Abstract
The intimate relationship between DNA double-strand break (DSB) repair and cancer susceptibility has sparked profound interest in how transactions on DNA and chromatin surrounding DNA damage influence genome integrity. Recent evidence implicates a substantial commitment of the cellular DNA damage response machinery to the synthesis, recognition, and hydrolysis of ubiquitin chains at DNA damage sites. In this review, we propose that, in order to accommodate parallel processes involved in DSB repair and checkpoint signaling, DSB-associated ubiquitin structures must be nonuniform, using different linkages for distinct functional outputs. We highlight recent advances in the study of nondegradative ubiquitin signaling at DSBs, and discuss how recognition of different ubiquitin structures may influence DNA damage responses.
Introduction
Ligand-inducible phosphorylation and phospho-protein binding enable the rapid assembly of macromolecular protein complexes and transmission of signals from the plasma membrane to the nucleus (Ptacek and Snyder, 2006; Pawson and Kofler, 2009). Phosphorylation is often coupled to substrate ubiquitylation, creating a diverse array of recognition platforms for association of these multicomponent protein complexes (Haglund and Dikic, 2005; Hunter, 2007; Schwartz and Ciechanover, 2009). There is now a plethora of molecular evidence to suggest that signaling events that are initiated in the nucleus invoke similar strategies to execute acute responses to DNA damage (Harper and Elledge, 2007; Greenberg, 2008; Huen and Chen, 2008). Phosphatidylinositol-3-kinase-related kinase (PIKK)-dependent phosphorylation mediates assembly of several different E3 ligase complexes at double-strand breaks (DSBs), each synthesizing ubiquitin chains on different substrates. In essence, a multitude of parallel signaling processes must rapidly develop at DSBs, necessitating different structural cues to coordinate repair and checkpoint events. Emerging evidence indicates that ubiquitin signaling is essential to this process. A substantial number of DNA repair proteins are dedicated to the synthesis, recognition, and breakdown of ubiquitin chains with specific topologies, supporting the concept that a diverse and dynamic ubiquitin landscape arises at DSBs to strongly influence both the efficiency and specificity of DNA repair.
Principles of ubiquitin
Numerous cellular processes are regulated by the posttranslational mark ubiquitin, including protein degradation, cell cycle regulation, DNA repair, transcription, and endocytosis. Specifically, the highly conserved 76–amino acid protein can alter the activity of its target in a variety of ways, from changing its localization or enzymatic activity to targeting it for degradation. Ubiquitylation, the process that involves the covalent attachment ubiquitin to the target protein, creates a covalent isopeptide linkage in a variety of different topologies to affect these diverse processes.
Ubiquitylation is a highly regulated process involving a specific cascade of activities performed by the E1, E2, and E3 series of enzymes (Hershko et al., 2000; Pickart, 2001). E1, or ubiquitin-activating enzyme, activates ubiquitin by forming a thiol ester link between the carboxy terminus of ubiquitin and the active site cysteine of E1 in an ATP-requiring step. The activated ubiquitin is then transferred to an E2 ubiquitin-conjugating enzyme, also through a thiol ester bond between ubiquitin and the active site cysteine of E2. E2, together with E3 or ubiquitin ligase, transfers the ubiquitin to its target, forming a covalent isopeptide linkage between the carboxyl terminus Gly-76 of ubiquitin to a primary amine (usually the ϵ-amino group of lysine) of the target protein.
Ubiquitylation is among the more unique forms of posttranslational modification in that a single ubiquitin monomer can be further ubiquitylated (polyubiquitylated) through one of seven lysines or through the amino terminus to create polyubiquitin chains (Fig. 1 A). Remarkably, different ubiquitin topologies or linkages between ubiquitin moieties can lead to vastly different biological outcomes. For example, the canonical lysine-48 (K48)-linked polyubiquitin targets the substrate protein for proteasomal degradation (Pickart and Cohen, 2004), whereas lysine-63 (K63)-linked polyubiquitin is often involved in localization or signaling events (Chen and Sun, 2009). Polyubiquitin can be linked through one residue to create a homogeneous chain, or through multiple residues, forming branched ubiquitin chains (Kim et al., 2007b).
Figure 1.
Structural topology of ubiquitin chains. (A) Surface representation of ubiquitin. All seven lysines (K6, K11, K27, K29, K33, K48, and K63) and the amino-terminus (M1), shown in blue, can be conjugated to the carboxy terminus of another ubiquitin molecule. The hydrophobic patch (L8, I44, and V70), shown in green, is recognized by several ubiquitin-binding proteins (PDB accession no. 1UBQ; Vijay-Kumar, et al., 1987). (B) Model of K63-tetraubiquitin (based on K63-diubiquitin, PDB accession no. 3A1Q; Sato et al., 2009). K63-tetraubiquitin forms long chains, exposing the I44 hydrophobic patch in green, to ubiquitin-binding proteins. (C) Model of K48-tetraubiquitin (based on K48-diubiquitin, PDB accession no. 1F9J; Phillips et al., 2001). K48-tetraubiquitin forms a compact structure, where the I44 hydrophobic patch in green is largely buried.
Even though polyubiquitin chains are made of the same basic ubiquitin unit, the structure of K63-linked chains is morphologically different than K48-linked chains (Fig. 1, B and C). The K48-linked isoform of ubiquitin adopts a compact superhelical structure in which a hydrophobic patch consisting of Leu8, Ile44, and Val70 is buried within a repeating tetramer (Fig. 1 C; Eddins et al., 2007). In contrast, there is very little interaction between ubiquitin monomers in K63-linked polyubiquitin, allowing for a high degree of torsional freedom about the carboxy-terminal glycine–glycine axis, as confirmed by nuclear magnetic resonance studies (Fig. 1 B; Varadan et al., 2004). This “beads-on-a-string” model of K63-linked polyubiquitin allows K63-specific ubiquitin binding proteins a great deal of flexibility for recognizing different surfaces on ubiquitin. Although no structures of other lysine-linked polyubiquitin chains have yet been structurally determined, mass spectrometry data suggest that these chains are formed through all lysine residues in vivo (Peng et al., 2003; Al-Hakim et al., 2008; Xu et al., 2009). Consistent with the structural diversity of ubiquitin chains, a myriad of ubiquitin-binding domains exists that recognizes different features and topologies of ubiquitin. The majority of ubiquitin-binding domains form α-helical structures that recognize the hydrophobic patch centered around Ile44 of ubiquitin (Harper and Schulman, 2006; Grabbe and Dikic, 2009). Though the affinities of individually isolated ubiquitin-binding domains are low to moderate, this affinity may increase in vivo due to multiple ubiquitin-binding members in a complex recognizing polyubiquitin chains in a synergistic fashion.
The hydrolysis of the isopeptide bond connecting ubiquitin to a substrate protein is performed by deubiquitinating enzymes (DUBs). DUBs act in a variety of regulatory processes, from “rescuing” previously targeted proteins from degradation to attenuating signaling pathways (Reyes-Turcu et al., 2009). Given the tight regulatory control of ubiquitin and the possibility for many different linkages between ubiquitin moieties, it is perhaps not surprising that many DUBs exhibit specificity for one type of ubiquitin chain isoform. There are generally two classes of DUBs that are distinguished on the basis of catalytic mechanism (Reyes-Turcu et al., 2009). The first utilizes an active site cysteine nucleophile to hydrolyze the ubiquitin isopeptide bond, which is remarkably similar to the papain family of cysteine proteases. The second class of DUB enzymes is the Jab1/MPN/Mov34 (JAMM) family of zinc-binding metalloproteases. JAMM domains function by binding a highly electrophilic metal (usually zinc) that delivers a hydroxide ion for nucleophilic attack on the isopeptide bond, performing isopeptide bond hydrolysis in a similar mechanism to the Zn2+-containing protease thermolysin.
Principles of DSB recognition
DSBs are potentially lethal lesions that can arise from endogenous and exogenous sources. To protect the genome, cells use a vast network of proteins to monitor DNA integrity and mount a response to DSBs. This DNA damage response usually includes cell cycle arrest and activation of the DNA repair pathways. In extreme cases, when cells are unable to properly repair DSBs, apoptosis or senescence pathways may be triggered.
Recognition and signaling at DSBs proceeds rapidly, with a distinct temporal and spatial order of association and dissociation of numerous DNA repair factors with the site of the break (Lisby et al., 2003). DNA repair protein recruitment and retention is conveniently visualized by fluorescence imaging of repair foci (often called ionizing radiation–induced foci [IRIF]). Foci are easily observed because many DNA repair proteins are retained at DSBs in suprastoichiometric ratios. It has been estimated that individual breaks contains at least 1,000 molecules of each DNA repair protein (Lisby et al., 2004), revealing that many repair proteins are concentrated at repair sites by a factor of 103. The reasons for this stoichiometry are unknown, although it is speculated that high local concentrations of repair factors are necessary to amplify signals for repair and checkpoint responses.
Robust foci formation occurs within minutes of DSB induction, which dictates that molecular recognition must rapidly develop at the site of DNA damage for repair protein recruitment. At the pinnacle of this chain of events is phosphorylation of the histone H2A variant H2AX. DNA damage activates cellular ataxia telangiectasia mutated (ATM) kinase and related PIKKs to phosphorylate H2AX at its C terminus on Ser139. Phospho H2AX (γ-H2AX) formation occurs within minutes after damage, and extends for up to a megabase from the site of the break in mammalian cells, providing a platform for subsequent DNA repair protein recruitment and amplification at DSBs. Indeed, H2AX-null cells demonstrate strongly reduced repair protein focus formation, which is consistent with H2AX being a master regulator of the recruitment of DNA repair proteins to chromatin at DSBs (Celeste et al., 2002). These findings provided a basis to understand repair factor–DSB stoichiometry. Thousands of repair protein molecules associate along chromatin that contains γ-H2AX in cis to the DSB, which explains the suprastoichiometric relationship of repair proteins to DSBs.
Although γ-H2AX is a master regulator of visible foci formation, γ-H2AX deficiency does not eliminate all DNA repair protein recruitment to DSBs, which indicates that foci are just part of the DSB repair puzzle. During DSB repair, there is a clearing of γ-H2AX and nucleosomes directly adjacent to the DSB (Shroff et al., 2004; Berkovich et al., 2007). These dechromatinized regions are thought to be the site of the majority of DSB repair chemistry. For example, proteins that perform nonhomologous end joining do not form foci, presumably because they are present only at the DSB termini and not along chromatin that contains γ-H2AX. Conversely, proteins dedicated to homologous recombination, such as the breast cancer early onset gene product BRCA1, are present at both non-nucleosomal regions and at chromatin that contains γ-H2AX (Celeste et al., 2003; Bekker-Jensen et al., 2006). Thus, DSB recognition at different locales on both DNA and chromatin flanking the break is essential for repair and the maintenance of genome integrity.
Connections between PIKK activity and ubiquitylation
Ubiquitylation directs repair proteins to DSBs.
Several prominent DNA repair pathways use a paired process in which PIKK-mediated phosphorylation is coupled to substrate ubiquitylation (Table I). In each instance, ubiquitylation of a protein associated with DNA repair is essential for its recruitment to DSBs. Notably, mutation in each of these DNA damage–associated ubiquitylation pathways is responsible for a human cancer susceptibility syndrome (Scully and Livingston, 2000; Chenevix-Trench et al., 2002; Wang, 2007).
Table I.
Connections between the DNA damage response and ubiquitin
| Substrate | PIKK | Ubiquitin linkage | E3 ligase | E2 ligase | DUB | Function |
| FancD2 | ATR | Mono | FancL | Unknown | USP1 | DSB localization |
| FancI | ATM/ATR | Mono | FancL | Ube2T | USP1 | DSB localization |
| BRCA1 | ATM/ATR | K6 (in vitro) | BRCA1-BARD1 | Ubch5c | Unknown | Unknown |
| CtIP | ATM | Unknown | BRCA1-BARD1 | Ubch5c | Unknown | DSB localization |
| H2A, H2AX | ATM/ATR/DNA-PK | Mono, K63 | RNF8, RNF168 | Ubc13 | USP3, BRCC36 | Recruitment of factors to DSB |
DNA-PK, DNA-dependent protein kinase.
Fanconi anemia is a recessive monogenic disease in which patients display developmental abnormalities, bone marrow failure, and cancer predisposition phenotypes. Fanconi syndrome is comprised of 13 different genetic complementation groups, and at least three pathways (Wang, 2007). All Fanconi mutant cell lines display the common characteristic of sensitivity to DNA cross-linking agents. The classical Fanconi pathway culminates in monoubiquitylation of the FancD2 and FancI proteins at a single lysine residue on each protein (Garcia-Higuera et al., 2001; Sims et al., 2007; Smogorzewska et al., 2007; Wang, 2007). FancD2 ubiquitylation occurs in S phase in response to phosphorylation by the Rad3-related PIKK, ataxia telangiectasia and Rad3 related (ATR; Andreassen et al., 2004). Monoubiquitylation is critical for FancD2 and FancI localization to DSBs and chromatin association (Meetei et al., 2004; Wang et al., 2004; Sims et al., 2007; Smogorzewska et al., 2007). Though it is not clear how ubiquitylation controls Fanconi pathway activity, a carboxy-terminal FancD2-ubiquitin fusion protein strongly associated with DSBs and chromatin, whereas a fusion containing the Ile44Ala mutation in ubiquitin did not. Because most ubiquitin-binding domains require direct binding to Ile44 on ubiquitin, this result raises the possibility that a chromatin-bound ubiquitin receptor mediates ubiquitylated FancD2 and FancI localization (Matsushita et al., 2005).
PIKK phosphorylation coupled to E3 ubiquitylation of tumor-suppressor repair proteins appears to be evolutionarily conserved. The breast cancer early onset 1 gene product, BRCA1, is a RING domain E3 ligase that pairs with its stoichiometric binding partner, the RING domain protein BARD1. Approximately 20% of the clinical missense mutations to BRCA1 occur in the RING domain and disrupt its E3 ligase activity (Brzovic et al., 2001). BRCA1-BARD1 heterodimers localize at DSBs and are necessary for a host of DNA damage response activities including homologous recombination and checkpoint activity. DNA damage induces an ATM and ATR kinase–dependent activation of BRCA1-BARD1 E3 ligase activity on chromatin in Caenorhabditis elegans and in human cells (Polanowska et al., 2006). The BRCA1 RING domain interaction with the E2 enzyme Ubch5c is necessary for autoubiquitylation via K6-linked ubiquitin in vitro (Wu-Baer et al., 2003; Nishikawa et al., 2004). K6-Ub foci are present at DSBs in both a BRCA1- and Ubch5c-dependent manner (Morris and Solomon, 2004; Polanowska et al., 2006), which suggests that BRCA1 deposits K6-Ub chains on proteins at repair sites to execute at least a portion of its myriad DNA repair activities. Notably, these experiments were primarily performed with ubiquitin mutants that contain a single lysine residue, and it is currently unclear how K6-Ub influences recognition and signaling processes. It will be important to readdress these findings upon development of K6-Ub–specific antibodies to more definitively understand the kinetics and relationship of K6-Ub to BRCA1 DNA repair activities.
The identification of BRCA1-BARD1 E3 substrates will be critical to understand how BRCA1 ligase activity contributes to the DNA damage response. In this regard, BRCA1 mediated, DNA damage–inducible ubiquitylation of the BRCA1 carboxy-terminal interacting partner, CtBP-interacting protein (CtIP), was reported in human cells (Yu et al., 2006). Interestingly, BRCA1 RING mutations that disrupt E3 ligase activity or CtIP deficiency failed to support CHK1 phosphorylation via ATR, resulting in a defective ionizing radiation–induced G2 checkpoint. BRCA1 E3 activity and interaction with CtIP was necessary for CtIP ubiquitylation and IRIF formation. As with the case of FancD2 and FancI ubiquitylation, mechanisms responsible for CtIP-Ub foci formation are not presently understood.
Evidence for ubiquitin recognition at DSBs.
The concept of retention of DNA repair proteins at DSBs by ubiquitin receptors was finally validated in studies that revealed a molecular basis for BRCA1 DSB recruitment. BRCA1 is targeted to DSBs via an interaction with a five-component complex containing RAP80, a DNA repair protein that contains tandem ubiquitin interaction motifs (UIMs; Kim et al., 2007a; Sobhian et al., 2007; Wang et al., 2007; Yan et al., 2007). These UIM domains preferentially recognize K63-linked ubiquitin over K48-linked structures, which suggests that BRCA1 would be targeted to nondegradative ubiquitin signals (Sobhian et al., 2007). Indeed, K63-linked structures accumulate in DSB foci, whereas K48-linked chains do not (Sobhian et al., 2007; Doil et al., 2009; Stewart et al., 2009). RAP80 DSB localization requires γ-H2AX and MDC1, both of which are necessary for polyubiquitylation at DSBs. Interestingly, an in-frame deletion in RAP80 UIM1 is associated with breast cancer in northern Finnish populations (Nikkilä et al., 2009). This RAP80 variant, RAP80ΔE81, demonstrates reduced ubiquitin binding and DSB localization while maintaining all other protein interactions. Because of these properties, RAP80ΔE81 functions as a dominant-negative allele by titrating BRCA1 away from DSBs.
RAP80 exists as a stable complex together with four other core components: BRCC36, BRCC45, Abraxas, and MERIT40/NBA1 (Feng et al., 2009; Shao et al., 2009b; Wang et al., 2009). Bioinformatic analysis suggests similarity of the RAP80 core complex to the lid domain within the 19S subunit of the 26S proteasome (Wang et al., 2009). BRCC36 is a member of the JAMM family of metalloprotease DUB enzymes, and mutation of the zinc-binding residues results in increased sensitivity to DSBs, an impaired G2 checkpoint, and elevated levels of conjugated ubiquitin at DSBs (Shao et al., 2009a,b). This JAMM domain DUB specifically deubiquitylates K63-Ub, which matches the ubiquitin-binding preference of the RAP80 UIM domains (Cooper et al., 2009; Shao et al., 2009a). Generally, knockdown of any of the members of the complex results in failure of the other constituents to form DSB-associated foci, and increased sensitivity to ionizing radiation. Although structural evidence is needed to determine the actual degree of similarity to the proteasome, it is intriguing that multiple ubiquitin-binding domains would be present within the RAP80 complex (Wang et al., 2009). Perhaps these domains regulate BRCC36 catalytic activity or, alternatively, modulate the avidity of the complex for ubiquitin chains at DSBs. In vitro reconstitution of this complex with recombinant proteins will be necessary to definitively address these possibilities.
PIKK activity has been linked to histone ubiquitylation, the putative DSB-associated ligand for the BRCA1–RAP80 complex (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Zhao et al., 2007). As mentioned earlier, γ-H2AX serves as a signal to initiate recruitment of additional factors to DNA repair foci. Chief among them is the mediator of DNA damage checkpoint 1 (MDC1). ATM phosphorylates MDC1 at an SQ/TQ-rich region, which is subsequently recognized by the E3 ubiquitin ligase RNF8, beginning the transition from primarily a phosphorylation cascade at DSBs to a series of ubiquitin-dependent signaling events. RNF8 works in conjunction with Ubc13 to ubiquitylate histones H2A and γ-H2AX. The RIDDLE syndrome E3 ligase RNF168 recognizes ubiquitin chains at DSBs synthesized by RNF8 through its two motif interacting with ubiquitin (MIU) domains which represent an inverted UIM (Doil et al., 2009; Stewart et al., 2009). RNF168, like RNF8, associates with Ubc13 to direct ubiquitylated H2A and K63-Ub synthesis at DSBs. RNF8 or RNF168 deficiency abrogates BRCA1–RAP80 foci formation, providing additional supportive evidence that ubiquitin is a DSB-targeting mechanism for the BRCA1–RAP80 complex.
RAP80 is not the only DNA repair protein that requires the combined efforts of RNF8 and RNF168 together with Ubc13 for DSB localization (Huang et al., 2009). The chromatin-bound DNA repair protein, p53-binding protein 1 (53BP1), also depends on RNF8 and RNF168 DSB-associated ubiquitylation, yet no evidence exists that 53BP1 directly binds ubiquitin. Instead, it appears to recognize methylated histones at DSBs (Huyen et al., 2004; Sanders et al., 2004; Botuyan et al., 2006). These results suggest that DSB-associated ubiquitin influences chromatin structure in a manner necessary to unveil modified histone epitopes. The current data are therefore consistent with ubiquitylation of DSB-associated proteins providing direct and indirect routes to damage site recognition for DNA repair proteins.
Structural basis for RAP80 K63-Ub specificity: a paradigm for DSB-associated ubiquitin recognition
The structural basis accounting for the K63-Ub specificity of the RAP80 tandem UIM domains was delineated in two recent papers (Sato et al., 2009; Sims and Cohen, 2009). Sims and Cohen (2009) measured the binding affinity for ubiquitin of the UIM1 and UIM2 to be quite modest (230 µM and 470 µM, respectively). The binding affinity increased >10-fold (17–22 µM) when the tandem UIM domains were measured for the ability to bind K63-linked diubiquitin. Mutational analysis showed that the linker region between the two UIMs was important for binding K63-linked polyubiquitin. Sato et al., (2009) recently described the mouse RAP80 tandem UIM domain structure bound to K63-linked diubiquitin (Fig. 2 A). The structure reveals that these domains consists of a single 60-Å long α-helix (Sato et al., 2009). Interestingly, the inter-UIM region adopts an α-helical secondary structure, as predicted by Sims and Cohen (2009). Each UIM domain binds to the hydrophobic patch surrounding Ile44 of the respective ubiquitin. The torsional freedom of K63-linked diubiquitin about the carboxy-terminal axis of the distal ubiquitin is evident when one compares the RAP80 bound and unbound structures. In the unbound structures (Datta et al., 2009; Komander et al., 2009), the Ile44 hydrophobic patches are rotated ∼100° from each other. Upon binding to RAP80, the hydrophobic patches of the two ubiquitins align along one face rotated just ∼10° from one another (Fig. 2 B; Sato et al., 2009). Combining the structural information with the mutational analysis reveals the inter-UIM region as more than just a linker between the two UIM domains. It serves as a molecular ruler measuring the distance between K63-linked ubiquitin and orients the hydrophobic regions to lie on one side. These findings led to a model of linkage-specific avidity, in which linker region orientation of the UIM domains can dictate specificity for different ubiquitin chain topologies (Sims and Cohen, 2009).
Figure 2.

Structural basis for the specificity of RAP80 binding to ubiquitin. (A) RAP80 UIM1 and UIM2 bound to K63-diubiquitin. The UIM domains of RAP80 (magenta) recognize the I44 hydrophobic patches (green) of ubiquitin. The inter-UIM region (pink) adopts an α-helical fold. (Sato et al., 2009) (B) Comparison of the RAP80 bound and unbound forms of K63-linked diubiquitin. The distal ubiquitin was superimposed to show differences in the proximal ubiquitin between bound (yellow) and unbound (pale blue) of K63-linked diubiquitin. RAP80 induces an ∼45° rotation about the carboxy-terminal region glycine–glycine axis of K63-linked diubiquitin (Komander et al., 2009; Sato et al., 2009).
Evidence for a heterogeneous ubiquitin landscape at DSBs
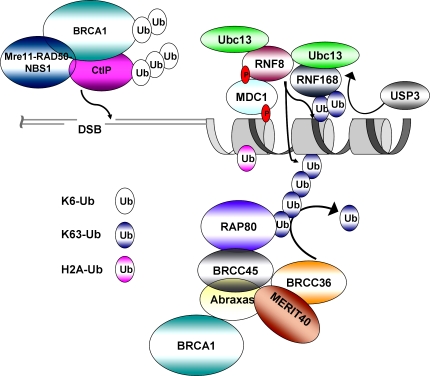
Several lines of evidence argue against a homogeneous ubiquitin response at DSBs (Fig. 3). The most compelling of which is that (1) different types of ubiquitin linkages have been detected at DSBs, and (2) that repair proteins that bind to ubiquitin display different kinetics of DSB foci formation. Comparison of RNF168 and RAP80 foci formation strongly suggests differential ubiquitin recognition at DSBs. Like RAP80, recruitment of RNF168 to DSBs is dependent on its ubiquitin-binding domains and RNF8. RNF168 DSB association appears before BRCA1–RAP80 and is necessary for both RAP80 and 53BP1 focus formation (Doil et al., 2009; Stewart et al., 2009). Expression of a RING domain RNF168 mutant revealed that it does not require its own catalytic activity for retention at DSBs (Doil et al., 2009). RNF8 E3 activity is thus sufficient for RNF168 DSB retention but insufficient for recruiting either RAP80 or 53BP1, again supporting the argument that the RAP80 UIM domains differ in their DSB recognition from RNF168 MIUs. The RAP80 UIMs selectively recognize K63- and K6-linked ubiquitin but exhibit much lower affinity for either monoubiquitin or K48-linked ubiquitin (Sobhian et al., 2007). It is thus unlikely that the increase in mono-ubiquitylated H2A is what attracts RAP80; the more likely culprit is K63-ubiquitin chains at DSBs. RAP80 specificity for K63-linked ubiquitin is encoded in part via a linker region between each UIM (Sims and Cohen, 2009). The RNF168 MIU domains have a much larger intervening sequence, and although in vitro ubiquitin binding to K63-Ub has been described (Penengo et al., 2006), the exact configuration of ubiquitin that facilitates RNF168 recruitment to sites of DNA damage still remains to be investigated.
Figure 3.
Model for differential ubiquitin-related recognition and repair activities at DSBs. Ubiquitin chains of differing topologies, as indicated, covalently linked to different substrate proteins create a varied ubiquitin landscape at DSBs. Differential recognition of this ubiquitin environment by DNA repair proteins targets repair and checkpoint activities to the appropriate location adjacent to DSBs.
The balance between E3 ligase and DUB activities has the potential to dynamically alter the ubiquitin landscape at DSBs (Fig. 3; Greenberg, 2008). BRCC36 demonstrates K63-specific DUB activity, failing to hydrolyze either K48 or K6-Ub chains, or monoubiquitylated proteins. BRCC36 is involved in a negative feedback pathway that controls the breakdown of K63-polyubiquitin at DSBs (Shao et al., 2009a). There are also several other DUBs that function at DSBs. Unlike BRCC36, these DUBs are cysteine proteases and do not accumulate in DSB foci. USP1 deubiquitylates FancD2-Ub, and USP3 is thought to act on uH2A (Nijman et al., 2005; Mailand et al., 2007; Nicassio et al., 2007). Other DUBs have been found to interact with proteins found at DSBs, including USP28 that stabilizes 53BP1, (Zhang et al., 2006) and STAMBP and OtuB1, which interact with Ubc13 (Sowa et al., 2009). Given the different substrate specificities, DUB regulation may have an impact on the relative ratios of different ubiquitin linkages at DNA lesions. This phenomenon is exemplified by the A20 protein, which remarkably contains both a DUB domain and an E3 ligase domain. With its DUB domain, A20 is able to cleave K63-linked polyubiquitin chains and, through its E3 ligase domain, is able to promote the K48 ubiquitylation of the same substrate (Newton et al., 2008). In other words, A20 possesses the ability to “edit” the topology of ubiquitin chains. Although A20 has not been reported at DSBs and may be unique in containing both activities in a single polypeptide chain, one can imagine that ubiquitin-editing activities may be performed by different members of the same complex. For example, the BRCA1–RAP80 complex contains both BRCC36-encoded K63-specific DUB activity and BRCA1-derived K6-specific E3 ligase activity, raising the possibility of ubiquitin editing activities at DSBs.
Ubiquitin dynamics and DNA repair mechanism
There is considerable evidence that ubiquitylation influences DNA repair mechanism. DSBs are primarily repaired by either homologous or nonhomologous methods. Each of these employs distinct DNA processing events. A pivotal step distinguishing the two repair mechanisms is the 5′-to-3′ end resection of DSB ends. Zhao et al. (2007) reported an interesting observation that Ubc13 is required for homologous recombination and that Ubc13-deficient cells lack wild-type levels of resection at a break, as detected by single-stranded binding replication protein A recruitment to laser-induced DSBs (Zhao et al., 2007). Ubc13 deficiency eliminated detectable ubiquitin DSB focus formation and conferred exquisite sensitivity to agents that induce DNA damage that requires homologous recombination for repair. These data implicate DSB-associated ubiquitin in the initial stages of repair.
Why Ubc13 is necessary for DSB end resection is not known; however, BRCA1 DSB association and E3 ligase activity were dramatically reduced in Ubc13-null cells (Zhao et al., 2007). This data raises the possibility of a connection between Ubc13 and BRCA1-dependent ubiquitylation and end resection. Central to the process of DSB end resection is the interaction of CtIP with BRCA1 and Mre11. The BRCA1–CtIP complex interacts with Mre11 in a DNA damage–inducible manner (Greenberg et al., 2006; Chen et al., 2008), and this supercomplex has been implicated in nucleolytic degradation of double-stranded termini (Fig. 3; Sartori et al., 2007; Yun and Hiom, 2009). As previously discussed, CtIP is polyubiquitylated after DNA damage in a manner dependent on its interaction with BRCA1 (Yu et al., 2006). CtIP ubiquitylation is correlated with its ability to form IRIF and to activate CHK1. Because CHK1 activation requires ATR activation on single-stranded DNA (Zou and Elledge, 2003), it is tempting to speculate that BRCA1 teams up with Ubc13 to ubiquitylate CtIP, thus activating CtIP–Mre11 complex nuclease activity for end resection. The creation of a clean, genetic system to investigate BRCA1 E3 activity has severely injured this hypothesis. BRCA1 RING domain I26A mutant knock-in embryonic stem cells have recently been made (Reid et al., 2008). This allele abrogates BRCA1 RING domain interaction with E2 enzymes (Brzovic et al., 2003; Christensen et al., 2007), strongly reducing BRCA1 E3 activity in vitro and autoubiquitylation in vivo (Reid et al., 2008). Surprisingly, BRCA1 I26A knock-in ES cells performed homologous recombination at similar levels to cells expressing wild-type BRCA1 (Reid et al., 2008). These findings are inconsistent with BRCA1 E3 activity being involved in end resection and instead are more supportive of Ubc13 working in conjunction with other E3 ligases (e.g., RNF8 and RNF168) to concentrate BRCA1–CtIP and other resection-promoting factors at DSBs. Perhaps CtIP ubiquitylation by BRCA1 is not essential for end resection and instead influences CHK1 phosphorylation by other means. An alternative explanation is that CtIP does not require BRCA1 in mouse ES cells for ubiquitylation and that a different E3 suffices in this cell type.
Concluding remarks
Nondegradative forms of ubiquitin have, quite literally, left their mark on the DNA damage response. A variety of repair protein substrates, E3 ligases, and DUBs, each with their own specificity for synthesizing, recognizing, or hydrolyzing ubiquitin chains, appear to make important contributions to DNA repair. These initial studies have created new opportunities to understand the DNA damage response, and perhaps additional pharmacologic opportunities for treating human disease.
Acknowledgments
R.A. Greenberg would like to gratefully acknowledge funding from: K08 award 1K08CA106597-01 from the National Cancer Institute, an American Cancer Society Research Scholar Grant, the Sidney Kimmel Foundation Scholar Award, the Mary Kay Ash Foundation Translational Innovation Award, and funds from the Abramson Family Cancer Research Institute. Troy E. Messick was supported by funds from the Center of Excellence in Environmental Toxicology and the University Research Foundation of the University of Pennsylvania.
Footnotes
Abbreviations used in this paper:
- ATM
- ataxia telangiectasia mutated
- ATR
- ataxia telangiectasia and Rad3 related
- CtIP
- CtBP-interacting protein
- DSB
- double-strand break
- DUB
- deubiquitinating enzyme
- IRIF
- ionizing radiation–induced foci
- MIU
- motif interacting with ubiquitin
- PIKK
- phosphatidylinositol-3-kinase-related kinase
- UIM
- ubiquitin interaction motif
References
- Al-Hakim A.K., Zagorska A., Chapman L., Deak M., Peggie M., Alessi D.R. 2008. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem. J. 411:249–260 10.1042/BJ20080067 [DOI] [PubMed] [Google Scholar]
- Andreassen P.R., D’Andrea A.D., Taniguchi T. 2004. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 18:1958–1963 10.1101/gad.1196104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M.B., Bartek J., Lukas J. 2006. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173:195–206 10.1083/jcb.200510130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E., Monnat R.J., Jr., Kastan M.B. 2007. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 9:683–690 10.1038/ncb1599 [DOI] [PubMed] [Google Scholar]
- Botuyan M.V., Lee J., Ward I.M., Kim J.E., Thompson J.R., Chen J., Mer G. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 127:1361–1373 10.1016/j.cell.2006.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic P.S., Meza J.E., King M.C., Klevit R.E. 2001. BRCA1 RING domain cancer-predisposing mutations. Structural consequences and effects on protein-protein interactions. J. Biol. Chem. 276:41399–41406 10.1074/jbc.M106551200 [DOI] [PubMed] [Google Scholar]
- Brzovic P.S., Keeffe J.R., Nishikawa H., Miyamoto K., Fox D., III, Fukuda M., Ohta T., Klevit R. 2003. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. USA. 100:5646–5651 10.1073/pnas.0836054100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A., Petersen S., Romanienko P.J., Fernandez-Capetillo O., Chen H.T., Sedelnikova O.A., Reina-San-Martin B., Coppola V., Meffre E., Difilippantonio M.J., et al. 2002. Genomic instability in mice lacking histone H2AX. Science. 296:922–927 10.1126/science.1069398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A., Fernandez-Capetillo O., Kruhlak M.J., Pilch D.R., Staudt D.W., Lee A., Bonner R.F., Bonner W.M., Nussenzweig A. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675–679 10.1038/ncb1004 [DOI] [PubMed] [Google Scholar]
- Chen Z.J., Sun L.J. 2009. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell. 33:275–286 10.1016/j.molcel.2009.01.014 [DOI] [PubMed] [Google Scholar]
- Chen L., Nievera C.J., Lee A.Y., Wu X. 2008. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 283:7713–7720 10.1074/jbc.M710245200 [DOI] [PubMed] [Google Scholar]
- Chenevix-Trench G., Spurdle A.B., Gatei M., Kelly H., Marsh A., Chen X., Donn K., Cummings M., Nyholt D., Jenkins M.A., et al. 2002. Dominant negative ATM mutations in breast cancer families. J. Natl. Cancer Inst. 94:205–215 [DOI] [PubMed] [Google Scholar]
- Christensen D.E., Brzovic P.S., Klevit R.E. 2007. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 14:941–948 10.1038/nsmb1295 [DOI] [PubMed] [Google Scholar]
- Cooper E.M., Cutcliffe C., Kristiansen T.Z., Pandey A., Pickart C.M., Cohen R.E. 2009. K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 28:621–631 10.1038/emboj.2009.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A.B., Hura G.L., Wolberger C. 2009. The structure and conformation of Lys63-linked tetraubiquitin. J. Mol. Biol. 392:1117–1124 10.1016/j.jmb.2009.07.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 136:435–446 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- Eddins M.J., Varadan R., Fushman D., Pickart C.M., Wolberger C. 2007. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J. Mol. Biol. 367:204–211 10.1016/j.jmb.2006.12.065 [DOI] [PubMed] [Google Scholar]
- Feng L., Huang J., Chen J. 2009. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 23:719–728 10.1101/gad.1770609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M.S., Timmers C., Hejna J., Grompe M., D’Andrea A.D. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 7:249–262 10.1016/S1097-2765(01)00173-3 [DOI] [PubMed] [Google Scholar]
- Grabbe C., Dikic I. 2009. Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem. Rev. 109:1481–1494 10.1021/cr800413p [DOI] [PubMed] [Google Scholar]
- Greenberg R.A. 2008. Recognition of DNA double strand breaks by the BRCA1 tumor suppressor network. Chromosoma. 117:305–317 10.1007/s00412-008-0154-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg R.A., Sobhian B., Pathania S., Cantor S.B., Nakatani Y., Livingston D.M. 2006. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 20:34–46 10.1101/gad.1381306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K., Dikic I. 2005. Ubiquitylation and cell signaling. EMBO J. 24:3353–3359 10.1038/sj.emboj.7600808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.W., Elledge S.J. 2007. The DNA damage response: ten years after. Mol. Cell. 28:739–745 10.1016/j.molcel.2007.11.015 [DOI] [PubMed] [Google Scholar]
- Harper J.W., Schulman B.A. 2006. Structural complexity in ubiquitin recognition. Cell. 124:1133–1136 10.1016/j.cell.2006.03.009 [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A., Varshavsky A. 2000. The ubiquitin system. Nat. Med. 6:1073–1081 10.1038/80384 [DOI] [PubMed] [Google Scholar]
- Huang J., Huen M.S., Kim H., Leung C.C., Glover J.N., Yu X., Chen J. 2009. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell Biol. 11:592–603 10.1038/ncb1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen M.S., Chen J. 2008. The DNA damage response pathways: at the crossroad of protein modifications. Cell Res. 18:8–16 10.1038/cr.2007.109 [DOI] [PubMed] [Google Scholar]
- Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 131:901–914 10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. 2007. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell. 28:730–738 10.1016/j.molcel.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Huyen Y., Zgheib O., Ditullio R.A., Jr., Gorgoulis V.G., Zacharatos P., Petty T.J., Sheston E.A., Mellert H.S., Stavridi E.S., Halazonetis T.D. 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 432:406–411 10.1038/nature03114 [DOI] [PubMed] [Google Scholar]
- Kim H., Chen J., Yu X. 2007a. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 316:1202–1205 10.1126/science.1139621 [DOI] [PubMed] [Google Scholar]
- Kim H.T., Kim K.P., Lledias F., Kisselev A.F., Scaglione K.M., Skowyra D., Gygi S.P., Goldberg A.L. 2007b. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282:17375–17386 10.1074/jbc.M609659200 [DOI] [PubMed] [Google Scholar]
- Kolas N.K., Chapman J.R., Nakada S., Ylanko J., Chahwan R., Sweeney F.D., Panier S., Mendez M., Wildenhain J., Thomson T.M., et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 318:1637–1640 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Reyes-Turcu F., Licchesi J.D., Odenwaelder P., Wilkinson K.D., Barford D. 2009. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 10:466–473 10.1038/embor.2009.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Mortensen U.H., Rothstein R. 2003. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 5:572–577 10.1038/ncb997 [DOI] [PubMed] [Google Scholar]
- Lisby M., Barlow J.H., Burgess R.C., Rothstein R. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 118:699–713 10.1016/j.cell.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 131:887–900 10.1016/j.cell.2007.09.040 [DOI] [PubMed] [Google Scholar]
- Matsushita N., Kitao H., Ishiai M., Nagashima N., Hirano S., Okawa K., Ohta T., Yu D.S., McHugh P.J., Hickson I.D., et al. 2005. A FancD2-monoubiquitin fusion reveals hidden functions of Fanconi anemia core complex in DNA repair. Mol. Cell. 19:841–847 10.1016/j.molcel.2005.08.018 [DOI] [PubMed] [Google Scholar]
- Meetei A.R., Yan Z., Wang W. 2004. FANCL replaces BRCA1 as the likely ubiquitin ligase responsible for FANCD2 monoubiquitination. Cell Cycle. 3:179–181 [PubMed] [Google Scholar]
- Morris J.R., Solomon E. 2004. BRCA1 : BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum. Mol. Genet. 13:807–817 10.1093/hmg/ddh095 [DOI] [PubMed] [Google Scholar]
- Newton K., Matsumoto M.L., Wertz I.E., Kirkpatrick D.S., Lill J.R., Tan J., Dugger D., Gordon N., Sidhu S.S., Fellouse F.A., et al. 2008. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 134:668–678 10.1016/j.cell.2008.07.039 [DOI] [PubMed] [Google Scholar]
- Nicassio F., Corrado N., Vissers J.H., Areces L.B., Bergink S., Marteijn J.A., Geverts B., Houtsmuller A.B., Vermeulen W., Di Fiore P.P., Citterio E. 2007. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 17:1972–1977 10.1016/j.cub.2007.10.034 [DOI] [PubMed] [Google Scholar]
- Nijman S.M., Huang T.T., Dirac A.M., Brummelkamp T.R., Kerkhoven R.M., D’Andrea A.D., Bernards R. 2005. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell. 17:331–339 10.1016/j.molcel.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Nikkilä J., Coleman K.A., Morrissey D., Pylkäs K., Erkko H., Messick T.E., Karppinen S.M., Amelina A., Winqvist R., Greenberg R.A. 2009. Familial breast cancer screening reveals an alteration in the RAP80 UIM domain that impairs DNA damage response function. Oncogene. 28:1843–1852 10.1038/onc.2009.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa H., Ooka S., Sato K., Arima K., Okamoto J., Klevit R.E., Fukuda M., Ohta T. 2004. Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J. Biol. Chem. 279:3916–3924 10.1074/jbc.M308540200 [DOI] [PubMed] [Google Scholar]
- Pawson T., Kofler M. 2009. Kinome signaling through regulated protein-protein interactions in normal and cancer cells. Curr. Opin. Cell Biol. 21:147–153 10.1016/j.ceb.2009.02.005 [DOI] [PubMed] [Google Scholar]
- Penengo L., Mapelli M., Murachelli A.G., Confalonieri S., Magri L., Musacchio A., Di Fiore P.P., Polo S., Schneider T.R. 2006. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 124:1183–1195 10.1016/j.cell.2006.02.020 [DOI] [PubMed] [Google Scholar]
- Peng J., Schwartz D., Elias J.E., Thoreen C.C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S.P. 2003. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21:921–926 10.1038/nbt849 [DOI] [PubMed] [Google Scholar]
- Phillips C.L., Thrower J., Pickart C.M., Hill C.P. 2001. Structure of a new crystal form of tetraubiquitin. Acta Crystallogr. D Biol. Crystallogr. 57:341–344 10.1107/S090744490001800X [DOI] [PubMed] [Google Scholar]
- Pickart C.M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503–533 10.1146/annurev.biochem.70.1.503 [DOI] [PubMed] [Google Scholar]
- Pickart C.M., Cohen R.E. 2004. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5:177–187 10.1038/nrm1336 [DOI] [PubMed] [Google Scholar]
- Polanowska J., Martin J.S., Garcia-Muse T., Petalcorin M.I., Boulton S.J. 2006. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. EMBO J. 25:2178–2188 10.1038/sj.emboj.7601102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J., Snyder M. 2006. Charging it up: global analysis of protein phosphorylation. Trends Genet. 22:545–554 10.1016/j.tig.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Reid L.J., Shakya R., Modi A.P., Lokshin M., Cheng J.T., Jasin M., Baer R., Ludwig T. 2008. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc. Natl. Acad. Sci. USA. 105:20876–20881 10.1073/pnas.0811203106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. 2009. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78:363–397 10.1146/annurev.biochem.78.082307.091526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S.L., Portoso M., Mata J., Bähler J., Allshire R.C., Kouzarides T. 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 119:603–614 10.1016/j.cell.2004.11.009 [DOI] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. 2007. Human CtIP promotes DNA end resection. Nature. 450:509–514 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Yoshikawa A., Mimura H., Yamashita M., Yamagata A., Fukai S. 2009. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 28:2461–2468 10.1038/emboj.2009.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz A.L., Ciechanover A. 2009. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu. Rev. Pharmacol. Toxicol. 49:73–96 10.1146/annurev.pharmtox.051208.165340 [DOI] [PubMed] [Google Scholar]
- Scully R., Livingston D.M. 2000. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 408:429–432 10.1038/35044000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao G., Lilli D.R., Patterson-Fortin J., Coleman K.A., Morrissey D.E., Greenberg R.A. 2009a. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc. Natl. Acad. Sci. USA. 106:3166–3171 10.1073/pnas.0807485106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao G., Patterson-Fortin J., Messick T.E., Feng D., Shanbhag N., Wang Y., Greenberg R.A. 2009b. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 23:740–754 10.1101/gad.1739609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R., Arbel-Eden A., Pilch D., Ira G., Bonner W.M., Petrini J.H., Haber J.E., Lichten M. 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 14:1703–1711 10.1016/j.cub.2004.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims A.E., Spiteri E., Sims R.J., III, Arita A.G., Lach F.P., Landers T., Wurm M., Freund M., Neveling K., Hanenberg H., et al. 2007. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 14:564–567 10.1038/nsmb1252 [DOI] [PubMed] [Google Scholar]
- Sims J.J., Cohen R.E. 2009. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol. Cell. 33:775–783 10.1016/j.molcel.2009.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E.R., III, Hurov K.E., Luo J., Ballif B.A., Gygi S.P., Hofmann K., D’Andrea A.D., Elledge S.J. 2007. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 129:289–301 10.1016/j.cell.2007.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B., Shao G., Lilli D.R., Culhane A.C., Moreau L.A., Xia B., Livingston D.M., Greenberg R.A. 2007. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 316:1198–1202 10.1126/science.1139516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa M.E., Bennett E.J., Gygi S.P., Harper J.W. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell. 138:389–403 10.1016/j.cell.2009.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G.S., Panier S., Townsend K., Al-Hakim A.K., Kolas N.K., Miller E.S., Nakada S., Ylanko J., Olivarius S., Mendez M., et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 136:420–434 10.1016/j.cell.2008.12.042 [DOI] [PubMed] [Google Scholar]
- Varadan R., Assfalg M., Haririnia A., Raasi S., Pickart C., Fushman D. 2004. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J. Biol. Chem. 279:7055–7063 10.1074/jbc.M309184200 [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar S., Bugg C.E., Cook W.J. 1987. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 194:531–544 10.1016/0022-2836(87)90679-6 [DOI] [PubMed] [Google Scholar]
- Wang B., Matsuoka S., Ballif B.A., Zhang D., Smogorzewska A., Gygi S.P., Elledge S.J. 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 316:1194–1198 10.1126/science.1139476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Hurov K., Hofmann K., Elledge S.J. 2009. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 23:729–739 10.1101/gad.1770309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. 2007. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 8:735–748 10.1038/nrg2159 [DOI] [PubMed] [Google Scholar]
- Wang X., Andreassen P.R., D’Andrea A.D. 2004. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell. Biol. 24:5850–5862 10.1128/MCB.24.13.5850-5862.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Baer F., Lagrazon K., Yuan W., Baer R. 2003. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J. Biol. Chem. 278:34743–34746 10.1074/jbc.C300249200 [DOI] [PubMed] [Google Scholar]
- Xu P., Duong D.M., Seyfried N.T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. 2009. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 137:133–145 10.1016/j.cell.2009.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Kim Y.S., Yang X.P., Li L.P., Liao G., Xia F., Jetten A.M. 2007. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 67:6647–6656 10.1158/0008-5472.CAN-07-0924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Fu S., Lai M., Baer R., Chen J. 2006. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 20:1721–1726 10.1101/gad.1431006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M.H., Hiom K. 2009. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 459:460–463 10.1038/nature07955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Zaugg K., Mak T.W., Elledge S.J. 2006. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 126:529–542 10.1016/j.cell.2006.06.039 [DOI] [PubMed] [Google Scholar]
- Zhao G.Y., Sonoda E., Barber L.J., Oka H., Murakawa Y., Yamada K., Ikura T., Wang X., Kobayashi M., Yamamoto K., et al. 2007. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell. 25:663–675 10.1016/j.molcel.2007.01.029 [DOI] [PubMed] [Google Scholar]
- Zou L., Elledge S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 300:1542–1548 10.1126/science.1083430 [DOI] [PubMed] [Google Scholar]