A screen identifies two more bouquet proteins required for meiotic telomere clustering: Bqt4 anchors the telomeres, whereas Bqt3 protects Bqt4 from degradation.
Abstract
In many organisms, telomeres cluster to form a bouquet arrangement of chromosomes during meiotic prophase. Previously, we reported that two meiotic proteins, Bqt1 and -2, are required for tethering telomeres to the spindle pole body (SPB) during meiotic prophase in fission yeast. This study has further identified two novel, ubiquitously expressed inner nuclear membrane (INM) proteins, Bqt3 and -4, which are required for bouquet formation. We found that in the absence of Bqt4, telomeres failed to associate with the nuclear membranes in vegetative cells and consequently failed to cluster to the SPB in meiotic prophase. In the absence of Bqt3, Bqt4 protein was degraded during meiosis, leading to a phenotype similar to that of the bqt4-null mutant. Collectively, these results show that Bqt4 anchors telomeres to the INM and that Bqt3 protects Bqt4 from protein degradation. Interestingly, the functional integrity of telomeres is maintained even when they are separated from the nuclear envelope in vegetative cells.
Introduction
Meiosis is a process required by all sexually reproducing eukaryotic organisms to generate haploid gametes from a diploid cell. Upon entering meiosis, chromosomes undergo significant movements within the nucleus to accomplish pairing and recombination of homologous chromosomes and subsequent chromosome segregation. During this process, telomeres dramatically change their organization: although telomeres are dispersed along the nuclear envelope (NE) during the mitotic cell cycle, during meiotic prophase, they form a cluster adjacent to the NE to produce a characteristic bundled configuration of chromosomes, which is called the bouquet arrangement (for reviews see Scherthan, 2001; Harper et al., 2004; Zickler, 2006; Chikashige et al., 2007). Bouquet formation is important for pairing and recombination of homologous chromosomes, which are essential for correct segregation of chromosomes during the meiotic divisions. Thus, understanding the mechanisms underlying meiotic chromosomal bouquet formation is clinically important because chromosome missegregation during meiosis is a major cause of human miscarriage and developmental abnormalities (Hassold et al., 1996).
A striking example of chromosomal bouquet formation has been demonstrated in the fission yeast Schizosaccharomyces pombe. In this organism, centromeres and telomeres show dramatic differences in organization upon entering meiosis. During the mitotic cell cycle, centromeres cluster near the spindle pole body (SPB; a centrosome-equivalent structure in fungi; Funabiki et al., 1993); however, during meiotic prophase, the centromeres detach from the SPB, and the telomeres cluster next to the SPB (Chikashige et al., 1994). Subsequently, the reorganized nucleus elongates and oscillates between the cell poles (known as the horsetail nucleus). This process is mediated by cytoplasmic microtubules and by cytoplasmic dynein, a microtubule motor protein complex (Ding et al., 1998; Yamamoto et al., 1999; for review see Yamamoto and Hiraoka, 2001). Loss of telomere clustering or horsetail nuclear movement results in reduced homologous recombination and increased missegregation (for reviews see Cooper, 2000; Yamamoto and Hiraoka, 2001). Direct observations of living cells have demonstrated that telomere clustering and horsetail nuclear movement facilitate the alignment of homologous chromosomes (Ding et al., 2004). Thus, chromosomal bouquet formation, or telomere clustering, is an important initial step in chromosome movements during meiosis.
Studies in S. pombe have revealed that bouquet formation involves interaction between a SUN domain protein, Sad1, and a putative KASH domain protein, Kms1. SUN and KASH domain proteins form complexes that span both membranes of the NE (Starr et al., 2001; for reviews see Starr and Han, 2003; Gruenbaum et al., 2005; Tzur et al., 2006). In S. pombe, Sad1 and Kms1 are components of the SPB (Hagan and Yanagida, 1995; Shimanuki et al., 1997). During meiosis, the Sad1–Kms1 protein complex links telomeres to microtubules and cytoplasmic dynein, which leads to telomere clustering at the SPB and telomere-led horsetail movement of the entire nucleus (for review see Chikashige et al., 2007).
Additional work from other organisms has indicated that the role of SUN–KASH domain pairs may be a conserved aspect of bouquet formation (for reviews see Chikashige et al., 2007; Starr, 2009). In the nematode Caenorhabditis elegans, a set of four zinc finger proteins, each specifically binding to one or two chromosomes, mediates pairing of homologous chromosomes in meiosis (Phillips et al., 2005; Phillips and Dernburg, 2006). These proteins associate with internal chromosome sites, which are known as pairing centers, and mediate their interactions with SUN-1 and ZYG-12 (Penkner et al., 2007), a SUN–KASH pair homologous to Sad1 and Kms1. ZYG-12 can interact with cytoplasmic dynein (Malone et al., 2003). In the budding yeast Saccharomyces cerevisiae, recent studies have shown that the SUN domain protein Mps3 is required for meiotic telomere clustering through interaction with Csm4 and Ndj1, and intranuclear movements of telomeres are mediated through actin filaments (Trelles-Sticken et al., 2005; Conrad et al., 2007, 2008; Scherthan et al., 2007; Kosaka et al., 2008; Koszul et al., 2008; Wanat et al., 2008). In mice, the SUN domain proteins Sun1 and -2 are dispersed along the envelope in somatic cells but relocalize to discrete foci where telomeres are attached to the NE in spermatocytes (Ding et al., 2007; Schmitt et al., 2007), suggesting that these proteins move within the NE during meiosis. Deletion of Sun1 causes defects in telomere attachment to the NE and in gametogenesis (Ding et al., 2007).
S. pombe Taz1 and Rap1 are constitutive telomere-associated proteins; loss of Taz1 or Rap1 does not affect mitotic cell viability but results in loss of telomere clustering during meiotic prophase (Cooper et al., 1997, 1998; Nimmo et al., 1998; Chikashige and Hiraoka, 2001; Kanoh and Ishikawa, 2001). As a prerequisite for telomere clustering, meiotically expressed Bqt1 and -2 connect the Rap1 telomere protein to the Sad1–Kms1 protein complex in the NE; Sad1–Kms1 complexes then tether telomeres to the SPB along the nuclear membranes using cytoplasmic microtubules through an interaction between Kms1 and dynein (for review see Chikashige et al., 2007).
In this study, further screening for factors that are required for meiotic telomere clustering in S. pombe has identified two inner nuclear membrane (INM) proteins, Bqt3 and -4. Our findings show that Bqt4 is required for anchoring telomeres to the nuclear membranes, which is mediated through its interaction with the Rap1 telomere protein in both vegetative and meiotic cells in the absence of Bqt1 and -2. Bqt3 is required during meiosis to protect Bqt4 from protein degradation by a yet-unknown mechanism. In the absence of Bqt3 or -4, telomeres fail to associate with the nuclear membrane and therefore usually fail to establish a connection to the Sad1–Kms1 complex once the Bqt1–Bqt2 complex is expressed during meiosis.
Results
Two novel nuclear membrane proteins required for bouquet formation
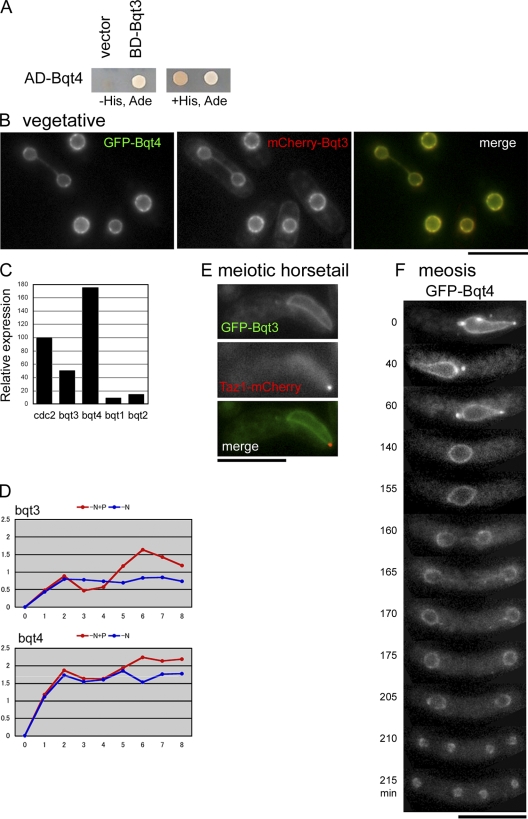
In a previous study, we generated 83 strains systematically deleted for each of the S. pombe genes that are induced by mating pheromone (Chikashige et al., 2006). We reported that one of the strains showed a complete loss in meiotic telomere clustering (bouquet formation), and named this gene bqt1 for bouquet (Chikashige et al., 2006). This was the only strain showing a complete loss of telomere clustering, but among the same 83 strains, we also observed one strain with a partial defect in telomere clustering. We named the corresponding gene bqt3 (S. Pombe GeneDB gene ID: SPCC594.07c). In a yeast two-hybrid screen using Bqt3 as the bait, we identified an interacting protein (S. Pombe GeneDB gene ID: SPBC19C7.10). We found that deletion of this gene resulted in loss of bouquet formation and have therefore named it bqt4. Two-hybrid interaction data for Bqt3 and -4 are shown in Fig. 1 A.
Figure 1.
Expression and localization of Bqt3 and -4. (A) Two-hybrid interaction of Bqt3 and -4. AD and BD indicate the constructs based on the pGADT7 and pGBKT7 plasmids, respectively (see Materials and methods). (B) Localization of GFP-Bqt4 (green) and mCherry-Bqt3 (red) in vegetatively growing cells. The observed strain is CRLt53 (h90 bqt3Δ::LEU2, bqt4Δ::ura4+, aur1::mCherry-bqt3, lys1::GFP-bqt4). (C) bqt3+ and -4+ genes were expressed in vegetatively growing cells. Relative levels of expression were determined by real-time PCR, under which the expression of cdc2+ was normalized to 100 as a standard. The mean of two experiments is shown. (D) The expression profiles of Bqt3 and -4 upon nitrogen starvation and mating pheromone (P-factor) treatment analyzed by DNA microarray experiments (as described in Chikashige et al. [2006]). Blue indicates nitrogen starvation. Red indicates P-factor treatment with nitrogen starvation. The vertical axis is the expression ratio relative to the vegetative level on a base 2 logarithmic scale, as described in Chikashige et al. (2006). The ratio of the expression levels of each gene is plotted for each time point (hours) after nitrogen starvation. (E) Localization of GFP-Bqt3 (green) and Taz1-mCherry (red) in a meiotic cell. The observed strain is CRLt73 (h90 aur1::GFP-bqt3, taz1-mCherry::kan). (F) Live observation of cells expressing GFP-Bqt4 in meiosis. The strain observed is CRLs53 (h90 bqt4Δ::ura4+, lys1::GFP-bqt4, taz1-mCherry::kan). Selected frames of time-lapse images are shown. The image at each time point is a projection of optical section images. Aggregates of Bqt4 often appear on the nuclear membranes. These aggregates sporadically detach from the trailing edge of the moving nucleus, as shown in Video 1. Numbers on the left of each image indicate the time in minutes. Bars, 10 µm.
Full-length cDNAs of these genes were isolated, and analysis revealed that the bqt3 and -4 genes encode proteins of 255 and 432 amino acids, respectively. We examined the amino acid sequences of Bqt3 and -4 with three prediction tools for identification of transmembrane sequences in proteins. According to the predictions, both Bqt3 and -4 were expected to be membrane proteins: Bqt3 contains six to eight transmembrane helices (TMHs) distributed throughout the protein, whereas Bqt4 contains a single TMH at its C terminus.
To test the prediction that Bqt3 and -4 are membrane proteins, we examined their subcellular localization using fluorescent protein fusion constructs. Although Bqt3 was originally identified by virtue of its up-regulation by mating pheromone (Fig. 1 D), it was found to be expressed in vegetatively growing cells (Fig. 1, B and C). We found that in both vegetative cells (Fig. 1 B) and meiotic cells (Fig. 1 E), Bqt3 was associated with the nuclear periphery. Bqt4 also localized to the nuclear periphery in both vegetative and meiotic cells (Fig. 1, B, C, and F; and Video 1). Together with the physical interaction detected by the yeast two-hybrid assay, these results suggest that Bqt3 and -4 form a complex that localizes to the NE.
Strains deleted for bqt3 and -4 show loss of bouquet formation
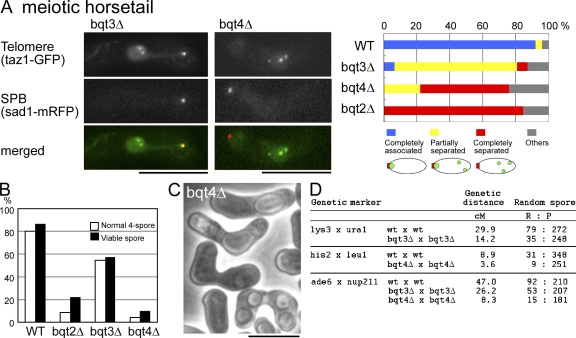
To characterize the functions of Bqt3 and -4, we examined the phenotypes of S. pombe cells deleted for either the bqt3 or -4 gene (bqt3Δ and -4Δ, respectively). Although both genes were expressed during vegetative growth, bqt3Δ and -4Δ strains did not perturb growth during the mitotic cell cycle (Fig. S1 B). Although both strains were defective in telomere clustering to the SPB during meiotic prophase, as mentioned in the previous section, the extent of the defects in each strain was different. In bqt3Δ cells, complete loss of telomere clustering was observed in only 6% of the cells examined (3 out of 47 cells). In the majority of bqt3Δ cells (74%; 35 out of 47 cells), partial loss of telomere clustering was observed: in these cells, telomeres were scattered, producing several spots, but one spot was associated with the SPB (Fig. 2 A). Normal, complete clustering of telomeres to the SPB was observed in 6% of the cells examined (3 out of 47 cells). In contrast, in bqt4Δ cells, there were no examples showing complete clustering of telomeres to the SPB. Partial loss of telomere clustering was observed in 22% of the cells (11 out of 50 cells), and complete loss of telomere clustering was observed in 54% of the cells (27 out of 50 cells; Fig. 2 A). These results contrast with bqt1Δ and -2Δ, which showed complete loss of telomere clustering in the majority of cells (Fig. 2 A, right). In addition, both bqt3Δ and -4Δ cells showed defects in viable spore formation and meiotic recombination frequency, as previously observed for other bouquet defective strains such as rap1Δ, taz1Δ, or bqt1Δ or -2Δ (Fig. 2, B–D; Chikashige et al., 2006). For each of these meiotic phenotypes, the defects in bqt3Δ cells were more variable than the bqt4Δ defects. Because deletion of bqt4 gave rise to stronger defects in meiosis than bqt3Δ, we expected that Bqt4 would play a major role in bouquet formation.
Figure 2.
Phenotypes of bqt3Δ and -4Δ cells. (A) Loss of telomere clustering in bqt3Δ and -4Δ cells. Cells expressing Taz1-GFP (green) and Sad1-mRFP (red) were observed after the induction of meiosis. The image at each wavelength is a projection of selected optical section images. The strains used are CRLs63 (h90 bqt3Δ::ura4+, taz1Δ::ura4+, lys1::taz1-GFP, sad1-mCherry::kan) and CRLp85 (h90 bqt4Δ::ura4+, taz1Δ::ura4+, lys1::taz1-GFP, sad1-mCherry::kan). (B) Spore formation and spore viability. Normal four-spore formation percentage = 100 x (number of asci containing four normal spores)/(number of randomly selected asci dissected). Spore viability percentage = 100 × (number of viable spores)/(4 × [number of asci dissected randomly]). The strains used are CRLs95 (wild type [WT]; 76 asci), CRLm03 (bqt2Δ::ura4+; 90 asci), CRLs57 (bqt3Δ::ura4+; 77 asci), and CRLp86 (bqt4Δ::ura4+; 93 asci). (C) Phase contrast image of asci of the bqt4Δ strain with defects in spore formation. (D) Random spore analysis and genetic linkage. Genetic distance according to Haldane's formula: d = −50 × ln(1 − 2R/[R + P]), where R is the number of recombinants and P is the number of colonies with parental genotype. Bars, 10 µm.
Bqt3 and -4 are embedded in the INM
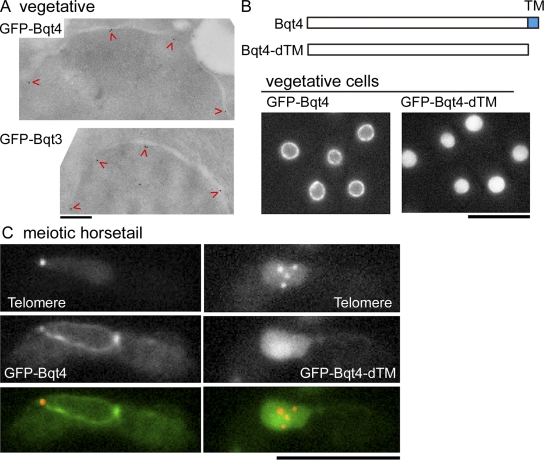
We investigated the precise localization of Bqt3 and -4 proteins using immunoelectron microscopy. As shown in Fig. 3 A, when GFP fusion proteins, in which GFP was fused to the N terminus of Bqt3 or -4, were detected with immunogold-conjugated antibodies, most of the gold signals were observed just interior to the INM. The Bqt4 protein is predicted to include one TMH at the C-terminal end. To test whether the TMH plays a role in the localization of Bqt4 to the INM, we constructed a Bqt4 protein deleted for the TMH (designated Bqt4-dTM) by removing 19 amino acid residues from the C-terminal end and examined its subcellular localization. As shown in Fig. 3 (B and C), the Bqt4-dTM protein localized uniformly throughout the nucleus. These results indicate that Bqt4 is embedded in the INM through its C-terminal TMH, with its N terminus oriented toward the nucleoplasm. As shown in Fig. 3 C, Bqt4-dTM expression did not suppress the defect of telomere clustering in the bqt4Δ strain. A normal telomere cluster was found in none of the GFP–Bqt4-dTM–expressing cells examined (n = 56), whereas 77% of GFP-Bqt4–expressing cells formed a complete telomere cluster (n = 124). Therefore, localization of Bqt4 to the INM must be essential for telomere clustering.
Figure 3.
Localization of Bqt3 and -4 to the INM. (A) Localization of Bqt4 and -3 proteins in vegetatively growing cells, as determined by immunoelectron microscopy. Arrowheads indicate GFP-Bqt4 or -Bqt3, in which GFP was fused to the N terminus of Bqt4 or -3. Samples were stained with polyclonal anti-GFP and immunogold-conjugated secondary antibodies. The observed strains are CRLr06 (h− bqt4Δ::ura4+, lys1::GFP-bqt4) and CRLs86 (h− bqt3Δ::LEU2, lys1::GFP-bqt3). (B) Localization of GFP-Bqt4 and –Bqt4-dTM in vegetatively growing cells. The observed strains are CRLt53 (left; h− bqt4Δ::ura4+, lys1::GFP-bqt4) and CRLt54 (right; h− bqt4Δ::ura4+, lys1::GFP–bqt4-dTM). (C) Expression of Bqt4-dTM protein did not suppress the loss of telomere clustering. Cells expressing both Taz1-mRFP (red) and GFP-Bqt4 or –Bqt4-dTM (green) were observed after the induction of meiosis. The observed strains are CRLs53 (h90 bqt4Δ::ura4+, lys1::GFP-bqt4, taz1-mCherry::kan) and CRLs38 (h90 bqt4Δ::ura4+, lys1::GFP–bqt4-dTM, taz1-mCherry::kan). Bars: (A) 200 nm; (B and C) 10 µm.
Telomeres are released from the NE in bqt4Δ
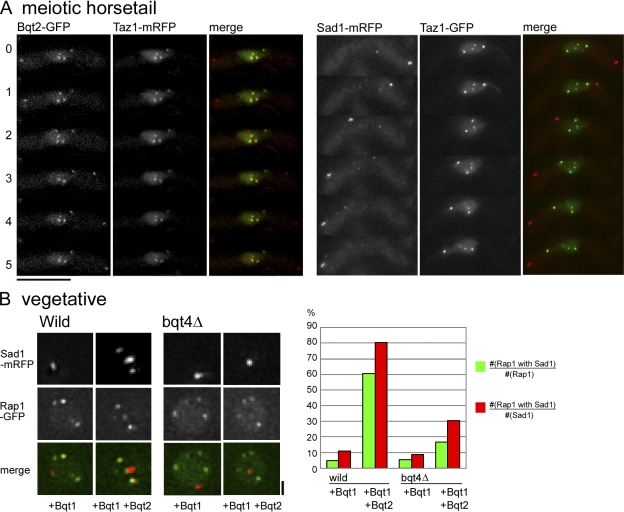
We examined the interdependence of telomere and SPB targeting of bouquet components. In bqt4Δ cells, Bqt2 protein colocalized with the scattered telomeres (Fig. 4 A, left). In contrast, Sad1 protein was localized primarily at the SPB and not with the scattered telomeres during meiotic prophase. Although weak foci of Sad1 could be occasionally seen away from the SPB (Fig. 4 A, right), these minor spots did not colocalize with telomeres, suggesting that they are unrelated to telomere dynamics. However, this observation may indicate that Bqt4 affects Sad1 mobility in the NE. We previously showed that ectopic expression of Bqt1 and -2 in vegetatively growing cells is sufficient to induce Sad1 colocalization with telomeres (Chikashige et al., 2006), and we found that this colocalization was strongly reduced in bqt4Δ cells (Fig. 4 B). These observations indicate that Sad1 cannot accumulate on telomeres in the absence of Bqt4 protein even if the connector Bqt2 is recruited normally to telomeres. Because Sad1p is a membrane protein containing one TMH, it is expected that its localization is confined to the NE. Thus, we hypothesized that telomeres are released from the NE in bqt4Δ cells, leading to the loss of interaction between telomeres and Sad1.
Figure 4.
Localization of Bqt2 and Sad1 in the absence of Bqt4. (A, left) Cells expressing Bqt2-GFP (red) and Taz1-mRFP (green) were observed after the induction of meiosis. The strain observed is CRLp83 (h90 bqt4Δ::ura4+, taz1Δ::ura4+, lys1::taz1-mRFP, bqt2-GFP::kan). (right) Cells expressing Sad1-mRFP (red) and Taz1-GFP (green) were observed after induction of meiosis. The strain observed is CRLp85 (h90 bqt4Δ::ura4+, taz1Δ::ura4+, lys1::taz1-GFP, sad1-mRFP::kan). Time-lapse images were recorded at intervals of 1 min. The image at each time point is a projection of optical section images. Numbers on the left of images indicate the time in minutes. (B) Localization of Sad1 (mRFP) and Rap1 (GFP) in vegetatively growing cells in which Bqt1 alone or both Bqt1 and -2 were ectopically expressed. In bqt4+ cells, Sad1 showed multiple foci, and many of these Sad1 foci colocalized with Rap1 when both bqt1 and -2 were expressed. The colocalization of Sad1 foci with Rap1 after ectopic expression was more extensive in bqt4+ than in bqt4Δ cells. Strains observed in each column are CRLs27 (bqt4+, CFP-bqt1), CRLs28 (bqt4+, CFP-bqt1, bqt2-myc), CRLs29 (bqt4Δ, CFP-bqt1), and CRLs30 (bqt4Δ, CFP-bqt1, bqt2-myc) from left to right. The graph shows the frequencies of colocalization of Sad1 and Rap1 in each strain. Numbers of Sad1 and Rap1 spots examined in each strain, from left to right, are 66 Sad1 spots and 153 Rap1 spots in 39 nuclei, 141 Sad1 spots and 187 Rap1 spots in 47 nuclei, 82 Sad1 spots and 141 Rap1 spots in 43 nuclei, and 89 Sad1 spots and 161 Rap1 spots in 51 nuclei. The green columns indicate the frequency with which Rap1 foci colocalized with Sad1 relative to total observed Rap1 foci. The red columns indicate the frequency with which Sad1 foci colocalized with Rap1 relative to total observed Sad1 foci. Bars: (A) 10 µm; (B) 1 µm.
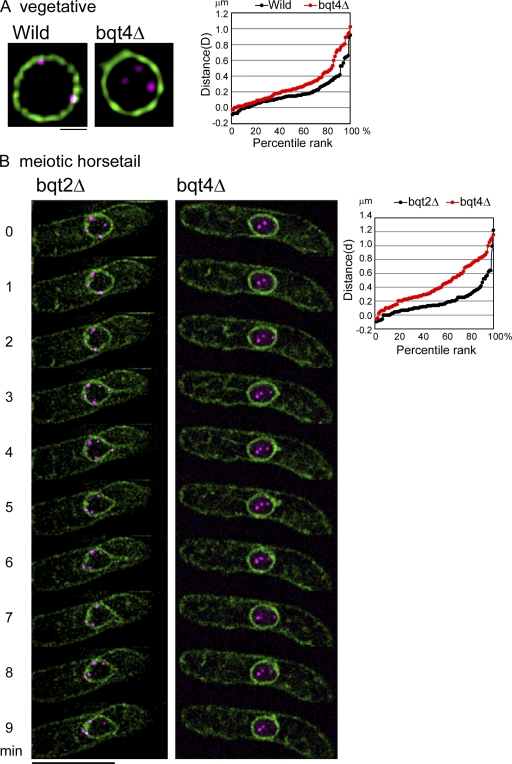
To test this hypothesis, we compared the distances between telomeres and the NE in bqt4Δ and wild-type cells (see Materials and methods). The results are shown for vegetative cells in Fig. 5 A and for meiotic prophase cells in Fig. 5 B. The telomere–NE distance distribution was clearly weighted toward longer distances in bqt4Δ than in bqt4+ vegetative cells (Fig. 5 A). A more pronounced difference was observed in meiotic prophase nuclei, which are elongated and moving. As shown in Fig. 5 B, scattered telomeres are observed along the NE in bqt2Δ meiotic prophase cells, which is consistent with our observations that telomeres are anchored to the NE but are not captured by Sad1 in the absence of the Bqt2 connector. In contrast, in the bqt4Δ cells, telomeres are not only dispersed but are not constrained to lie near the NE. Thus, we conclude that the association of telomeres with Sad1 requires their association with the NE, which is mediated by Bqt4.
Figure 5.
Telomeres are released from the NE in bqt4Δ cells. (A and B) Cells expressing Taz1-mCherry (magenta) and Ccr1N-GFP (green) were observed during mitosis (A) and meiosis (B). Numbers on the left of each image in B indicate the time in minutes. The image at each time point is a projection of three optical sections around the nuclear midline taken in three dimensions. The strains observed are CRLr75 (wild type), CRLr81 (bqt4Δ), and CRLr82 (bqt2Δ). Percentile ranks of the distance between the telomere and the NE are shown in the right panels. The vertical axis is the 3D radial distance D for mitotic cells and is the 2D in-focus distance d for meiotic cells (see Fig. 9 and Materials and methods). In a percentile rank plot, the percentage of telomeres in which the distance to the NE is equal or less than a given distance is plotted as a function of the distance. Bars: (A) 1 µm; (B) 10 µm.
Bqt4 anchors telomeres to the NE through association with Rap1
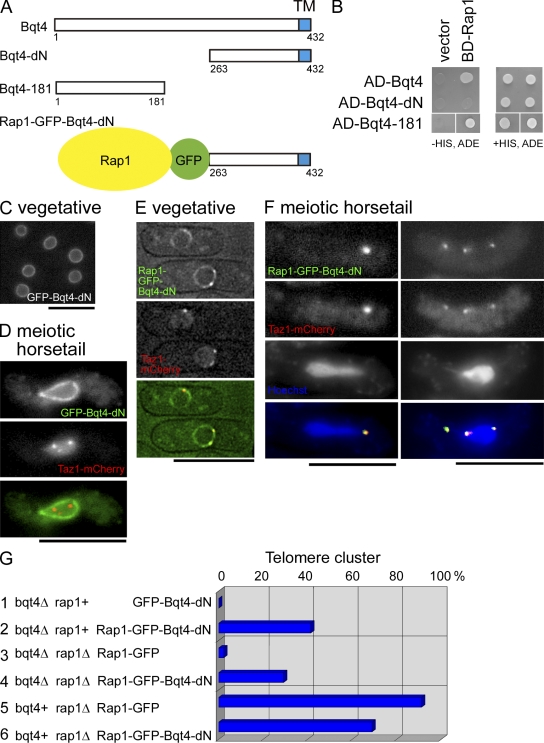
As described in the previous sections, Bqt4 localizes at the nucleoplasmic surface of the INM, and telomeres do not associate with the NE in the absence of Bqt4, therefore being unable to form the bouquet. These results suggest that Bqt4 provides an attachment surface for telomeres to the NE. Using a yeast two-hybrid assay, we found that Bqt4 protein interacts with Rap1, a constitutively telomere-associated protein (Chikashige and Hiraoka, 2001; Kanoh and Ishikawa, 2001). Further analysis showed that the N-terminal portion of Bqt4 is necessary and sufficient for interaction with Rap1: a truncated protein lacking the N-terminal 262 amino acid residues (Bqt4-dN) did not bind to Rap1, whereas a fragment containing the N-terminal 181 amino acid residues of Bqt4 (Bqt4-181) did bind (Fig. 6 B). However, Bqt4-dN localized at the NE (Fig. 6, C and D) but did not rescue the defects in bouquet formation exhibited by bqt4Δ cells (Fig. 6, D and G). These results indicate that the binding of Rap1 to the N-terminal domain of Bqt4 is necessary for telomere clustering.
Figure 6.
The interaction between Bqt4 and Rap1 is essential for telomere clustering. (A) Diagram of Bqt4 constructs. (B) Yeast two-hybrid assay between Bqt4 constructs and the full-length Rap1. AD and BD indicate the constructs based on the pGADT7 and pGBKT7 plasmids, respectively (see Materials and methods). (C) NE localization of GFP–Bqt4-dN in vegetative bqt4Δ cells. (D) NE localization of GFP–Bqt4-dN and scattered telomeres (Taz1-mCherry) in the horsetail nucleus in a bqt4Δ cell. (E and F) Localization of Rap1-GFP–Bqt4-dN in vegetative cells (E) and in meiotic horsetail (F) in bqt4Δ. (G) Percentages of nuclei bearing a single cluster of telomeres in each strain as indicated. Strains observed are row 1, CRLs25 (n = 103); row 2, CRLs33 (n = 128); row 3, CRLx02 (n = 86); row 4, CRLx01 (n = 113); row 5, CRLx03 (n = 57); and row 6, CRLx04 (n = 74). Bars, 10 µm.
To further investigate this possibility, we constructed a Bqt4-dN protein fused directly with Rap1 (Rap1-GFP–Bqt4-dN) and examined whether expression of the fusion construct could suppress the bqt4Δ mutation. The fusion protein localized to the NE and telomeres during the vegetative growth, reflecting the localization of Bqt4 and Rap1 (Fig. 6 E). During meiotic prophase, Rap1-GFP–Bqt4-dN partially restored meiotic telomere clustering in bqt4Δ cells. The frequency of telomere clustering increased to 29% in bqt4Δ rap1Δ double-deletion cells expressing Rap1-GFP–Bqt4-dN (Fig. 6 G, row 4; compare with row 3 as a control). The fusion protein had reduced functionality of Rap1, as shown in Fig. 6 G (compare row 6 with row 5). In the presence of wild-type Rap1, expression of Rap1-GFP–Bqt4-dN further increased the telomere clustering frequency to 41% in bqt4Δ cells (Fig. 6 G, row 2); the rest of the cells showed scattered telomeres. Rap1-GFP–Bqt4-dN colocalized with both clustered and scattered telomeres (Fig. 6 F, left and right, respectively). Although the partial rescue suggests that the fusion protein cannot fully substitute for wild-type Bqt4, these results indicate that the N-terminal domain of Bqt4 anchors telomeres to the INM through an interaction with Rap1, which is necessary for bouquet formation. Collectively, telomeres attach to the NE through the interaction between Rap1 and Bqt4 extending from the INM, and this attachment ensures that Sad1 encounters telomeres on the NE to form the bouquet arrangement in meiosis.
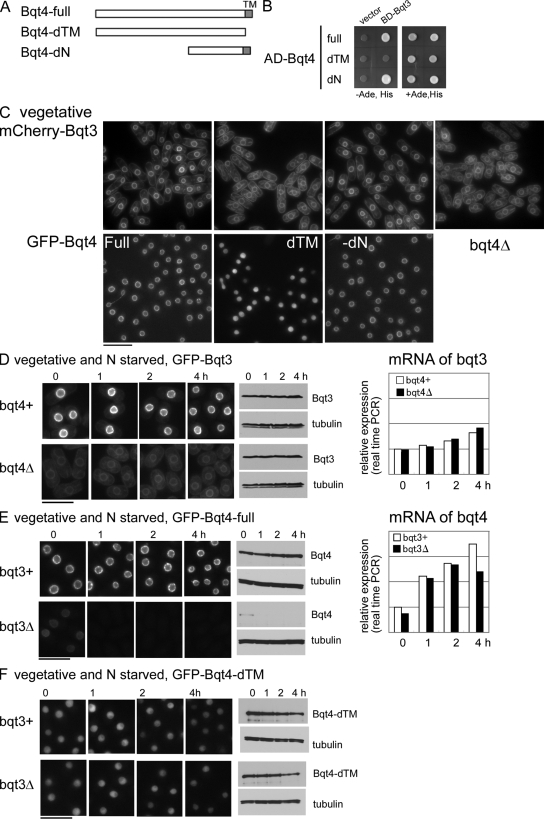
Bqt3 stabilizes Bqt4 in the INM
Bqt3 and -4 displayed interdependent localization to the NE. In the absence of Bqt4, levels of Bqt3 mRNA and protein remained unchanged (Fig. 7 D). However, Bqt3 protein was localized to various membrane-bound compartments such as the ER (Fig. 7 C, rightmost panel) rather than being restricted to the NE. Similar mislocalization of Bqt3 was also observed in cells expressing Bqt4-dTM, which does not bind to Bqt3 (Fig. 7 C, dTM). Expression of the Bqt4-dN protein, which interacts with Bqt3 but not with Rap1, rescued the diffuse localization of Bqt3 (Fig. 7 C, dN), although it did not restore telomere cluster formation (Fig. 6 D).
Figure 7.
Bqt3 stabilizes Bqt4 in the INM. (A) Bqt4-dTM represents Bqt4 deleted for the TMH by removal of 19 amino acid residues from the C-terminal. Bqt4-dN represents Bqt4 deleted of the N-terminal 262 amino acid residues. (B) Two-hybrid interactions of Bqt4 and its deletion derivatives with Bqt3. AD and BD indicate the constructs based on the pGADT7 and pGBKT7 plasmids, respectively (see Materials and methods). (C) Localization of mCherry-Bqt3 in bqt4-full (CRLu61; h− bqt3Δ::LEU2, aur1::mCherry-bqt3, bqt4Δ::ura4+ lys1::GFP-bqt4), bqt4-dTM (CRLu62; h− bqt3Δ::LEU2, aur1::mCherry-bqt3, bqt4Δ::ura4+ lys1::GFP–bqt4-dTM), bqt4-dN (CRLu63; h− bqt3Δ::LEU2, aur1::mCherry-bqt3, bqt4Δ::ura4+ lys1::GFP–bqt4-dN), and bqt4Δ (CRLu69; h− bqt3Δ::LEU2, aur1::mCherry-bqt3, bqt4Δ::ura4+) was observed in mitotic cells. (D–F) Heterothallic h− haploid cells expressing GFP-Bqt3 (D), -Bqt4 (E), and –Bqt4-dTM (F) were observed in the vegetative phase and after nitrogen starvation. Numbers on the top of each image indicate the time in hours after nitrogen starvation. (D) In bqt4Δ cells, levels of Bqt3 mRNA and protein remained unchanged. The strains examined are CRLt01 (h− bqt3Δ::LEU2, aur1::GFP-bqt3, bqt4+) and CRLs87 (h− bqt3Δ::LEU2, aur1::GFP-bqt3, bqt4Δ::ura4+). The graph shows the bqt3 mRNA level measured by real time PCR (mean of two experiments). (E) In bqt3Δ cells, Bqt4 protein was detected at low levels on the NE but disappeared after nitrogen starvation. Levels of Bqt4 protein measured by Western blotting were markedly decreased in bqt3Δ cells. The graph shows the bqt4 mRNA level, as measured by real-time PCR (mean of two experiments). The amount of bqt4 mRNA in bqt3Δ is comparable with that in bqt3+ in vegetative cells as well as after nitrogen starvation. The strains examined are CRLr06 (h− bqt3+, bqt4Δ::ura4+, lys1::GFP-bqt4) and CRLp93 (h− bqt3Δ::LEU2, bqt4Δ::ura4+, lys1::GFP-bqt4). (F) The Bqt4-dTM protein, which does not localize to the NE, escaped degradation in bqt3Δ cells. The strains examined are CRLr24 (h− bqt3+, bqt4Δ::ura4+, lys1::GFP–bqt4-dTM) and CRLr25 (h− bqt3Δ::LEU2, bqt4Δ::ura4+ lys1::GFP–bqt4-dTM). Bars, 10 µm.
Conversely, in bqt3Δ cells, Bqt4 protein was faintly observed at the NE but disappeared upon nitrogen starvation (Fig. 7 E). The disappearance of Bqt4 did not reflect its mislocalization but rather its degradation. In the absence of Bqt3, levels of Bqt4 protein, as measured by Western blotting, markedly decreased, whereas the level of bqt4 mRNA remained unchanged both during vegetative growth and after nitrogen starvation (Fig. 7 E). Intriguingly, the Bqt4-dTM protein, which does not localize to the NE, escaped degradation in bqt3Δ cells (Fig. 7 F), suggesting that the degradation of Bqt4 may occur either at the NE or through the secretory pathway. These results indicate that Bqt3 protein is essential during meiotic prophase to stabilize Bqt4 protein in the nuclear membrane and suggest that the “leaky” bouquet formation defects in bqt3Δ cells may reflect variable residual levels of Bqt4 protein.
Discussion
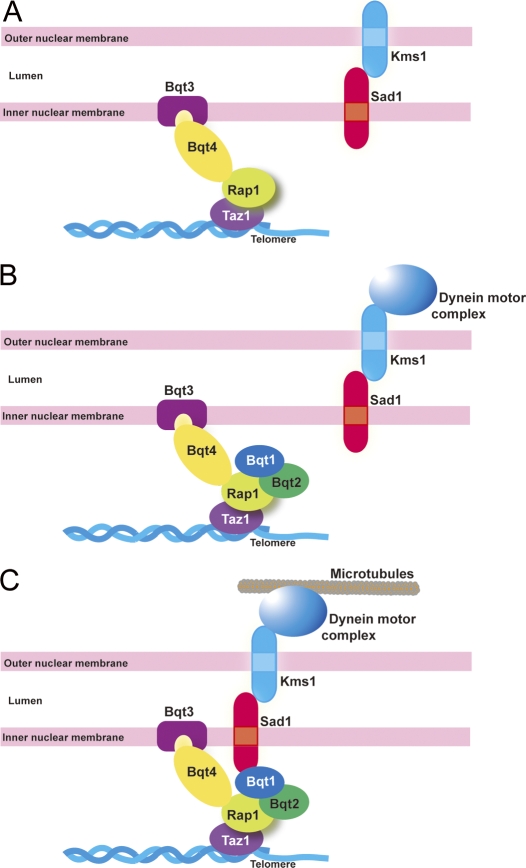
We have identified Bqt3 and -4 as two new INM proteins that are necessary to connect telomeres to the NE during both vegetative growth and meiosis. This connection ensures clustering of telomeres to the SPB when cells enter meiotic prophase, as schematically summarized in Fig. 8. Bqt4 provides an attachment surface for telomeres to the NE through direct interaction between Rap1 and Bqt4 (Fig. 8 A). During meiosis, Bqt3 protects Bqt4 from degradation. In response to mating pheromone signaling during meiosis, Bqt1 and -2 are expressed and accumulate at telomeres through their interaction with Rap1 (Fig. 8 B). An interaction between Sad1 and Bqt1 mediates the association of the Sad1–Kms1 complex and associated microtubule motor proteins with NE-bound telomeres, culminating with telomere clustering at the SPB and dramatic horsetail movement (Fig. 8 C).
Figure 8.
A group of proteins required for telomere positioning. (A) The Bqt3–Bqt4 complex provides an attachment surface for telomeres to the NE in vegetative cells. (B and C) The Bqt1–Bqt2 complex binds to Rap1 at the telomere upon mating pheromone signaling during meiosis, followed by accumulation of the Sad1–Kms1 complex to the telomere through interaction with Bqt1.
Anchoring telomeres to the NE
Bqt4 is an INM protein required to anchor telomeres to the NE. The distribution of the measured telomere–envelope distances is larger in bqt4Δ than in wild-type or bqt2Δ cells (as shown in Fig. 5). This is the first demonstration of the INM protein that directly interacts with telomere-associated proteins in S. pombe. When the telomere–envelope connection is lost in S. cerevisiae, telomeres are randomly distributed throughout the nucleus (Hediger et al., 2002; Taddei et al., 2004). However, in S. pombe, telomeres tend to reside near the NE in the absence of Bqt4, but their separation from the envelope can be frequently seen in live cell observations and becomes obvious during nuclear movement, indicating that the telomere–envelope connection may be weakened rather than eliminated. Thus, Bqt4 is a major component that anchors telomeres to the NE; however, we consider it possible that other components are involved.
Clustering of telomeres to the SPB
During telomere clustering, Sad1 accumulates at telomeres on the NE. Although Sad1 appears to be localized at the SPB all of the time, analysis by FRAP has shown that Sad1 actually diffuses within the NE (unpublished data). Because Sad1 is confined to the NE, telomeres must have contact with the NE to be captured by Sad1. In the absence of Bqt4, the chance of contact between telomeres and Sad1 on the NE is reduced, leading to a pronounced defect in telomere clustering. Bqt4 is degraded in the absence of Bqt3, but residual protein may account for the variable phenotype of bqt3Δ cells.
Once the Sad1–Kms1 complex becomes associated with telomeres, the dynein motor complex is involved in clustering the telomeres to the SPB. However, it is likely that other motor proteins are also involved because telomere clustering is not abolished by disruption of the dynein heavy chain dhc1 gene (Yamamoto et al., 1999; Tomita and Cooper, 2007). Conversely, oscillatory movement of the entire nucleus, which follows telomere clustering, is completely eliminated by disruption of the dhc1 gene (Yamamoto et al., 1999). Thus, these two types of movements are regulated by overlapping but separate mechanisms.
Possible additional roles for Bqt4
Although Bqt3 and -4 are expressed during mitotic cell cycles, no obvious roles for Bqt4 in mitosis have been found. Cells disrupted for bqt4 are viable, and grow at a rate similar to wild-type cells with no obvious defects in chromosome or spindle dynamics. Growth curves and doubling times are shown in Fig. S1 B. Telomere length and telomere silencing were not affected by the loss of Bqt4 (Fig. S1, A and C). Thus, the chromatin environment of the telomere is likely maintained even when separated from the NE. However, the loss of Bqt4 causes additional distinct meiotic phenotypes. It has been previously reported that cells defective in meiotic telomere association with Sad1 and Kms1, including bqt1Δ and taz1Δ mutants, show defects in meiotic spindle formation (Tomita and Cooper, 2007). Consistently, bqt4Δ cells also showed aberrant meiotic spindles (Fig. S1, D and E). In addition, bqt4Δ cells show more pronounced defects in spore viability than bqt2Δ cells. This contrasts with the observed defects in telomere clustering, which are less severe in bqt4Δ cells than in bqt2Δ cells. These observations may suggest additional roles for Bqt4 in the progression of meiosis or sporulation. Bqt4 has been annotated as a putative transcription factor containing a DNA-binding APSES domain (http://www.genedb.org/genedb/pombe/). However, such a function seems unlikely, as its localization is restricted to the NE and our analysis of Bqt4 gave no indication of direct DNA binding.
Evolutional conservation of Bqt proteins
To date, no obvious sequence homologues of the Bqt proteins, Bqt1–4, have been identified in other organisms, with the exceptions of another fission yeast, Schizosaccharomyces japonicus, and some other fungi (e.g., Neurospora crassa and Gibberella zeae, which have a possible homologue of Bqt4). In S. cerevisiae, the Sir (silent information regulation) proteins are involved in anchoring telomeres to the NE (Gotta et al., 1996; Hediger et al., 2002). Sir4 interacts directly with SUN domain protein Mps3 (Bupp et al., 2007); Sir4 also binds to the telomere-associated protein Rap1 and attaches to the NE through interaction with Esc1 (Taddei et al., 2004). Esc1 is localized along the inner face of the NE, like Bqt4, which is localized in the INM through its transmembrane domain. Esc1 has no transmembrane domain but bears posttranslational lipid modifications, which may mediate its membrane association (Taddei et al., 2004). It is curious that Sir4 and Esc1 are not found in S. pombe and, in turn, that Bqt4 is not found in S. cerevisiae. S. cerevisiae Esc1 may be functionally related to S. pombe Bqt4 even though they do not share sequence homology.
Materials and methods
Strains, culture media, and induction of meiosis
The genotypes of the strains used in this paper are shown in Table I. The swi6Δ and taz1Δ strains were gifts from J. Kanoh (Osaka University, Suita, Osaka, Japan), and the TAS-ura4+-tel2(L) strain was a gift from R.C. Allshire (Medical Research Council, Edinburgh, Scotland, UK).YE, YES, or EMM2 were used for routine mitotic culture of S. pombe cells. ME agar plates were used for the induction of meiosis and spore formation. S. pombe h90 cells grown in a liquid medium were transferred to an ME plate at 26°C to induce meiosis. Chemical compositions of YE, YES, EMM2, and ME media are described in Moreno et al. (1991).
Table I.
List of strains used
| Strain name | Mating type | Genotype |
| CRLg48 | h+ | his2-245-245 leu1-32 lys1+::LEU2 |
| CRLh20 | h− | leu1-32 lys1+::LEU2 lys3 ura1 |
| CRLi50 | h90 | leu1-32 ura4-D18 ade6-210 sad1-mRFP::kan lys1+::taz1-GFP taz1Δ::ura4+ |
| CRLk22 | h90 | leu1-32 ura4-D18 ade6-216 sad1-mRFP::kan lys1+::taz1-GFP taz1Δ::ura4+ bqt2Δ::ura4+ |
| CRLl02 | h+ | his2-245 leu1-32 |
| CRLl12 | h+ | his2-245 leu1-32 bqt3Δ::LEU2 |
| CRLl58 | h+ | his2-245 leu1-32 bqt3Δ::LEU2 lys3 ura1 |
| CRLl88 | h+ | his2-245 leu1-32 lys1-131 ade6-216 |
| CRLm03 | h90 | ura4-D18 bqt2Δ::ura4+ lys1+ |
| CRLp44 | h− | ura4-D18 bqt4Δ::ura4+ |
| CRLp67 | h+ | his2-245 leu1-32 lys1-131 ura4-D18 ade6-216 bqt4Δ::ura4+ |
| CRLp83 | h90 | leu1-32 ura4-D18 ade6-216 lys1+::taz1-mRFP bqt2-GFP::kan taz1Δ::ura4+ bqt4Δ::ura4+ |
| CRLp85 | h90 | leu1-32 ura4-D18 ade6-216 sad1-mRFP::kan lys1+::taz1-GFP taz1Δ::ura4+ bqt4Δ::ura4+ |
| CRLp86 | h90 | ura4-D18 bqt4Δ::ura4+ |
| CRLp93 | h− | leu1-32 ura4-D18 bqt4Δ::ura4+ bqt3Δ::LEU2 lys1+::GFP-bqt4 |
| CRLr06 | h− | ura4-D18 bqt4Δ::ura4+ lys1+::GFP-bqt4 |
| CRLr24 | h− | ura4-D18 bqt4Δ::ura4+ lys1+::GFP–bqt4-dTM |
| CRLr25 | h− | leu1-32 ura4-D18 bqt4Δ::ura4+ lys1+::GFP–bqt4-dTM bqt3Δ::LEU2 |
| CRLr75 | h90 | leu1-32 lys1-131 ura4-D18 ade6-210 hus5+::ccr1N-GFP::LEU2 taz1-mCherry::kan |
| CRLr81 | h90 | leu1-32 lys1-131 ura4-D18 ade6-210 bqt4Δ::ura4+ hus5+::ccr1N-GFP::LEU2 taz1-mCherry::kan |
| CRLr82 | h90 | leu1-32 lys1-131 ura4-D18 ade6-216 bqt2Δ::ura4+ hus5+::ccr1N-GFP::LEU2 taz1-mCherry::kan |
| CRLs25 | h90 | leu1-32 ura4-D18 ade6-210 bqt4Δ::ura4+ lys1+::GFP–bqt4-dN taz1-mCherry::kan |
| CRLs27 | h−/h− | leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-210/ade6-216 sad1-mRFP::kan/sad1-mRFP::kan lys1+::CFP-bqt1/lys1-131 rap1-GFP::LEU2/rap1+ |
| CRLs28 | h−/h− | leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-210/ade6-216 sad1-mRFP::kan/sad1-mRFP::kan lys1+::CFP-bqt1/lys1+::bqt2-myc rap1-GFP::LEU2/rap1+ |
| CRLs29 | h−/h− | leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-210/ade6-216 sad1-mRFP::kan/sad1-mRFP::kan lys1+::CFP-bqt1/lys1-131 rap1-GFP::LEU2/rap1+ bqt4D::ura4/bqt4D::ura4+ |
| CRLs30 | h−/h− | leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-210/ade6-216 sad1-mRFP::kan/sad1-mRFP::kan lys1+::CFP-bqt1/lys1+::bqt2-myc rap1-GFP::LEU2/rap1+ bqt4Δ::ura4/bqt4Δ::ura4+ |
| CRLs33 | h90 | leu1-32 ura4-D18 ade6-210 bqt4Δ::ura4+ taz1-mCherry::kan lys1+::rap1-GFP–bqt4-dN |
| CRLs38 | h90 | leu1-32 ura4-D18 ade6-210 bqt4Δ::ura4+ taz1-mCherry::kan lys1+::GFP–bqt4-dTM |
| CRLs53 | h90 | leu1-32 ura4-D18 ade6-210 bqt4Δ::ura4+ taz1-mCherry::kan lys1+::GFP-bqt4 |
| CRLs57 | h90 | ura4-D18 bqt3Δ::ura4+ |
| CRLs63 | h90 | leu1-32 ura4-D18 ade6-210 sad1-mRFP::kan lys1+::taz1-GFP taz1Δ::ura4+ bqt3Δ::ura4+ |
| CRLs87 | h− | leu1-32 ura4-D18 bqt3Δ::LEU2 aur1::GFP-bqt3 bqt4Δ::ura4+ |
| CRLs95 | h90 | ura4-D18 lys1+::ura4+ |
| CRLt01 | h− | leu1-32 lys1-131 ura4-D18 bqt3Δ::LEU2 aur1::GFP-bqt3 lys1::ura4+ |
| CRLt07 | h+ | his2-245 leu1-32 ura4-D18 bqt4Δ::ura4+ |
| CRLt39 | h− | leu1-32 bqt3Δ::LEU2 |
| CRLt53 | h− | leu1-32 ura4-D18 bqt3Δ::LEU2 bqt4Δ::ura4+ aur1r::mCherry-bqt3 lys1+::GFP-bqt4 |
| CRLt54 | h− | leu1-32 ura4-D18 bqt3Δ::LEU2 bqt4Δ::ura4+ aur1r::mCherry-bqt3 lys1+::GFP–bqt4-dTM |
| CRLt55 | h− | leu1-32 ura4-D18 bqt3Δ::LEU2 bqt4Δ::ura4+ aur1r::mCherry-bqt3 lys1+::GFP–bqt4-dN |
| CRLt61 | h− | leu1-32 lys1-131 ura4-D18 ade6-210 bqt4Δ::ura4+ nup211-40 |
| CRLt62 | h− | leu1-32 lys1-131 ura4-D18 ade6-210 nup211-40 |
| CRLt63 | h− | leu1-32 lys1-131 ura4-D18 ade6-210 bqt3Δ::LEU2 nup211-40 |
| CRLt73 | h90 | leu1-32 lys1-131 ura4-D18 ade6-216 taz1-mCherry::kan aur1r::GFP-bqt3 |
| CRLu61 | h− | leu1-32 ura4-D18 bqt4Δ::ura4+ bqt3Δ::LEU2 lys1+::GFP-bqt4 aur1r::mCherry-bqt3 |
| CRLu62 | h− | leu1-32 ura4-D18 bqt4Δ::ura4+ bqt3Δ::LEU2 lys1+::GFP–bqt4-dN aur1r::mCherry-bqt3 |
| CRLu63 | h− | leu1-32 ura4-D18 bqt4Δ::ura4+ bqt3Δ::LEU2 lys1+::GFP–bqt4-dTM aur1r::mCherry-bqt3 |
| CRLu69 | h− | leu1-32 lys1-131 ura4-D18 bqt4Δ::ura4+ bqt3Δ::LEU2 aur1r::mCherry-bqt3 |
| CRLw70 | h− | leu1-32 ura4-swi6Δ::kan TAS-ura4+-tel2(L) |
| CRLw71 | h− | leu1-32 ura4-TAS-ura4+-tel2(L) |
| CRLw73 | h− | leu1-32 ura4-taz1Δ::kan TAS-ura4+-tel2(L) |
| CRLw95 | h− | leu1-32 ura4-bqt4Δ::hyg TAS-ura4+-tel2(L) |
| CRLx01 | h90 | leu1-32 lys1-131 ura4-D18 ade6-210 bqt4Δ::ura4+ rap1Δ::ura4+ taz1-mCherry::kan lys1::rap1-GFP–bqt4-dN |
| CRLx02 | h90 | leu1-32 lys1-131 ura4-D18 ade6-210 bqt4Δ::ura4+ rap1Δ::ura4+ taz1-mCherry::kan lys1::rap1-GFP |
| CRLx03 | h90 | leu1-32 lys1-131 ura4-D18 ade6-210 rap1Δ::ura4+ taz1-mCherry::kan lys1::rap1-GFP–bqt4-dN |
| CRLx04 | h90 | leu1-32 lys1-131 ura4-D18 ade6-210 rap1Δ::ura4+ taz1-mCherry::kan lys1::rap1-GFP–bqt4-dN |
Prediction of transmembrane sequences
The prediction tools that we used to identify transmembrane sequences are found at http://bp.nuap.nagoya-u.ac.jp/sosui/, http://www.cbs.dtu.dk/services/TMHMM-2.0/, and http://www.ch.embnet.org/software/TMPRED_form.html.
Fluorescent fusion constructs
GFP-Bqt4, GFP–Bqt4-dTM, GFP–Bqt4-dN, Rap1-GFP, and Rap1-GFP–Bqt4-dN fusion constructs were made by integration of the following plasmids at the lys1-131 gene locus: pCSS18, pCSS23, pCSS42, pCSS62, and pCSS44, respectively. Monomeric Cherry (mCherry)–Bqt3 and GFP-Bqt3 fusion constructs were made by integration of the following plasmids at the aur1 gene locus: pCSS46 and pCSS48, respectively. Maps of these plasmids are shown in Fig. S2. Integration was confirmed by PCR. Constructs for Taz1-GFP, Sad1-mRFP, and Bqt2-GFP were described previously (Chikashige et al., 2006). Strains carrying Taz1-mCherry were constructed by replacing the taz1+ gene with a selection marker kanr by a PCR-based gene-targeting method (Bähler et al., 1998).
Image acquisition and processing
Fluorescence microscope images were obtained using a computer-controlled fluorescence microscope system (DeltaVision; Applied Precision); for imaging of live cells, a DeltaVision microscope system set up in a temperature-controlled room was used (Haraguchi et al., 1999). This microscope system is based on an inverted fluorescence microscope (IX70; Olympus) equipped with a charge-coupled device (CoolSNAP HQ; Photometrics). The objective lens used was an oil immersion Plan-Apochromat 60× NA 1.4 lens (Olympus). For time-lapse observation, living cells were mounted in a 35-mm glass-bottom culture dish (MatTek Corp.) coated with concanavalin A or lectin and observed in EMM2 medium at 26°C for meiotic cells or at 30°C for vegetative cells. Images were acquired using SoftWoRx software (Applied Precision), which was provided as part of the DeltaVision system. A 3D stack of images spanning 9–15 focal planes at 0.3-µm increments was recorded at each time point. A single section of unprocessed 3D images is displayed in Figs. 1 (B and E), 3 B, 6 C, and 7 (C–F). Image deconvolution was performed using SoftWoRx software on the DeltaVision system (Figs. 4 B and 5, A and B; and Fig. 6 E). Projection images were generated using a maximum intensity method (Figs. 1 F, 2 A, 3 C, and 4, A and B; Fig. 5, A and B; Fig. 6, D and F; and Fig. S1, D and E). Photoshop 10.0 (Adobe) was used to adjust the brightness and contrast for figure production.
Measurement of the distance between telomeres and the NE
Telomeres and the NE were visualized with Taz1-mCherry and Ccr1N-GFP, respectively (Ccr1N-GFP was described as D817 in Ding et al. [1998] and Tange et al. [1998]). Optical section data (15 focal planes at 0.3-µm spacing) were collected on a Peltier-cooled charge-coupled device using an oil immersion Plan-Apochromat 60× NA 1.4 objective lens on the DeltaVision microscope system and computationally processed by a 3D deconvolution method (Agard et al., 1989). Distances were measured after pixel interpolation using SoftWoRx functions.
We measured distances (designated by d) from the center of intensity of the telomere spot to the middle of the NE signal in each focal plane (Fig. 9 A). We selected only telomeres that showed their brightest signal in the three optical sections near the nuclear midline. The position of the telomere spot is (r, z) in the cylindrical coordinate system. D represents the 3D radial distance from the telomere to the NE (Fig. 9 A, blue line). d represents the telomere–envelope distance measured in the focal plane at z = z1 (Fig. 9 A, red line). R represents the radius of the nucleus in three dimensions. r1 represents the radius of the nucleus in the focal plane at z = z1, and r2 represents the radius of the nucleus in the focal plane at z = z2.
Figure 9.
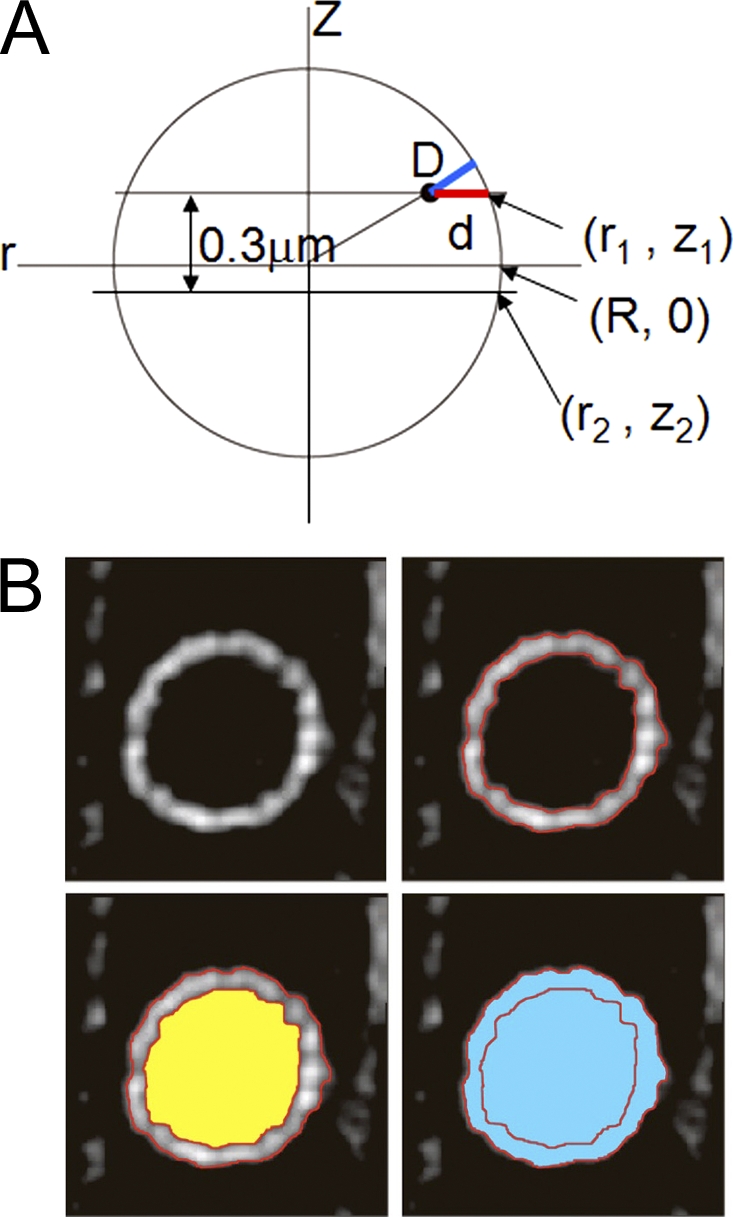
Measurement of the distance between telomeres and the NE. (A and B) A schematic diagram of the nucleus on the side view (A) and on the top view (B).
When the nucleus is considered to be a sphere, the 3D telomere–envelope distance is obtained as
| (1) |
R and z1 are obtained as solutions of the following equations: Δz = z1 − z2 and Therefore,
| (2) |
| (3) |
Δz = 0.3 µm in this series of experiments.
The radiuses r1 and r2 were obtained from the area as follows: the areas outside (S1 and S2; shown in blue) and inside (s1 and s2; shown in yellow) the NE were measured as shown in Fig. 9 B, and the radius (r1 and r2) was determined from the area:
D was obtained by inserting the r1 and r2 values into Eqs. 1, 2, and 3.
Because the nucleus in a vegetative cell can be considered a sphere, 3D radial distance D was measured. However, meiotic nuclei do not show a constant shape because of nuclear movement; thus, the 2D distance d in a focal plane was measured in those nuclei. This is a good approximation as the difference between d and D is small when the observed focal planes are close to the equatorial plane of the nucleus.
Immunoelectron microscopy
Cells were fixed with 0.5% glutaraldehyde and 3% paraformaldehyde in 100 mM phosphate buffer, pH 7.2, and 150 mM NaCl (PBS) for 2 h at 4°C, washed with PBS, treated with 1% Na metaperiodate for 15 min and 50 mM ammonium chloride for 30 min, washed again with PBS, and placed into 2% low-melt agarose. The specimens were dehydrated through a graded ethanol series (50–99.5% vol/vol), embedded in LR White resin (London Resin Co.), and polymerized under ultraviolet irradiation for 48 h at −20°C. Immunostaining with anti-GFP polyclonal antibody (1:20 dilution; gift from R.J. McIntosh, University of Colorado, Boulder, CO) was performed on ultrathin sections and picked up on nickel grids after blocking with 3% normal goat serum in TBS (50 mM Tris-HCl buffer containing 150 mM NaCl, pH 7.6) containing 0.5% BSA for 30 min at room temperature. 10-nm colloidal gold-conjugated goat anti–rabbit IgG (1:50 dilution; BB International) was used as the secondary antibody. After immunostaining, sections were fixed with 1% glutaraldehyde in 100 mM TBS. The grids were stained in 4% uranyl acetate and viewed with a JEM 1200EXS (JEOL) at 80 kV. Cryofixation by high-pressure freezing was performed as described in Humbel et al. (2001).
Yeast two-hybrid assays
Yeast two-hybrid screening was performed using the BD Matchmaker library construction and screening kits. The BD Matchmaker cDNA library for two-hybrid screening was constructed from S. pombe cells treated with mating pheromone for 5–6 h (Chikashige et al., 2006). The diploid cells carrying both pGADT7 and pGBKT7 plasmids were tested for expression of the two reporter genes (ADE2 and HIS3) according to the BD Matchmaker system manual.
Real-time RT PCR
Real-time PCR with TaqMan technology (Applied Biosystems) was used to measure levels of mRNA. For measurement of mRNA levels, RNA was reverse transcribed to cDNA by MultiScribe Reverse transcription (Applied Biosystems). Specific primers and TaqMan probes were designed with Primer Express software (Applied Biosystems). Sequences of the primers and TaqMan probes are listed in Table S1. A comparative threshold cycle (Ct) was used to determine gene expression relative to the vegetative growing phase using the 2-ΔΔCt technique (User Bulletin #2; Applied Biosystems), in which cdc2+ or act1+ was used as an endogenous reference.
Preparation of cell extracts and immunoblotting
Cell cultures of ∼5–8 × 106 cells/ml in YES or EMM2-N medium were incubated at 30°C. Cells were collected at appropriate times, washed once with ice-cold water, suspended in 25 µl of ice-cold water, and boiled for 5 min. After the cell suspension was frozen in liquid nitrogen, 35 µl of 2× Laemmli buffer (2% SDS, 20% glycerol, and 0.12 M Tris-HCl, pH 6.6) was added. Cells were disrupted with 0.5-mm glass beads using a Multibeads Shocker (Yasui Kikai). The resulting cell extracts were centrifuged at 15,000 rpm for 15 min. Aliquots of the cell extracts, each containing 80 µg of total protein, were separated on an SDS-PAGE gel. GFP-tagged proteins were detected using JL8 mouse monoclonal anti-GFP antibody (Takara Bio Inc.). To confirm equal loading, tubulin protein was detected with TAT1 (Woods et al., 1989).
Online supplemental material
Fig. S1 shows the phenotypes of bqt4Δ cells. Fig. S2 shows maps of the fusion plasmid constructs made in this study. Video 1 shows the live observation of S. pombe meiotic cells expressing GFP-Bqt4. Table S1 lists the primers used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200902122/DC1.
Acknowledgments
We thank Akio Matsukage and Shou Waga for providing their electron microscope facilities, Richard J. McIntosh for providing anti-GFP antibody, Junko Kanoh and Robin C. Allshire for providing S. pombe strains, and Abby F. Dernburg for critical reading of the manuscript.
This work was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology, the Japan Society for the Promotion of Science, and the Japan Science and Technology Agency to Y. Chikashige, T. Haraguchi, and Y. Hiraoka.
Footnotes
Abbreviations used in this paper:
- INM
- inner nuclear membrane
- NE
- nuclear envelope
- SPB
- spindle pole body
- TMH
- transmembrane helix
References
- Agard D.A., Hiraoka Y., Shaw P., Sedat J.W. 1989. Fluorescence microscopy in three dimensions. Methods Cell Biol. 30:353–377 10.1016/S0091-679X(08)60986-3 [DOI] [PubMed] [Google Scholar]
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951 [DOI] [PubMed] [Google Scholar]
- Bupp J.M., Martin A.E., Stensrud E.S., Jaspersen S.L. 2007. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 179:845–854 10.1083/jcb.200706040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y., Hiraoka Y. 2001. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr. Biol. 11:1618–1623 10.1016/S0960-9822(01)00457-2 [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Ding D.Q., Funabiki H., Haraguchi T., Mashiko S., Yanagida M., Hiraoka Y. 1994. Telomere-led premeiotic chromosome movement in fission yeast. Science. 264:270–273 10.1126/science.8146661 [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Tsutsumi C., Yamane M., Okamasa K., Haraguchi T., Hiraoka Y. 2006. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 125:59–69 10.1016/j.cell.2006.01.048 [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Haraguchi T., Hiraoka Y. 2007. Another way to move chromosomes. Chromosoma. 116:497–505 10.1007/s00412-007-0114-8 [DOI] [PubMed] [Google Scholar]
- Conrad M.N., Lee C.Y., Wilkerson J.L., Dresser M.E. 2007. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 104:8863–8868 10.1073/pnas.0606165104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M.N., Lee C.Y., Chao G., Shinohara M., Kosaka H., Shinohara A., Conchello J.A., Dresser M.E. 2008. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 133:1175–1187 10.1016/j.cell.2008.04.047 [DOI] [PubMed] [Google Scholar]
- Cooper J.P. 2000. Telomere transitions in yeast: the end of the chromosome as we know it. Curr. Opin. Genet. Dev. 10:169–177 10.1016/S0959-437X(00)00070-8 [DOI] [PubMed] [Google Scholar]
- Cooper J.P., Nimmo E.R., Allshire R.C., Cech T.R. 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 385:744–747 10.1038/385744a0 [DOI] [PubMed] [Google Scholar]
- Cooper J.P., Watanabe Y., Nurse P. 1998. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 392:828–831 10.1038/33947 [DOI] [PubMed] [Google Scholar]
- Ding D.Q., Chikashige Y., Haraguchi T., Hiraoka Y. 1998. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci. 111:701–712 [DOI] [PubMed] [Google Scholar]
- Ding D.Q., Yamamoto A., Haraguchi T., Hiraoka Y. 2004. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell. 6:329–341 10.1016/S1534-5807(04)00059-0 [DOI] [PubMed] [Google Scholar]
- Ding X., Xu R., Yu J., Xu T., Zhuang Y., Han M. 2007. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell. 12:863–872 10.1016/j.devcel.2007.03.018 [DOI] [PubMed] [Google Scholar]
- Funabiki H., Hagan I., Uzawa S., Yanagida M. 1993. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121:961–976 10.1083/jcb.121.5.961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M., Laroche T., Formenton A., Maillet L., Scherthan H., Gasser S.M. 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134:1349–1363 10.1083/jcb.134.6.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Margalit A., Goldman R.D., Shumaker D.K., Wilson K.L. 2005. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 6:21–31 10.1038/nrm1550 [DOI] [PubMed] [Google Scholar]
- Hagan I., Yanagida M. 1995. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129:1033–1047 10.1083/jcb.129.4.1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T., Ding D.Q., Yamamoto A., Kaneda T., Koujin T., Hiraoka Y. 1999. Multiple-color fluorescence imaging of chromosomes and microtubules in living cells. Cell Struct. Funct. 24:291–298 10.1247/csf.24.291 [DOI] [PubMed] [Google Scholar]
- Harper L., Golubovskaya I., Cande W.Z. 2004. A bouquet of chromosomes. J. Cell Sci. 117:4025–4032 10.1242/jcs.01363 [DOI] [PubMed] [Google Scholar]
- Hassold T., Abruzzo M., Adkins K., Griffin D., Merrill M., Millie E., Saker D., Shen J., Zaragoza M. 1996. Human aneuploidy: incidence, origin, and etiology. Environ. Mol. Mutagen. 28:167–175 [DOI] [PubMed] [Google Scholar]
- Hediger F., Neumann F.R., Van Houwe G., Dubrana K., Gasser S.M. 2002. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 12:2076–2089 10.1016/S0960-9822(02)01338-6 [DOI] [PubMed] [Google Scholar]
- Humbel B.M., Konomi M., Takagi T., Kamasawa N., Ishijima S.A., Osumi M. 2001. In situ localization of beta-glucans in the cell wall of Schizosaccharomyces pombe. Yeast. 18:433–444 10.1002/yea.694 [DOI] [PubMed] [Google Scholar]
- Kanoh J., Ishikawa F. 2001. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 11:1624–1630 10.1016/S0960-9822(01)00503-6 [DOI] [PubMed] [Google Scholar]
- Kosaka H., Shinohara M., Shinohara A. 2008. Csm4-dependent telomere movement on nuclear envelope promotes meiotic recombination. PLoS Genet. 4:e1000196 10.1371/journal.pgen.1000196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Kim K.P., Prentiss M., Kleckner N., Kameoka S. 2008. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 133:1188–1201 10.1016/j.cell.2008.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.J., Misner L., Le Bot N., Tsai M.C., Campbell J.M., Ahringer J., White J.G. 2003. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 115:825–836 10.1016/S0092-8674(03)00985-1 [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Nimmo E.R., Pidoux A.L., Perry P.E., Allshire R.C. 1998. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 392:825–828 10.1038/33941 [DOI] [PubMed] [Google Scholar]
- Penkner A., Tang L., Novatchkova M., Ladurner M., Fridkin A., Gruenbaum Y., Schweizer D., Loidl J., Jantsch V. 2007. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev. Cell. 12:873–885 10.1016/j.devcel.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Phillips C.M., Dernburg A.F. 2006. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell. 11:817–829 10.1016/j.devcel.2006.09.020 [DOI] [PubMed] [Google Scholar]
- Phillips C.M., Wong C., Bhalla N., Carlton P.M., Weiser P., Meneely P.M., Dernburg A.F. 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell. 123:1051–1063 10.1016/j.cell.2005.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H. 2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2:621–627 10.1038/35085086 [DOI] [PubMed] [Google Scholar]
- Scherthan H., Wang H., Adelfalk C., White E.J., Cowan C., Cande W.Z., Kaback D.B. 2007. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 104:16934–16939 10.1073/pnas.0704860104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J., Benavente R., Hodzic D., Höög C., Stewart C.L., Alsheimer M. 2007. Transmembrane protein Sun2 is involved in tethering mammalian meiotic telomeres to the nuclear envelope. Proc. Natl. Acad. Sci. USA. 104:7426–7431 10.1073/pnas.0609198104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanuki M., Miki F., Ding D.Q., Chikashige Y., Hiraoka Y., Horio T., Niwa O. 1997. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol. Gen. Genet. 254:238–249 10.1007/s004380050412 [DOI] [PubMed] [Google Scholar]
- Starr D.A. 2009. A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 122:577–586 10.1242/jcs.037622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Han M. 2003. ANChors away: an actin based mechanism of nuclear positioning. J. Cell Sci. 116:211–216 10.1242/jcs.00248 [DOI] [PubMed] [Google Scholar]
- Starr D.A., Hermann G.J., Malone C.J., Fixsen W., Priess J.R., Horvitz H.R., Han M. 2001. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 128:5039–5050 [DOI] [PubMed] [Google Scholar]
- Taddei A., Hediger F., Neumann F.R., Bauer C., Gasser S.M. 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 23:1301–1312 10.1038/sj.emboj.7600144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange Y., Horio T., Shimanuki M., Ding D.-Q., Hiraoka Y., Niwa O. 1998. A novel fission yeast gene, tht1+, is required for the fusion of nuclear envelopes during karyogamy. J. Cell Biol. 140:247–258 10.1083/jcb.140.2.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K., Cooper J.P. 2007. The telomere bouquet controls the meiotic spindle. Cell. 130:113–126 10.1016/j.cell.2007.05.024 [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken E., Adelfalk C., Loidl J., Scherthan H. 2005. Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J. Cell Biol. 170:213–223 10.1083/jcb.200501042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur Y.B., Wilson K.L., Gruenbaum Y. 2006. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Natl. Rev. 7:782–788 10.1038/nrm2003 [DOI] [PubMed] [Google Scholar]
- Wanat J.J., Kim K.P., Koszul R., Zanders S., Weiner B., Kleckner N., Alani E. 2008. Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet. 4:e1000188 10.1371/journal.pgen.1000188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A., Sherwin T., Sasse R., MacRae T.H., Baines A.J., Gull K. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491–500 [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Hiraoka Y. 2001. How do meiotic chromosomes meet their homologous partners?: lessons from fission yeast. Bioessays. 23:526–533 10.1002/bies.1072 [DOI] [PubMed] [Google Scholar]
- Yamamoto A., West R.R., McIntosh J.R., Hiraoka Y. 1999. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol. 145:1233–1249 10.1083/jcb.145.6.1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D. 2006. From early homologue recognition to synaptonemal complex formation. Chromosoma. 115:158–174 10.1007/s00412-006-0048-6 [DOI] [PubMed] [Google Scholar]