Multifunctional protein Rif1 accumulates at stalled replication forks to facilitate DNA repair during S phase.
Abstract
Rif1, originally recognized for its role at telomeres in budding yeast, has been implicated in a wide variety of cellular processes in mammals, including pluripotency of stem cells, response to double-strand breaks, and breast cancer development. As the molecular function of Rif1 is not known, we examined the consequences of Rif1 deficiency in mouse cells. Rif1 deficiency leads to failure in embryonic development, and conditional deletion of Rif1 from mouse embryo fibroblasts affects S-phase progression, rendering cells hypersensitive to replication poisons. Rif1 deficiency does not alter the activation of the DNA replication checkpoint but rather affects the execution of repair. RNA interference to human Rif1 decreases the efficiency of homology-directed repair (HDR), and Rif1 deficiency results in aberrant aggregates of the HDR factor Rad51. Consistent with a role in S-phase progression, Rif1 accumulates at stalled replication forks, preferentially around pericentromeric heterochromatin. Collectively, these findings reveal a function for Rif1 in the repair of stalled forks by facilitating HDR.
Introduction
In mammalian cells, two major checkpoints ensure the fidelity of DNA replication. The intra–S phase checkpoint is elicited by exogenously inflicted DNA damage that creates double-strand breaks (DSBs) and is controlled by the PI3-like kinase ataxia telangiectasia mutated (ATM). ATM can be activated in every phase of the cell cycle, including S phase, and the main signal transducer is Chk2 (for review see Cimprich and Cortez, 2008). Neither Chk2 nor ATM are essential genes (Barlow et al., 1996; Elson et al., 1996; Xu et al., 1996; Hirao et al., 2002; Takai et al., 2002). The second and essential checkpoint (hitherto referred as the DNA replication checkpoint) monitors the process of DNA replication itself and is activated by single-stranded DNA (ssDNA) accumulated at stalled replication forks. The activation of the checkpoint depends on another PI3-like kinase, ataxia telangiectasia and Rad3 related (ATR), and its main signal transducer is Chk1 (for review see Cimprich and Cortez, 2008). Errors in DNA replication are the main source of endogenous DNA damage, and genes required for the DNA replication checkpoint are essential (Liu et al., 1994; Brown and Baltimore, 2000; Takai et al., 2000; Weiss et al., 2000; Yamane et al., 2002; Budzowska et al., 2004; Hopkins et al., 2004). Activation of the DNA replication checkpoint arrests the cell cycle and activates the homologous recombination pathway, which mediates the restart of the arrested replication fork (for review see Lambert et al., 2007).
We have previously shown that Rif1 participates in the intra–S phase checkpoint, contributing to the inhibition of DNA replication associated with the activation of ATM (Silverman et al., 2004). Human Rif1 localizes to DSBs induced by a variety of clastogenic agents but not to DNA lesions generated by UV radiation. This association with DSBs is dependent on the activation of the ATM kinase and the DNA damage response factor 53BP1 (Schultz et al., 2000; Silverman et al., 2004).
Rif1 was originally identified in budding yeast based on its ability to interact with Rap1, a protein which plays a key role at telomeres (Hardy et al., 1992). By tethering Rif1 and a second Rap1-interacting factor, Rif2, Rap1 can limit the action of telomerase in cis and thus establish telomere length homeostasis (Marcand et al., 1997; Levy and Blackburn, 2004; Bianchi and Shore, 2008). Rif2 is a diverged version of ORC4 that can bind to Rif1 as well as Rap1 (Wotton and Shore, 1997; Marcand et al., 2008). No other Rif1 interacting partners have been identified in yeast, and the mechanism by which Rif1 enforces the inhibition of telomerase has not been established. Orthologues of Rif1 and Rap1 (but not Rif2) have been recognized in Schizosaccharomyces pombe and in vertebrates (Li et al., 2000; Kanoh and Ishikawa, 2001; Li and de Lange, 2003; Adams and McLaren, 2004; Silverman et al., 2004). Although fission yeast Rif1 does not bind Rap1, it does interact with telomeres and contribute to telomere length homeostasis (Kanoh and Ishikawa, 2001).
So far there has been no indication of a conserved role for yeast and mammalian Rif1 orthologues. Mammalian Rif1 does not appear to bind normal telomeres nor to have a role in telomere homeostasis (Silverman et al., 2004; Xu and Blackburn, 2004; this study). Although rif1Δ budding yeast looses chromosomes at a slightly increased rate (Wotton and Shore, 1997; Banerjee and Myung, 2004), Rif1 deficiency does not affect the rate of gross chromosomal rearrangements (Myung et al., 2001), which are largely caused by the repair of S-phase damage. Similarly, there is no data indicating that fission yeast Rif1 plays a prominent role in the response to S-phase damage.
In addition to regulation of telomere maintenance in budding yeast and response to DSBs in human cells, Rif1 has also been implicated in transcriptional silencing at yeast telomeres and ribosomal DNA (Hardy et al., 1992; Smith et al., 1999; Teng et al., 2000; Chan et al., 2001; Silverman et al., 2004; Teixeira et al., 2004) and in controlling mouse embryonic stem (ES) cells’ identity (Loh et al., 2006; Wang et al., 2006; Fazzio et al., 2008; Liang et al., 2008). Rif1 has no informative protein motifs or interacting partners that could point to the molecular mechanism by which Rif1 contributes to these different pathways.
To gain further insight into the function of this enigmatic protein, we targeted Rif1 in the mouse, generating a setting in which the consequences of the absence of Rif1 could be studied. Our data reveal a new role for mammalian Rif1 in the context of the repair machinery that is mobilized by the DNA replication checkpoint.
Results
Rif1 deficiency impairs embryonic development
To determine the consequences of Rif1 deficiency, we initially used a mouse ES cell line from a gene trap collection (Hansen et al., 2003) in which a pT1β-GEO vector was integrated between exons 21 and 22 of the Rif1 gene (referred to as Rif1GT; Fig. S1, A–D). This insertion generates a truncated allele, coding for a Rif1/β-GEO fusion protein which lacks a nuclear localization signal (Fig. S1 E). Immunoblots of Rif1GT/+ mouse embryonic fibroblasts (MEFs) showed diminished expression of the Rif1/β-GEO protein compared with wild type, suggesting that the truncated protein is unstable (Fig. S1 E). Furthermore, as expected from the lack of a nuclear localization signal, Rif1/β-GEO failed to show the specific nuclear signals detected for wild-type Rif1 (unpublished data). Intercrosses of Rif1GT/+ mice yielded homozygous Rif1GT/GT offspring at a severely reduced frequency (Fig. S1 F), suggesting a critical role for Rif1 during mouse development. In addition, examination of the genotypes of embryonic day (E) 12.5 embryos showed only one third of the expected number of Rif1GT/GT embryos (Fig. S1 F), indicating that Rif1 contributes to embryonic development at early as well as late stages. In addition, a conventional knockout allele of Rif1, in which exons 8 and 9 were replaced with a Neomycin resistance (NeoR) cassette, resulted in embryonic lethality on a pure C57BL/6J background (unpublished data). These results are in contrast with the findings with ATM and 53BP1 knockout mice (Barlow et al., 1996; Elson et al., 1996; Xu et al., 1996; Morales et al., 2003; Ward et al., 2003), which are born at the expected Mendelian ratios and are fully viable. Therefore, the embryonic lethality indicates that Rif1 has functions in addition to those downstream of ATM and 53BP1. Furthermore, the recovery of the Rif1-deficient embryos at E12.5 (Fig. S1 F) argues that Rif1 is not essential for the function of Nanog, whose absence results in lethality around E5.5 (Mitsui et al., 2003).
The few Rif1GT/GT mice born survived for at least 1 yr but appeared slightly retarded in their growth and were less fertile than their littermates (unpublished data). Because these rare Rif1GT/GT survivors are likely reflecting some type of compensation for the essential function of Rif1, we did not study them further.
Conditional deletion of Rif1 induces an S-phase defect
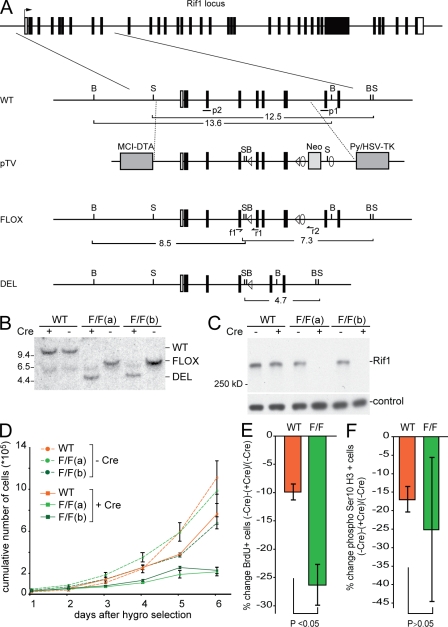
Given the diminished viability of Rif1-deficient embryos and the associated possibility of selection for compensation, we generated a conditional allele of Rif1. In the conditional allele (Rif1F), LoxP target sites for the Cre recombinase were positioned 5′ of exon 5 and 3′ of exon 7 (Fig. 1 A). Mice homozygous for the Rif1F/F allele were born at the expected frequency, viable, healthy, and fertile (unpublished data). Fibroblasts were prepared from E12.5 Rif1F/F, and wild-type littermate embryos were derived from Rif1F/+ intercrosses. Cells were either used as primary cultures (pMEFs) or immortalized with SV40 large T antigen (SV40LT; MEFs). Infection with a Cre-expressing retrovirus efficiently generated the Rif1-null allele (Fig. 1, B and C). In the absence of Rif1, primary and SV40LT immortalized cells proliferated significantly slower than the controls (Fig. 1 D and not depicted), which is consistent with the findings from the Rif1GT/GT cells (Fig. S1 G). Rif1-deficient cells, like Rif1GT/GT pMEFs, also showed reduced uptake of BrdU during a 30-min pulse, indicating that the proliferation phenotype is (at least in part) caused by a defect in entry and/or progression through S phase (Fig. 1 E, Fig. S2 A, and not depicted). We could not detect any significant effect of Rif1 deletion on the amount of cells in G2/M based on FACS analysis of phosphorylated Ser10 histone H3 (Fig. 1 F). Consistent with previous data on human Rif1 (Silverman et al., 2004), Rif1−/− cells revealed no obvious telomere defects such as telomere fusions or loss of telomeric DNA (Fig. S1 H and not depicted), arguing that their diminished proliferation is not telomere related. Thus, unlike the fungal Rif1 genes, mammalian Rif1 is required for normal cell proliferation.
Figure 1.
Conditional deletion of mouse Rif1 affects cell growth. (A) Schematic diagram of the mouse Rif1 locus, the targeting vector (pTV), the conditional Rif1 allele (FLOX), and the null allele (DEL). LoxP sites (triangles) flank exons 5, 6, and 7. FRT sites (circles) flank the NeoR gene. SacI (S) and BamHI (B) fragment sizes are indicated for each genotype, and the probes p1 and p2 are shown. f1, r1, and r2 are primers for genomic PCR. (B) Southern blots from MEF clones. Genomic DNA digested with SacI and probed with p1. The MEFs were infected with retrovirus carrying either an empty vector (−Cre) or pWZL-Cre (+Cre). Length is indicated in kilobases. (C) Western blot of extracts from MEFs of the indicated genotypes probed with mouse Rif1 antibody 1240. mTOR was used as a loading control. (D) Growth curves of Rif1 wild type (WT) and two independent Rif1F/F (F/F) MEF lines with and without treatment with Cre. (E) Changes in the S-phase index upon infection with Cre determined based on BrdU uptake during a 30-min pulse of asynchronous populations of Rif1-proficient and -deficient cells. The changes were quantified by calculating the percentage of BrdU-positive cells in (−Cre) − (+Cre)/(−Cre). (F) The same quantification as in E was performed for the G2/M cells by staining for histone H3 phospho-Ser10 and FACS analysis. (D–F) Error bars represent SD.
Rif1 deficiency exacerbates sensitivity to aphidicolin
The reduced proliferation of Rif1-null cells and their diminished BrdU uptake suggested a role for Rif1 in S phase that is distinct from its ATM-dependent response to DSBs. Therefore, we asked whether Rif1 deficiency affected the response to the DNA polymerase inhibitor aphidicolin. The effect of aphidicolin treatment was evaluated on metaphase chromosomes from Rif1-deficient immortalized MEFs or Cre-treated wild-type control cells (Fig. 2, A and B). The standard dose of aphidicolin used for this experiment does not inhibit cell cycle progression but slows replication fork progression and renders it more prone to stalling, especially at fragile sites (Glover et al., 1984). In wild-type cells, this treatment induces chromatid breaks and chromosomal fragments. The frequency of these anomalies was significantly elevated (threefold) in Rif1-deficient cells, suggesting an increase in fork-stalling frequency, a decrease in the stability of stalled forks, or a defective restart of the stalled replication fork. Other aberrations such as translocations and radial chromosomes were not significantly altered in the Rif1-null setting (Fig. 2 B). In the absence of aphidicolin treatment, Rif1-null cells showed a spontaneous elevation of chromatid breaks and chromosomal fragments over the negligible background detected in Cre-treated wild-type cells. A similar breakage phenotype occurred in Rif1GT/GT MEFs (unpublished data).
Figure 2.
Rif1-null cells are hypersensitive to aphidicolin. (A) Representative examples of metaphase chromosomes from wild-type (left) or Rif1F/F (right) cells treated with Cre and aphidicolin. Open arrows indicate chromatid breaks, the closed arrow indicates a fragment, and arrowheads indicate radial chromosomes. (B) Table summarizing chromosome anomalies scored by SKY analysis of metaphase spreads from MEFs of the indicated genotypes. Values in parentheses indicate percentages of chromosomes with the relevant abnormality. aph, aphidicolin. (C and D) Graphs of colony survival of MEFs with the indicated genotypes treated with aphidicolin (C) or MMC (D). Error bars represent SD. Bar, 10 µm.
Because chromosome spreads showed that replication forks in Rif1-null cells are more sensitive to low doses of aphidicolin, we examined how Rif1 deficiency influenced the ability of cells to survive aphidicolin treatment (Fig. 2 C). Two independent Rif1F/F MEF lines and a control wild-type line were treated with Cre recombinase and examined for their sensitivity to aphidicolin. Rif1 deletion resulted in significantly diminished survival compared with Rif1-proficient cells. The sensitivity to agents interfering directly with DNA polymerases appeared to be specific. Rif1-null cells also showed an approximate sixfold increase in cell death after low doses of hydroxyurea (HU; Fig. S2 B). In contrast, treatment with the interstrand cross-linking agent mitomycin C (MMC; Fig. 2 D) or the topoisomerase inhibitor ICRF-193 (Fig. S2 C) did not affect cells in a Rif1-dependent manner. Ionizing radiation (IR)–induced DSBs only marginally affected the survival of Rif1-null cells (1.3- and 2.5-fold reduced survival compared with Rif1-proficient cells when irradiated with 1 Gy or 5 Gy, respectively, as shown in Fig. S2 D), which is consistent with previous findings with RNAi to Rif1 in human cells (Silverman et al., 2004).
Rif1-null cells accumulate S phase–induced DNA damage
Aphidicolin treatment causes the activation of the DNA replication checkpoint, which ensures both the stability of the stalled forks and the activation of the DNA repair pathways responsible for fork restart (Casper et al., 2002; Cha and Kleckner, 2002; Sogo et al., 2002; Zachos et al., 2003; Calzada et al., 2005). The hypersensitivity to the aphidicolin treatment of the Rif1-null cells could be explained either by a defect in some aspect of the ATR kinase cascade or by a problem during repair. To test the first possibility, we monitored the phosphorylation state of the ATR target Chk1. Aphidicolin treatment of Rif1-null cells induced the same level of Chk1 phosphorylation as in the Rif1-proficient controls (Fig. 3 A), indicating that the main signaling cascade of the DNA replication checkpoint does not require Rif1. In addition, Rif1 deletion itself, without additional treatment, elicited a low level of Chk1 phosphorylation, whereas the phosphorylation of Chk2 was not obviously elevated compared with wild-type cells (Fig. 3 B). The specific phosphorylation of Chk1 and not Chk2 in Rif1-null cells is consistent with a defect in the repair of DNA damage in S phase. Rif1-null cells did not have a defect in their initial response to replication stress, as measured based on the level of γ-H2AX in cells treated with aphidicolin (Fig. 3 C). However, the aphidicolin-induced γ-H2AX response persisted in Rif1-null cells, which is consistent with a defect in the repair of aphidicolin-induced DNA damage (Fig. 3 D). To test more directly the requirements for Rif1 during normal S-phase progression, we synchronized in G0 Rif1GT/GT pMEFs and analyzed their progression through one cell cycle. Rif1-null cells appeared to transit through S phase reproducibly slower than wild-type controls (Fig. 3 E and Fig. S2 E), which is consistent with the presence of replication-induced DNA damage.
Figure 3.
Rif1 deletion causes accumulation of DNA damage. (A and B) Western blotting for the phosphorylation state of Chk1 and -2. MEFs of the indicated genotypes were treated with aphidicolin (A) or analyzed untreated (B). Proteins were extracted and analyzed for total levels of Chk1, Chk1 phosphorylation on Ser345, Chk2 phosphorylation, and levels of cyclin A. As a control, NIH3T3 cells were treated with aphidicolin (aph) or γ-irradiated (IR). (C and D) Cells were treated or not (no APH) with aphidicolin for 3 h and 30 min. The presence of the DNA damage is identified by IF for γ-H2AX and is quantified as the mean intensity of specific signal in the nuclei. The percentage of each cell population that showed a value of mean intensity signal >50 was calculated for each time point. Data represent the mean ± SEM of two independent Rif1F/F + Cre and the mean ± SD of three wild type (WT; Rif1 WT, Rif WT + Cre, and Rif1F/F). (C) Mean intensity of γ-H2AX signal after 3 h and 30 min of aphidicolin (APH) treatment is comparable between wild-type and Rif1-null cells. (D) Time course monitoring the variation of γ-H2AX signal intensity after release from aphidicolin-induced S-phase arrest. Data were plotted as fold induction over time 0 mean value for each clone. Each time point represents the mean value of two independent Rif1F/F + Cre clones and three wild type (Rif1 WT, Rif WT + Cre, and Rif1F/F). The breaks in the lines indicate that the “no APH” is not a time point in a time course but the basal signal level. (E) Rif1GT/GT pMEF and wild-type littermate controls were synchronized in G0 and released in BrdU to monitor S-phase progression. At the indicated time point, cells were collected and analyzed for BrdU content by FACS. One representative experiment out of two is shown.
Rif1 stimulates homology-directed repair (HDR)
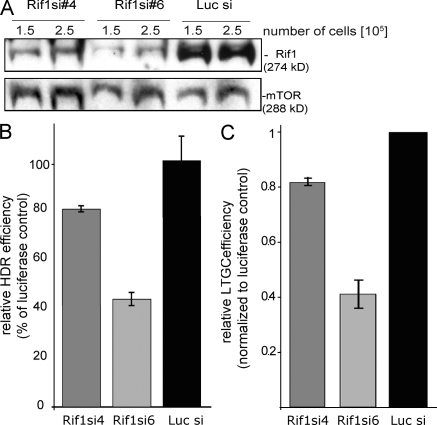
To further explore the role of Rif1 in the repair of S-phase damage, we used a previously established assay for HDR, the main pathway for the repair of replication-associated DNA damage (Johnson and Jasin, 2000; Puget et al., 2005; for reviews see Cromie et al., 2001; Helleday, 2003). The HDR assay is based on a human U2OS cell line containing two tandem mutant copies of the gene encoding GFP. The first copy is promoterless and contains a small deletion in the 5′ end; the second has a promoter but has an I-SceI site interrupting the GFP ORF. Upon introduction of I-SceI, an intact GFP gene can be generated through DSB repair by unequal sister or intrachromatid recombination. The same reporter allows for assaying the efficiency of long tract gene conversion (LTGC). It contains the two halves of the blasticidin resistance gene, such that I-SceI–stimulated LTGC leads to the reconstitution of the wild-type gene. To reduce Rif1 levels, we used two Rif1-specific siRNAs and an irrelevant siRNA to luciferase as a control (Fig. 4 A). Quantitative immunoblots indicated that the siRNAs reduced Rif1 levels by 30–60% compared with the level in the luciferase siRNA control cells (Fig. 4 A). Despite the modest knockdown of Rif1, we detected a significant effect on the efficiency of HDR. Cells transfected with siRNA #6 showed a threefold decrease in HDR, whereas siRNA #4 had a lesser effect (Fig. 4 B). As it has been shown for two other genes involved in HDR, BRCA1 and -2, depletion of Rif1 leads to a reduced S phase (Fig. S3 A; Patel et al., 1998; Kumaraswamy and Shiekhattar, 2007). Diminished Rif1 levels also affected the efficiency of LTGC to an extent proportional to the magnitude of the knockdown achieved by the different siRNAs.
Figure 4.
Rif1 promotes HDR. (A and B) Knockdown of Rif1 decreases HDR efficiency. (A) Rif1 levels were reduced by two specific siRNAs, #4 and #6, in U2OS#18. siRNA against luciferase (Luc si) was used as a control. Western blot showing a titration to validate the reduction of Rif1 levels upon treatment with specific siRNAs. mTOR was used as a loading control. (B) Upon DSB induction by I-SceI transfection, HDR was evaluated by quantifying the amount of GFP-positive cells by FACS. The effect on HDR is expressed as the percentage of the luciferase siRNA. P-values are <0.05 for both siRNAs. (C) LTGC efficiency was calculated by counting the number of colonies growing in the presence of blasticidin and normalized for plating efficiency. The effect of LTGC is expressed relative to the luciferase siRNA control. (B and C) Error bars represent SD.
Rif1 localizes at sites of replication fork stalling, mostly at pericentromeric heterochromatin
Given the possibility of a role for Rif1 at stalled replication forks, we examined the distribution of Rif1 during an unperturbed cell cycle. Cells were briefly labeled with BrdU such that sites of DNA replication could be detected with anti-BrdU antibodies under denaturing conditions. This technique allows the identification of cells in different stages of S phase (Fig. 5; Nakamura et al., 1986; Dimitrova and Berezney, 2002). Immunofluorescence (IF) for Rif1 showed a punctuate nuclear staining pattern in late G1 with more prominent signals at the pericentromeric heterochromatin. In early and mid S phase, Rif1 progressively concentrated at pericentromeric heterochromatin. The Rif1 signal never overlapped with BrdU or proliferating cell nuclear antigen (unpublished data) except when pericentromeric heterochromatin started to be replicated in mid S phase. At that stage, some BrdU foci colocalized with Rif1, again mostly at pericentromeric heterochromatin. In mid–late and late S phase, Rif1 appeared to have left the chromatin, showing a diffuse nuclear staining pattern. These data raised the possibility that Rif1 has a specific function in the replication of certain regions of the genome, including the pericentromeric heterochromatin.
Figure 5.
Rif1 binds chromatin before and during DNA replication but only seldom colocalizes with the replication fork in mid–late S phase and around pericentromeric heterochromatin. IF on wild-type MEFs for Rif1 (red) and BrdU visualized by denaturing BrdU staining (green). Late G1, early, mid, mid–late, and late S were identified by the BrdU staining pattern. Insets contain enlarged details of the indicated areas.
To determine whether the sites of BrdU–Rif1 colocalization represent sites of fork stalling, we visualized stalled replication forks in aphidicolin-treated cells based on the presence of ssDNA using BrdU staining without denaturation (Fig. 6, A and B). Rif1 colocalized with ssDNA in only ∼12% of the untreated and 27% of the aphidicolin-treated BrdU-positive cells (Fig. 6 A and Fig. S5 B). In these cells, only a subset of the stalled forks showed the presence of Rif1 (Fig. 6 A), mostly, but not exclusively, at or near the pericentromeric heterochromatin (Fig. 6 A, bottom). Control experiments with Rif1−/− cells did not show foci of Rif1 coinciding with BrdU, establishing the specificity of the antibody (Fig. S3 B). To further verify the localization pattern of Rif1, we executed dual IF for the ssDNA-binding protein RPA34 and Rif1 (Fig. S3 C). Consistent with the BrdU data, there was significant colocalization of Rif1 with a subset of the RPA34 foci. These data support the idea that the Rif1–BrdU colocalization at pericentromeric heterochromatin of unperturbed cells reflects sites of fork stalling.
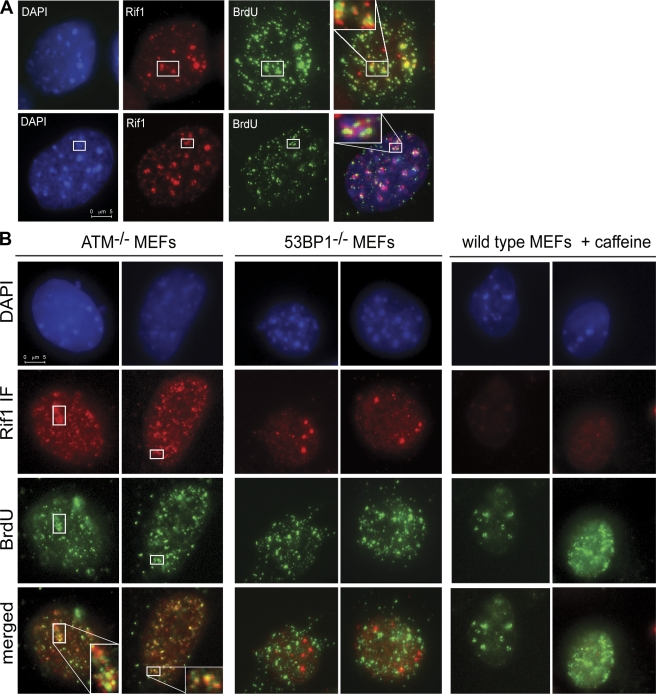
Figure 6.
Rif1 localizes to sites of replication stress. (A) IF on wild-type MEFs for Rif1 (red) and ssDNA visualized by nondenaturing BrdU staining (green). The bottom row shows an example of Rif1 localized at stalled replication forks near heterochromatin. (B) Rif1 localization at aphidicolin-induced sites of ssDNA is abolished in 53BP1-null cells and upon caffeine treatment. IF for Rif1 (red) and ssDNA (green) on ATM−/−, 53BP1−/−, and caffeine-treated wild-type MEFs. All cells were treated with aphidicolin. (A and B) Insets contain enlarged details of the indicated areas.
Rif1 localization at stalled replication forks requires ATR and 53BP1
To further characterize Rif1 foci, we analyzed the presence of other fork-stalling response factors. The Rif1 foci at ssDNA always contained 53BP1 (Fig. S4 A), although there were ssDNA sites containing 53BP1 but no Rif1. Similarly, Rif1 foci at stalled forks colocalized with the BLM RecQ helicase, but not all BLM foci coincided with Rif1 signals (Fig. S4 B).
The localization of Rif1 at IR-induced DSBs is dependent on ATM and 53BP1 (Silverman et al., 2004). In contrast, ATM−/− MEFs retained the ability to accumulate Rif1 at aphidicolin-induced ssDNA, whereas the absence of 53BP1 abrogated this response (Fig. 6 B). This suggested that the ATR kinase might be involved in providing the signal responsible for the localization of 53BP1 and Rif1 at the stalled forks. Consistent with this, the Rif1 pattern was significantly altered by treating the cells with the PI3-like kinase inhibitor caffeine (Fig. 6 B) or by inhibiting ATR with a specific short hairpin RNA (shRNA; Fig. S5, A–C). Together, the data suggest that Rif1 requires ATR and 53BP1 to localize to a subset of stalled replication forks, very often located in the proximity of pericentromeric heterochromatin. On the contrary, Rif1 is not required for 53BP1 or BLM localization at stalled replication forks (Fig. S5 D). This finding is in agreement with the fact that Rif1-deficient cells do not show an increase in sister chromatid exchanges (unpublished data).
Aberrant Rad51 aggregates in Rif1-deficient cells
Because we found that Rif1 contributes to the efficiency of HDR, we examined the behavior of one of the main effectors of homologous recombination, the protein Rad51, in Rif1-deficient MEFs. Co-IF for endogenous Rad51 and 53BP1 revealed that 15% of the Rad51-positive cells accumulated large aberrant Rad51 aggregates during S phase in the absence of Rif1 (Fig. 7 A). These types of structures were present exclusively in cells that were in S phase (Fig. 7 B; Starborg et al., 1996) and were virtually absent from wild-type cells (Fig. 7, A and D). In contrast, the overall percentage of Rad51-positive cells was not significantly different between wild-type and Rif1-null cells (Fig. 7 C). The presence of these aggregates could be explained if Rif1 had a direct or indirect role in promoting late steps of Rad51 turnover at the sites of recombination. Alternatively, their presence could be an indirect consequence of the accumulation of damage during the replication of specific regions of the genome that cannot be repaired in the absence of Rif1.
Figure 7.
Aberrant Rad51 aggregates in Rif1-deficient cells. (A) IF for Rad51 (green) and 53BP1 (red) in Rif1F/F (left) and wild-type (right) MEFs treated with Cre and aphidicolin. Arrowheads point to aberrant Rad51 aggregates. Insets contain enlarged details of the indicated areas. (B) IF for Rad51 (green), Ki67 (red), and DAPI staining (blue) on Rif1F/F MEFs treated with Cre and aphidicolin (aph). (C and D) Quantification of the percentage of Rad51-positive cells (C) and of the percentage of Rad51-positive (pos) cells containing Rad51 large aggregates (D). One representative experiment out of two is shown. wt, wild type.
Discussion
Our results identify Rif1 as a novel component of the DNA replication checkpoint, essential for embryonic development and normal DNA replication. We had previously shown that Rif1 was contributing to the inhibition of DNA replication in response to ATM activation by DSBs (Silverman et al., 2004). However, Rif1 is essential, whereas ATM is not. The requirement for Rif1 for cellular and embryonic survival can be explained from the S-phase function of Rif1 documented in this study. We show that upon ATR activation and 53BP1 recruitment, Rif1 localizes at a subset of stalled replication forks, where it could be important to regulate HDR during replication. Indeed, Rif1 deletion induces accumulation of chromatid-type genome instability and, as a consequence, renders cells hypersensitive to aphidicolin and HU. We have also shown that Rif1 localizes at stalled replication forks preferentially during mid–late S phase, often at/around replicating pericentromeric heterochromatin. Centromeres, like telomeres, are highly repetitive sequences and are highly recombinogenic (Jaco et al., 2008; Nakamura et al., 2008). In particular, it was recently shown that Rad51 function is essential to suppress gross chromosomal rearrangement at centromeres in fission yeast (Nakamura et al., 2008). Therefore, we hypothesize that Rif1 regulates Rad51-dependent homologous recombination at replication forks stalled in regions of the genome that are difficult to replicate like repetitive sequences or tight protein–DNA complexes.
These findings can be reconciled with observations in yeast, in which Rif1 has a telomere-specific function. Budding yeast strains that survive without telomerase maintain their telomeres either through Rad51-dependent HDR (type I survivors) or through a less understood Rad50-dependent pathway (type II survivors; Lundblad and Blackburn, 1993; Lendvay et al., 1996; Le et al., 1999; Teng and Zakian, 1999). In the absence of Rif1, the frequency of type I survivors is diminished (Teng et al., 2000), which is consistent with a role for Rif1 in the Rad51 pathway. Data from fission yeast Rif1 can also be explained from an HDR perspective. When the telomeric protein Taz1 is absent, fission yeast telomeres can become entangled, presumably reflecting a pathological product of an HDR event taking place after replication fork arrest (Miller and Cooper, 2003; Miller et al., 2006). Rif1 deficiency suppresses the lethality associated with this process, which is consistent with a role in HDR (Miller et al., 2005). Therefore, modulation of Rad51-dependent HDR could be Rif1’s function conserved during evolution. The finding that Rif1 depends on 53BP1 and positively contributes to HDR execution is in apparent conflict with observations on 53BP1-null MEFs. Deletion of 53BP1 does not sensitize cells to aphidicolin (unpublished data) and promotes HDR (Adams et al., 2005; Xie et al., 2007). This discrepancy could be explained if Rif1 is one of several factors that act downstream of 53BP1 to navigate replication problems such that its deletion creates an imbalance in HDR that leads to pathological products. One interesting possibility is that Rif1 inhibits (directly or not) BLM, which has been proposed to have a 53BP1-regulated role at stalled forks (Sengupta et al., 2004; Bugreev, 2007; Tripathi et al., 2007, 2008). Inhibition of BLM by Rif1 would be consistent with genetic data on the survivor pathway in yeast, which allows budding yeast to proliferate without telomerase (McEachern and Haber, 2006). The yeast orthologue of BLM, Sgs1, is required for the type II (Rad50 dependent) survivor pathway (Johnson et al., 2001), but this requirement is alleviated by the deletion of Rif1 (Tsai et al., 2006). Because Rif1 localizes only to a subset of the stalled replication forks, the interplay between BLM and Rif1 could direct the choice between different types of HDR at single forks, depending on the DNA sequence and chromatin context.
Finally, the finding that Rif1 plays a role in HDR is relevant to the recent identification of Rif1 mutations in human breast cancer. One breast cancer cell line (HCC2218) was found to have two mutations in Rif1 (E1784K and D1955H; Sjöblom et al., 2006), and a second breast cancer cell line (HCC1806) carries a translocation that truncates Rif1 after exon 21 (Howarth et al., 2008). According to our data on Rif1/β-GEO, this truncation is expected to be nonfunctional. The possible inactivation of Rif1 in sporadic cases of breast cancer is of interest with regard to the BRCA2 gene, whose inactivation predisposes to breast and ovarian cancer. Like Rif1, BRCA2 is implicated in the Rad51 pathway (for review see Thorslund and West, 2007). Given the current interest in treating BRCA2-deficient breast cancer with therapies based on their inability to execute HDR efficiently (Bryant et al., 2005; Farmer et al., 2005), it may be useful to consider sporadic tumors with Rif1 mutations for this treatment approach.
Materials and methods
Gene targeting
The mouse Rif1 locus contains 36 exons, with the second exon containing the ATG for the 2,426–amino acid ORF. The targeting strategy allows Cre-mediated deletion of exons 5, 6, and 7; the splicing of exon 4 to exon 8 generates a frame shift. The targeting vector was generated by recombineering (Lee et al., 2001) using bacterial artificial chromosome RCPI-23 395O2 (American Type Culture Collection) in Escherichia coli strain EL350. The first LoxP site was introduced in the unique EcoRV site upstream of exon 4 by inserting an oligonucleotide that also introduced SacI and BamHI restriction sites used for the analysis of targeting in ES cells. The unique EcoRI site 3′ of exon 7 was used to insert a LoxP-FRT-NeomycinR(NeoR)-FRT cassette (gift from D. O’Carroll, European Molecular Biology Laboratory Monterotondo, Monterotondo, Italy). The construct was subcloned into the pDTA-TKIII vector, which allows double negative selection (DTA and TK genes), linearized with NotI, and electroporated into Bruce4 C57BL/6 ES cells. Four independent ES cell clones were injected by standard techniques into albino C57BL/6J blastocysts, and chimeras were evaluated based on coat color. Chimeric founders were crossed to C57BL/6J females, and the mice were kept in a pure C57BL/6J background. The NeoR cassette was removed by crossing the F1 mice with an FLPe deleter mouse strain (Jackson ImmunoResearch Laboratories, Inc.). Genotyping of mice and derived cells was performed by PCR with the following primers: f1, 5′-TTAGAGGAACTGAGGGGTAGGTAG-3′; r1, 5′-AACTGCAACTCTACTGAGGGAAG-3′; and r2, 5′-TGAAACCGTAGCCAGAAACTG-3′. PCR generated a 78-bp product for the wild-type allele, a 138-bp product for the LoxP-modified (FLOX) allele, and a 246-bp product for the deleted (DEL) allele. The southern probes p1 and p2 were generated by PCR from genomic DNA with the following primers: p1 E3s, 5′-GAATTGTAGTTAATGATGGGCC-3′; and E3a 5′-TCTATGGATGTAGCTTAGTGGTACA-3′; and p2 E8a, 5′-TGGTTCACATCTACGATCCCA-3′; and E8s 5′-GAATTGTAGTTAATGATGGGCC-3′.
Rif1GT mice were generated from an ES cell clone carrying a trapped Rif1 allele (A045A01) available from the German Gene Trap Consortium (http://genetrap.gsf.de/). Chimeric founders were crossed to C57BL/6J females, and the mice were kept in a mixed 129/C57BL/6J background. Genotyping of mice and derived cells was performed by PCR with the following primers: f2, 5′-GCTAGTCTAGGCACCAGGAACC-3′; f3, 5′-AGGAGAAAATACCGCATCAGG-3′; and r3, 5′-TCCTCAGCATCAACCAAAGAG-3′. The PCR products were 135 bp and 190 bp for the wild-type and GT allele, respectively. The southern probe p3 was generated by PCR from genomic DNA with the following primers: E22s, 5′-TCAGATTGGTCCAGAAAGAA-3′; and E24a, 5′-CATTCCACAACAGAGCACTCT-3′. The Neo probe was obtained by digesting the plasmid pZero2-LoxP-Neo-LoxP with PstI and NcoI and isolating the fragment corresponding to part of the NeoR ORF.
Generation of MEFs, conditional deletion of Rif1, and cell manipulation
MEFs were generated from E12.5 embryos according to standard protocols and propagated in DME high glucose, 15% heat-inactivated FBS (Invitrogen), 100 U/ml penicillin, 0.1 mg/ml streptomycin, 0.2 mM l-Gln, 0.1 mM nonessential amino acids, and 50 µM β-mercaptoethanol. To immortalize primary MEFs, early passage (P2) cells were infected with pBabeSV40LT (gift from G. Hannon, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY).
ATM−/− MEFs were prepared from heterozygous intercrosses of ATM+/− mice (129S6/SvEvTac-Atmtm1Awb/J; Jackson ImmunoResearch Laboratories, Inc.). 53BP1−/− MEFs were prepared from heterozygous intercrosses of mice provided by J. Chen (Yale University School of Medicine, New Haven, CT). MEFs were immortalized with SV40LT as described in the previous paragraph.
For Cre-mediated deletion of the Rif1 gene, cells were infected six times at 6–8-h intervals with the supernatant from Phoenix packaging cells (American Type Culture Collection) transfected by standard calcium phosphate protocol with either empty pWZL-hygro (gift from S. Lowe, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) or pWZL-hygro-CRE. Cells were split into medium containing hygromycin B (Sigma-Aldrich) at 90 µg/ml and grown for 3 d.
Growth curves were determined by plating 34,000 or 68,000 cells per well of a 6-well plate in triplicate for each day of the experiment, for immortalized and primary cells, respectively. Cells were trypsinized and counted with a Z2 counter (Beckman Coulter).
Aphidicolin sensitivity was measured by plating 1,400 and 1,000 cells per well of a 6-well plate for Rif1F/F and wild-type immortalized MEFs, respectively, in triplicate. After 24 h, cells were either left untreated or treated for 24 h with the different doses of aphidicolin, washed three times with PBS, and incubated in growth media for about 1 wk. After rinsing with PBS, cells were fixed for 10 min in a solution containing 50% methanol, 7% glacial acetic acid, and 0.1% Coomassie brillant blue. The number of colonies with >20 cells was counted. MMC sensitivity was tested in the same conditions, except that the drug treatment was performed only for 2 h and 30 min.
G0 synchronization of pMEFs was obtained by 5 d of contact inhibition and 48 h of serum starvation (0.1% FBS). Cells were pulsed with 10 µM BrdU for 1 h and subsequently released in 15% FBS medium containing 10 µM BrdU by plating 1.5 × 106 cells per 10 cm.
BrdU staining for FACS analysis
Asynchronous cell populations were pulsed for 30 min with 10 µM BrdU. After fixation in 70% ethanol at −20°C, 3 × 105 cells were stained according to the BD protocol, with 10–15 µl per sample of anti-BrdU FITC conjugated (BD). The samples were analyzed by FACS on a FACSCalibur System One laser (488 nm; BD), and the data were analyzed with FlowJo software (Tree Star, Inc.).
Aphidicolin treatment, metaphase spreads, and spectral karyotyping analysis (SKY)
Cells were treated for 24 h with or without 0.3 µM aphidicolin, followed by 4 h with 0.2 µg/ml colcemid, and then collected by trypsinization and treated according to standard protocols to prepare metaphase chromosomes. Metaphase spreads were analyzed either by DAPI staining or by SKY (Liyanage et al., 1996) using paint mixtures from Applied Spectral Imaging according to the manufacturer’s protocol. Imaging was performed on a microscope (BX-61; Olympus) equipped with an interferometer driven by a desktop computer and SpectraView software (Applied Spectral Imaging).
IF
Cells grown on coverslips were rinsed with PBS and extracted for 10 min at 4°C in Triton buffer (0.5% Triton X-100, 20 mM Hepes, pH 7.9, 50 mM NaCl, 3 mM MgCl2, and 300 mM sucrose), fixed for 10 min in 3% paraformaldehyde/2% sucrose at room temperature, and permeabilized again for 10 min at 4°C in Triton buffer. IF was performed as described previously (Zhu et al., 2000), with minor modifications. In brief, cells were blocked in PBG buffer (0.2% cold fish gelatin [Sigma-Aldrich] and 0.5% BSA) for 30 min at room temperature. The primary antibodies were diluted in PBG and incubated for 1 h at room temperature. The fluorescent-conjugated secondary antibodies were also diluted in PBG and incubated for 45 min.
Nondenaturing BrdU IF was performed according to Raderschall et al. (1999). In brief, cells were labeled with 3 µM BrdU for 25 h. After harvesting, IF was performed as described (Raderschall et al., 1999), using the BD anti-BrdU antibody at 1:1,000. An appropriate secondary fluorescent-conjugated anti–mouse antibody was used to detect the BrdU.
Denaturing BrdU IF was performed by pulsing cells for 30 min with 10 µM BrdU. Cells were pre-extracted and fixed as described in the previous paragraph. 100 U/ml DNaseI and 3 mM MgCl2 were added to the primary antibody mix to denature the DNA.
For the triple staining, either one or both primary rabbit antibodies were directly conjugated to the fluorophore using the Zenon kit (Invitrogen) according to the manufacturer’s instructions. Anti-BrdU antibody was detected using a Cy5-conjugated anti–mouse secondary antibody (Millipore). To assay the effect of PI3-like kinase’s inhibition on Rif1 localization at stalled replication forks, wild-type MEFs were incubated for 3 h with 10 mM caffeine before aphidicolin treatment. Caffeine was maintained during the aphidicolin treatment. Slides were mounted in embedding medium (1 mg/ml p-phenylene diamine [Sigma-Aldrich] in 1× PBS and 90% glycerol). Images were acquired with a microscope (Axioplan 2 [Carl Zeiss, Inc.], DM6000 [Leica], or confocal TCS SP5 [Leica]) with a 63× 1.4 NA oil objective. The camera used was either a C4742-95 (Hamamatsu Photonics) or a DFC340FX (Leica). The acquisition software used was either OpenLab (PerkinElmer) or LAS AF (Leica). Contrast adjustment and cropping was performed in Photoshop (Adobe), and figures were composed in Illustrator (Adobe). For the confocal images (Fig. 7), maximum projection is shown.
In the case of the quantification of the γ-H2AX signal over the time course (Fig. 3, C and D), cells from two Rif1F/F + Cre and three wild type were imaged using a wide-field DM6000 microscope with a 63× objective (30 fields from each time point). Two channels were imaged: channel 1 to find the nuclei (DAPI) and channel 2 to image the γ-H2AX signal. The mean intensity of channel 2 was quantified by using MetaMorph software (MDS Analytical Technologies). After application of the threshold in channel 1 (DAPI), objects were identified and masked. The images and masks were visually inspected for accuracy. Then, the percentage of cells with a mean intensity signal of channel 2 >50 was calculated (n > 100) for each condition and normalized to the percentage of the same cell line at time 0.
Western blotting
To monitor Chk1 Ser345 phosphorylation in response to aphidicolin, cells were treated for 3 h and 30 min with 5 µM aphidicolin and collected. Control irradiated NIH3T3 were treated with 5 Gy and allowed to recover for 30 min before preparing protein extracts. Cells were collected by trypsinization, rinsed in PBS, directly lysed in 2× Laemmli buffer, and passed through a 29-gauge insulin syringe. Alternatively, cells were lysed in 50 mM Tris-HCl, pH 7.4, 1% Triton X-100, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, phosphatase, and protease inhibitors. Chromatin-bound proteins were extracted by increasing the NaCl concentration to 500 mM. The extract was centrifuged for 10 min at 13,200 rpm at 4°C, and the supernatant was recovered. Proteins were separated by gel electrophoresis and blotted according to standard protocols. All of the Western blots, with the exception of phospho–Ser345 Chk1, were blocked in 5% milk/PBS/0.1% Tween, and the primary antibody was incubated in 1% milk/PBS/0.1% Tween. For the anti-phospho–Ser345 Chk1 antibody (Cell Signaling Technology), the manufacturer’s instructions were followed.
shRNA, siRNA, and sister chromatid recombination assay
Knockdown of ATR in wild-type or ATM−/− MEFs was performed as described previously (Denchi and de Lange, 2007). In brief, cells plated on coverslips were infected four times in 24 h with retroviruses carrying either the ATR or the control luciferase shRNA. After 24 h in 2 µg/ml puromycin and 3 µM BrdU, cells were incubated for 3 h in 5 µM aphidicolin and processed for IF as described in the IF section. Sister chromatid recombination assay was performed using U2OS#18 (gift from R. Scully, Harvard University, Cambridge, MA) as described previously, with minor modifications (Puget et al., 2005). In brief, 50,000 cells were plated per well of a 12-well plate in triplicate for each siRNA. siRNA transfections were performed as described previously (Silverman et al., 2004), except for the use of DharmaFECT Duo (Thermo Fisher Scientific) instead of Oligofectamine (Invitrogen). The sequences of siRNA #4 and #6 and luciferase were published previously (Silverman et al., 2004). The siRNAs were synthesized either by Thermo Fisher Scientific or Mycrosynth. 1 d after the second transfection, the medium was changed, and cells were collected for Western blot analysis and for blasticidin selection. 250,000 cells were plated in duplicate in a 10-cm dish and, after 24 h, were switched to full medium containing 5 µg/ml blasticidin. 200 cells were also plated in a 6-cm dish in triplicate to calculate plating efficiency. Cells were collected for GFP FACS at 48 h after the second transfection. During the FACS analysis, at least 30,000 events per sample were acquired. Statistical analysis was performed using a standard t test with a two-tailed distribution and equal variance.
Antibodies
Rabbit anti–mouse Rif1 serum 1240 was raised against a keyhole limpet hemocyanin–conjugated peptide spanning amino acids 401–424 of the mouse Rif1 sequence. The antibody was affinity purified and used at a 1:10,000 dilution for IF and Western blotting. Other antibodies used in this study were anti-53BP1 (1:1,000; Novus Biologicals), a mouse anti-53BP1 serum generated against the C-terminal 312 amino acids of human 53BP1 and fused to GST (1:2,000), anti-BLM (1:200; Abcam), anti-RPA (1:500; GeneTex, Inc.), anti-Ki67 (1:1,000; BD), anti–γ-H2AX (1:1,000; Bethyl Laboratories, Inc.), and anti-Rad51 (1:2,500 for IF; gift from R. Kanaar, Erasmus Medical Center, Rotterdam, Netherlands; van Veelen et al., 2005). For Western blotting, anti-Chk2 (BD) was used at 1:1,000, anti-Chk1 (Santa Cruz Biotechnology, Inc.) was used at 1:200; anti-phospho–Ser345 Chk1 (Cell Signaling Technology) was used at 1:1,000, anti-ATR (Santa Cruz Biotechnology, Inc.) was used at 1:500, anti–mammalian target of rapamycin (mTOR; Cell Signaling Technology) was used at 1:1,000, and anti–cyclin A (Cell Sciences) was used at 1:1,000.
Online supplemental material
Fig. S1 shows that insertional inactivation of Rif1 leads to embryonic lethality and reduced proliferation of MEFs. Fig. S2 exemplifies that Rif1-null cells exhibit a reduced S-phase index and are hypersensitive to HU, as sensitive as wild-type cells to ICRF-193, and only mildly sensitive to γ-irradiation. Fig. S3 shows a decreased S-phase index in cells treated with Rif1-specific siRNAs and demonstrates the specificity of IF with anti–mouse Rif1 antibody 1240 and Rif1–RPA34 colocalization. Fig. S4 illustrates that Rif1 colocalizes with BLM and 53BP1 at stalled replication forks. Fig. S5 shows that the Rif1 localization at stalled replication forks depends on ATR and that Rif1 is not required for 53BP1 and BLM localization at stalled replication forks. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200902039/DC1.
Acknowledgments
We gratefully acknowledge Devon White for mouse husbandry, The Rockefeller University (RU) and Memorial Sloan-Kettering Cancer Center Transgenic facilities for ES cell injections, the RU FACS facility, Daniel Bilbao at the European Molecular Biology Laboratory (EMBL) FACS facility, and Valeria Berno at EMBL Monterotondo for confocal images and precious support in the quantification of the images. We thank Donal O’Carroll and Alexander Tarakhovsky for invaluable help with mouse protocols and scientific input. We greatly appreciate Eros Lazzerini Denchi, Hiro Takai, and Agnel Sfeir for their support with helpful discussions and protocols. R. Scully provided the U2OS#18 line, R. Kanaar provided the Rad51 antibody, and J. Chen provided the 53BP1-null mice.
This work was supported by the Breast Cancer Research Foundation and National Institutes of Health grant RO1 CA076027 to T. de Lange. S.B.C. Buonomo was supported by the William D. Hassett Fellowship of the Damon Runyon Cancer Research Foundation. D. Ferguson was supported by a grant from the Sidney Kimmel Foundation for Cancer Research.
Note added in review. While this paper was under review, a paper was published showing, with an independent assay, a role for Rif1 in HDR during DSBs response (Wang et al. 2009. Carcinogenesis. 30:1314–1319).
Footnotes
Abbreviations list:
- ATM
- ataxia telangiectasia mutated
- ATR
- ataxia telangiectasia and Rad3 related
- DSB
- double-strand break
- ES
- embryonic stem
- HDR
- homology-directed repair
- HU
- hydroxyurea
- IF
- immunofluorescence
- IR
- ionizing radiation
- LTGC
- long tract gene conversion
- MEF
- mouse embryonic fibroblast
- MMC
- mitomycin C
- mTOR
- mammalian target of rapamycin
- NeoR
- Neomycin resistance
- shRNA
- short hairpin RNA
- SKY
- spectral karyotyping analysis
- ssDNA
- single-stranded DNA
- SV40LT
- SV40 large T antigen
References
- Adams I.R., McLaren A. 2004. Identification and characterisation of mRif1: a mouse telomere-associated protein highly expressed in germ cells and embryo-derived pluripotent stem cells. Dev. Dyn. 229:733–744 10.1002/dvdy.10471 [DOI] [PubMed] [Google Scholar]
- Adams M.M., Wang B., Xia Z., Morales J.C., Lu X., Donehower L.A., Bochar D.A., Elledge S.J., Carpenter P.B. 2005. 53BP1 oligomerization is independent of its methylation by PRMT1. Cell Cycle. 4:1854–1861 [DOI] [PubMed] [Google Scholar]
- Banerjee S., Myung K. 2004. Increased genome instability and telomere length in the elg1-deficient Saccharomyces cerevisiae mutant are regulated by S-phase checkpoints. Eukaryot. Cell. 3:1557–1566 10.1128/EC.3.6.1557-1566.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D., Wynshaw-Boris A. 1996. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 86:159–171 10.1016/S0092-8674(00)80086-0 [DOI] [PubMed] [Google Scholar]
- Bianchi A., Shore D. 2008. Molecular biology. Refined view of the ends. Science. 320:1301–1302 10.1126/science.1159104 [DOI] [PubMed] [Google Scholar]
- Brown E.J., Baltimore D. 2000. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 14:397–402 [PMC free article] [PubMed] [Google Scholar]
- Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 434:913–917 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- Budzowska M., Jaspers I., Essers J., de Waard H., van Drunen E., Hanada K., Beverloo B., Hendriks R.W., de Klein A., Kanaar R., et al. 2004. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J. 23:3548–3558 10.1038/sj.emboj.7600353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev D.V., Yu X., Egelman E.H., Mazin A.V. 2007. Novel pro- and antirecombination activities of the Bloom's syndrome helicase. Genes Dev. 21:3085–3094 10.1101/gad.1609007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzada A., Hodgson B., Kanemaki M., Bueno A., Labib K. 2005. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 19:1905–1919 10.1101/gad.337205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper A.M., Nghiem P., Arlt M.F., Glover T.W. 2002. ATR regulates fragile site stability. Cell. 111:779–789 10.1016/S0092-8674(02)01113-3 [DOI] [PubMed] [Google Scholar]
- Cha R.S., Kleckner N. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 297:602–606 10.1126/science.1071398 [DOI] [PubMed] [Google Scholar]
- Chan S.W., Chang J., Prescott J., Blackburn E.H. 2001. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr. Biol. 11:1240–1250 10.1016/S0960-9822(01)00391-8 [DOI] [PubMed] [Google Scholar]
- Cimprich K.A., Cortez D. 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9:616–627 10.1038/nrm2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie G.A., Connelly J.C., Leach D.R. 2001. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell. 8:1163–1174 10.1016/S1097-2765(01)00419-1 [DOI] [PubMed] [Google Scholar]
- Denchi E.L., de Lange T. 2007. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 448:1068–1071 10.1038/nature06065 [DOI] [PubMed] [Google Scholar]
- Dimitrova D.S., Berezney R. 2002. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell Sci. 115:4037–4051 10.1242/jcs.00087 [DOI] [PubMed] [Google Scholar]
- Elson A., Wang Y., Daugherty C.J., Morton C.C., Zhou F., Campos-Torres J., Leder P. 1996. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl. Acad. Sci. USA. 93:13084–13089 10.1073/pnas.93.23.13084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 434:917–921 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- Fazzio T.G., Huff J.T., Panning B. 2008. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 134:162–174 10.1016/j.cell.2008.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover T.W., Berger C., Coyle J., Echo B. 1984. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 67:136–142 10.1007/BF00272988 [DOI] [PubMed] [Google Scholar]
- Hansen J., Floss T., Van Sloun P., Füchtbauer E.M., Vauti F., Arnold H.H., Schnütgen F., Wurst W., von Melchner H., Ruiz P. 2003. A large-scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc. Natl. Acad. Sci. USA. 100:9918–9922 10.1073/pnas.1633296100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C.F., Sussel L., Shore D. 1992. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 6:801–814 10.1101/gad.6.5.801 [DOI] [PubMed] [Google Scholar]
- Helleday T. 2003. Pathways for mitotic homologous recombination in mammalian cells. Mutat. Res. 532:103–115 [DOI] [PubMed] [Google Scholar]
- Hirao A., Cheung A., Duncan G., Girard P.M., Elia A.J., Wakeham A., Okada H., Sarkissian T., Wong J.A., Sakai T., et al. 2002. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol. Cell. Biol. 22:6521–6532 10.1128/MCB.22.18.6521-6532.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins K.M., Auerbach W., Wang X.Y., Hande M.P., Hang H., Wolgemuth D.J., Joyner A.L., Lieberman H.B. 2004. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol. Cell. Biol. 24:7235–7248 10.1128/MCB.24.16.7235-7248.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth K.D., Blood K.A., Ng B.L., Beavis J.C., Chua Y., Cooke S.L., Raby S., Ichimura K., Collins V.P., Carter N.P., Edwards P.A. 2008. Array painting reveals a high frequency of balanced translocations in breast cancer cell lines that break in cancer-relevant genes. Oncogene. 27:3345–3359 10.1038/sj.onc.1210993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaco I., Canela A., Vera E., Blasco M.A. 2008. Centromere mitotic recombination in mammalian cells. J. Cell Biol. 181:885–892 10.1083/jcb.200803042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.D., Jasin M. 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19:3398–3407 10.1093/emboj/19.13.3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F.B., Marciniak R.A., McVey M., Stewart S.A., Hahn W.C., Guarente L. 2001. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 20:905–913 10.1093/emboj/20.4.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J., Ishikawa F. 2001. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 11:1624–1630 10.1016/S0960-9822(01)00503-6 [DOI] [PubMed] [Google Scholar]
- Kumaraswamy E., Shiekhattar R. 2007. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol. Cell. Biol. 27:6733–6741 10.1128/MCB.00961-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S., Froget B., Carr A.M. 2007. Arrested replication fork processing: interplay between checkpoints and recombination. DNA Repair (Amst.). 6:1042–1061 10.1016/j.dnarep.2007.02.024 [DOI] [PubMed] [Google Scholar]
- Le S., Moore J.K., Haber J.E., Greider C.W. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 152:143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D.A., Court D.L., Jenkins N.A., Copeland N.G. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 73:56–65 10.1006/geno.2000.6451 [DOI] [PubMed] [Google Scholar]
- Lendvay T.S., Morris D.K., Sah J., Balasubramanian B., Lundblad V. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 144:1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.L., Blackburn E.H. 2004. Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol. Cell. Biol. 24:10857–10867 10.1128/MCB.24.24.10857-10867.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., de Lange T. 2003. Rap1 affects the length and heterogeneity of human telomeres. Mol. Biol. Cell. 14:5060–5068 10.1091/mbc.E03-06-0403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Oestreich S., de Lange T. 2000. Identification of human Rap1: implications for telomere evolution. Cell. 101:471–483 10.1016/S0092-8674(00)80858-2 [DOI] [PubMed] [Google Scholar]
- Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S.Y., Qin J., Wong J., Cooney A.J., Liu D., Songyang Z. 2008. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 10:731–739 10.1038/ncb1736 [DOI] [PubMed] [Google Scholar]
- Liu C., Mao X., Lustig A.J. 1994. Mutational analysis defines a C-terminal tail domain of RAP1 essential for Telomeric silencing in Saccharomyces cerevisiae. Genetics. 138:1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyanage M., Coleman A., du Manoir S., Veldman T., McCormack S., Dickson R.B., Barlow C., Wynshaw-Boris A., Janz S., Wienberg J., et al. 1996. Multicolour spectral karyotyping of mouse chromosomes. Nat. Genet. 14:312–315 10.1038/ng1196-312 [DOI] [PubMed] [Google Scholar]
- Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38:431–440 10.1038/ng1760 [DOI] [PubMed] [Google Scholar]
- Lundblad V., Blackburn E.H. 1993. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell. 73:347–360 10.1016/0092-8674(93)90234-H [DOI] [PubMed] [Google Scholar]
- Marcand S., Gilson E., Shore D. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science. 275:986–990 10.1126/science.275.5302.986 [DOI] [PubMed] [Google Scholar]
- Marcand S., Pardo B., Gratias A., Cahun S., Callebaut I. 2008. Multiple pathways inhibit NHEJ at telomeres. Genes Dev. 22:1153–1158 10.1101/gad.455108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M.J., Haber J.E. 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 75:111–135 10.1146/annurev.biochem.74.082803.133234 [DOI] [PubMed] [Google Scholar]
- Miller K.M., Cooper J.P. 2003. The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Mol. Cell. 11:303–313 10.1016/S1097-2765(03)00041-8 [DOI] [PubMed] [Google Scholar]
- Miller K.M., Ferreira M.G., Cooper J.P. 2005. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 24:3128–3135 10.1038/sj.emboj.7600779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K.M., Rog O., Cooper J.P. 2006. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 440:824–828 10.1038/nature04638 [DOI] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 113:631–642 10.1016/S0092-8674(03)00393-3 [DOI] [PubMed] [Google Scholar]
- Morales J.C., Xia Z., Lu T., Aldrich M.B., Wang B., Rosales C., Kellems R.E., Hittelman W.N., Elledge S.J., Carpenter P.B. 2003. Role for the BRCA1 C-terminal repeats (BRCT) protein 53BP1 in maintaining genomic stability. J. Biol. Chem. 278:14971–14977 10.1074/jbc.M212484200 [DOI] [PubMed] [Google Scholar]
- Myung K., Chen C., Kolodner R.D. 2001. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 411:1073–1076 10.1038/35082608 [DOI] [PubMed] [Google Scholar]
- Nakamura H., Morita T., Sato C. 1986. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp. Cell Res. 165:291–297 10.1016/0014-4827(86)90583-5 [DOI] [PubMed] [Google Scholar]
- Nakamura K., Okamoto A., Katou Y., Yadani C., Shitanda T., Kaweeteerawat C., Takahashi T.S., Itoh T., Shirahige K., Masukata H., Nakagawa T. 2008. Rad51 suppresses gross chromosomal rearrangement at centromere in Schizosaccharomyces pombe. EMBO J. 27:3036–3046 10.1038/emboj.2008.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K.J., Yu V.P., Lee H., Corcoran A., Thistlethwaite F.C., Evans M.J., Colledge W.H., Friedman L.S., Ponder B.A., Venkitaraman A.R. 1998. Involvement of Brca2 in DNA repair. Mol. Cell. 1:347–357 10.1016/S1097-2765(00)80035-0 [DOI] [PubMed] [Google Scholar]
- Puget N., Knowlton M., Scully R. 2005. Molecular analysis of sister chromatid recombination in mammalian cells. DNA Repair (Amst.). 4:149–161 10.1016/j.dnarep.2004.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raderschall E., Golub E.I., Haaf T. 1999. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. USA. 96:1921–1926 10.1073/pnas.96.5.1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz L.B., Chehab N.H., Malikzay A., Halazonetis T.D. 2000. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151:1381–1390 10.1083/jcb.151.7.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S., Robles A.I., Linke S.P., Sinogeeva N.I., Zhang R., Pedeux R., Ward I.M., Celeste A., Nussenzweig A., Chen J., et al. 2004. Functional interaction between BLM helicase and 53BP1 in a Chk1-mediated pathway during S-phase arrest. J. Cell Biol. 166:801–813 10.1083/jcb.200405128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman J., Takai H., Buonomo S.B., Eisenhaber F., de Lange T. 2004. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev. 18:2108–2119 10.1101/gad.1216004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science. 314:268–274 10.1126/science.1133427 [DOI] [PubMed] [Google Scholar]
- Smith J.S., Caputo E., Boeke J.D. 1999. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19:3184–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J.M., Lopes M., Foiani M. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 297:599–602 10.1126/science.1074023 [DOI] [PubMed] [Google Scholar]
- Starborg M., Gell K., Brundell E., Höög C. 1996. The murine Ki-67 cell proliferation antigen accumulates in the nucleolar and heterochromatic regions of interphase cells and at the periphery of the mitotic chromosomes in a process essential for cell cycle progression. J. Cell Sci. 109:143–153 [DOI] [PubMed] [Google Scholar]
- Takai H., Tominaga K., Motoyama N., Minamishima Y.A., Nagahama H., Tsukiyama T., Ikeda K., Nakayama K., Nakanishi M., Nakayama K. 2000. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(-/-) mice. Genes Dev. 14:1439–1447 [PMC free article] [PubMed] [Google Scholar]
- Takai H., Naka K., Okada Y., Watanabe M., Harada N., Saito S., Anderson C.W., Appella E., Nakanishi M., Suzuki H., et al. 2002. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 21:5195–5205 10.1093/emboj/cdf506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M.T., Arneric M., Sperisen P., Lingner J. 2004. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 117:323–335 10.1016/S0092-8674(04)00334-4 [DOI] [PubMed] [Google Scholar]
- Teng S.C., Zakian V.A. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083–8093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S.C., Chang J., McCowan B., Zakian V.A. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell. 6:947–952 10.1016/S1097-2765(05)00094-8 [DOI] [PubMed] [Google Scholar]
- Thorslund T., West S.C. 2007. BRCA2: a universal recombinase regulator. Oncogene. 26:7720–7730 10.1038/sj.onc.1210870 [DOI] [PubMed] [Google Scholar]
- Tripathi V., Nagarjuna T., Sengupta S. 2007. BLM helicase–dependent and –independent roles of 53BP1 during replication stress–mediated homologous recombination. J. Cell Biol. 178:9–14 10.1083/jcb.200610051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V., Kaur S., Sengupta S. 2008. Phosphorylation-dependent interactions of BLM and 53BP1 are required for their anti-recombinogenic roles during homologous recombination. Carcinogenesis. 29:52–61 10.1093/carcin/bgm238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H.J., Huang W.H., Li T.K., Tsai Y.L., Wu K.J., Tseng S.F., Teng S.C. 2006. Involvement of topoisomerase III in telomere-telomere recombination. J. Biol. Chem. 281:13717–13723 10.1074/jbc.M600649200 [DOI] [PubMed] [Google Scholar]
- van Veelen L.R., Essers J., van de Rakt M.W., Odijk H., Pastink A., Zdzienicka M.Z., Paulusma C.C., Kanaar R. 2005. Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52. Mutat. Res. 574:34–49 [DOI] [PubMed] [Google Scholar]
- Wang J., Rao S., Chu J., Shen X., Levasseur D.N., Theunissen T.W., Orkin S.H. 2006. A protein interaction network for pluripotency of embryonic stem cells. Nature. 444:364–368 10.1038/nature05284 [DOI] [PubMed] [Google Scholar]
- Ward I.M., Minn K., van Deursen J., Chen J. 2003. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23:2556–2563 10.1128/MCB.23.7.2556-2563.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R.S., Enoch T., Leder P. 2000. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 14:1886–1898 [PMC free article] [PubMed] [Google Scholar]
- Wotton D., Shore D. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11:748–760 10.1101/gad.11.6.748 [DOI] [PubMed] [Google Scholar]
- Xie A., Hartlerode A., Stucki M., Odate S., Puget N., Kwok A., Nagaraju G., Yan C., Alt F.W., Chen J., et al. 2007. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 28:1045–1057 10.1016/j.molcel.2007.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Blackburn E.H. 2004. Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J. Cell Biol. 167:819–830 10.1083/jcb.200408181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Ashley T., Brainerd E.E., Bronson R.T., Meyn M.S., Baltimore D. 1996. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10:2411–2422 10.1101/gad.10.19.2411 [DOI] [PubMed] [Google Scholar]
- Yamane K., Wu X., Chen J. 2002. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol. Cell. Biol. 22:555–566 10.1128/MCB.22.2.555-566.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos G., Rainey M.D., Gillespie D.A. 2003. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J. 22:713–723 10.1093/emboj/cdg060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.D., Küster B., Mann M., Petrini J.H., de Lange T. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25:347–352 10.1038/77139 [DOI] [PubMed] [Google Scholar]