Abstract
Here we use single-molecule imaging to determine coarse-grained intrinsic energy landscapes for nucleosome deposition on model DNA substrates. Our results reveal distributions that are correlated with recent in silico predictions, reinforcing the hypothesis that DNA contains some intrinsic positioning information. We also show that cis-regulatory sequences in human DNA coincide with peaks in the intrinsic landscape, whereas valleys correspond to non-regulatory regions, and we present evidence arguing that nucleosome deposition in vertebrates is influenced by factors not accounted for by current theory. Finally, we demonstrate that intrinsic landscapes of nucleosomes containing the centromere-specific variant CenH3 are correlated with patterns observed for canonical nucleosomes, arguing that CenH3 does not alter sequence preferences of centromeric nucleosomes. However, the non-histone protein Scm3 alters the intrinsic landscape of CenH3-containing nucleosomes, enabling them to overcome the otherwise exclusionary effects of poly(dA–dT) tracts, which are enriched in centromeric DNA.
The distribution of nucleosomes throughout the genome has profound consequences for DNA transcription, repair, and chromosome segregation1–4. Canonical nucleosomes consist of ~147-base pairs (bp) of DNA wrapped in ~1.7 turns around an octamer containing two of each histone H2A, H2B, H3, and H43,5. There has been tremendous interest in developing in silico models of genome-wide nucleosome positions, the first of which came from Segal et al.,6 and calculated probabilistic nucleosome distributions based upon the preference of nucleosomes for bendable DNA containing AA/TT/AT dinucleotides with 10-bp periodicities in counter-phase with GC dinucleotides1,7. This supported the hypothesis of a “second genetic code”, which asserts that genomes intrinsically encode information dictating nucleosome distributions, and posits that extrinsic factors, such as chromatin remodeling proteins, play a limited role in establishing steady-state positions. However, accumulating evidence suggests stiff DNA sequences that resist bending, including poly(dA–dT) tracts8, play a dominant role in influencing nucleosome distributions9–13, yet these sequences were unaccounted for in the original algorithms. Two more recent theoretical models from Segal and colleagues included exclusion effects from poly(dA–dT) tracts along with other sequence motifs found enriched in linker DNA14,15. These modeling efforts together with other in vivo mapping studies of nucleosome positions are starting to yield unprecedented details of chromatin structure and its relationship to gene regulation.
The information content within eukaryotic genomes is also enriched through deposition of histone variants4, such as CenH3, which is an H3-variant that serves as a universally conserved centromere-specific epigenetic marker necessary for directing kinetochore assembly for chromosome segregation during meiosis and mitosis 16–18. CenH3 (called Cse4 in S. cerevisiae) contains a conserved fold resembling histone H3 along with a unique N-terminal extension that distinguishes it from a canonical histone. Biophysical studies have shown that CenH3/H4 tetramers are more rigid and compact than canonical H3/H4 tetramers due to an amino acid sequence called the CATD (CENP-A centromeres targeting domain)16,19,20. Yeast centromeres also contain a non-histone protein called Scm3, which was identified as a high-copy suppressor of Cse4 mutations21,22. Deletion of Scm3 is lethal and Scm3 is required for the binding of other inner kinetochore proteins, indicating that it plays an early role in kinetochore assembly18,21–24. Scm3 is found in the point centromeres of S. cerevisiae and the regional centromeres of S. pombe, and homologs have been identified in higher eukaryotes 25–29. The roles of Scm3 remain unclear, but it is known to form a stable complex with H4 and Cse4, yet does not interact with a conventional H3/H4 tetramer18. Interestingly, H2A and H2B are excluded by Scm3, leading to the hypothesis that H4/Cse4/Scm3 forms an atypical hexameric nucleosome that is somehow restricted to centromeres18. One feature of centromeric DNA is that it is often highly A/T-rich2,30. For example the point centromeres (CEN sequences) of S. cerevisiae range from 86–98% A/T, and contain numerous tracts of poly(dA–dT), suggesting that mechanisms must exist enabling centromeric nucleosomes to bind to these otherwise unfavorable sequences31–34. Despite the importance of CenH3 and Scm3, relatively little is known about how they are deposited and maintained at centromeres, or whether they might alter the intrinsic sequence preferences of nucleosomes, enabling them to bind to poly(dA–dT)-rich DNA.
To begin addressing these and other questions in chromatin biology we have established a unique single molecule microscopy assay to directly visualize individual fluorescent nucleosomes bound to aligned curtains of DNA. Here we use this assay to determine the coarse-grained energy landscapes for nucleosome deposition using phage λ DNA as a model substrate. The observed patterns for canonical nucleosomes and nucleosomes containing H2AZ are well correlated with recent theoretical predictions, and we confirm that exclusionary effects of poly(dA–dT) tracts play a dominant role in dictating the landscape. We also map the intrinsic landscape for nucleosome deposition on a substrate derived from the human β-globin locus and show that all intrinsically preferred binding sites within this 23-kb fragment are correlated with transcriptional regulatory regions. However, the intrinsic landscape for the β-globin locus does not match reported in vivo nucleosome positions, suggesting that sequence preferences alone are insufficient to predict nucleosome distributions in vertebrate genomes. Finally, we demonstrate that canonical nucleosomes and nucleosomes containing Cse4 display similar distribution patterns, indicating that the DNA binding properties of both are subject to similar thermodynamic principles, despite the fact that Cse4 is targeted to poly(dA–dT)-rich centromeric DNA in vivo. In striking contrast, nucleosomes bearing both Cse4 and Scm3 display a distinct binding distribution, revealing an unanticipated role for Scm3 in overcoming the aversion of nucleosomes for poly(dA–dT) tracts.
RESULTS
A DNA curtain assay for imaging individual nucleosomes
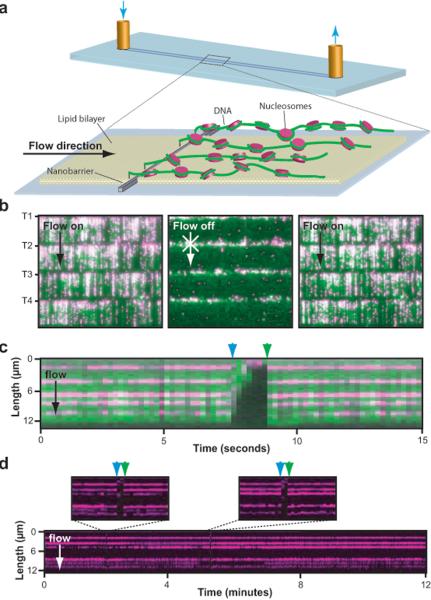
For our assay DNA molecules are anchored via a biotin-neutravidin interaction to a fluid lipid bilayer on the surface of a fused silica microfluidic sample chamber and hydrodynamic force is used to push the molecules towards the leading edges of nanofabricated barriers35,36. The DNA molecules align along these barriers, enabling us to visualize thousands of individual molecules in real time using total internal reflection fluorescence microscopy (TIRFM; Fig. 1a, and Supplementary Fig. 1 and Supplementary Video 1)35,36.
Fig. 1. Visualizing fluorescently tagged recombinant nucleosomes on DNA curtains.
Panel (a) depicts the experimental design; full details of the nanofabrication and DNA curtain assembly can be found in Fazio et al35. YOYO1-stained DNA curtains (green) bound by nucleosomes (magenta) that were tagged with anti-FLAG QDs are shown in (b). The tethered end of each curtain is indicated as T1–T4, and arrows indicate the direction of flow. A kymogram illustrating five nucleosomes on one DNA molecule is shown in (c). All nucleosome reconstitution reactions were performed at 4°C, the samples were shifted to 37°C for a minimum of 15-minutes after injection into the microfluidic sample chamber, and all single molecule distribution measurements were made at 37°C. The nucleosomes disappear when flow is temporarily interrupted (blue arrowheads), and reappear when flow is resumed (green arrowheads), verifying they are bound to the DNA and do not interact with the lipid bilayer. A longer kymogram collected without YOYO1 to avoid laser-induced photocleavage of the DNA is shown in (d). Insets highlight 30-second windows where flow was transiently paused. Other signal gaps result from QD blinking. During a 25-minute observation at 37°C 0.9% of the nucleosomes (5 of 580) moved and/or dissociated from the DNA, and the remaining 99.1% were stationary.
To visualize nucleosomes, we expressed and purified recombinant S. cerevisiae histones from E. coli. Histone octamers were assembled in vitro and purified by gel filtration, and purified octamers containing FLAG-H2B were deposited onto biotinylated λ-DNA by salt dialysis (Supplementary Fig. 2), which recapitulates thermodynamically favorable binding distributions3,37–39. The nucleosomes were labeled in situ with anti-FLAG quantum dots (QDs) and visualized by TIRFM (Fig. 1b, Supplementary Video 2). Time courses confirmed that ≥99% of the fluorescent nucleosomes did not move or dissociate from the DNA during the experiments (Fig. 1b–d). Transient termination of hydrodynamic force provoked entropic collapse of the DNA, causing the molecules to drift outside the detection volume defined by the penetration depth of the evanescent field, verifying that they were anchored only via the biotin tag (Fig. 1b–d). For the position distribution measurements described below, conditions were selected to yield ~5 nucleosomes per DNA, equivalent to ~1.5% coverage of the 48.5-kb substrate (Fig. 1). In addition, only full-length DNA molecules well resolved from adjacent molecules were used for distribution measurements. Any DNA molecules that did not meet these criteria were discarded from further analysis, and we were able to typically obtain ~200–300 data points per experiment.
Visualizing intrinsic landscapes for nucleosome deposition
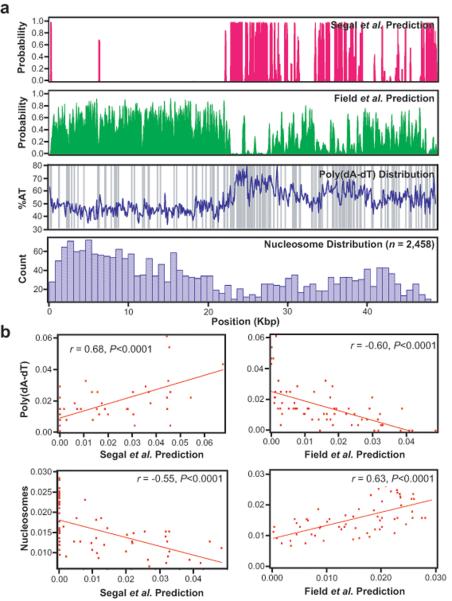
We sought to explore the relative performance of recent theoretical models as general tools for predicting intrinsic energy landscapes for nucleosome deposition using phage λ-DNA as a simple model substrate. λ-DNA is not subject to evolutionary pressure to position nucleosomes, but analysis of the phage genome revealed an unexpected advantage: the original Segal et al.6 model yielded surprisingly dissimilar predictions for the nucleosome distributions compared to the more recent models of both Field et al.14 and Kaplan et al.15 (Fig. 2a, Supplementary Fig. 3–5). Pearson correlation analysis revealed that the theoretical predictions of Segal et al.6 were actually anticorrelated with the two more recent models (r = −0.76, P < 0.0001 and r = −0.27, P = 0.031, for the Field and Kaplan models, respectively; Supplementary Fig. 3 and 5, and data not shown). The strikingly dissimilar predictions arise due to the natural asymmetric division of the phage genome into A/T- and G/C-rich halves (Fig. 2a), and can be further attributed to the abundance of exclusionary poly(dA–dT) tracts (Fig. 2a), which are greatly enriched near the center of the λ phage genome. The effects of the poly(dA–dT) tracts were further evidenced by the observation that the Segal et al.6 model was correlated with poly(dA–dT) tracts (r = 0.68, P < 0.0001), whereas the Field et al.14 and Kaplan et al.15 models were anticorrelated with these same sequence features (r = −0.60, P < 0.0001, and r = −0.65, P < 0.0001, for the Field and Kaplan models respectively; Fig. 2b and data not shown). The drastically different predicted distributions generated by the theoretical algorithms provided a means for evaluating their relative performance, and the differences in the theoretical models were readily evident even when the 1-bp resolution in silico predictions were binned to match our experimental data (Supplementary Fig. 1 and Fig. 4).
Fig. 2. Predicted and observed nucleosome distribution patterns on bacteriophage λ-DNA.
The nucleosome distributions predicted by the Segal et al.6 (magenta) and the Field et al.14 (green) models for λ-DNA are shown in (a). The AT-content of the λ is shown (blue line; calculated with a 100-bp window) superimposed with the distribution of poly(dA–dT) tracts ≥5-mers (gray bars), which are asymmetrically distributed and comprise 3.2% of the phage genome. The observed nucleosome distribution is shown in the lower panel, and is comprised of data from both canonical nucleosomes and nucleosomes bearing H2AZ (Supplementary Fig. 7). The theoretical data is shown at 1-bp resolution, and the observed data is compiled into 758-bp bins. Supplementary Fig. 4 shows examples where the predicted locations are binned to the same resolution as the experimental data. Panel (b) shows Pearson correlation analysis of the poly(dA–dT) distribution and the observed nucleosome distribution compared to the theoretical predictions.
To test the theoretical predictions we measured the locations of 1,248 canonical nucleosomes and 1,210 nucleosomes containing the histone variant H2AZ, which is a conserved histone H2A variant that influences gene regulation40–45 (n = 2,458 total; Fig. 2, Supplementary Fig. 6). A histogram built from these experiments represents a coarse-grained profile of the thermodynamically favored intrinsic energy landscape for nucleosome deposition (Fig. 2a). As shown in figure 2, the observed nucleosome distribution was anticorrelated with both the Segal et al.6 prediction (r = −0.55, P < 0.0001; Fig. 2b) and the poly(dA–dT) distribution (r = −0.32, P = 0.01), but bore a remarkable resemblance to the Field et al.14 prediction (r = 0.63, P < 0.0001; Fig. 2a, b, and Supplementary Fig. 4) and also to the prediction of Kaplan et al.15 (r = 0.63, P < 0.0001, not shown). The positions of nucleosomes that were heated to 37°C for a period of 10 hours were even more strongly correlated with the Field et al.14 and Kaplan et al.15 models (n = 1,247; r = 0.74, P < 0.0001, and r = 0.75, P < 0.0001, respectively; Supplementary Fig. 6).
Our data represent a coarse-grained profile of the global energy landscape, and we can not confirm the accuracy of the algorithms at base pair resolution. Nevertheless, because the predictions of the different algorithms are so dissimilar we can test their performance relative to one another, and our results are most consistent with predictions from the recent Field et al.14, and Kaplan et al.15 algorithms. These findings support the Field et al.14 and Kaplan et al.15 algorithms as general means for predicting intrinsic energy landscapes of nucleosome binding sites in vitro, but do not address their performance for predicting in vivo landscapes (see discussion). Our results also show that nucleosomes bearing H2AZ displayed the same distribution trends as canonical nucleosomes, demonstrating that H2AZ does not drastically alter thermodynamic sequence preferences, and these conclusions are supported by independent comparison of the two separate data sets (Supplementary Fig. 7). Finally, the non-random nucleosome distributions observed in our assay, and the agreement between the newer theoretical models and our single molecule data reinforces the validity of our unique experimental system, and confirms that the experimentally observed distributions reflect the actual intrinsic energy landscape for nucleosome sequence preferences.
Intrinsic landscapes in human DNA
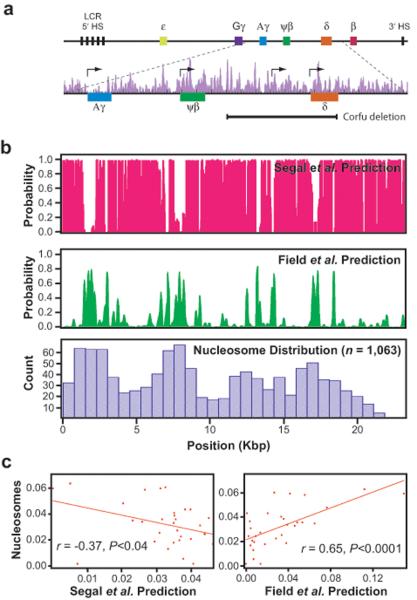
We next sought to determine the intrinsic landscape of a eukaryotic substrate, and for this we selected a 23-kb PCR fragment derived from the human β-globin locus (Fig. 3a). Comparison of the original Segal et al.6 model with the more recent Field et al.14 and Kaplan et al.15 models revealed that the theoretical distributions for this substrate were again anticorrelated (r = −0.74, P < 0.0001, and r = −0.70, P < 0.0001 for the Field et al., and Kaplan et al., models respectively; Fig. 3b,c; Supplementary Fig. 3). The models were also anticorrelated for several regions of the yeast genome (Supplementary Fig. 8). While β-globin DNA lacks the fortuitous A/T and G/C asymmetry and high poly(dA–dT) content found in λ-phage, its base composition is similar to the yeast genome (61% versus 62% A/T), and the Field et al.14 and Kaplan et al.15 models still predict a non-random landscape with four particularly prominent clusters of positioned nucleosomes that should be discernable in the coarse-grained landscape obtained from the single molecule in vitro data (Fig. 3b, Supplementary Fig. 4).
Fig. 3. Predicted and observed nucleosome distribution patterns on human β-globin DNA.
Panel (a) shows an overview of the 82-kb human β-globin locus, highlighting details of the 23-kb fragment (chr11 5,205,941–5,229,259) used in this study. Also shown are the in vivo nucleosome positions (purple) from Schones et al.53, and the ~7.2-kb deletion spanning most of the δ-globin gene and ~6-kb of upstream sequence found in Corfu δβ-thalassemia patients. Major ticks are at 1-kb intervals, and minor ticks are in 200-bp subdivisions. The nucleosome distribution patterns predicted by the Segal et al.6 (magenta) and Field et al.14 (green) models for the 23-kb fragment are shown in (b), along with the observed patterns of nucleosome deposition determined by single-molecule imaging (blue). Panel (c) shows Pearson correlation analysis of the observed data for the β-globin locus compared results from the Segal et al.6 (left panel) and Field et al.14 models (right panel).
As shown in Fig. 3b, the distribution of nucleosomes (n = 1,063) bound to the β-globin DNA was different from that found for the λ-DNA, indicating that the substrate sequence contributed to the overall patterns. Analysis of the nucleosome distribution for the β-globin substrate revealed that it was weakly anticorrelated with the Segal et al.6 prediction (r = −0.37, P = 0.04), but was well correlated with the Field et al.14 (r = 0.65, P < 0.0001) and the Kaplan et al.15 (r = 0.64, P < 0.0001) predictions (Fig. 3c, and not shown). The agreement between our data and the predictions, indicates that nucleosomes obey similar thermodynamic rules, regardless of the origin of the DNA.
Inspection of the β-globin data suggested that the intrinsic landscape reflected underlying organizational features of the DNA. Every peak within the intrinsic landscape for the human DNA substrate coincided with regulatory sequences, including the promoter-proximal regions of the Aγ- and δ-globin genes, and the non-coding ψβ-globin gene. An additional peak in the intrinsic landscape encompassed an intergenic developmental stage-specific promoter located ~2.6-kb upstream from the δ-globin gene within a regulatory region necessary for silencing the fetal γ-globin genes that is deleted in patients with Corfu δβ-thalassemia (Fig. 3a,b)46–48. In contrast, all valleys within the intrinsic landscape corresponded to non-transcribed and non-regulatory DNA. This data reveals that cis-regulatory regions within the human β-globin locus are poised with thermodynamically preferred nucleosome-binding sites. This evolutionarily-driven architecture has likely arisen not because more or more tightly bound nucleosomes are required in these regions, but rather because precise nucleosome positioning within regulatory sequences may be much more critical compared to other areas of the genome40,49–51. It remains to be determined whether this organization is a general trend for human DNA, but our interpretation is consistent with the finding that eukaryotic transcriptional start sites (TSS) are typically flanked on either side by well-positioned nucleosomes4,40,49,50,52.
The locations of nucleosomes across the β-globin locus have been mapped in CD4+ T cells53, but there was no obvious correlation between these positions and any of the theoretical predictions (Fig. 3a,b). The in vivo nucleosome distribution also did not fully coincide with the coarse-grain features of the experimentally observed intrinsic landscape, although the promoter proximal regions of the ψβ- and δ-globin genes were occupied by positioned nucleosomes as expected based on the intrinsic landscape (Fig. 3a,b). However, the in vivo data were dissimilar with the experimentally defined intrinsic landscape in the region encompassing the Aγ-globin gene, illustrating that thermodynamically preferred sequences alone can not predict nucleosome occupancy within the human genome.
Scm3 alters the intrinsic energy landscape
How centromeric nucleosomes are targeted to centromeres is an unanswered question in chromatin biology, and it remains unclear whether they are subject to the same energetic landscapes that dictate favorable binding by canonical nucleosomes. If centromeric nucleosomes obey the same principles as canonical nucleosomes, and bind to intrinsically bendable DNA, then the in vitro distributions of centromeric versus canonical nucleosomes should be similar. Alternatively, if centromeric nucleosomes have different sequence preferences, then they should exhibit distribution patterns that are distinct from canonical nucleosomes. The intrinsic landscape is particularly intriguing for Cse4, which is targeted to poly(dA–dT)-rich CEN DNA in vivo, and can form an unusual hexameric nucleosome wherein H2A/H2B is replaced with the non-histone protein Scm318. Interestingly, the algorithms of Field et al.14 and Kaplan et al.15 both predict exceptionally low probabilities of nucleosome occupancy at yeast CEN sequences (P = 7.5×10−7 – 4.9×10−4) and also at A/T-rich human α-satellite repeats (P = 0.015 – 0.039), suggesting that mechanisms must exist enabling centromeric nucleosomes to overcome the exclusionary effects of these sequences.
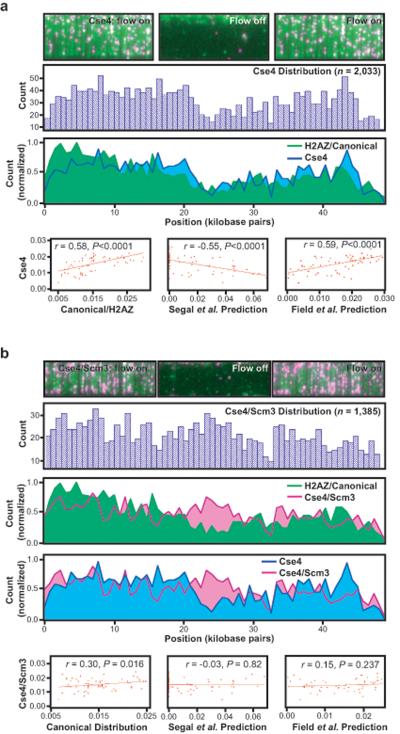
To determine whether centromere-specific nucleosomes have unique DNA binding properties that might contribute to their targeting specificity recombinant nucleosomes were assembled and deposited onto λ-DNA as described above, with the exception that H3 was replaced with Cse4 (Supplementary Fig. 2). As shown in Fig. 4a, replacement of H3 with Cse4 did cause ~8% of the total nucleosomes to redistribute away from the G/C-rich left 10-kb of the λ-DNA towards more A/T-rich regions (Fig. 4a), reflecting a subtle perturbation of the intrinsic landscape. However, the overall distribution of Cse4-nucleosomes (n = 2,033) was still correlated with that of the canonical nucleosomes (r = 0.58, P < 0.0001), was correlated with both the Field et al.14 (r = 0.59, P < 0.0001) and Kaplan et al.15 predictions (r = 0.63, P < 0.0001), and was anticorrelated with the Segal et al.6 prediction (r = −0.55, P < 0.0001). Moreover, comparison of the canonical, H2AZ, and Cse4 data sets reveals similar patterns, and the combined data from all of these octameric nucleosomes (n = 4,491) shows even better correlation with the Field et al.14 (r = 0.69, p < 0.0001) and Kaplan et al.15 (r = 0.74, P < 0.0001) theoretical predictions (Supplementary Fig. 8). These findings demonstrate that Cse4 does not drastically alter the intrinsic DNA binding landscape. This conclusion is reasonable from a physical perspective given that MNase footprinting reveals that ~1.7 turns of DNA still wrap around histone octamers harboring the centromeric H3-variant (Supplemental Fig. 2). We conclude that octameric nucleosomes harboring Cse4 are subject to the same physical principles that dictate preferential binding by canonical nucleosomes and exhibit a preference for deposition onto intrinsically bendable DNA.
Fig. 4. Thermodynamic landscapes for centromeric nucleosomes.
Panels (a and b) show images in the presence and absence of buffer flow, population distribution histograms, overlays with the canonical/H2AZ distributions, and correlation analysis for nucleosomes bearing Cse4 or Cse4/Scm3, respectively.
We next tested the effect of Scm3 on the intrinsic landscape of nucleosomes harboring Cse4. For this, nucleosomes were assembled from Scm3, Cse4, and FLAG tagged H4, and these purified nucleosomes were then deposited onto DNA via salt dialysis, as described (Supplementary Fig. 2)18. As previously shown, Scm3, Cse4, and H4 form a stable complex that can be isolated by gel filtration and bulk biochemical assays verified that these centromeric nucleosomes could bind to DNA (Supplementary Fig. 2)18. Affinity pull-down assays confirmed that Scm3, Cse4, and H4 were all present in the DNA-bound complexes (Supplementary Fig. 9). Consistent with the findings of Miziguchi et al.18, our bulk experiments argue that Scm3 is bound to DNA, and argue against the possibility that Scm3 is readily displaced after deposition of H4 and Cse4 onto DNA. Single molecule imaging assays revealed that nucleosomes containing both Cse4 and Scm3 displayed an altered DNA-binding landscape relative to the octameric nucleosomes (Fig. 4b). The profile observed for the Scm3/Cse4/H4 nucleosomes was not significantly correlated with any of the theoretical models (Fig. 4b) and was only weakly correlated with the canonical/H2AZ distribution (r = 0.30, P = 0.016; Fig. 4b). Nor was the distribution of the Scm3/Cse4/H4 nucleosomes significantly correlated with the distribution observed for the nucleosomes harboring just Cse4 (r = 0.31, P = 0.013; not shown) or the poly(dA–dT) tracts (r = 0.11, P = 0.387; not shown). All other nucleosomes tested displayed reduced occupancy within the poly(dA–dT)-rich center of λ-DNA (Fig. 2a and Fig. 4a), but this aversion was fully relieved by Scm3 (Fig. 4b). Consistent with these findings, in vitro reconstitution and gel shift experiments have revealed that DNA-protein complexes containing Cse4, H4 and Scm3 form on A/T-rich CEN sequences with greater efficiency than do Cse4/H4/H2A/H2B nucleosomes (Carl Wu, personal communication). Taken together, these results suggest that Scm3 alters centromeric nucleosomes enabling them to more readily tolerate stiff DNA sequences. These results argue that the poly(dA–dT)-rich CEN sequences found in S. cerevisiae may have evolved in part to help exclude octameric nucleosomes, irrespective of whether they contain histone H3 or Cse4, and that Scm3 abrogates this inhibition, thus ensuring a unique landmark for kinetocore assembly.
DISCUSSION
We have established a unique system capable of visualizing thousands of individual fluorescently tagged nucleosomes in real time under conditions compatible with many types of biochemical reactions. This experimental platform offers the potential for much higher throughput than other comparable single molecule imaging approaches, and opens new experimental paths for studying nucleosomes, chromatin, and chromatin remodeling. Here we use this approach to demonstrate that recombinant nucleosomes assembled onto model DNA substrates reveal global distribution patterns reflecting intrinsic properties of the underlying DNA sequence.
Intrinsic nucleosome positioning and chromatin organization
Our data with the human β-globin locus is the first measurement of an intrinsic nucleosome binding profile for a vertebrate DNA substrate, and the agreement between our data and the theoretically predicted landscape suggests that the observed nucleosome distribution reflects sequence-dependent intrinsic properties of the DNA. Moreover, our finding that all of the peaks identified in the experimentally measured intrinsic profile coincided with promoters and regulatory regions argues that the intrinsic profile reflects the underlying organization of this DNA locus. There are at least two potential explanations for this pattern: (1) incorrectly positioned nucleosomes near promoters could result in a loss of regulatory capacity through inadvertent occlusion of RNA polymerase or other factors, suggesting an evolutionary advantage to ensure correct nucleosome positioning in these regions through the use of intrinsically preferred sequences; and/or (2) strongly positioned nucleosomes at or near promoters and regulatory regions may be necessary to dictate the positions of downstream nucleosomes within a gene through a steric occlusion mechanism called statistical packing that has been likened to a can of tennis balls1,4,51. The absence of intrinsic peaks outside of regulatory regions would facilitate the establishment of nucleosome positions via statistical packing by ensuring that the most tightly bound nucleosomes were restricted to a small subset of available sites, allowing for more flexible positioning of the adjacent nucleosomes.
For S. cerevisiae, the theoretical intrinsic profiles provided by the newest generation of predictive algorithms have been reported to match the observed in vivo nucleosome positions4,14,15. However, the Field et al.14, and Kaplan et al.15 algorithms do not fully agree with recently measured in vivo nucleosome positions from Mavrich et al.51 The reasons for the discrepancies remain unclear, and should be the subject of future studies, nevertheless our data show that the Field et al.14 and Kaplan et al.15 algorithms can predict the general features of in vitro intrinsic nucleosome landscapes. Importantly, there does not appear to be a clear relationship between intrinsically favored positions and actual nucleosome locations for more complex vertebrate genomes4, and neither our experimentally derived intrinsic landscape nor the corresponding theoretical predictions were sufficient to describe the distributions of nucleosome positions mapped across the β-globin locus in human CD4+ T cells. The discrepancy between the intrinsically preferred nucleosome landscape and in vivo data may arise from several sources, including competition between nucleosomes and other binding factors, steric occlusion at high nucleosome densities, the effects of nucleosome remodeling proteins or nucleosome modifications, higher-order folding of chromatin, and/or developmentally regulated changes in gene expression. Ultimately the disagreement between the intrinsically favored landscape for the β-globin DNA and the in vivo positions argues that future predictive algorithms will need to account for these added levels of complexity.
Effects of Scm3 and implications for centromere biology
We are just beginning to understand the molecular determinants that are essential for centromere organization and function2,30. The universal marker for centromere function is the variant histone CenH3, but little is known about how it is targeted to centromeric DNA. Interestingly, S. cerevisiae Cse4 can replace CenH3 in human centromeres, indicating a high degree of functional conservation54. This is despite the fact that S. cerevisiae has well-defined point centromeres, whereas humans have regional centromeres comprised of repetitive A/T-rich units of 171-bp α-satellite sequences that span megabases of DNA. Our findings demonstrate that Cse4 by itself does not impart centromeric nucleosomes with unique DNA binding properties that might influence targeting to centromeric DNA. This begs the question of what mechanisms might contribute to Cse4 targeting. Known mechanisms that contribute to Cse4 targeting in S. cerevisiae include protein-protein interactions between Scm3 and the Ndc10 subunit of the CBF3 complex that binds the CDEIII-site within yeast centromeres21, and proteolytic degradation of any Cse4 that is mistargeted to chromosome arms55. Our results suggest a potential third level of regulation for the Cse4 centromere-targeting mechanism involving intrinsic exclusion of normal octameric nucleosomes from centromeric DNA, coupled with the positive targeting effects of Scm3, which enables nucleosomes containing Cse4 to overcome the exclusionary barrier presented by poly(dA–dT) tracts found in yeast CEN sequences. To our knowledge, this is the first example of a nucleosome with such drastically altered DNA binding characteristics, and the first direct evidence that centromeric nucleosomes have distinct DNA binding properties that might facilitate targeting to centromeric DNA. This finding further suggests that DNA bendability does not dominate the intrinsic binding landscape experienced by centromeric nucleosomes harboring Scm3, implying that these nucleosomes may be bound to DNA in an altered configuration distinct from canonical nucleosomes.
Methods
Protein expression, purification and nucleosome reconstitution
Histones were expressed in E. coli, purified from inclusion bodies and reconstituted as described (Supplementary Fig. 2)56. H2AZ was cloned into pET-100/D-TOPO, purified and reconstituted using the same procedure (Supplementary Fig. 2)56. We purified and reconstituted Cse4 as described (Supplementary Fig. 2)18. Scm3 was also expressed in E. coli, but remained soluble after cell lysis and was ammonium sulfate precipitated (45% saturation), resuspended in unfolding buffer (7 M guanidinium-HCl, 1 M NaCl, 50 mM Tris-HCl [pH 7.8], 1 mM EDTA, 1 mM DTT) and dialyzed against urea buffer (7 M urea, 1 M NaCl, 10 mM Tris-HCl [pH 7.8], 1 mM EDTA, 5 mM β-mercaptoethanol). Scm3 was purified on Ni-NTA agarose in urea buffer, eluted with 200 mM imidazole, and dialysed against 10 mM Tris-HCl [pH 7.8] plus 5 mM β-mercaptoethanol, followed by 10 mM Tris-HCl [pH 7.8], then lyophilized and stored at −20°C. All reconstituted complexes were purified by gel filtration and deposited onto DNA by salt dialysis (Supplementary Fig. 2)38,39,57, and the DNA was assembled into curtains as described35. All reconstitution reactions were performed at 4°C, the samples were shifted to 37°C for a minimum of 15-minutes after injection into the microfluidic sample chamber, and all single molecule measurements were made at 37°C.
Single molecule assays
Nucleosomes were labeled with 2 nM QDs (Invitrogen, 705 nm emission) conjugated to anti-FLAG antibodies (Sigma). Illumination was provided by a 488-nm laser (Coherent, Sapphire-CDHR), images were collected using a water immersion objective (Nikon, 1.2 NA Plan Apo, 60x). YOYO1 and QD emission spectra were separated using a dichroic mirror (630 nm DCXR, Chroma Technologies) and recorded onto separate halves an EMCCD (Photometrics, Cascade 512B). Images for position analysis were acquired at a flow rate of 0.4 ml min−1 in 40 mM Tris-HCl [pH 7.8], 1 mM DTT, and 0.2 mg ml−1 BSA. For all experimental data sets flow was transiently paused to exclude non-specifically surface bound QDs from analysis (Fig. 1b–d). Only full-length DNA molecules well resolved from adjacent neighboring molecules and bound by ~5 nucleosomes or fewer were used for distribution measurements.
Statistical analysis
Data were collected using NIS-Elements software and processed using ImageJ. Dual color images were aligned, cropped at the DNA tether points, and contrast was adjusted to improve upon the signal-to-noise ratio. The QD images were then imported into Igor Pro for position analysis, as previously described58. In brief, a 2D Gaussian fit was applied to each manually selected nucleosome particle to determine its location in microns relative to the DNA tether point based on the centroid position of the fluorescent QDs58. The position data from all of the images was divided into bins based on the experimentally measured variation of QDs attached to DNA curtains at single fixed positions (Supplementary Fig. 1), and for convenience the size of the bins was set at 758-bp, which divides the 48,502-bp λ-DNA substrate into 64 equivalent sections. For correlation analysis, the theoretically predicted distributions were also averaged over 758-bp bins, so that they matched the resolution of the experimental data (Supplementary Fig. 4). The normalized values of the experimental and theoretical data sets were then graphed against each other and the Pearson linear correlation coefficient (r) was calculated using Igor Pro by dividing the covariance between the two variables by the product of their standard deviations. The P value for n-2 degrees of freedom, where n is the total number of bins, was calculated using GraphPad software (http://www.graphpad.com/quickcalcs/pvalue1.cfm). All correlation trends were cross-validated using randomly selected subsets of the experimental data (Supplementary Fig. 10).
Supplementary Material
Acknowledgments
We thank Richard Axel, Harmen Bussemaker, Ruben Gonzalez, Alla Grishok, Richard Mann, and members of the Greene laboratory for critically reading the manuscript. We thank Brad Cairns (University of Utah & HHMI) for S. cerevisiae H2A, H2B, H3, and H4 expression plasmids. We thank Carl Wu and Hua Xiao (National Institutes of Health) for the Cse4 and Scm3 expression plasmids and for sharing unpublished results. We thank Jonathan Widom (Northwestern University) for the plasmid bearing the 601 nucleosome binding sequence, and for communicating unpublished results. We thank Luke Kaplan for cloning FLAG-H4 and purifying Scm3, Jason Gorman for assisting with data analysis, and YoungHo Kwon (Yale University) for assistance with the salt-dialysis protocol. This work was supported by a Basil O'Connor Starter Scholar Award from the March of Dimes and a grant from the National Institutes of Health (GM082848) to E.C.G., and was partially funded by the Initiatives in Science and Engineering grant program through Columbia University, and the Nanoscale Science and Engineering Initiative of the National Science Foundation under NSF Award Number CHE-0641523 and by the New York State Office of Science, Technology, and Academic Research (NYSTAR). Dr. Greene is an HHMI Early Career Scientist.
References
- 1.Rando OJ, Ahmad K. Rules and regulation in the primary structure of chromatin. Curr Opin Cell Biol. 2007;19:250–256. doi: 10.1016/j.ceb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Morris CA, Moazed D. Centromere assembly and propagation. Cell. 2007;128:647–650. doi: 10.1016/j.cell.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Widom J. Structure, dynamics, and function of chromatin in vitro. Annu Rev Biophys Biomol Struct. 1998;27:285–327. doi: 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]
- 4.Rando OJ, Chang HY. Genome-Wide Views of Chromatin Structure. Annu Rev Biochem. 2009 doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 6.Segal E, Fondufe-Mittendorf Y, Chen L, ThÂstr^m A, Field Y, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 8.Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 9.Struhl K. Naturally occurring poly(dA–dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985;82:8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 11.Lee W, Tillo D, Bray N, Morse RH, Davis RW, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 12.Whitehouse I, Tsukiyama T. Antagonistic forces that position nucleosomes in vivo. Nat Struct Mol Biol. 2006;13:633–640. doi: 10.1038/nsmb1111. [DOI] [PubMed] [Google Scholar]
- 13.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, et al. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 17.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 18.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 19.Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 20.Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Camahort R, Li B, Florens L, Swanson SK, Washburn MP, et al. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, et al. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci U S A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker RE. CENP-A targeting moves a step back. Mol Cell. 2009;33:411–413. doi: 10.1016/j.molcel.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Aravind L, Iyer LM, Wu C. Domain architectures of the Scm3p protein provide insights into centromere function and evolution. Cell Cycle. 2007;6:2511–2515. doi: 10.4161/cc.6.20.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 29.Foltz DR, Jansen LE, Bailey AO, Yates JR, Bassett EA, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalal Y, Furuyama T, Vermaak D, Henikoff S. Structure, dynamics, and evolution of centromeric nucleosomes. Proc Natl Acad Sci U S A. 2007;104:15974–15981. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cumberledge S, Carbon J. Mutational Analysis of Meiotic and Mitotic Centromere Function in Saccharomyces cerevisiae. Genetics. 1987;117:203–212. doi: 10.1093/genetics/117.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker RE, Rogers K. Genetic and Genomic Analysis of the AT-rich Centromere DNA Element II of Saccharomyces cerevisiae. Genetics. 2005;171:1463–1475. doi: 10.1534/genetics.105.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malik HS, Henikoff S. Conflict begets complexity: the evolution of centromeres. Current Opinion in Genetics & Development. 2002;12:711–718. doi: 10.1016/s0959-437x(02)00351-9. [DOI] [PubMed] [Google Scholar]
- 34.Espelin CW, Simons KT, Harrison SC, Sorger PK. Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10p to CDEII. Mol Biol Cell. 2003;14:4557–4568. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fazio T, Visnapuu ML, Wind S, Greene EC. DNA curtains and nanoscale curtain rods: high-throughput tools for single molecule imaging. Langmuir. 2008;24:10524–10531. doi: 10.1021/la801762h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Visnapuu ML, Fazio T, Wind S, Greene EC. Parallel arrays of geometric nanowells for assembling curtains of DNA with controlled lateral dispersion. Langmuir. 2008;24:11293–11299. doi: 10.1021/la8017634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Thästrôm A, Bingham LM, Widom J. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J Mol Biol. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 39.Thästrôm A, Lowary PT, Widom J. Measurement of histone-DNA interaction free energy in nucleosomes. Methods. 2004;33:33–44. doi: 10.1016/j.ymeth.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 40.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 41.Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, et al. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 43.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, et al. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- 47.Bank A. Regulation of human fetal hemoglobin: new players, new complexities. Blood. 2006;107:435–443. doi: 10.1182/blood-2005-05-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 49.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 53.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P. Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:6620–6630. doi: 10.1128/MCB.24.15.6620-6630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 56.Wittmeyer J, Saha A, Cairns B. DNA translocation and nucleosome remodeling assays by the RSC chromatin remodeling complex. Methods Enzymol. 2004;377:322–343. doi: 10.1016/S0076-6879(03)77020-7. [DOI] [PubMed] [Google Scholar]
- 57.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 58.Prasad TK, Robertson RB, Visnapuu ML, Chi P, Sung P, et al. A DNA-translocating Snf2 molecular motor: Saccharomyces cerevisiae Rdh54 displays processive translocation and extrudes DNA loops. J Mol Biol. 2007;369:940–953. doi: 10.1016/j.jmb.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.