are, respectively, the lifetime histogram and the total number of pixels at time point t = i.
Statistical analysis
To interpret the distribution of the percent FRET efficiency data, box and whisker plots were used. The box and whisker plot is a histogram-like method for displaying upper and lower quartiles and maximum and minimum values in addition to median. We used a nonparametric Mann-Whitney test to compare the medians of each dataset for the two-photon FLIM measurements using Prism software (GraphPad Software, Inc.).
FRAP
HeLaH2B-2FP cells were cultured in glass-bottomed dishes (WILCO; Intracel). Before imaging, growth medium was replaced with phenol red–free CO2-independent medium (Invitrogen). In all experiments, a 60× oil immersion NA 1.4 Plan-Apochromat objective (Olympus) was used. HeLaH2B-2FP cells were photobleached by using the photokinetic experiment function of the DeltaVision Spectris wide-field fluorescence microscope (Phair and Misteli, 2001). In brief, a small region in the cells was photobleached with a 488-nm laser (100% laser power for the duration of 0.15 s), and time-lapse sequences of single optical sections for imaging EGFP fluorescence were collected with an exposure time of 0.1 s for each image. SoftWoRx software was used for image acquisition. The fluorescence intensities in the bleached and nonbleached area before and after laser photobleaching were quantitated with the SoftWoRx software. These fluorescence intensity measurements were corrected from the total fluorescence intensity before and after laser photobleaching and from the background intensity level. The mobile fraction of the proteins was calculated using Excel (Microsoft) and Prism software.
TEM
After fixation in 4% paraformaldehyde/2.5% glutaraldehyde in 0.2 M Pipes, pH 7.3, cells were scraped and then pelleted in Eppendorf tubes and rinsed in 0.2 M Pipes. Samples were postfixed in 1% aqueous osmium tetroxide, rinsed in water, dehydrated in graded alcohol-propylene oxide, and then embedded in Durapan resin (Sigma-Aldrich). Serial sections (60–70 nm) were cut on an ultramicrotome (Ultracut UCT; Leica) and collected on 100 mesh copper grids coated with 1% Pioloform (Agar) in chloroform. Sections were stained with uranyl acetate and lead citrated and examined with a transmission electron microscope (Tecnai 12; FEI) at 6,000 magnification. Images were collected on digital imaging plates, which were read out with a micrometer imaging plate scanner (Ditabis; Digital Biomedical Imaging Systems AG).
Online supplemental material
Fig. S1 shows the structural model organization of a nucleosome core particle labeled with both H2B-EGFP and mCherry-H2B fluorescent-tagged histones and the biochemical characterization of the HeLaH2B-2FP stable cell line. Fig. S2 shows the effect of ATP depletion on the mean fluorescence lifetime of H2B-GFP (donor) and the effect on the localization of H3K9triMe staining. Fig. S3 shows the mobility by FRAP of H2B-EGFP upon ATP depletion. Fig. S4 shows the statistical analysis of the FRET efficiency percentage upon TSA treatment and the relocalization of CENP-C and HP1-α proteins after TSA treatment. Fig. S5 shows time-lapse FLIM–FRET measurements of a HeLaH2B-2FP cell progressing along mitosis from prometaphase to telophase and the FRET distributions of the control HeLaH2B-GFP stable cell line. Videos 1 and 2 show time-lapse videos of HeLaH2B-2FP cells either untreated or upon ATP depletion treatment, respectively. Tables S1 and S2 show DNA sequences of the H2B-EGFP and mCherry-H2B histone genes, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200907029/DC1.
Acknowledgments
We thank Drs. T. Kanda, G. Wahl, R.Y. Tsien, H. Kimura, P.R. Cook, and J. Swedlow for providing reagents and cell lines. We are grateful to the members of the Lamond laboratory for helpful discussions and suggestions and thank Dr. P. Appleton and C. Thomson of the College of Life Sciences Light Microscopy Facility for advice and technical assistance. We thank Dr. A. Prescott for advice on TEM experiments.
This work was supported by the Wellcome Trust (programme grant 073980/Z/03/Z and strategic award WT083524MA). A.I. Lamond is a Wellcome Trust Principal Research Fellow. D.G. Norman was supported by the Biotechnology and Biological Sciences Research Council (programme grant BB/E022286/1).
Footnotes
Abbreviations used in this paper:
- 2-DG
- 2-deoxyglucose
- 5-FU
- 5-fluorouracil
- CENP-C
- centromere protein C
- FLIM
- fluorescence lifetime imaging microscopy
- FRET
- Förster resonance energy transfer
- H3K9triMe
- histone H3 lysine trimethylation
- MNase
- micrococcal nuclease
- MPLSM
- multiphoton laser-scanning microscopy
- PB
- physiological buffer
- ROI
- region of interest
- TCSPC
- time-correlated single-photon counting
- TEM
- transmission EM
- TSA
- trichostatin A
References
- Bastiaens P.I., Squire A. 1999. Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol. 9:48–52 10.1016/S0962-8924(98)01410-X [DOI] [PubMed] [Google Scholar]
- Becker W. 2005. The bh TCSPC handbook. Becker and Hickl GmbH. http://www.becker-hickl.de/handbook.htm(accessed November 27, 2007)
- Belmont A.S., Dietzel S., Nye A.C., Strukov Y.G., Tumbar T. 1999. Large-scale chromatin structure and function. Curr. Opin. Cell Biol. 11:307–311 10.1016/S0955-0674(99)80041-6 [DOI] [PubMed] [Google Scholar]
- Choi J.K., Howe L.J. 2009. Histone acetylation: truth of consequences? Biochem. Cell Biol. 87:139–150 10.1139/O08-112 [DOI] [PubMed] [Google Scholar]
- Cushman I., Stenoien D., Moore M.S. 2004. The dynamic association of RCC1 with chromatin is modulated by Ran-dependent nuclear transport. Mol. Biol. Cell. 15:245–255 10.1091/mbc.E03-06-0409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S.C. 1996. Heterochromatin and gene regulation in Drosophila. Curr. Opin. Genet. Dev. 6:193–202 10.1016/S0959-437X(96)80050-5 [DOI] [PubMed] [Google Scholar]
- Förster T. 1949. Experimental and theoretical investigation of the intermolecular transfer of electronic excitation energy. Z. Naturforsch. [A]. 4:321–327 [Google Scholar]
- Gerlich D., Hirota T., Koch B., Peters J.M., Ellenberg J. 2006. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr. Biol. 16:333–344 10.1016/j.cub.2005.12.040 [DOI] [PubMed] [Google Scholar]
- Gilbert N., Boyle S., Fiegler H., Woodfine K., Carter N.P., Bickmore W.A. 2004. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 118:555–566 10.1016/j.cell.2004.08.011 [DOI] [PubMed] [Google Scholar]
- Grewal S.I., Elgin S.C. 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12:178–187 10.1016/S0959-437X(02)00284-8 [DOI] [PubMed] [Google Scholar]
- Grunstein M. 1998. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 93:325–328 10.1016/S0092-8674(00)81160-5 [DOI] [PubMed] [Google Scholar]
- Huisinga K.L., Brower-Toland B., Elgin S.C. 2006. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma. 115:110–122 10.1007/s00412-006-0052-x [DOI] [PubMed] [Google Scholar]
- Huynh V.A., Robinson P.J., Rhodes D. 2005. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J. Mol. Biol. 345:957–968 10.1016/j.jmb.2004.10.075 [DOI] [PubMed] [Google Scholar]
- Kanda T., Sullivan K.F., Wahl G.M. 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8:377–385 10.1016/S0960-9822(98)70156-3 [DOI] [PubMed] [Google Scholar]
- Keppler A., Arrivoli C., Sironi L., Ellenberg J. 2006. Fluorophores for live cell imaging of AGT fusion proteins across the visible spectrum. Biotechniques. 41:167–170: 172: 174–175 10.2144/000112216 [DOI] [PubMed] [Google Scholar]
- Kimura H., Cook P.R. 2001. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341–1353 10.1083/jcb.153.7.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Tao Y., Roeder R.G., Cook P.R. 1999. Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol. Cell. Biol. 19:5383–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M.J., Celeste A., Dellaire G., Fernandez-Capetillo O., Müller W.G., McNally J.G., Bazett-Jones D.P., Nussenzweig A. 2006. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 172:823–834 10.1083/jcb.200510015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz J.R. 2006. Principles of Fluorescence Spectroscopy. Third edition Springer, New York: 954 pp [Google Scholar]
- Lippincott-Schwartz J., Patterson G.H. 2003. Development and use of fluorescent protein markers in living cells. Science. 300:87–91 10.1126/science.1082520 [DOI] [PubMed] [Google Scholar]
- Llères D., Swift S., Lamond A.I. 2007. Detecting protein-protein interactions in vivo with FRET using multiphoton fluorescence lifetime imaging microscopy (FLIM). Curr. Protoc. Cytom. Chapter 12:Unit12.10. [DOI] [PubMed] [Google Scholar]
- Luger K. 2003. Structure and dynamic behavior of nucleosomes. Curr. Opin. Genet. Dev. 13:127–135 10.1016/S0959-437X(03)00026-1 [DOI] [PubMed] [Google Scholar]
- Luger K., Hansen J.C. 2005. Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol. 15:188–196 10.1016/j.sbi.2005.03.006 [DOI] [PubMed] [Google Scholar]
- Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 389:251–260 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Misteli T., Gunjan A., Hock R., Bustin M., Brown D.T. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature. 408:877–881 10.1038/35048610 [DOI] [PubMed] [Google Scholar]
- Mora-Bermúdez F., Ellenberg J. 2007. Measuring structural dynamics of chromosomes in living cells by fluorescence microscopy. Methods. 41:158–167 10.1016/j.ymeth.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Mora-Bermúdez F., Gerlich D., Ellenberg J. 2007. Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat. Cell Biol. 9:822–831 10.1038/ncb1606 [DOI] [PubMed] [Google Scholar]
- Perche P.Y., Vourc’h C., Konecny L., Souchier C., Robert-Nicoud M., Dimitrov S., Khochbin S. 2000. Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr. Biol. 10:1531–1534 10.1016/S0960-9822(00)00832-0 [DOI] [PubMed] [Google Scholar]
- Phair R.D., Misteli T. 2001. Kinetic modelling approaches to in vivo imaging. Nat. Rev. Mol. Cell Biol. 2:898–907 10.1038/35103000 [DOI] [PubMed] [Google Scholar]
- Platani M., Goldberg I., Lamond A.I., Swedlow J.R. 2002. Cajal body dynamics and association with chromatin are ATP-dependent. Nat. Cell Biol. 4:502–508 10.1038/ncb809 [DOI] [PubMed] [Google Scholar]
- Rhodes D., Laskey R.A. 1989. Assembly of nucleosomes and chromatin in vitro. Methods Enzymol. 170:575–585 10.1016/0076-6879(89)70065-3 [DOI] [PubMed] [Google Scholar]
- Robinett C.C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685–1700 10.1083/jcb.135.6.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Steinbach P.A., Tsien R.Y. 2005. A guide to choosing fluorescent proteins. Nat. Methods. 2:905–909 10.1038/nmeth819 [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y., Darzacq X., Shenoy S.M., Fusco D., Janicki S.M., Spector D.L., Singer R.H. 2004. Dynamics of single mRNPs in nuclei of living cells. Science. 304:1797–1800 10.1126/science.1099754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strukov Y.G., Belmont A.S. 2009. Mitotic chromosome structure: reproducibility of folding and symmetry between sister chromatids. Biophys. J. 96:1617–1628 10.1016/j.bpj.2008.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedlow J.R., Sedat J.W., Agard D.A. 1993. Multiple chromosomal populations of topoisomerase II detected in vivo by time-lapse, three-dimensional wide-field microscopy. Cell. 73:97–108 10.1016/0092-8674(93)90163-K [DOI] [PubMed] [Google Scholar]
- Taddei A., Maison C., Roche D., Almouzni G. 2001. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3:114–120 10.1038/35055010 [DOI] [PubMed] [Google Scholar]
- Telford D.J., Stewart B.W. 1989. Micrococcal nuclease: its specificity and use for chromatin analysis. Int. J. Biochem. 21:127–137 10.1016/0020-711X(89)90100-6 [DOI] [PubMed] [Google Scholar]
- Tóth K.F., Knoch T.A., Wachsmuth M., Frank-Stöhr M., Stöhr M., Bacher C.P., Müller G., Rippe K. 2004. Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. J. Cell Sci. 117:4277–4287 10.1242/jcs.01293 [DOI] [PubMed] [Google Scholar]
- Treanor B., Lanigan P.M., Kumar S., Dunsby C., Munro I., Auksorius E., Culley F.J., Purbhoo M.A., Phillips D., Neil M.A., et al. 2006. Microclusters of inhibitory killer immunoglobulin-like receptor signaling at natural killer cell immunological synapses. J. Cell Biol. 174:153–161 10.1083/jcb.200601108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremethick D.J. 2007. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 128:651–654 10.1016/j.cell.2007.02.008 [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Andrews P.D., Wickramasinghe S., Sleeman J., Prescott A., Lam Y.W., Lyon C., Swedlow J.R., Lamond A.I. 2003. Time-lapse imaging reveals dynamic relocalization of PP1gamma throughout the mammalian cell cycle. Mol. Biol. Cell. 14:107–117 10.1091/mbc.E02-07-0376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.M. 1998. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell. Mol. Life Sci. 54:21–31 10.1007/s000180050122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P., Hudson D.F., Ribeiro S.A., Trinkle-Mulcahy L., Spence J.M., Lai F., Farr C.J., Lamond A.I., Earnshaw W.C. 2006. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat. Cell Biol. 8:1133–1142 10.1038/ncb1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Munster E.B., Gadella T.W. 2005. Fluorescence lifetime imaging microscopy (FLIM). Adv. Biochem. Eng. Biotechnol. 95:143–175 [DOI] [PubMed] [Google Scholar]
- Wallrabe H., Periasamy A. 2005. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 16:19–27 10.1016/j.copbio.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Wolffe A.P., Kurumizaka H. 1998. The nucleosome: a powerful regulator of transcription. Prog. Nucleic Acid Res. Mol. Biol. 61:379–422 10.1016/S0079-6603(08)60832-6 [DOI] [PubMed] [Google Scholar]
- Wouters F.S., Bastiaens P.I. 2001. Imaging protein-protein interactions by fluorescence resonance energy transfer (FRET) microscopy. Curr. Protoc. Cell Biol. Chapter 17:Unit 17.1. [DOI] [PubMed] [Google Scholar]