Abstract
The identification and structural characterization of a series of ornithine lipids extracted from the cell membranes of wild-type Rhodobacter sphaeroides, as well as from a glycerophosphocholine-deficient strain, has been achieved by multistage tandem mass spectrometry of their protonated and deprotonated precursor ions in a linear quadrupole ion trap. Systematic examination of the multistage gas-phase fragmentation reactions of these ions, combined with the use of hydrogen/deuterium exchange, has enabled the pathways responsible for sequential losses of the 3-hydroxy linked fatty acyl chain and the amide linked 3-OH fatty acid chain from these lipids, as well as for formation of the previously reported ornithine specific positively charged ‘fingerprint’ ion at m/z 115, to be determined. Additionally, the fragmentation pathways responsible for formation of a previously unreported ornithine lipid head group specific product ion at m/z 131 in negative ion mode have been examined. Based on these results, and by comparison with the fragmentation behavior of model lipoamino acid standard compounds, a series of novel glutamine containing lipids have also been identified, with analogous structures but with masses 14 Da higher than several of the ornithine lipids observed in this study. Characteristic ‘fingerprint’ ions indicative of these glutamine lipids were found at m/z 147, 130 and 129 in positive ion mode, and at m/z 145 and 127 in negative ion mode. The results from this study establish an experimental basis for future efforts aimed at the sensitive identification, characterization and quantitative analysis of ornithine and glutamine lipids in complex unfractionated cellular extracts.
Introduction
Ornithine lipids (OL) belong to a class of fatty acylated amino acids that do not contain either glycerol or phosphate. Since their discovery over 40 years ago [1–3], ornithine lipids have been found to be broadly distributed among a number of gram-negative bacteria and several gram-positive bacteria [4]. However, the function of the OL species in these bacteria remains unclear. Ornithine lipids constitute different proportions of the total membrane lipids depending on the individual bacterium and growth conditions, ranging from around 2 % in many bacteria grown under normal conditions, to being nearly the sole polar lipid present under phosphate-limited growth conditions in Pseudomonas fluorescens [5]. An increase in the abundance of OL under phosphate-limiting growth conditions, along with sulfoquinovosyldiacylglyceride (SQDG) and diacylglyceryl–N, N, N–trimethylhomoserine (DGTS) lipids, has also been observed in Rhodobacter sphaeroides (R. sphaeroides), as revealed by TLC using 14C labeling quantification [6]. In Sinorhizobium meliloti, either OL or DGTS are required under phosphate-limiting conditions to ensure normal cell yields [7]. Also in Sinorhizobium meliloti, an OL genetic deficiency has been shown to be compensated for by increases in glycerophosphatidylethanolamine (GPEtn)/N-methyl GPEtn, glycerophosphatidylcholine (GPCho) and glycerophosphatidylglycerol (GPGro), with wild-type growth behavior obtained [8]. OL’s have also been implicated in high temperature tolerance in Burkholderia cepacia [9] and in acid tolerance in Rhizobium tropici [10]. OL’s from Bordetella pertussis and Flavobacterium meningosepticum have been shown to have a number of biological functions, including hemagglutination, and stimulation of macrophages [11–13]. It is also known that OL is critical to obtaining optimal yields of cytochrome c in Rhodobacter capsulatus [14]. Finally, we have recently noted that OL’s are one of the lipid species that remain associated to cytochrome c oxidase (CcO), when purified and crystallized from R. sphaeroides [15].
To date, ornithine is the only type of non-glycerol and non-phosphate containing lipoamino acid reported to be present in R. sphaeroides [6,16,17]. However, lipids containing lysine or serine amino acids, or serine-glycine dipeptides, with analogous backbone structures to OL have previously been observed in Agrobacterium tumefaciens [18] and Flavobacterium meningosepticum [19]. Unfortunately, little detail regarding the molecular structures of OL in R. sphaeroides is available, with prior work limited to their identification based on their migration position on TLC plates using ninhydrin (amino-positive) and molybdate (phosphate-negative) spray results [6,16,17].
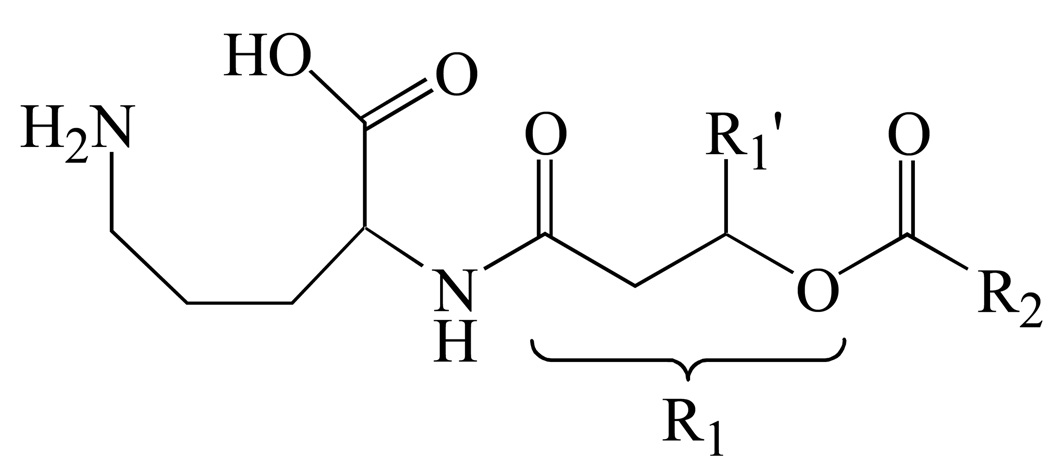
Ornithine lipids typically contain an ornithine head group that is connected via its Nα–amino group to a 3–OH fatty acid (R1), with a second fatty acid chain (R2) esterified to the 3–OH group of the first fatty acid [3] (Scheme 1). However, an increase in growth temperature from 25 to 40 °C has also been shown to promote the incorporation of a 2–OH fatty acyl group to the second fatty acid of OL in Burkholderia cepacia [9]. Furthermore, the incorporation of 14C-labeled ornithine in Rhizobium tropici revealed four different forms of 14C-labeled lipid products, two of which were ninhydrin-negative [10].
Scheme 1.
Schematic structure of an Ornithine lipid (OL).
Mass spectrometry (MS) coupled with electrospray ionization (ESI) or matrix assisted laser desorption ionization (MALDI) has emerged as an attractive method to analyze intact, non-volatile, polar lipids, and is being increasingly applied to a wide range of lipid studies [20]. However, the precursor ions for most membrane lipids fall into a relatively narrow m/z range between 600 – 1600, resulting in significant crowding and potential overlapping of peaks in the resultant mass spectra, particularly in the region of m/z from 600 – 900. The mass spectra are further complicated by the presence of various cationic or anionic adducts from the lipids that may be present in the mixture (e.g., [M+H]+ and [M+Na]+ in positive ion mode, and [M−H]−, [M+Cl]− or [M+CH3OCO2]− in negative ion mode) [20,21]. Therefore, the use of tandem mass spectrometry (MS/MS) or multistage tandem mass spectrometry (MSn) is generally required to unambiguously identify and characterize the individual lipid species that may be present, based on the characteristic product ions that are formed upon fragmentation.
Numerous mass spectrometry studies have previously been carried out to examine the gas-phase fragmentation reactions of OL [14, 22–27]. From these studies, a characteristic low mass product ion at m/z 115, resulting from the dissociation of protonated OL in positive ion mode MS/MS, and corresponding to the formation of a 3–amino–2–oxopiperidinium ion, has been consistently observed [14, 22–25]. This ion may therefore be considered a ‘fingerprint’ ion for ornithine lipid identification. However, this product is not generally observed due to the inherent low mass cutoff (LMCO) imposed on product ion detection when performing collision induced dissociation (CID)–MS/MS in quadrupole ion trap mass spectrometers under typical operating conditions. In this case, further dissociation (i.e., MSn) of the high mass product ions that are initially formed during MS/MS may be required to provide information regarding the presence and identity of the OL species. Furthermore, with the exception of a single report in the literature [24], little information is available regarding the negative ion mode CID–MS/MS fragmentation behavior of OL species.
An improved understanding of the fundamental gas-phase fragmentation behavior of ornithine lipids using modern mass spectrometry instrumentation, in both positive and negative ion modes, is therefore critical to the development of sensitive, unequivocal identification, characterization and quantitative analysis strategies for this class of lipid molecule, and for subsequently determining their functional roles. The results from these studies could also be applied to elucidation of the presence and structures of other lipoamino acid species that may be present.
Materials and Methods
Materials
L–ornithine, L–lysine, L–glutamine, Nα–acetyl L–ornithine, Nα–acetyl L–lysine, Nε–acetyl L–lysine, methanol (HPLC grade) were purchased from Sigma-Aldrich (St. Louis, MO). Propionic anhydride and iodomethane were purchased from EMD Chemicals (Gibbstown, NJ). Acetic anhydride, ammonium bicarbonate, glacial acetic acid, chloroform, KCl and KH2PO4 were purchased from Mallinckrodt Baker (Phillipsburg, NJ). Ethyl ether was from Jade Scientific (Canton, MI). Ammonium hydroxide (28.0–30.0 %, m/v) and hydrogen peroxide (30%, m/v) were purchased from Columbus Chemical Industries (Columbus, WI).. EDTA and Tris (≥99%) were purchased from Invitrogen Life Technologies (Carlsbad, CA). DNase I and RNase A (bovine pancrease) were from EMD Biosciences (La Jolla, CA). CD3OD (D, 99.8%) and CDCl3 (D, 99.8%) were purchased from Cambridge Isotope Laboratories (Andover, MA). Solvents were filtered with a sintered glass funnel (pore size 4–8 µm) prior to use. Nα–acetylation and Nα–propylation of L–glutamine and O–methylation of Nα–acetyl L–ornithine were performed as previously described [28]. Nδ–methylation of Nα–acetyl L–ornithine was performed by reacting Nα–acetyl L–ornithine with a 100 fold excess of CH3I in 50 mM NH4HCO3 aqueous buffer at room temperature for up to 48 hours.
Construction and growth of wild type (2.4.1 + CcO) and GPCho deficient (CHB20 + CcO) R. sphaeroides
A pRK-pYJ123H plasmid carrying the genes coding for a wild-type CcO [29] was transferred by conjugation from Escherichia coli S17-1 into the wild type (2.4.1) and GPCho-deficient (CHB20) [17] strains of R. sphaeroides, creating the 2.4.1 + CcO and CHB20 + CcO strains, respectively. These strains were grown aerobically on plates in Sistrom's media containing 50 µg/mL spectinomycin, 50 µg/mL streptomycin and 1 µg/mL tetracycline pH 7.0 at 30 °C for 3–5 days, then the cells were picked and inoculated into small flasks with 100 mL of Sistrom's media containing the same antibiotics and grown for two days at 30 °C with vigorous shaking at 250 rpm. Then 50 mL of starter culture was used to inoculate a 2.8 L Fernbach flask with 800 mL Sistrom's media containing 1 µg/mL tetracycline, 25 µg/mL streptomycin and 25 µg/mL spectinomycin. The Fernbach flasks were shaken at 250 rpm at 30 °C for 2–3 days until the UV-Vis absorbance at 660 nm exceeded 1.2 and the pH was greater than 8.5.
Membrane isolation and lipid extraction
R. sphaeroides cells were harvested by centrifugation in a GS-3 rotor at 14,000 g for 20 minutes and re-suspended in pH 6.5 buffer containing 50 mM KH2PO4 and 1 mM EDTA. After addition of DNase I and RNase, the re-suspended cells were homogenized and lysed by passing through French press twice under 20,000 psi at 4 °C. The mixture was then centrifuged at 30,000 g for 30 minutes at 4 °C to remove the precipitated unbroken cells and debris, and the supernatant was then subjected to ultracentrifugation at 200,000 g for 90 minutes at 4 °C to precipitate the membrane pellets [29]. The membrane pellets were then resuspended in the buffer containing 10 mM Tris, pH 8.0 and 40 mM KCl, quick-frozen in liquid N2 and stored at −80 °C prior to further use. 10 µL each of the suspended membranes from either wild type or mutant R. sphaeroides cells (protein concentrations of approximately 1.4 mg/mL) were then freeze-dried, and lipids extracted by the sequential addition of 2 × 50 µL chloroform: methanol (2:1,v/v), 1 × 50 µL ethyl ether: ethanol (1:1,v/v), and 2 × 50 µL chloroform: methanol: ammonium hydroxide (100:50:2,v/v/v) followed by vortexing and removal of the solvent at each step [30]. The extracts were combined, then dried under vacuum and re-dissolved in 50 µL CHCl3:CH3OH (1:1, v/v) prior to mass spectrometry analysis.
Identification of lipids using ESI MSn (n=1, 2, 3, 4, 5)
Mass spectrometry analysis of lipid extracts was performed using a linear quadrupole ion trap mass spectrometer (Thermo Fisher Scientific, model LTQ, San Jose, CA), equipped with electrospray ionization (ESI) or nanoelectrospray ionization (nanoESI) sources. Total lipid extracts from each membrane sample were diluted 4 fold using CHCl3:CH3OH (1:1, v/v) containing 40 mM ammonium hydroxide, immediately prior to introduction to the mass spectrometer by direct nanoESI infusion through non-coated silica tips with internal diameters of 30 µm (New Objective, Inc. Woburn, MA), at a flow rate of 0.3 µL/min. Amino acid and N–acetylated amino acid standards were analyzed as 100 µM solutions in H2O:CH3OH (1:1, v/v) containing 40 mM ammonium hydroxide. NanoESI conditions were optimized to maximize the sensitivity and stability of the precursor ions of interest while minimizing ‘in-source’ fragmentation. Typical nanoESI conditions were: heated capillary temperature 180 °C, spray voltage 1.8 kV, capillary voltage 20 V and tube lens voltage 75 V. H/D exchange of lipid extracts was achieved by drying the original lipid extracts under vacuum and redissolving in CDCl3:CD3OD (1:1, v/v) solvent. This process was repeated twice, then the sample was redissolved in CDCl3:CD3OD (1:1, v/v) containing 40 mM ammonium hydroxide. The sample was then introduced to the mass spectrometer via infusion using the ESI source at a flow rate of 1 uL/min. Typical ESI conditions, optimized to minimize back exchange, were: heated capillary temperature 180 °C, spray voltage 4.0 kV, sheath gas (N2) flow rate 12 arbitrary units, auxiliary gas (N2) flow rate 15 arbitrary units and sweep gas (N2) flow rate 10 arbitrary units.
Mass spectra (MS) were acquired from m/z 150 to 2000 in both positive ion and negative ion modes. Lipid precursor ions that had normalized relative abundances higher than 1% in the mass spectra were further examined by CID MSn using helium as the collision gas and an activation time of 30 ms. Collision energies were optimized (ranging from 20–30% normalized collision energy) for each precursor ion of interest. Typically, an activation q value of 0.2 was used in order to achieve a reasonable low-mass-cutoff while still maintaining good sensitivity. The spectra shown were typically the average of 50–300 scans.
Results and Discussion
ESI-Mass spectrometry analysis of wild type and GPCho-deficient R. sphaeroides
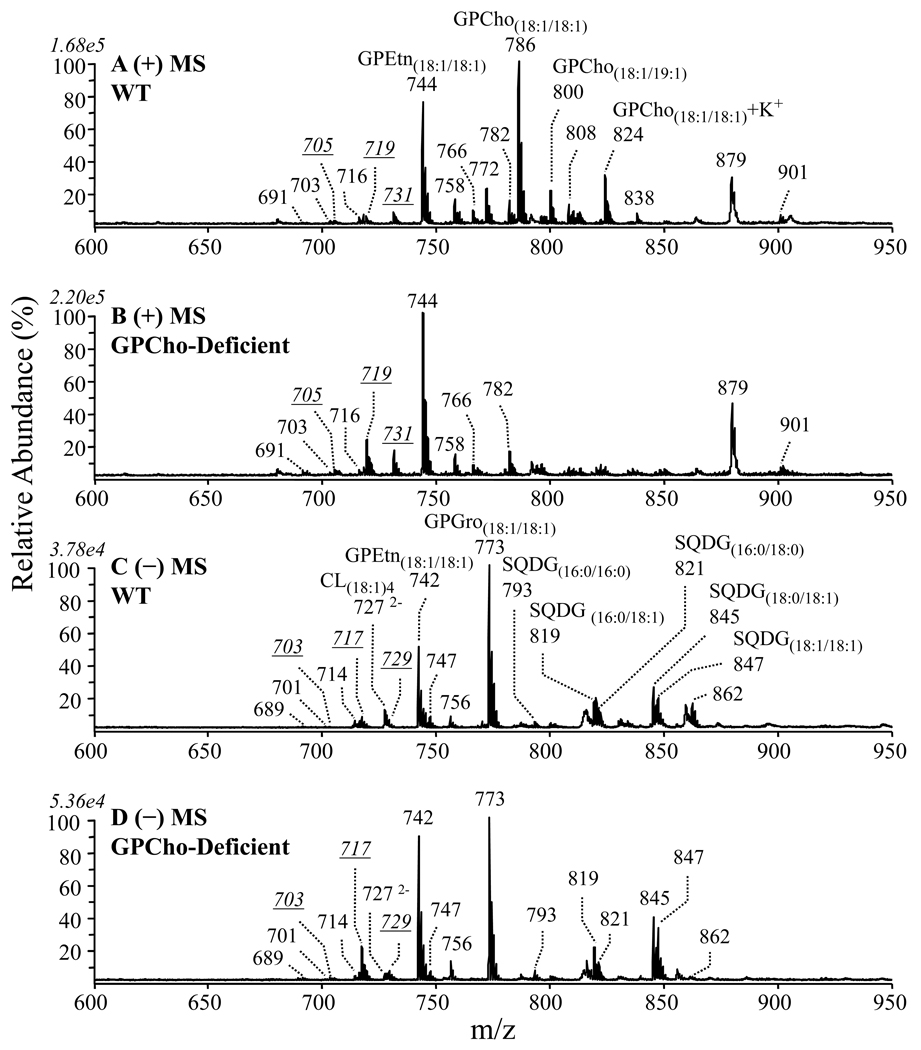
The mass spectra of membrane lipid extracts obtained from wild type R. sphaeroides, as well as from a GPCho-deficient mutant of R. sphaeroides, acquired in both positive and negative ion modes are shown in Figures 1A and 1B, and in Figures 1C and 1D, respectively. As the lipid extracts examined here were comprised of complicated mixtures, and were introduced to the mass spectrometer without prior separation, the m/z values of individual ions could correspond to a number of different molecular compositions.
Figure 1.
Positive ion ESI MS of crude lipid extracts isolated from the cell membranes of (A) wild type and (B) GPCho-deficient mutant R. sphaeroides (both with CcO over-expression). The negative ion ESI MS spectra are shown in panels (C) and (D), respectively. Only those ions within the selected m/z range of interest (600 – 950) are displayed. The italic underlined numbers represent ions of interest to this study.
Thus, all lipids whose precursor ions had normalized relative abundances higher than 1% in the mass spectrum were subjected to analysis by using CID MSn in order to determine their identities. The identities of several of the major lipids observed in the most abundant ions in these spectra are labeled in Figure 1. The nomenclature employed for lipid assignments is that recommended by Fahy et al [31]. The focus of the study described here however, was a series of odd m/z ions, including the italicized and underlined ions at m/z 705, m/z 719 and m/z 731 in positive ion mode, and at m/z 703, m/z 717 and m/z 729 in negative ion mode. According to the nitrogen rule, these ions should contain zero or an even number of nitrogen atoms, and therefore could correspond to putative protonated and deprotonated ions for various OL species, respectively. A comparison of the spectra obtained from the wild type and GPCho deficient mutant lipid extracts indicated an apparent increase in the relative abundances of various ions in the GPCho mutant, including the putative ornithine lipid ions. Previously, in the absence of overexpression of CcO, the GPCho-deficient mutant showed no increase in OL content, as quantified by TLC with 14C labeling [17]. However, similar studies have not yet been carried out to compare the wild type and GPCho strains when CcO is overexpressed in these systems. Here, it was not possible to determine if the observed changes in abundances for the putative ornithine lipid ions between the wild type and GPCho strains, obtained with CcO overexpression, was reflective of real changes in their concentrations, or whether the apparent increases were simply due to the removal of ionization suppression effects in the GPCho deficient mutant, due to the unavailability of authentic ornithine lipid internal standards. Regardless of this limitation, we were particularly interested to determine the presence and structural composition of the ornithine lipids in these samples, and to develop a comprehensive understanding of their fragmentation behaviors using MSn for use in further identification and quantification applications. The higher ion abundances of these ions in the GPCho-deficient mutant made it the sample of choice in which to investigate these species. However, the corresponding ions in the wild type lipid extract were also examined in parallel.
Identification and characterization of ornithine lipids in R. sphaeroides by using positive and negative ion mode MSn
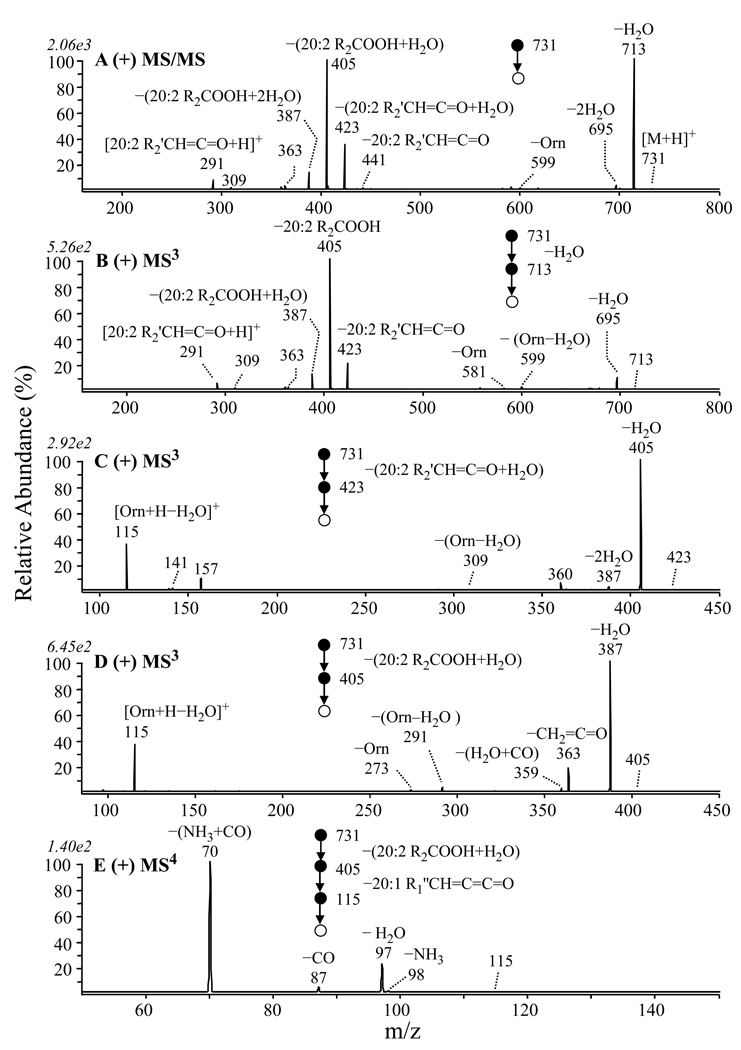
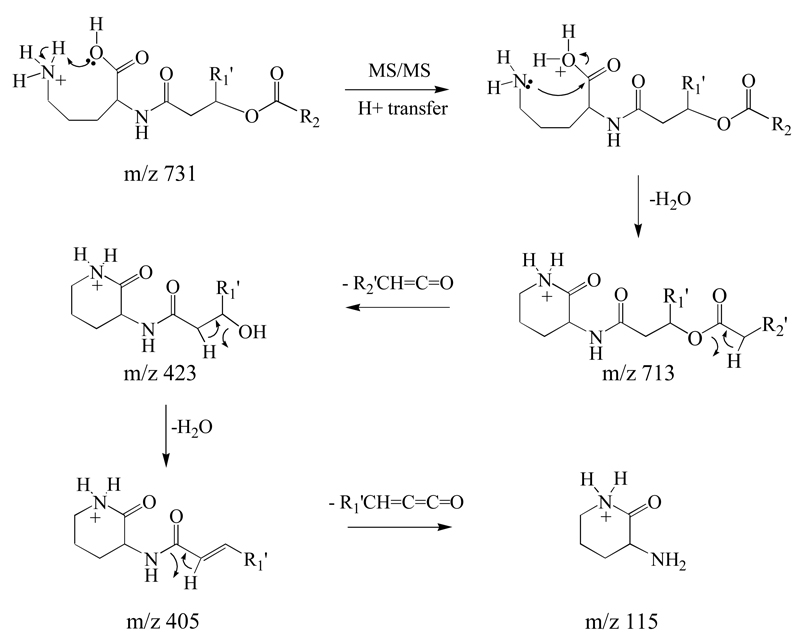
The MS/MS and MSn spectra for the ion at m/z 731 in positive ion mode, and for the corresponding ion at m/z 729 in negative ion mode observed in the membrane lipid extract of the GPCho deficient strain of R. sphaeroides are shown in Figure 2 and Figure 3, respectively. Figure 2A shows the positive ion mode MS/MS for m/z 731. In this spectrum, the most abundant product ion observed above the low mass cutoff of m/z 160 (at an applied activation q value of 0.2) was m/z 713, formed via the loss of water from the protonated precursor ion (Scheme 2). The other abundant product ions in this spectrum provided information on the fatty acid composition of the precursor ion, that were more clearly revealed by MS3 analysis of the [M+H−H2O]+ product ion. This ion underwent further fragmentation via (i) the loss of a 20:2 R2 fatty acyl chain as a neutral ketene (−R2'CH=C=O, m/z 423), where 20:2 represents the total number of carbons and the number of unsaturations, respectively. Note that ornithine lipid species containing a cyclopropane group within a fatty acyl chain have previously been described [2] (ii) the combined loss of R2'CH=C=O+H2O or the loss of a neutral fatty acid (R2COOH) (m/z 405), and (iii) the combined loss of R2COOH+H2O (m/z 387) (Figure 2B). Further dissociation of the [M+H−H2O−R2'CH=C=O]+ (m/z 423) product ion from Figure 2A led to the dominant loss of H2O to yield the product ion at m/z 405 (Figure 2C). MS3 of the m/z 405 product ion observed in Figure 2A was then used to demonstrate that this ion underwent further dissociation via the loss of a neutral R1 ketene (20:1 R1’CH=C=C=O) to yield the characteristic ‘fingerprint’ ion for the ornithine lipid at m/z 115 (i.e., [ornithine−H2O+H]+) (Figure 2D). Thus, the structure of the ornithine lipid ion at m/z 731 is proposed to correspond to OL 3–OH 20:1/20:2 (note that the acyl chain designations for the OL (e.g., OL 3-OH 20:1/20:2) are listed in order of R1 and R2).
Figure 2.
Identification of ornithine lipid (OL 3–OH 20:1/20:2) at m/z 731 from Figure 1B by positive ion CID-MS/MS and −MSn. (A) MS/MS of m/z 731, (B) MS3 of m/z 713 from panel A, (C) MS3 of m/z 423 from panel A, (D) MS3 of m/z 405 from panel A, and (E) MS4 of m/z 115 from panel D.
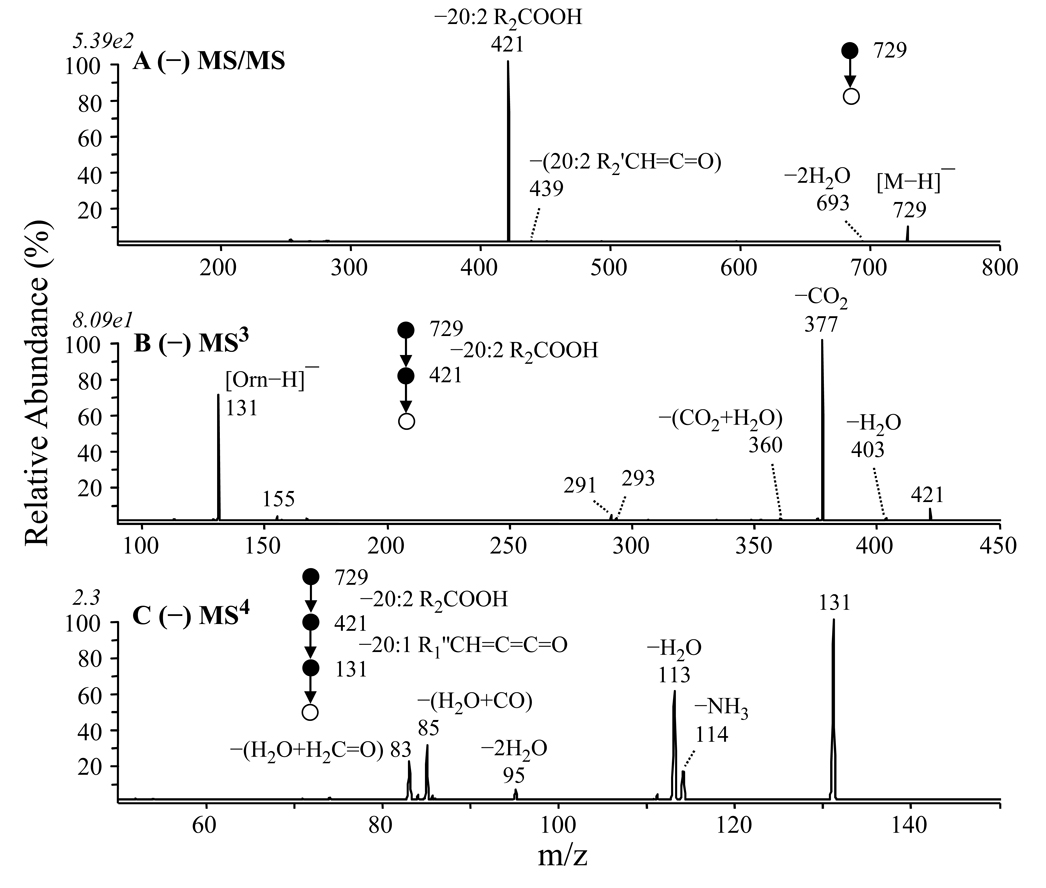
Figure 3.
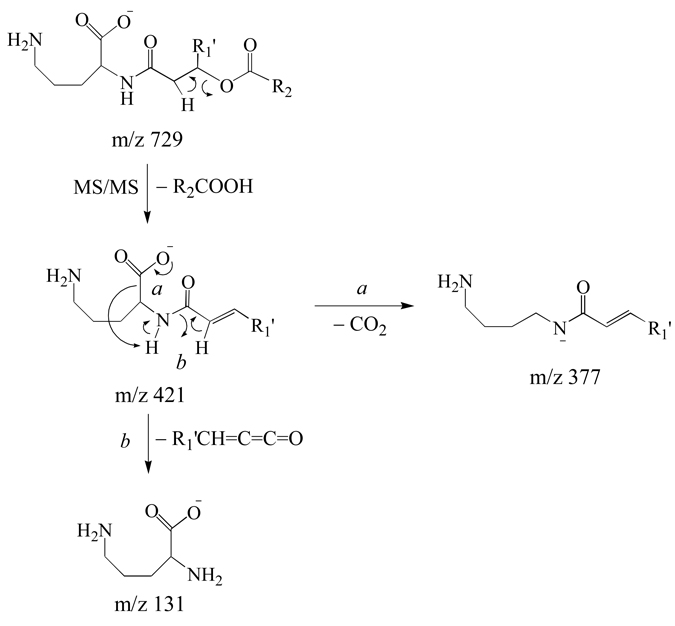
Identification of ornithine lipid (OL 3–OH 20:1/20:2) (m/z 729) from Figure 1D by negative ion CID-MS/MS and −MSn. (A) MS/MS of m/z 729, (B) MS3 of m/z 421 from panel A, and (C) MS4 of m/z 131 from panel B.
Scheme 2.
Proposed pathways for the multistage gas-phase fragmentation reactions of protonated ornithine lipids.
To confirm its identity, the product ion at m/z 115 was subjected to further dissociation by MS4 (Figure 2E), and was compared to the fragmentation behavior of the [M+H−H2O]+ or [M+H−H2O−CH2CO]+ product ions formed by MS/MS or MS3 of the [M+H]+ precursor ions of authentic ornithine or Nα–acetyl–ornithine standards, respectively (data not shown). The resultant spectra, revealing the losses of NH3 (m/z 98), H2O (m/z 97), CO (m/z 87) and NH3+CO (m/z 70), were identical, thereby providing compelling evidence for its structure. Further evidence for the identity of this ornithine lipid was provided by observing the incorporation of five deuterium atoms upon H/D exchange and MS analysis of this ion (i.e. m/z 731 was observed to shift to m/z 736). However, as extensive deuterium scrambling of the deuterated precursor ion upon CID MS/MS, presumably resulting from exchange between the deuterium atoms in the amino and amide groups with the Cα hydrogen atoms in the backbone, was observed, as evidenced by the losses of H2O, HOD, and D2O, further mechanistic details regarding these losses could not be obtained. Thus, while Scheme 2 shows a potential pathway for formation of the ion at m/z 115, via the ‘charge-directed’ loss of water from the [M+H]+ precursor ion, followed by the ‘charge-remote’ sequential losses of ketene (−R2'CH=C=O), water and ketene (R1’CH=C=C=O), we note that alternate charge-remote and charge-directed pathways cannot be ruled out.
Figure 3 shows the MS/MS and MS3 spectra obtained by dissociation of the deprotonated [M−H]− precursor ion at m/z 729 from the GPCho deficient strain of R. sphaeroides. The MS/MS spectrum (Figure 3A) was dominated by the loss of a neutral 20:2 fatty acid (R2COOH, m/z 421), consistent with that observed from the positive ion mode data. This loss is proposed in Scheme 3 to occur via a cis-1, 2 elimination reaction. This proposed mechanism was supported by the results from MS/MS of the fully H/D exchanged precursor ion (3 exchanges observed), where the loss of R2COOH was predominantly observed, rather than R2COOD (data not shown). Subsequent MS3 of the m/z 421 product ion from the protonated precursor ion resulted in the loss of CO2 (m/z 377), as well as the loss of a neutral 20:1 ketene (−R1’CH=C=C=O), thereby yielding a product ion at m/z 131 that corresponds to deprotonated ornithine (Figure 3B). A suggested pathway for these fragmentation reactions, consistent with previous reports in the literature describing the fragmentation reactions of deprotonated amino acids [32–34], is shown in Scheme 3. The formation of the m/z 131 product ion in negative ion mode is also consistent with the sequential fragmentation reactions observed for the corresponding precursor ion in positive ion mode analysis. Also, MS4 of the m/z 131 product ion (Figure 3C) resulted in identical spectra to those obtained by MS/MS or MS3 of protonated ornithine or Nα–acetyl ornithine standards, respectively (data not shown), thereby confirming its identity.
Scheme 3.
Proposed pathways for the multistage gas-phase fragmentation reactions of deprotonated ornithine lipids.
Analysis of the complementary pair of precursor ions observed at m/z 731 and m/z 729 in positive and negative ion modes, as well as comparison of the fragmentation behavior of their characteristic ‘fingerprint’ ions observed at m/z 115 and m/z 131, respectively, with those of authentic ornithine or Nα–acetyl ornithine standards, provide insights into the multistage gas-phase fragmentation behavior of ornithine lipids, thereby making possible the rapid and unambiguous assignment of these species in an unseparated complex lipid mixture, without requirement for independent synthesis of an entire OL lipid standard.
The ornithine head group in the structures shown in Scheme 2 and Scheme 3 are linked to the fatty acid chain via a Nα-amide bond, consistent with the structures of OL previously proposed for other bacteria. The linkage of the ornithine head group to the fatty acids could also potentially occur by forming an Nδ-amide bond. However, dissociation of this species would not result in the same product ions observed here by MS/MS and MS3 in either positive or negative ion modes. For example, CID MS/MS of protonated amino acids containing a free α-amino group are well known to result in the combined losses of H2O and CO (46 Da)), thereby allowing it to be disregarded.
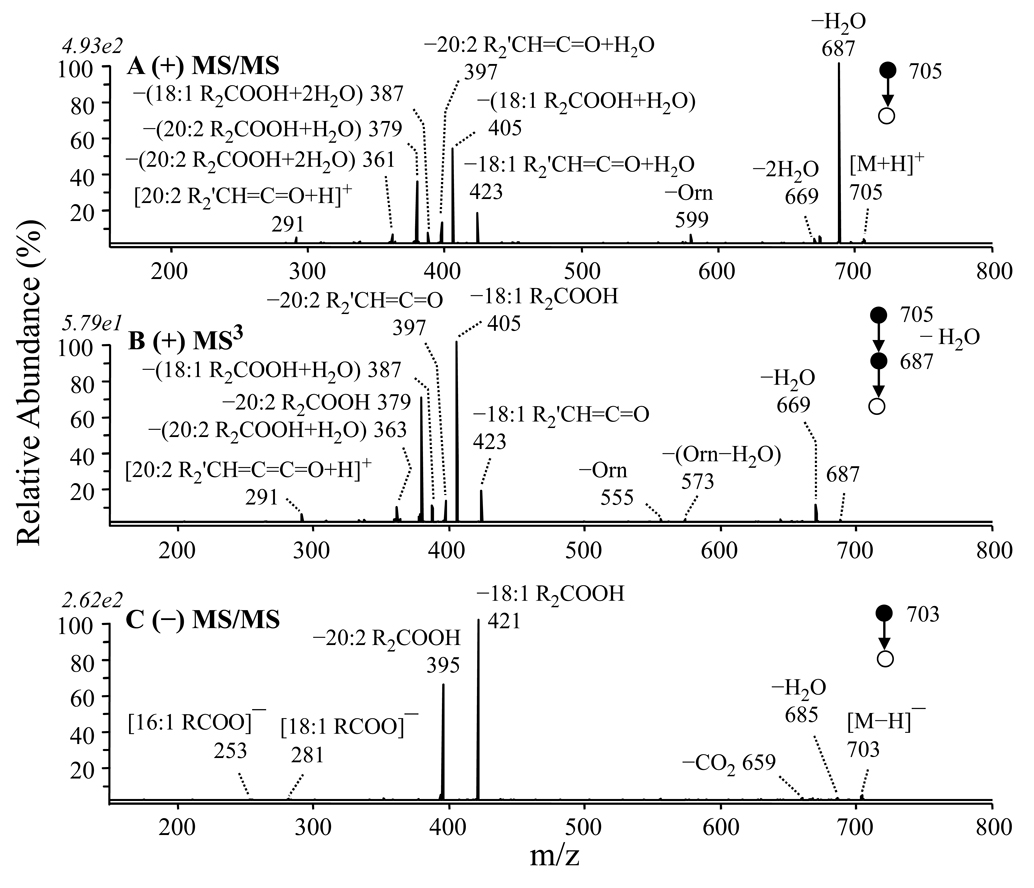
Elucidation of the fragmentation characteristics of different types of lipids is particularly useful when dealing with complicated lipid mixtures, since overlapping of multiple precursor ions at the same m/z frequently occurs. Figure 4A and 4B show the MS/MS and MS3 spectra of the positive ion mode precursor ion observed at m/z 705 in Figure 1B and its [M+H−H2O]+ product ion, respectively. Similar to that observed in Figure 2A, the loss of water was observed as a dominant fragmentation pathway in the positive ion mode MS/MS spectrum, along with a seemingly complicated set of other product ions. However, MS3 of the [M+H−H2O]+ ion at m/z 687 (Figure 4B) confirmed that these ions originated from the initial water loss product ion, indicative of the presence of 18:1 and 20:2 fatty acid chains. The product ions at m/z 423, 405 and 387 correspond to loss of the 18:1 fatty acid chain as a neutral ketene (18:1 R2'CH=C=O), a neutral fatty acid (18:1 R2COOH or 18:1 R2'CH=C=O+H2O) and a fatty acid + water (18:1 R2COOH+H2O), respectively, while m/z 397, 379 and 361 all correspond to the loss of the 20:2 fatty acid chain. This suggested the presence of two separate ornithine lipid species at similar relative abundances. Subsequent MS3 analysis of the m/z 405 product ion (the resultant spectrum was identical to Figure 2D), as well as the m/z 379 product ion (data not shown), both led to formation of the m/z 115 ornithine ‘fingerprint’ ion via the loss of a 20:1 and 18:0 fatty acid, respectively, indicating that the identity of the two lipids present at m/z 705 corresponded to OL 3–OH 20:1/18:1 and OL 3–OH 18:0/20:2.
Figure 4.
Identification of ornithine lipids (OL 3–OH 18:0/20:2 and OL 3–OH 20:1/18:0) at m/z 705 from Figure 1B by (A) CID–MS/MS and by (B) CID-MSn of the m/z 687 ion from panel A. (C) Negative ion CID-MS/MS of the corresponding ion at m/z 703 from Figure 1D.
Confirmation of these lipids were obtained by MS/MS of the corresponding negative ion mode precursor observed at m/z 703 (Figure 4C). From this spectrum, two major product ions were formed, corresponding to the neutral losses of 18:1 R2COOH (m/z 421) and 20:2 R2COOH (m/z 395) fatty acid chains, respectively. MS3 of these products both resulted in formation of the characteristic m/z 131 product ion via the loss of neutral 20:1 and 18:0 ketene molecules (R1’CH=C=C=O), respectively (data not shown).
Identification and characterization of glutamine lipids in R. sphaeroides by using positive and negative ion mode MSn
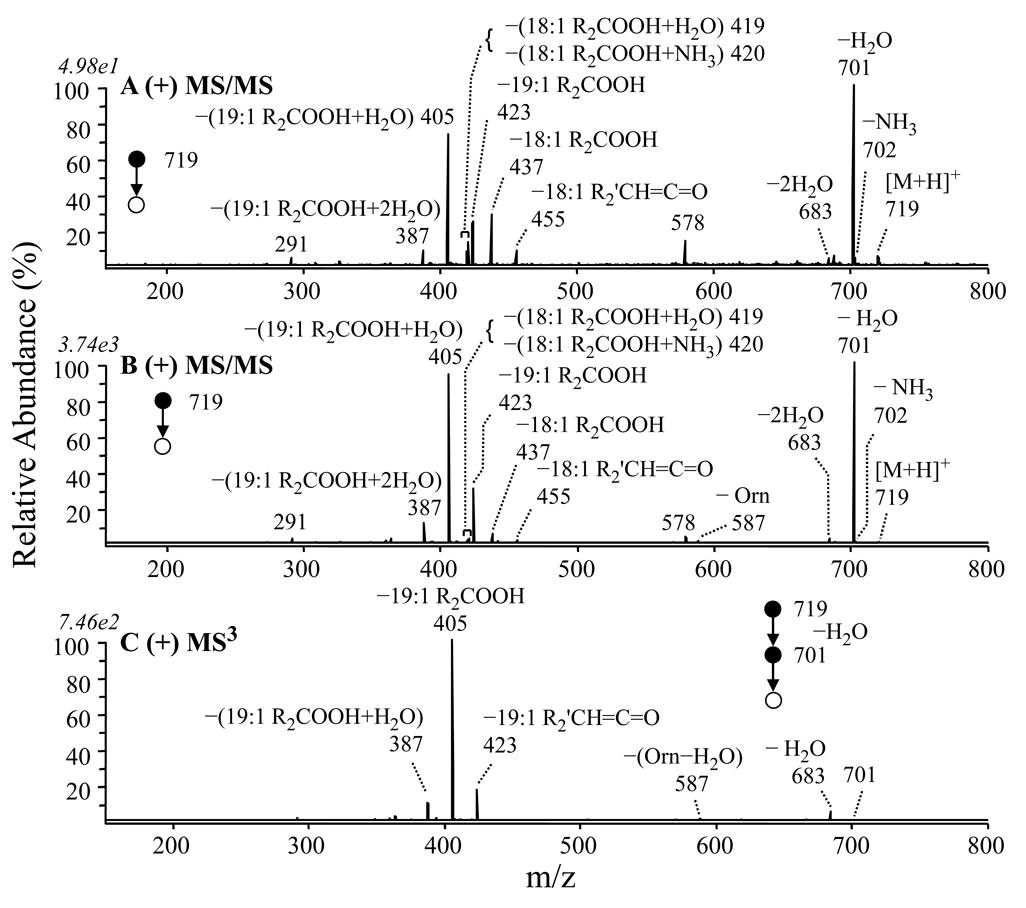
Similar to that seen in Figure 4A, MS/MS of the protonated precursor at m/z 719 from the membrane lipid extracts of both the wild type and GPCho deficient strains of R. sphaeroides resulted in the dominant loss of water (m/z 701), along with multiple products corresponding to the loss of either a 19:1 (m/z 423, 405 and 387) or an 18:1 (m/z 455, 437, 420 and 419) fatty acyl chain (Figures 5A and 5B, respectively). However, MS3 of the [M+H−H2O]+ product ion (Figure 5B) resulted in formation of only the 19:1 loss product ion series, suggesting that a non-ornithine containing lipid species was responsible for the 18:1 loss products. Subsequent fragmentation of the m/z 405 product ion in Figure 5C (the resultant spectrum was again identical to Figure 2D) resulted in the loss of a 20:1 fatty acid, allowing confirmation of the m/z 719 ornithine lipid that was present as being OL 3–OH 20:1/19:1. According to the nitrogen rule, the unknown lipid species that was also present at m/z 719 in addition to the OL 3–OH 20:1/19:1 lipid ion should contain an even number of nitrogen atoms.
Figure 5.
Identification of overlapping ornithine (OL 3–OH 20:1/19:1) and glutamine (QL 3–OH 20:1/18:1) lipids at m/z 719 by positive ion ESI CID-MS/MS and −MSn. (A) MS/MS of m/z 719 from the wild type membrane lipid extract in Figure 1A, (B) MS/MS of m/z 719 from the GPCho-deficient mutant membrane extract in Figure 2B. (C) MS3 of m/z 701 from panel B.
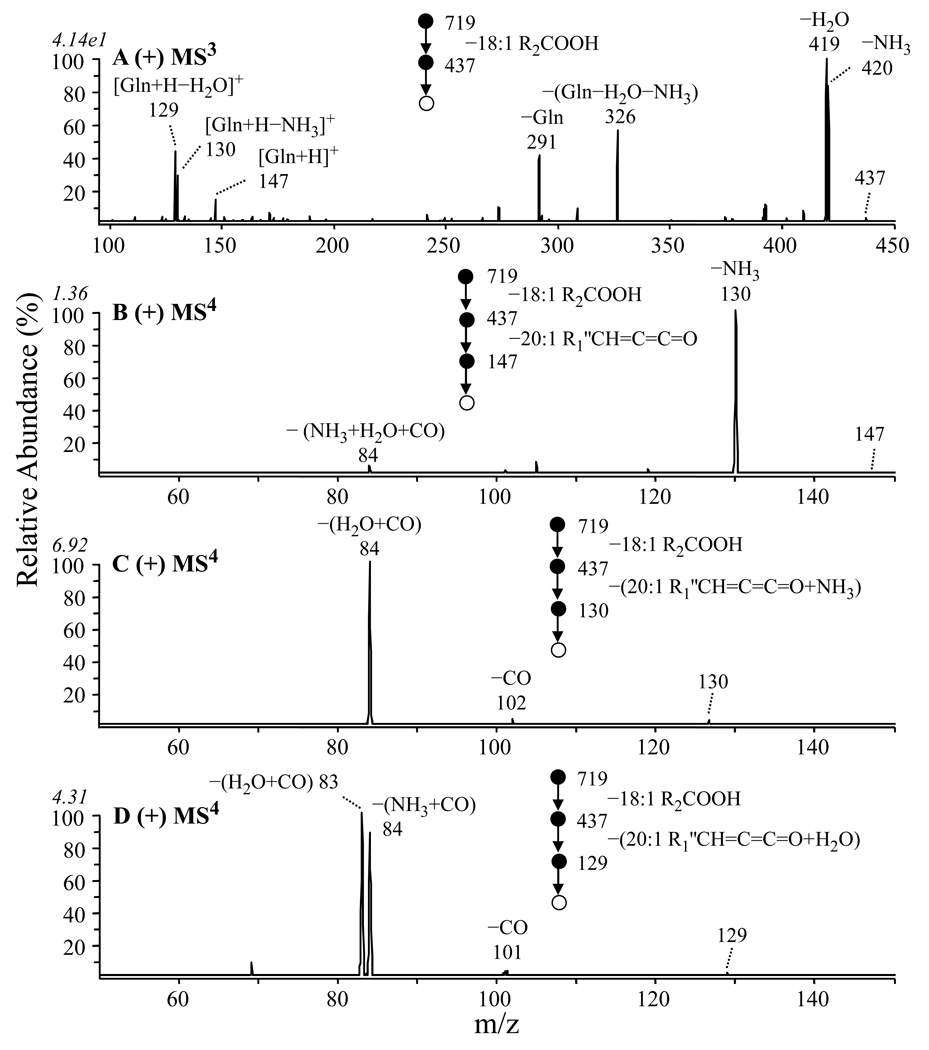
Figure 6A shows the MS3 spectrum of the m/z 437 ion from Figure 5B. The most abundant product ions in this spectra were seen at m/z 420 and m/z 419, corresponding to the loss of water and ammonia, respectively. Notably, although the characteristic ornithine specific ion at m/z 115 was not observed, a series of products at m/z 147, 130 and 129 were formed, via the neutral loss of a 20:1 ketene (−R1’CH=C=C=O) from the m/z 437 precursor ion or from the m/z 420 and 419 product ions, respectively. Importantly, the ion at m/z 147 corresponds to the m/z of protonated ornithine with the addition of +14 Da, while the ions at m/z 130 and 129 correspond to the m/z of protonated ornithine with the addition of +14 Da, minus ammonia and water, respectively.
Figure 6.
Identification of overlapping ornithine (OL 3–OH 20:1/19:1) and glutamine (QL 3–OH 20:1/18:1) lipids at m/z 719 by positive ion ESI CID-MSn. (A) MS3 of m/z 437 from Figure 5B, (B) MS4 of m/z 147 from panel B, (C) MS4 of m/z 130 from panel B, and (D) MS4 of m/z 129 from panel B.
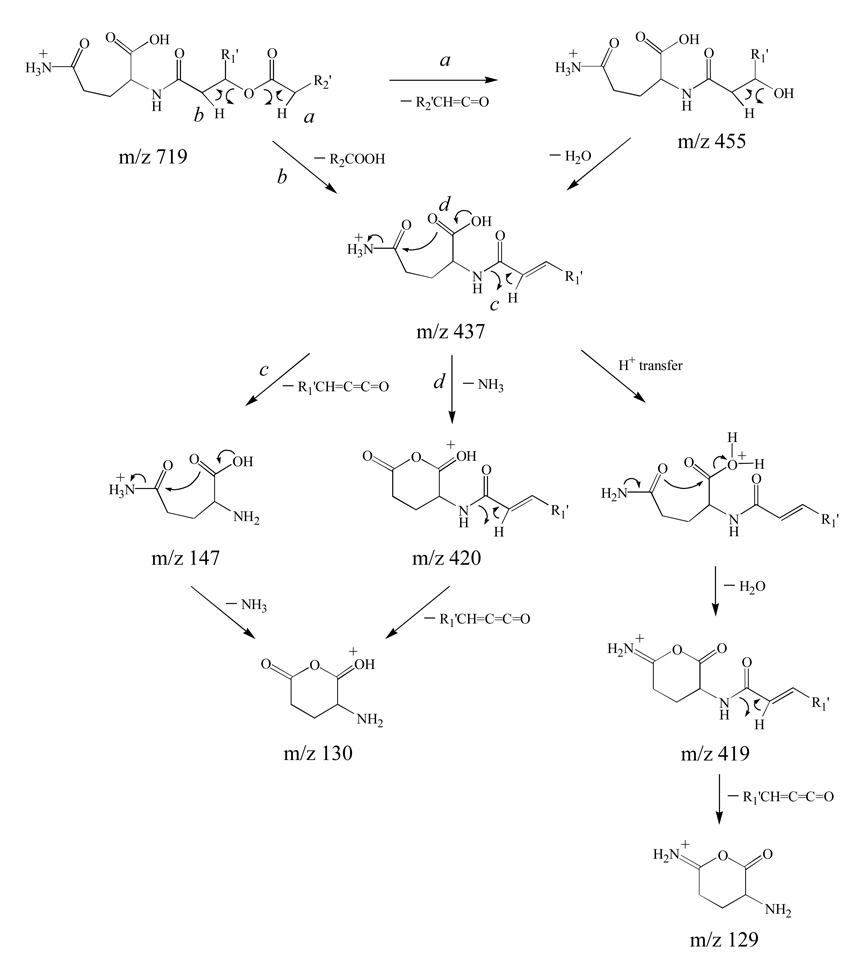
This +14 Da mass shift could potentially result from substitution of ornithine by lysine (analogous to incorporation of a methylene group into the ornithine amino acid side chain), or via N– or O–methylation of ornithine to yield N–Me ornithine or ornithine–OMe, respectively. Alternatively, the +14 Da mass shift could result from substitution of the ornithine lipid by glutamine (analogous to oxidation of the δ methylene group of the ornithine amino acid side chain). N–methylation is a commonly observed process in lipid biosynthesis in R. sphaeroides. For instance, GPCho can be synthesized by consecutive N–methylation of GPEtn by the action of GPEtn N–methyl transferase [17], and both mono N–Me–GPEtn (14 Da shift) and N,N–diMe–GPEtn (+28 Da shift) intermediates have been previously identified [17]. Similarly, the amino group in diacylglycerolhomoserine (DGHS) can be consecutively N-methylated to yield diacylglycerol–N,N,N–trimethyl homoserine (DGTS), a substitute for GPCho under phosphate-limiting growth conditions [35]. Stark differences were observed however, between the product ions observed by MS3 of m/z 437 (Figure 6A) with those formed by MS/MS of protonated Nα–acetyl lysine, Nε–acetyl lysine, Nα–acetyl Nδ–Me ornithine and Nα–acetyl ornithine–OMe, as well as between the MS4 spectra obtained from the m/z 147, 130 and 129 product ions from Figure 6A (Figures 6B, 6C and 6D, respectively), and the MS/MS or MS3 spectra obtained by dissociation of protonated lysine, Nδ–Me–ornithine and ornithine–OMe (data not shown), thereby allowing us to rule out these possible structures. In contrast, the product ions observed by MS3 of m/z 437 (Figure 6A) closely resembled those formed by MS/MS of Nα–acetyl or Nα–propyl glutamine, while the spectra shown in Figures 6B, 6C and 6D were found to be essentially identical to the spectra obtained by dissociation of the [M+H]+, [M+H−NH3]+ and [M+H−H2O]+ ions from protonated glutamine (data not shown), indicating that the +14 Da mass shift had resulted from this substitution. The structure of the proposed glutamine lipid (QL) ion at m/z 719 therefore corresponds to QL 3–OH 20:1/18:1. Potential mechanisms to rationalize the product ion fragmentation pathways observed for this glutamine-containing lipid, and for formation of the m/z 147, 130 and 129 ‘fingerprint’ ions, are shown in Scheme 4. Note that the pathway leading to the m/z 129 product ion in Scheme 4 is shown to occur via nucleophilic attack from the carbonyl oxygen of the glutamine amide side chain. This proposed pathway is consistent with previous mechanistic studies on the fragmentation reactions of protonated peptide ions [36]. However, an alternative pathway involving nucleophilic attack from the amide nitrogen, resulting in the formation of a protonated 3-aminohexahydro-2,6-pyridinedione product ion, can not be ruled out.
Scheme 4.
Proposed pathways for the multistage gas-phase fragmentation reactions of protonated glutamine lipids.
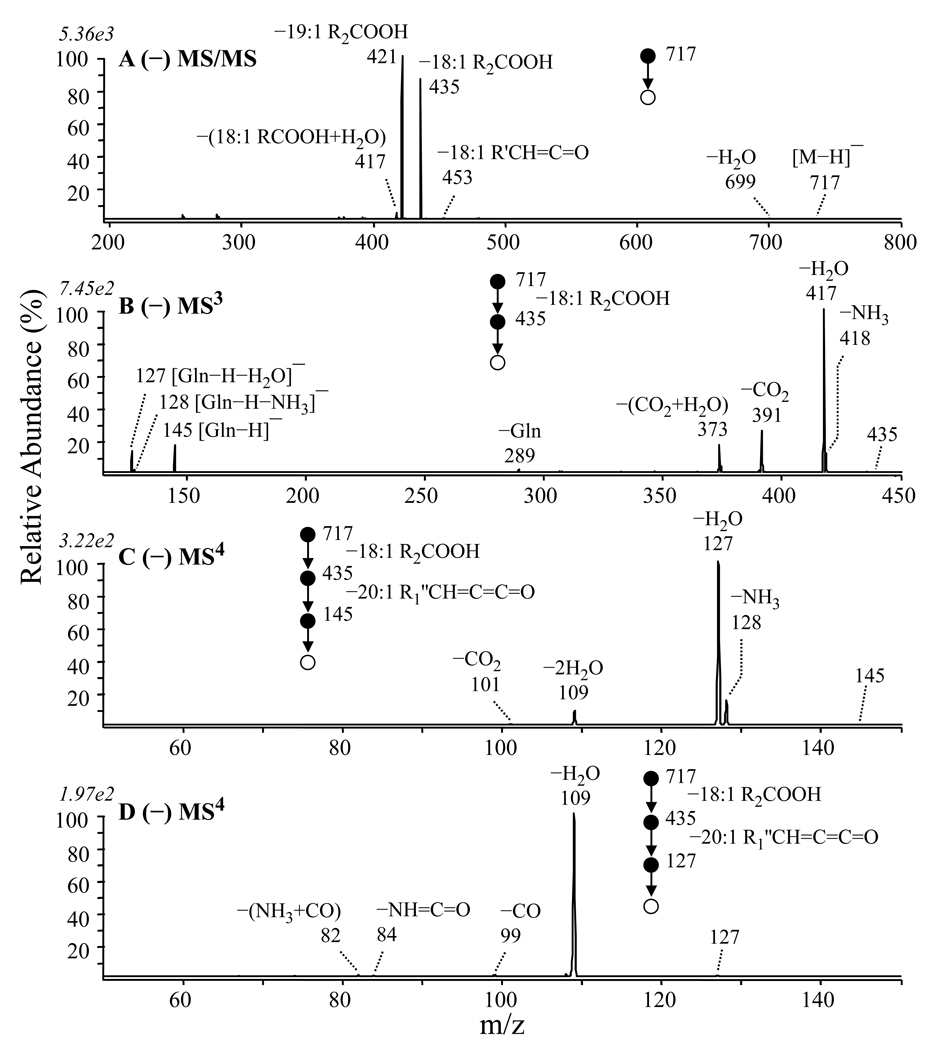
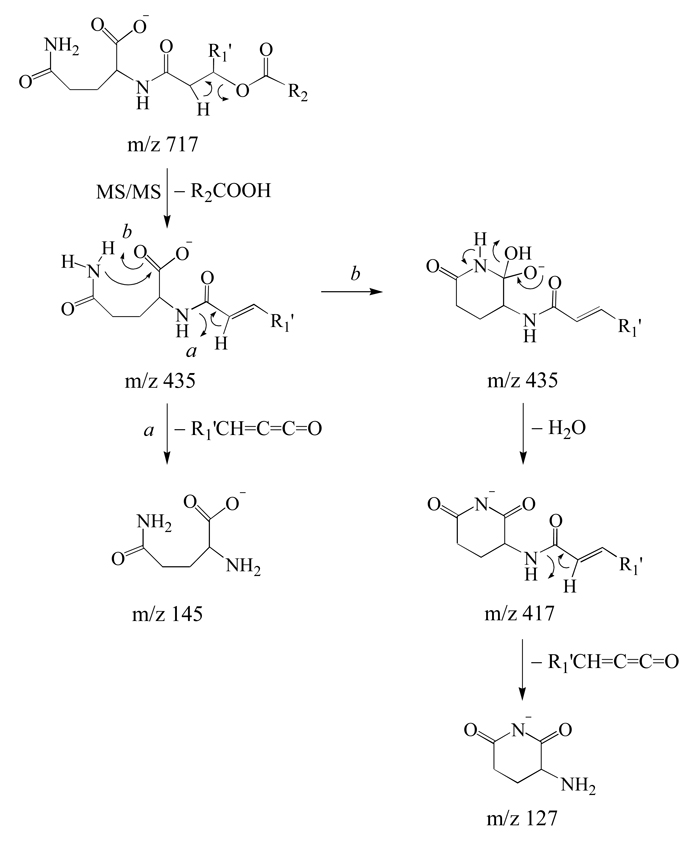
Further evidence for the presence of the QL 3–OH 20:1/18:1 glutamine containing lipid was obtained by inspection of the MS/MS, MS3,MS4 and MS5 spectra obtained from the negative ion precursor at m/z 717 in Figure 1D. (Figures 7A–7D). Similar to that observed in the positive ion mode spectra, MS/MS of the negative ion precursor led to the neutral loss of either a 19:1 (m/z 421) or 18:1 (m/z 435) fatty acid (−R2COOH) (Figure 7A). MS3 of the m/z 421 ion resulted in loss of CO2, as well as the loss of a 20:2 ketene to yield the ‘fingerprint’ ornithine specific product ion at m/z 131, confirming the presence of the OL 3–OH 20:1/19:1 ornithine lipid (data not shown). MS3 fragmentation of the m/z 435 ion led to the losses of NH3 (m/z 418), H2O (m/z 417), CO2 (m/z 391) and CO2+H2O (m/z 373), as well as the formation of product ions at m/z 145, 128 and 127 (Figure 7B). Similar to that observed in positive ion mode therefore, these ions correspond to the [M−H]−, [M−H−NH3]− and [M−H−H2O]− ions of deprotonated glutamine. Indeed, the MS4 spectra of the m/z 145 and 127 product ions from Figure 7B (Figures 7C and 7D, respectively) (the m/z 128 product ion was too low in abundance to allow its isolation and fragmentation) were found to be identical to the MS/MS and MS3 spectra acquired from an authentic deprotonated glutamine standard (data not shown). Potential mechanisms to rationalize the formation of the m/z 145 and 127 glutamine specific ‘fingerprint’ ions are shown in Scheme 5. Note that the pathway for formation of the m/z 127 product ions from the m/z 435 product could also potentially occur via nucleophilic attack from the carbonyl oxygen of the glutamine amide side chain following initial proton transfer from the amide nitrogen to the carboxylate anion, analogous to that shown in Scheme 4. Importantly, these proposed mechanisms are supported by the results from H/D exchange experiments. For example, MS/MS of the deuterated precursor ions at m/z 720 resulted in loss of the 18:1 fatty acid as R2COOH rather than R2COOD, indicating that this loss mainly occurs by a cis-1,2 elimination mechanism. MS3 of m/z 435 resulted in formation of m/z 145 via loss of R1’CH=C=C=O, consistent with the charge-directed fragmentation mechanism shown in the left branch of Scheme 5. In contrast, the loss of water was observed at m/z 418 (D2O) and m/z 419 (HOD), at similar abundances, indicating that this loss could involve both exchangeable protons and the non-exchangeable α-proton. Interestingly, the significantly increased abundance of the m/z 435 glutamine containing product ion in the negative ion mode MS/MS spectrum in Figure 7A relative to that of the m/z 421 ornithine containing product ion, compared to the relative abundance of the corresponding ions in the positive ion mode spectrum (compare Figure 7A and Figure 5B) is consistent with an expected improvement in the ionization efficiency for the glutamine lipid precursor ion in negative mode, due to the presence of its less basic amide functional group, compared to a primary amine in the ornithine lipid. In contrast, OL are expected to exhibit some preferential ionization in positive mode due to the presence of the more basic primary amine compared to the amide functional group.
Figure 7.
Identification of overlapping ornithine (OL 3-OH 20:1/19:1) and glutamine (QL 3–OH 20:1/18:1) lipids at m/z 717 by negative ion ESI CID-MS/MS and −MSn. (A) MS/MS of m/z 717 from Figure 1D, (B) MS3 of m/z 435 from panel A, (C) MS4 of m/z 145 from panel B, and (D) MS5 of m/z 127 from panel C.
Scheme 5.
Proposed pathways for the multistage gas-phase fragmentation reactions of deprotonated glutamine lipids.
Summary of the ornithine and glutamine lipid species identified from wild type and GPCho deficient R. sphaeroides cell membranes
Using the characteristic multistage gas-phase fragmentation behaviors of the OL and QL lipids described above, we have examined all of the ions appearing above 1% normalized relative abundance in the positive and negative ion mode mass spectra for the wild type and GPCho deficient R. sphaeroides cell membranes. The identities of all the lipids identified from this study are listed in Table 1. Interestingly, none of the major OL molecular species reported previously in bacteria Sinorhizobium meliloti [25] and Rhodobacter capsulatus [14] were found to be abundant in R. sphaeroides. The R1 3–OH fatty acyl chains connected to the amino acid head group predominantly consist of 18:0, 18:1, 20:0 and 20:1 chains. The R2 fatty acyl chain attached via the 3–OH group were found to primarily consist of 18:0, 18:1, 19:1, 20:2, 20:3 and 21:1 chains. Based on prior reports in the literature [37], the 19:1 and 21:1 chains are likely to contain a cyclopropane group, formed via the addition of CH2 onto a double bond of an 18:1 or 20:1 fatty acid, rather than containing an odd number of carbons in its backbone.
Table 1.
Summary of the ornithine (OL) and glutamine (QL) lipid species extracted from the cell membranes of wild type and GPCho deficient R. sphaeroidesa
| [M+H]+ m/z |
[M−H]− m/z |
Ornithine Lipids (OL) (wild type) |
Ornithine Lipids (OL) (GPCho deficient) |
Glutamine Lipids (QL) (wild type) |
Glutamine Lipids (QL) (GPCho deficient) |
|---|---|---|---|---|---|
| 677 | 675 | OL 3-OH 18:1/18:1 OL 3-OH 20:1/16:1 OL 3-OH 16:0/20:2 |
|||
| 691 | 689 | OL 3-OH 18:1/19:1 | OL 3-OH 18:1/19:1 | ||
| 693 | 691 | OL 3-OH 18:0/19:1 | OL 3-OH 18:0/19:1 | QL 3-OH 20:1/16:0 QL 3-OH 18:0/18:1 |
|
| 703 | 701 | OL 3-OH 18:1/20:2 OL 3-OH 18:0/20:3 |
OL 3-OH 18:1/20:2 OL 3-OH 18:0/20:3 |
||
| 705 | 703 | OL 3-OH 18:0/20:2 OL 3-OH 20:1/18:1 |
OL 3-OH 18:0/20:2 OL 3-OH 20:1/18:1 |
||
| 707 | 705 | OL 3-OH 20:0/18:1 OL 3-OH 20:1/18:0 |
OL 3-OH 20:0/18:1 OL 3-OH 20:1/18:0 |
||
| 719 | 717 | OL 3-OH 20:1/19:1 | OL 3-OH 20:1/19:1 | QL 3-OH 20:1/18:1 | QL 3-OH 20:1/18:1 |
| 721 | 719 | OL 3-OH 20:0/19:1 | OL 3-OH 20:0/19:1 | QL 3-OH 20:0/18:1 | QL 3-OH 20:0/18:1 |
| 731 | 729 | OL 3-OH 20:1/20:2 OL 3-OH 20:0/20:3 |
OL 3-OH 20:1/20:2 | ||
| 733 | 731 | OL 3-OH 20:0/20:2 | QL 3-OH 20:1/19:1 | ||
| 747 | 745 | OL 3-OH 20:1/21:1 | OL 3-OH 20:1/21:1 | ||
| 749 | 747 | OL 3-OH 20:0/21:1 | OL 3-OH 20:0/21:1 |
Only those ions with normalized relative abundances > 1% in the MS spectra were examined.
Glutamine-containing lipids have not previously been reported in the literature, and their origin remains unknown. The presence of a side chain carbonyl group in the glutamine lipids should make this lipid unreactive with the ninhydrin reagent (Here, Nα–acetyl glutamine was found to give a negative ninhydrin test result (data not shown)). Thus, although evidence is lacking, it is intriguing to speculate that the ninhydrin-negative phosphate-free lipid species observed following incorporation of 14C-labeled ornithine as a substrate for a potential homolog of aspartyl/aspariginal β–hydroxylase in Rhizobium tropici [10], correspond to glutamine lipids.
The possibility of these glutamine lipids appearing as artifacts due to oxidation of ornithine lipid species during sample preparation cannot be ruled out. However, if this had occurred, it would be expected that the more susceptible double bonds in the fatty acid chains of the ornithine lipids would not be found intact. Efforts to chemically oxidize the δ methylene group of N–acetyl–ornithine, using conditions previously demonstrated to result in the effective oxidation of methionine, cysteine, S–alkyl cysteine and tryptophan [38], did not yield N–acetyl–glutamine, indicating the stability of the ornithine head group to oxidative modifications. Furthermore, despite the wide variety of ornithine lipids observed here, only a limited number of glutamine lipids were observed, and the relative abundances of these lipids did not reflect those of the ornithine lipids. We would not expect artificial oxidation to display such selectivity.
Conclusions
The molecular structures of ornithine lipids (OL) and novel glutamine lipid (QL) species extracted from the membranes of wild type R. sphaeroides, as well as from a GPCho-deficient mutant, have been determined by using multistage tandem mass spectrometry in a linear quadrupole ion trap. The glutamine lipids were found to share an analogous structure with the ornithine lipids, where the amino acid head group is connected to the first fatty acid via an amide bond at the Nα–position, and the second fatty acid is incorporated via an ester bond with a 3–OH group within the first fatty acid. Characteristic ‘fingerprint’ product ions for OL’s are observed at m/z 115 in positive ion mode and at m/z 131 in negative ion mode, whereas analogous ‘fingerprint’ ions for QL’s are observed at m/z 147, 130 and 129 in positive ion mode, and at m/z 145 and 127 in negative ion mode. The fundamental knowledge obtained here regarding the multistage gas-phase fragmentation behavior of both ornithine and glutamine lipids establishes the basis for future applications aimed toward the sensitive identification and quantification of these lipid species in complex lipidomic mixtures.
Acknowledgements
We thank Dr. Christoph Benning and Ms. Banita Tamot for providing the original R. sphaeroides 2.4.1 and CHB20 strains used in this study and Dr. Carrie Hiser for the construction and growth of the cytochrome oxidase over-producing versions of these strains. This work was supported by NIH GM26916 (SF-M), and a MSU Research Excellence Fund grant (SF-M, GER).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gorchein A. Distribution and metabolism of ornithine in Rhodopseudomonas spheroides. Proc. Roy. Soc., Ser. B. 1968;170:265–278. doi: 10.1098/rspb.1968.0038. [DOI] [PubMed] [Google Scholar]
- 2.Shively JM, Knoche HW. Isolation of an ornithine-containing lipid from Thiobacillus thiooxidans. J. Bacteriol. 1969;98:829–830. doi: 10.1128/jb.98.2.829-830.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knoche HW, Shively JM. The structure of an ornithine-containing lipid from Thiobacillus thiooxidans. J. Biol. Chem. 1972;247:170–178. [PubMed] [Google Scholar]
- 4.Lopez-Lara IM, Sohlenkamp C, Geiger O. Membrane lipids in plant-associated bacteria: Their biosyntheses and possible functions. Mol. Plant-Microbe Interact. 2003;16:567–579. doi: 10.1094/MPMI.2003.16.7.567. [DOI] [PubMed] [Google Scholar]
- 5.Minnikin DE, Abdolrahimzadeh H. Replacement of phosphatidylethanolamine and acidic phospholipids by an ornithine-amide lipid and a minor phosphorus-free lipid in Pseudomonas fluorescens NCMB 129. FEBS (Fed. Eur. Biochem. Soc.) Lett. 1974;43:257–260. doi: 10.1016/0014-5793(74)80655-1. [DOI] [PubMed] [Google Scholar]
- 6.Benning C, Huang Z-H, Gage DA. Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch. Biochem. Biophys. 1995;317:103–111. doi: 10.1006/abbi.1995.1141. [DOI] [PubMed] [Google Scholar]
- 7.Lopez-Lara IM, Gao J-L, Soto MJ, Solares-Perez A, Weissenmayer B, Sohlenkamp C, Verroios GP, Thomas-Oates J, Geiger O. Phosphorus-free membrane lipids of Sinorhizobium meliloti are not required for the symbiosis with alfalfa but contribute to increased cell yields under phosphorus-limiting conditions of growth. Mol. Plant-Microbe Interact. 2005;18:973–982. doi: 10.1094/MPMI-18-0973. [DOI] [PubMed] [Google Scholar]
- 8.Weissenmayer B, Gao J-L, Lopez-Lara IM, Geiger O. Identification of a gene required for the biosynthesis of ornithine-derived lipids. Mol. Microbiol. 2002;45:721–733. doi: 10.1046/j.1365-2958.2002.03043.x. [DOI] [PubMed] [Google Scholar]
- 9.Taylor CJ, Anderson AJ, Wilkinson SG. Phenotypic variation of lipid composition in Burkholderia cepacia: a response to increased growth temperature is a greater content of 2-hydroxy acids in phosphatidylethanolamine and ornithine amide lipid. Microbiology (Reading, U. K.) 1998;144:1737–1745. doi: 10.1099/00221287-144-7-1737. [DOI] [PubMed] [Google Scholar]
- 10.Rojas-Jimenez K, Sohlenkamp C, Geiger O, Martinez-Romero E, Werner D, Vinuesa P. A CIC chloride channel homolog and ornithine-containing membrane lipids of Rhizobium tropici CIAT899 are involved in symbiotic efficiency and acid tolerance. Mol. Plant-Microbe Interact. 2005;18:1175–1185. doi: 10.1094/MPMI-18-1175. [DOI] [PubMed] [Google Scholar]
- 11.Kawai Y, Yano I. Ornithine-containing lipid of Bordetella pertussis, a new type of hemagglutinin. Eur. J. Biochem. 1983;136:531–538. doi: 10.1111/j.1432-1033.1983.tb07773.x. [DOI] [PubMed] [Google Scholar]
- 12.Kawai Y, Akagawa K. Macrophage activation of an ornithine-containing lipid or a serine-containing lipid. Infect. Immun. 1989;57:2086–2091. doi: 10.1128/iai.57.7.2086-2091.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawai Y, Nakagawa Y, Matuyama T, Akagawa K, Itagawa K, Fukase K, Kusumoto S, Nishijima M, Yano I. A typical bacterial ornithine-containing lipid Nalpha-(D)-[3-(hexadecanoyloxy)hexadecanoyl]-ornithine is a strong stimulant for macrophages and a useful adjuvant. FEMS Immunol. Med. Microbiol. 1999;23:67–73. doi: 10.1111/j.1574-695X.1999.tb01718.x. [DOI] [PubMed] [Google Scholar]
- 14.Aygun-Sunar S, Mandaci S, Koch H-G, Murray IVJ, Goldfine H, Daldal F. Ornithine lipid is required for optimal steady-state amounts of c-type cytochromes in Rhodobacter capsulatus. Mol. Microbiol. 2006;61:418–435. doi: 10.1111/j.1365-2958.2006.05253.x. [DOI] [PubMed] [Google Scholar]
- 15.Tamot B, Zhang X, Hiser C, Reid GE, Ferguson-Miller SM, Benning C. Cytochrome c oxidase produced in a cardiolipin-deficient mutant of Rhodobacter sphaeroides is fully functional and crystallizable. In Submission. 2008 [Google Scholar]
- 16.Benning C, Beatty JT, Prince RC, Somerville CR. The sulfolipid sulfoquinovosyldiacylglycerol is not required for photosynthetic electron transport in Rhodobacter sphaeroides but enhances growth under phosphate limitation. Proc. Natl. Acad. Sci. U. S. A. 1993;90:1561–1565. doi: 10.1073/pnas.90.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arondel V, Benning C, Somerville CR. Isolation and functional expression in Escherichia coli of a gene encoding phosphatidylethanolamine methyltransferase (EC 2.1.1.17) from Rhodobacter sphaeroides. J. Biol. Chem. 1993;268:16002–16008. [PubMed] [Google Scholar]
- 18.Yasutaka T, Yuzi Y, Keiji K. A new lysine-containing lipids isolated from Agrobacterium tumefaciens. Agric. Biol. Chem. 1976;40:1449–1450. [Google Scholar]
- 19.Kawai Y, Ishida Okawara A, Okuyama H, Kura F, Suzuki K. Modulation of chemotaxis, O2− production and myeloperoxidase release from human polymorphonuclear leukocytes by the ornithine-containing lipid and the serine-glycine-containing lipid of Flavobacterium. FEMS Immunol. Med. Microbiol. 2000;28:205–209. doi: 10.1111/j.1574-695X.2000.tb01478.x. [DOI] [PubMed] [Google Scholar]
- 20.Pulfer M, Murphy RC. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 2003;22:332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Reid GE. Multistage tandem mass spectrometry of anionic phosphatidylcholine lipid adducts reveals novel dissociation pathways. Int. J. Mass Spectrom. 2006;252:242–255. [Google Scholar]
- 22.Linscheid M, Diehl BWK, Oevermoehle M, Riedl I, Heinz E. Membrane lipids of Rhodopseudomonas viridis. Biochim. Biophys. Acta, Lipids Lipid Metab. 1997;1347:151–163. doi: 10.1016/s0005-2760(97)00065-9. [DOI] [PubMed] [Google Scholar]
- 23.Hilker DR, Gross ML, Knocke HW, Shively JM. The interpretation of the mass spectrum of an ornithine-containing lipid from Thiobacillus thiooxidans. Biomed. Mass Spectrom. 1978;5:64–71. doi: 10.1002/bms.1200050112. [DOI] [PubMed] [Google Scholar]
- 24.Tomer KB, Crow FW, Knoche HW, Gross ML. Fast atom bombardment and mass spectrometry/mass spectrometry for analysis of a mixture of ornithine-containing lipids from Thiobacillus thiooxidans. Anal. Chem. 1983;55:1033–1036. [Google Scholar]
- 25.Geiger O, Rohrs V, Weissenmayer B, Finan TM, Thomas-Oates JE. The regulator gene phoB mediates phosphate stress-controlled synthesis of the membrane lipid diacylglyceryl-N,N,N-trimethylhomoserine in Rhizobium (Sinorhizobium) meliloti. Mol. Microbiol. 1999;32:63–73. doi: 10.1046/j.1365-2958.1999.01325.x. [DOI] [PubMed] [Google Scholar]
- 26.Hilker DR, Knoche HW, Gross ML. Thermolysis chemical ionization of a complex polar lipid. Biomed. Mass Spectrom. 1979;6:356–358. doi: 10.1002/bms.1200060810. [DOI] [PubMed] [Google Scholar]
- 27.Cerny RL, Tomer KB, Gross ML. Desorption ionization combined with tandem mass spectrometry: advantages for investigating complex lipids, disaccharides and organometallic complexes. Org. Mass Spectrom. 1986;21:655–660. [Google Scholar]
- 28.Reid GE, Simpson RJ, O'Hair RAJ. A mass spectrometric and ab initio study of the pathways for dehydration of simple glycine and cysteine-containing peptide [M+H]+ ions. J. Am. Soc. Mass Spectrom. 1998;9:945–956. [Google Scholar]
- 29.Zhen Y, Qian J, Follmann K, Hayward T, Nilsson T, Dahn M, Hilmi Y, Hamer AG, Hosler JP, Ferguson-Miller S. Overexpression and purification of cytochrome c oxidase from Rhodobacter sphaeroides. Protein Expr. and Purif. 1998;13:326–336. doi: 10.1006/prep.1998.0903. [DOI] [PubMed] [Google Scholar]
- 30.Awasthi YC, Chuang TF, Keenan TW, Crane FL. Tightly bound cardiolipin in cytochrome oxidase. Biochim. Biophys. Acta, Bioenerg. 1971;226:42–52. doi: 10.1016/0005-2728(71)90176-9. [DOI] [PubMed] [Google Scholar]
- 31.Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CRH, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA. A comprehensive classification system for lipids. J. Lipid Res. 2005;46:839–862. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Bowie JH, Brinkworth CS, Dua S. Collision-induced fragmentations of the (M−H)− parent anions of underivatized peptides: an aid to structure determination and some unusual negative ion cleavages. Mass Spectrom. Rev. 2002;21:87–107. doi: 10.1002/mas.10022. [DOI] [PubMed] [Google Scholar]
- 33.Bowie JH. The fragmentations of even-electron organic negative ions. Mass Spectrom. Rev. 1990;9:349–379. [Google Scholar]
- 34.Eckersley M, Bowie JH, Hayes RN. Collision-induced dissociations of deprotonated alpha -amino acids. The occurrence of specific proton transfers preceding fragmentation. Int. J. Mass Spectrom. Ion Processes. 1989;93:199–213. [Google Scholar]
- 35.Klug RM, Benning C. Two enzymes of diacylglyceryl-O-4'-(N,N,N,-trimethyl)-homoserine biosynthesis are encoded by btaA and btaB in the purple bacterium Rhodobacter sphaeroides. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5910–5915. doi: 10.1073/pnas.101037998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Hair RAJ. The role of nucleophile-electrophile interactions in the unimolecular and bimolecular gas-phase ion chemistry of peptides and related systems. J. Mass Spectrom. 2000;35:1377–1381. doi: 10.1002/1096-9888(200012)35:12<1377::AID-JMS83>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 37.Grogan DW, Cronan JEJ. Cyclopropane ring formation in membrane lipids of bacteria Microbiol. Mol. Biol. Rev. 1997;61:429–441. doi: 10.1128/mmbr.61.4.429-441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Froelich JM, Reid GE. The origin and control of ex vivo oxidative peptide modifications prior to mass spectrometry analysis. Proteomics. 2008;8:1334–1345. doi: 10.1002/pmic.200700792. [DOI] [PubMed] [Google Scholar]