Abstract
A novel custom microarray for largemouth bass (Micropterus salmoides) was designed with sequences obtained from a normalized cDNA library using the 454 Life Sciences GS-20 pyrosequencer. This approach yielded in excess of 58 million bases of high-quality sequence. The sequence information was combined with 2,616 reads obtained by traditional suppressive subtractive hybridizations to derive a total of 31,391 unique sequences. Annotation and coding sequences were predicted for these transcripts where possible. 16,350 annotated transcripts were selected as target sequences for the design of the custom largemouth bass oligonucleotide microarray. The microarray was validated by examining the transcriptomic response in male largemouth bass exposed to 17β-œstradiol. Transcriptomic responses were assessed in liver and gonad, and indicated gene expression profiles typical of exposure to œstradiol. The results demonstrate the potential to rapidly create the tools necessary to assess large scale transcriptional responses in non-model species, paving the way for expanded impact of toxicogenomics in ecotoxicology.
Keywords: pyrosequencing technology, largemouth bass, microarrays, 17β-œstradiol
INTRODUCTION
In recent years, toxicogenomics approaches have been widely used in environmental toxicology to identify altered gene transcriptional profiles following exposure to environmental xenobiotics. Microarrays are the most commonly used method for transcriptional profiling as they allow precise and accurate quantification of large scale gene transcriptional activity. Fish are commonly affected by environmental contaminants in aquatic ecosystems, making them important models for ecotoxicology. There are commercially available microarrays from manufacturers such as Agilent or Affymetrix for well-developed model species such as zebrafish (Danio rerio) (van der Meer et al., 2005; Hook et al., 2006; van der Ven et al., 2006; Santos et al., 2007) or fathead minnow (Pimephales promelas) (Larkin et al., 2007; Garcia-Reyero et al., 2008; Perkins et al., 2008). (See Miracle et al., 2003; Douglas, 2006; Denslow et al., 2007 for reviews covering the use of microarrays for gene expression profiling in fish). While useful, these tools are largely lacking for non-model species that are often more interesting from an environmental or toxicological viewpoint.
Researchers have made enormous efforts to develop custom-made microarrays of ecologically relevant species in order to get a better understanding of the adverse effects of pollutants. Successful examples include a custom cDNA microarray constructed for European flounder (Platichthys flesus) to detect toxic stress responses in flounder (Williams et al., 2003; Sheader et al., 2006), a membrane-based macroarray to study the effects of xenobiotic exposure in largemouth bass (LMB, Micropterus salmoides) (Larkin et al., 2003), and custom arrays for carp (Cyprinus carpio) (Moens et al., 2006; Moens et al., 2007a; Moens et al., 2007b) and goldfish brain (Carassius auratus) (Martyniuk et al., 2006).
A primary factor limiting toxicogenomic investigations of non-model species is the large amount of sequence knowledge required to construct a high-quality array for a given species (Ju et al., 2007). Attempts to work around this limitation include using techniques such as Differential Display-RT-PCR (DD-RTPCR) (Denslow et al., 2001a; Denslow et al., 2001b) or Suppressive Subtractive Hybridization (SSH) (Diatchenko et al., 1996; Blum et al., 2004) to identify likely candidate genes for spotting on arrays. Unfortunately, DD-RTPCR and SSH are imperfect at correctly identifying differentially expressed genes, making it possible to miss important genes. Additionally, due to the lack of sequence identity among genes of even closely related species (Yang et al., 2005), microarrays have less than ideal utility across species. Alternative techniques such as Massively Parallel Signature Sequencing (MPSS) (Reinartz et al., 2002), or Serial Analysis of Gene Expression (SAGE) (Velculescu et al., 1995; Lorenz and Dean, 2002; Griffitt et al., 2007), are useful, but can be technically difficult and incur significant sequencing costs.
Clearly there is a need for researchers working on non-model species with potentially limited resources to quickly and cheaply generate large amounts of sequence necessary to construct a high-quality microarray. Recently introduced ultra-high throughput sequencing techniques such as the 454 Life Sciences GS-FLX model (454 Life Sciences, Branford, CT), can generate 80 to 100 million bp of sequence data per run and have allowed the cost of large scale sequencing efforts (price per base pair (bp)) to drop considerably. Our goal was to use pyrosequencing technology to quickly generate a large amount of sequence information to create a transcriptome-wide microarray from a non-model species. In this study, we utilized LMB, a member of the sunfish family (Centrarchidae) that is widely distributed in fisheries throughout the Continental United States. LMB are generally considered apex predators and bioaccumulate lipophilic xenobiotics such as organochlorine pesticides (Marburger et al., 2002). In this report, we validate the use of pyrosequencing normalized cDNA libraries to construct a high-quality microarray on the Agilent platform using LMB exposed to œstradiol.
MATERIAL AND METHODS
SSH SEQUENCING
Liver, brain and gonad from female and male LMB controls and fish treated with either p,p’-DDE (45.9 µg/g) or dieldrin (0.81 µg/g) in the diet as previously described (Garcia-Reyero et al., 2006) were used to perform SSH. Briefly, PolyA RNA was isolated from the different tissues and different combinations of tissues and treatments were used to perform SSH reactions using the PCR-Select™ cDNA Subtraction Kit (Clontech, Palo Alto, CA), following the manufacturer’s protocol. The subtracted gene pools were then cloned into pGEM T-Easy (Promega, Madison, WI) and sequenced at the Interdisciplinary Center for Biotechnology Research (ICBR, University of Florida, Gainesville, FL). This effort generated 2,616 different reads corresponding to 758 genes. These genes were included in the microarray.
CONSTRUCTION OF THE LMB NORMALIZED cDNA LIBRARY
Two female LMB were anesthetized with 100 ppm tricaine methanesulfonate (MS-222) buffered with sodium bicarbonate and euthanized in accordance with the University of Florida’s Institutional Animal Care and Use Committee (IACUC) guidelines. Total RNA was isolated from 100 mg samples of brain, liver and gonad using Trizol (Invitrogen, Carlsbad, CA) following the manufacturer’s protocols. RNA was analyzed for quality by denaturing gel electrophoresis and for quantity by spectrophotometry using a Nanodrop ND100 (Nanodrop Technologies, Wilmington, DE). Equal masses of total RNA from the six individual samples were mixed to create a pooled sample with a final concentration of 200 ng/µl. A cDNA library was constructed from three microliters (600 ng) of this material using the SMART kit from Clontech (Mountain View, CA). Briefly, the RNA was used as template to synthesize cDNA, which then was amplified using an Advantage 2 PCR kit (Clontech, Mountain View, CA) for a total of 16 cycles, as determined from an optimization experiment. PCR products were purified (Wizard SV, Promega, Madison, WI), and eluted in 75 µl milliQ H20. Twenty µl of this product was split into two tubes and used as a template for a second PCR for 16 cycles. The product was pooled, purified on a Wizard SV spin column, eluted in H20 and then finally reconstituted to a concentration of 100 ng/µl.
The generated cDNA library was normalized using the Trimmer kit from Evrogen (Evrogen Joint Stock Company, Moscow, Russia) following the provided protocols, starting with 1200 ng of cDNA. After one round of normalization, significant levels of non-normalized cDNA remained, so the normalization steps were repeated. Normalization efficiency was assessed by amplifying several genes that are expressed at widely different levels in LMB, including β-actin and GAPDH (Glyceraldehyde 3-phosphate dehydrogenase), expected to be expressed highly in the tissue, and œstrogen receptor alpha (ERα), normally expressed at low levels in male LMB. Amplifications were performed for 15 cycles and visualized on a 1.5% agarose gel.
SEQUENCING OF THE LMB LIBRARY
DNA sequencing was performed on a 454 Life Sciences GS-20 pyrosequencer by ICBR. Sequencing was performed as described in the supplementary material and methods in (Margulies et al., 2005) with slight modifications as specified by 454 Life Sciences. Briefly, high molecular weight DNA, amplified using the rolling circle amplification (RCA) reaction was sheared by nebulization to a size range of 300 to 800 bp. DNA fragment ends were repaired and phosphorylated using T4 DNA polymerase and T4 polynucleotide kinase. Adaptor oligonucleotides “A” and “B,” supplied with the 454 Life Sciences sequencing reagent kit, were ligated to the DNA fragments using T4 DNA ligase. Purified DNA fragments were hybridized to DNA capture beads and clonally amplified by emulsion PCR (emPCR). DNA capture beads containing amplified DNA were deposited in individual wells of a 70 × 75mm PicoTiter plate and DNA sequences were determined using the GS-20 instrument.
BIOINFORMATICS AND ARRAY CONSTRUCTION
DNA sequence data from the titration and the production runs were combined in a single assembly using version 1.0.52.06 of the GS 20 Newbler sequence assembly software. Paracel Transcript Assembler (Paracel Inc., Pasadena, CA) was then used to combine identified contiguous sequences (contigs) with the reads obtained by traditional suppressive subtractive hybridizations, where a series of sequence cleaning, chimera identification, clustering and assembly steps were performed.
All contigs and singlets were annotated by BLAST search against the NCBI NR and NT databases where the e-value threshold was set at e-4. For each query sequence, the top 100 BLAST hits were obtained and stored in BlastQuest (Farmerie et al., 2005), a SQL database developed by the ICBR that facilitates similarity-based sequence annotation with GeneOntology information. The NCBI Gene database was used to map LMB sequences to homologs from zebrafish, human, mouse and other organisms.
In cases where more than one LMB sequence mapped to the same gene, the contig with best e- value or highest abundance was selected as the representative target sequence for probe design. For sequences where we were not able to identify the genes because the BLAST hits were above the e−4 threshold, ESTScan (Iseli et al., 1999) was used to predict CDS (coding sequence) regions. The final set of target sequences includes contigs and singlets that have either similarity-based annotations or for which there are predicted CDS.
The “GE Probe Design” tool on eArray (Agilent, Palo Alto, CA) was used to design the 60-mer probes. Because it was often not possible to determine directionality of the sequences, two probes were selected from the sense strand and one probe from the antisense strand for each target sequence. These probes were synthesized directly on the array in a random layout by Agilent, using their proprietary method. The glass slide contained 4 separate arrays, each with 45,220 elements (4 × 44K format). In addition to the LMB-specific probes, the arrays contained 1,417 internal quality control features.
EXPOSURE OF LMB TO E2
Adult male LMB received a single intraperitoneal (IP) dose of E2 (1 mg/kg) dissolved in sesame oil. Control fish received an IP injection of sesame oil without any chemical. Four fish were used per treatment group. After 48h, fish were euthanized in accordance with the University of Florida’s IACUC guidelines. Liver and gonadal tissues were excised, immediately flash frozen in liquid nitrogen, and stored at −80°C until RNA was isolated.
RNA EXTRACTION FROM TEST LARGEMOUTH BASS
Total RNA was isolated from LMB liver and gonad with the RNA Stat-60 reagent (Tel-test, Friendswood, TX), as described previously (Garcia-Reyero et al., 2006; Garcia-Reyero et al., 2008). Briefly, 30–50 mg of tissue was homogenized and the extraction process was repeated. The RNA pellet was resuspended in an appropriate volume (50–150 µl) of RNA secure (Ambion, Austin, TX) to inactivate RNases following the manufacturer’s protocol. A total of 10 µg of RNA was treated with DNase to remove contaminating DNA using DNA-free (Ambion, Austin, TX) following the manufacturer’s protocol. The quality of total RNA was assessed with the Agilent 2100 BioAnalyzer using the RNA 6000 Nanochip assay kit (Agilent, Palo Alto, CA) and the quantity was determined on a nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE). RNA was stored at −80°C until further use.
MICROARRAY VALIDATION
Array hybridizations were performed using a reference design, where each sample was compared to a reference sample. This design does not require dye-swapping since each of the biological samples is compared to the same reference sample, as a normalizer. The reference sample consisted of equal amounts of RNA from control LMB female and male liver and gonad. Four biological replicates were analyzed for each of the treatments (control and E2). cDNA synthesis, cRNA labelling, amplification and hybridization were performed following the manufacturer’s kits and protocols (Agilent Low RNA Input Fluorescent Linear Amplification Kit and Agilent 60-mer oligo microarray processing protocol; Agilent, Palo Alto, CA). Briefly, a primer containing poly dT and a T7 polymerase promoter was added to 1 µg of total RNA. Reverse transcriptase was added to the reaction to synthesize the first and second strands of cDNA. Next, cRNA was synthesized from the double-stranded cDNA using T7 RNA polymerase, which simultaneously incorporates cyanine 3- (Cy3) or cyanine 5- (Cy5) labelled CTP into the product. The gonad or liver samples were labelled with Cy5, while the reference sample was labelled with Cy3. Once the labelling was complete, samples were hybridized to the microarray for 17 hours. The microarrays were washed and scanned with a laser-based detection system (Agilent, Palo Alto, CA). In order to fulfil the MIAME standards (Brazma et al., 2001), text versions of the processed scanner output have been deposited at the Gene Expression Omnibus website (GEO: http://www.ncbi.nlm.nih.gov/geo/; and are available under Series Accession number: GSE10693.
Microarray image processing and data pre-processing were performed using Agilent’s Feature Extraction software v 9.5. The intensity of each spot was summarized by the median pixel intensity. A log2 transformed signal ratio between the experimental channel and the reference channel was calculated for each spot, followed by within-array lowess transformation and between-array scale normalization on median intensities (Zahurak et al., 2007). Probes that did not hybridize in any sample were removed from consideration.
Two-way ANOVA was performed on normalized log2 transformed signal ratios of each probe individually, followed by Tukey-HSD pair-wise comparisons to determine genes whose expression was significantly regulated by the treatment compared to the untreated samples. A P value of < 0.05 was used as the cut-off. After testing for significance we also eliminated from consideration genes whose fold-expression changes were less than 1.5 fold.
GeneOntology (GO) annotations were derived from similarity search of the NCBI Gene database. Over-representation of differentially expressed genes in the biological process GO category was determined by Fisher Exact Test (P ≤ 0.05) and the false discovery rate was determined (Benjamini and Hochberg, 1995). PathwayStudio® software from Ariadne Genomics (Nikitin et al., 2003) was used to determine the list of common regulators among the genes that were differentially expressed in the experiment.
REAL-TIME PCR
To validate the changes in gene expression observed with the microarray, several critical genes were quantified by real-time quantitative reverse transcriptase PCR (QPCR). RNA was extracted as described above from the samples used for microarray analysis, and 2 µg total RNA was reverse transcribed with random decamer primers using the Retroscript kit (Ambion, Austin, TX). Following reverse transcription, the samples were diluted 1:5 with DEPC-treated water, and subjected to QPCR. The following genes were analyzed: ERα, œstrogen receptor beta b (ERβb), œstrogen receptor beta a (ERβa), vitellogenin (Vtg), zona pellucida protein 4 (ZPC-4), steroidogenic acute regulatory protein (StAR) and androgen receptor (AR). Primer details were published previously (Garcia-Reyero et al., 2006). Reactions were performed as duplicate 20 µl reactions on an iCycler (BioRAD, Hercules, CA) using 10µl iQ SYBR Green supermix (BioRAD, Hercules, CA), 0.25 µM each primer (final concentration), and 2 µl diluted cDNA in each reaction. Gene expression levels for each gene were calculated using the provided iQ5 software, and expressed as log2 of the fold change from control levels (normalized to 18S expression levels) using a ΔΔCT approximation.
RESULTS
cDNA LIBRARY CONSTRUCTION AND NORMALIZATION
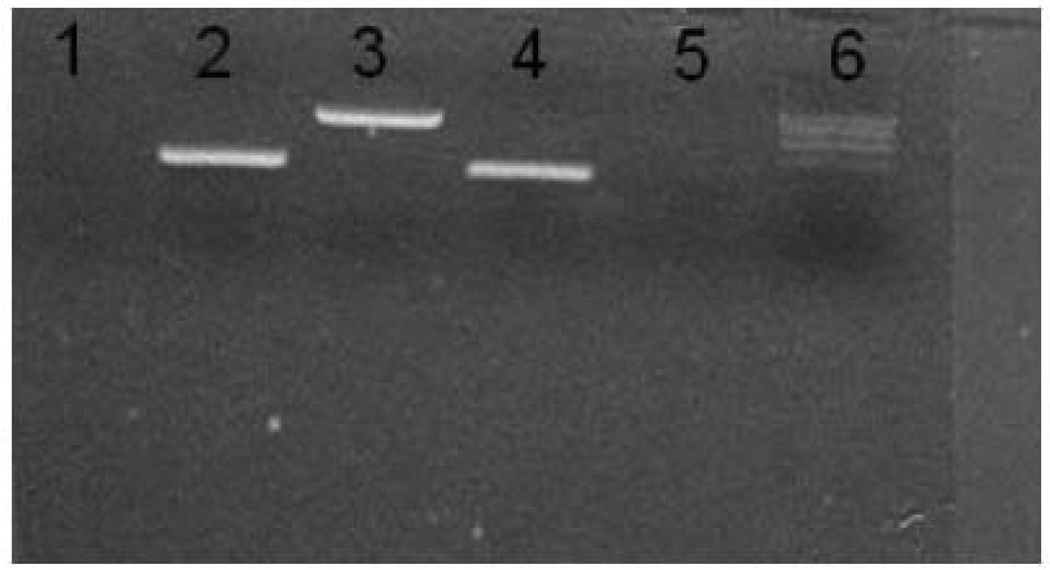
To assess cDNA library normalization, three genes were amplified from the normalized cDNA pool, GAPDH, ERα, and β-actin. GAPDH and β-actin are highly expressed in LMB tissues, while ERα is normally expressed at much lower levels. As a negative control we also used primers for human GST theta in a region that has low homology with the LMB homolog. After two rounds of normalization, band intensities of the three LMB genes were similar (Fig 1), indicating that the transcripts were present at roughly the same concentration range in the cDNA library and that the library was normalized.
Figure 1.
PCR products to test LMB cDNA library normalization. Equal volumes of the normalized library were subjected to 15 cycles of PCR amplification with gene specific primers and a 10µl aliquot of each product was placed in adjacent wells in a 1.5 % agarose gel. Lane 1, human GST theta, Lane 2, LMB beta-actin, Lane 3, LMB GAPDH, Lane 4, LMB ERα , Lane 5, no template control and Lane 6, 100-bp ladder. Human GST theta primers were included to test for contamination from a human library constructed concurrently as a positive control.
GS-20 SEQUENCING
Three separate sequencing runs were performed from the normalized LMB cDNA library (Table 1). The initial titration run (used to identify the appropriate density of beads on the plate) resulted in 711,648 bases. Two production runs resulted in 21,581,763 and 36,139,922 bases respectively. Assembly of sequence from these three runs resulted in 32,882 contigs.
Table 1.
Sequencing results from a titration run and two production runs on the 454 GS20.
| File | Type | Bases | Sequences | Avg Length | Min Length | Max Length |
|---|---|---|---|---|---|---|
| Production-1 | Fasta | 21,581,763 | 205,793 | 104.87 | 35 | 288 |
| Production-2 | Fasta | 36,139,922 | 343,685 | 105.15 | 38 | 299 |
| Titration | Fasta | 711,648 | 6,845 | 103.97 | 41 | 257 |
| Total | 58,433,333 | 556,323 | 105.03 | 35 | 299 |
BIOINFORMATICS AND MICROARRAY DESIGN
The final assembly produced 31,391 non-redundant sequences. Among these, 29,632 were GS-20-454 assemblies only, 78 were SSH assemblies only, and 1,681 were SSH-454 combined assemblies. In total we had 7,395 annotated genes and 8,555 un-annotated segments for a total of 15,950 sequences that were spotted on the array. The array format was the Agilent 4×44K. For each array, there were 45,220 total features which included 1,417 Agilent control probes and 43,803 user probes. To occupy the 43,803 user features, the four probe groups were printed as specified below:
-- Annotated sequences (sense strand): 2 probes per target sequence if available; 10,289 probes representing 7,395 targets. And these were spotted in duplicate on the array for a total of 20,578 probes.
-- Annotated sequences (antisense strand): 1 probe per target sequence; 7,395 probes representing 7,395 targets.
-- Un-annotated sequences with open reading frames (sense strand): 2 probes per target sequence if available; 10,146 probes representing 8,555 targets.
-- Un-annotated sequences with open reading frames (antisense strand): 2 probes per target sequence if available; 8,555 probes representing 8,555 targets. After printing the above three probe groups, there were only 5,684 features left on the array. This probe group was used to fill these spots. Out of the total 8,555 probes in this group, only 5,684 were printed.
MICROARRAY ANALYSIS
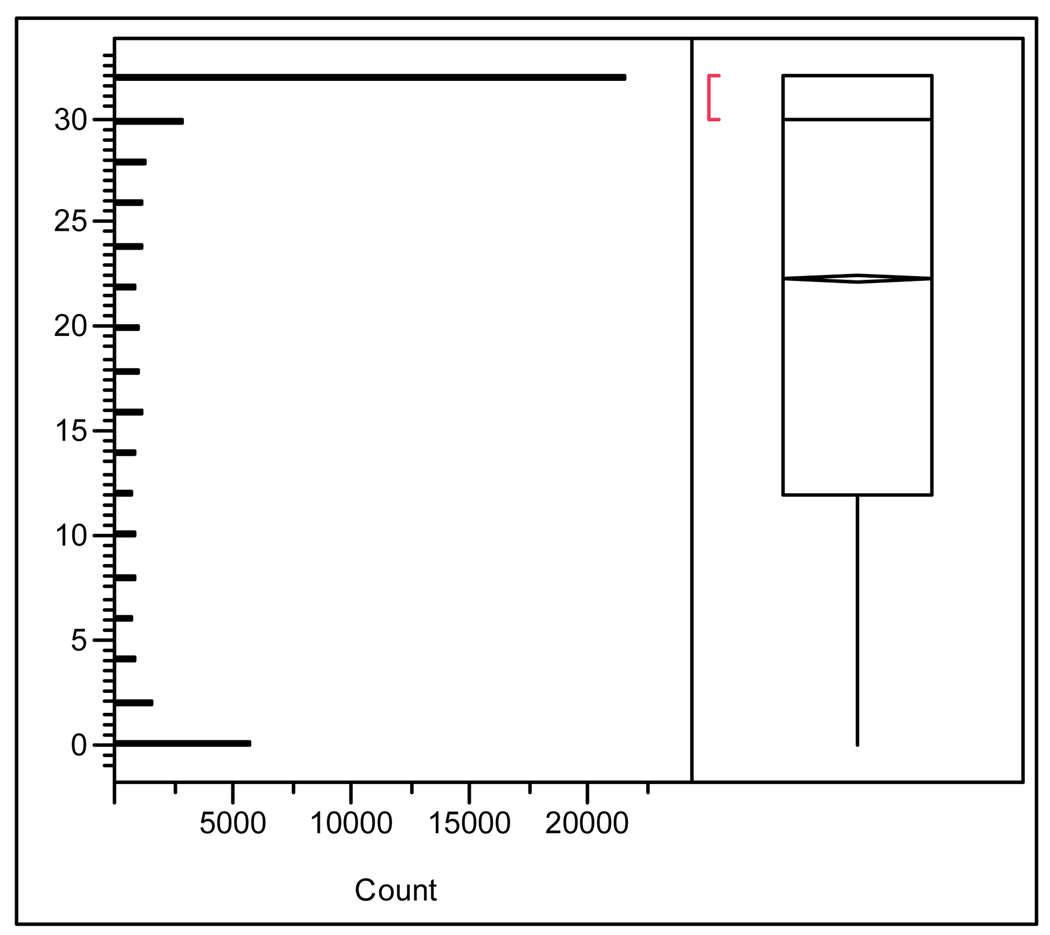
Custom oligo microarrays were used to analyze the transcriptional responses in liver and gonad of male LMB treated with œstradiol. To assess the quality of individual probes included on the array, each probe was assessed for hybridization success. Using JMP Genomics 3.0 (SAS, Cary, NC) the number of times each probe hybridized successfully on any array was plotted (Fig. 2). Values ranged from 0 (spot never hybridized) to 32 (spot always hybridized on both channels across 16 arrays). Only hybridizations resulting in a “present” call using the Agilent feature extraction software were considered successful. There were 21,620 probes that successfully hybridized in at least one sample across all arrays on both channels and 5,761 that were never successful, likely representing antisense probes. Probes that never hybridized were removed from further analysis.
Figure 2.
Distribution analysis of hybridization success across 16 microarrays. For each spot, the number of times it was called “Present” for both channels by Agilent’s feature extraction software was calculated. Distribution (Y-axis) ranges from 0 (spot never hybridized) to 32 (spot was called “Present” for both channels on all 16 microarrays. Mean, median, and IQR values for hybridization success are plotted also.
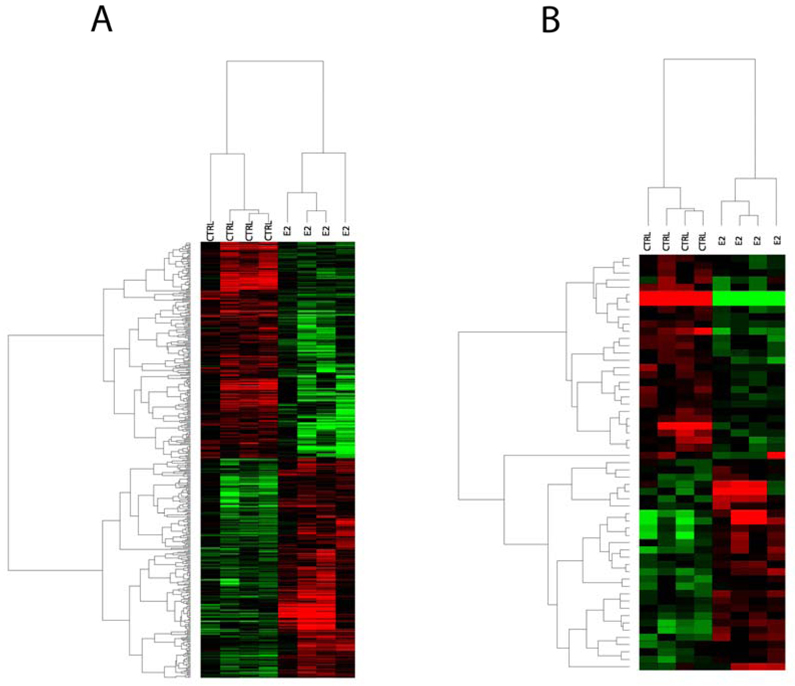
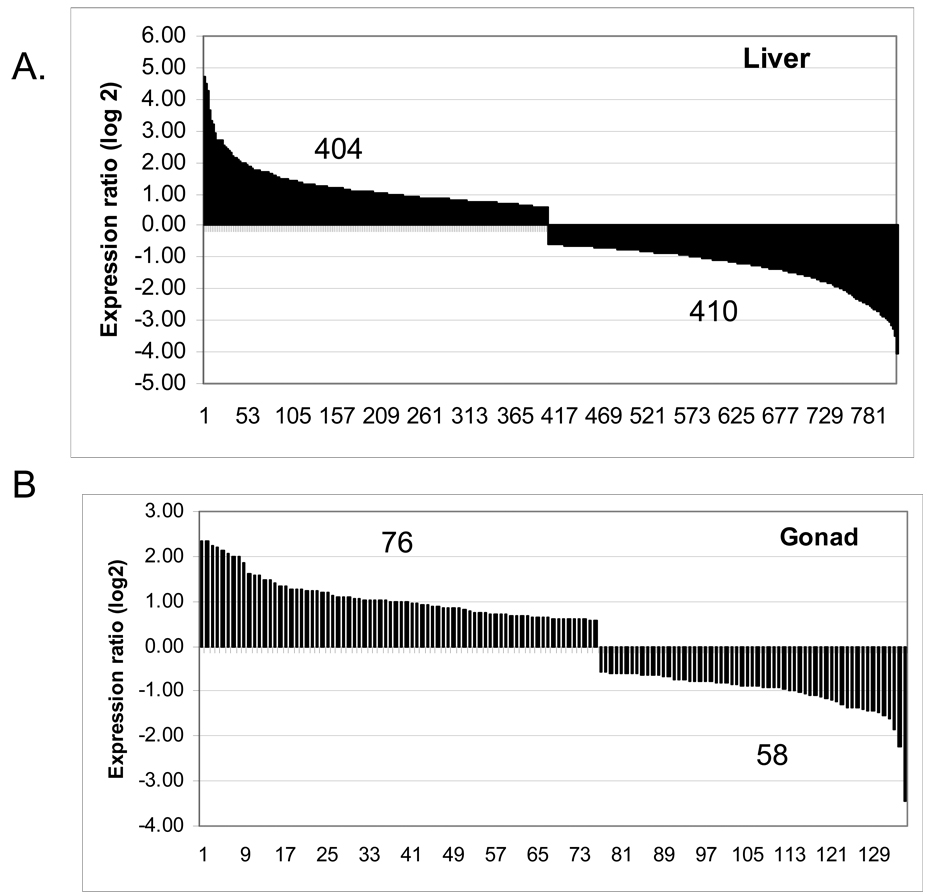
Genes that were significantly differentially expressed (p < 0.05) are represented in the heat maps (Fig. 3). The hierarchical clusters depict the actual expression level for genes in each sample compared to the reference sample, as described in the Materials and Methods section. To obtain fold differences between treated and control samples, the mean expression level for each gene was determined for the four treated biological replicates and divided by the mean of the controls for the same gene. At the P < 0.01 level, 814 probes were regulated in the liver (404 were up-regulated and 410 were down-regulated, Fig 4A). Among the most up-regulated genes were ZRP and Vtg, which are expected to be affected by œstrogenic exposure of male fish. In the gonad, only 134 probes were altered, with 76 being up-regulated and 58 being down-regulated (Fig 4B).
Figure 3.
Two-way hierarchical clustering for genes found to be differentially expressed following E2 treatment. Expression data was analyzed by ANOVA and then z-transformed. Genes used in the cluster were significant at P < 0.05. Represented are genes that are up-regulated (red) or down-regulated (green) by the treatments compared to the reference sample. (A) Cluster for genes changed in the liver and (B) cluster for genes changed in the gonad
Figure 4.
Overall gene regulation by oestradiol for genes that are significantly regulated (P < 0.01) as determined by ANOVA. (A) Gene regulation in the liver; (B) gene regulation in the gonad.
In the liver, at the P<0.05 1,394 probes were up-regulated and 1,388 probes were down-regulated. Of these, 1,130 genes were annotated using BLAST (roughly 41%) and human homologs were assigned to 389 genes (14%) that were used for the Pathway Studio® analysis below (Supplemental Table S1). In the gonad, 555 probes were up-regulated and 586 were down-regulated. Of these, we were able to annotate 477 genes by BLAST (roughly 42%). We were able to assign human homologs to only 355 genes (31% of the original group) that were used for the Pathway Studio® analysis below (Supplemental Table S2).
REAL TIME REVERSE TRANSCRIPTASE-PCR RESULTS
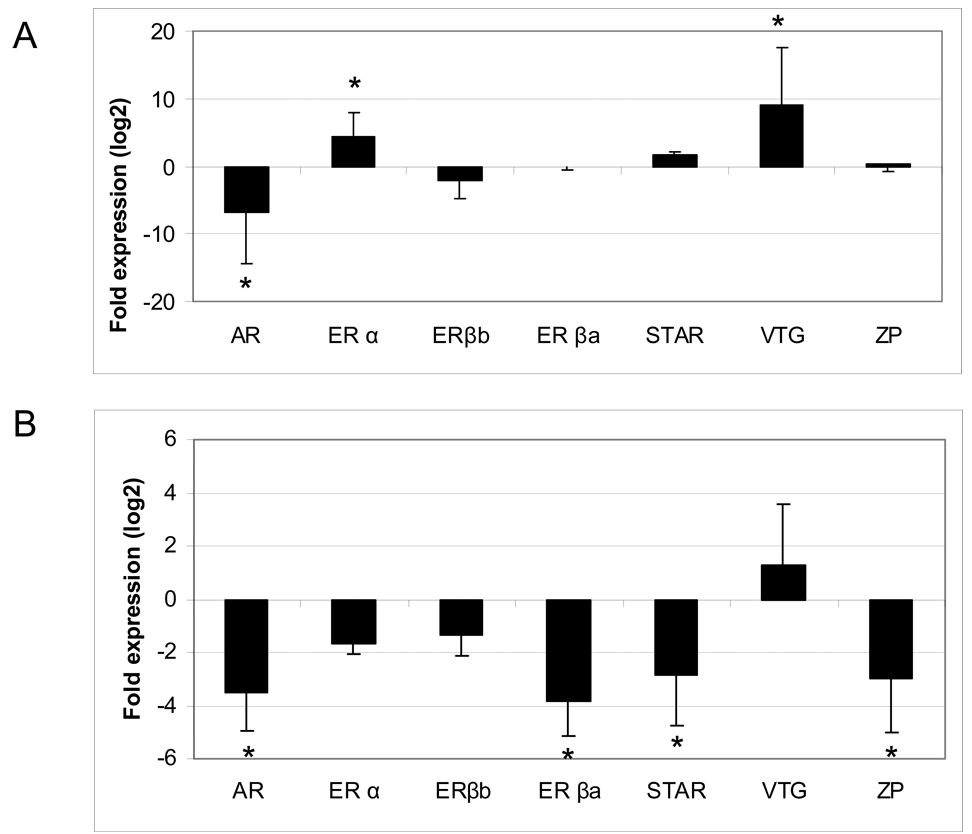
To validate the microarray results, relative expression of several critical genes was measured by Q-PCR. The results fit a common gene expression profile for LMB exposed to E2, and indicate that the microarray results are responsive to E2 exposure. In LMB liver (Fig. 5a), Vtg and ERα were significantly up-regulated by 528-fold and 20-fold, respectively, and AR was significantly down-regulated by 116 fold, while ERβa, ERβb, and ZPC-4 were largely unaffected. By microarray analysis, Vtg and ERα were up-regulated by 26-fold and 15-fold, respectively. AR was down-regulated by 1.5-fold but it was not significant. ERβa, ERβb and ZPC-4 were not affected.
Figure 5.
Real time quantitative (Q)-PCR analysis of critical genes involved in reproduction. (A) Liver and (B) Gonad. Significance at (P < 0.05) is denoted by an asterisk.
In LMB testis (Fig 5b), Q-PCR results for AR, StAR, ERβa and ZPC-4 showed that they were down-regulated by 11.5-fold, 7-fold, 14-fold, and 7-fold respectively. Other genes tested by Q-PCR were not significantly changed. Probes for all of these genes were present on the microarray, but only StAR was significantly down-regulated by 4.4-fold.
GO ANALYSES
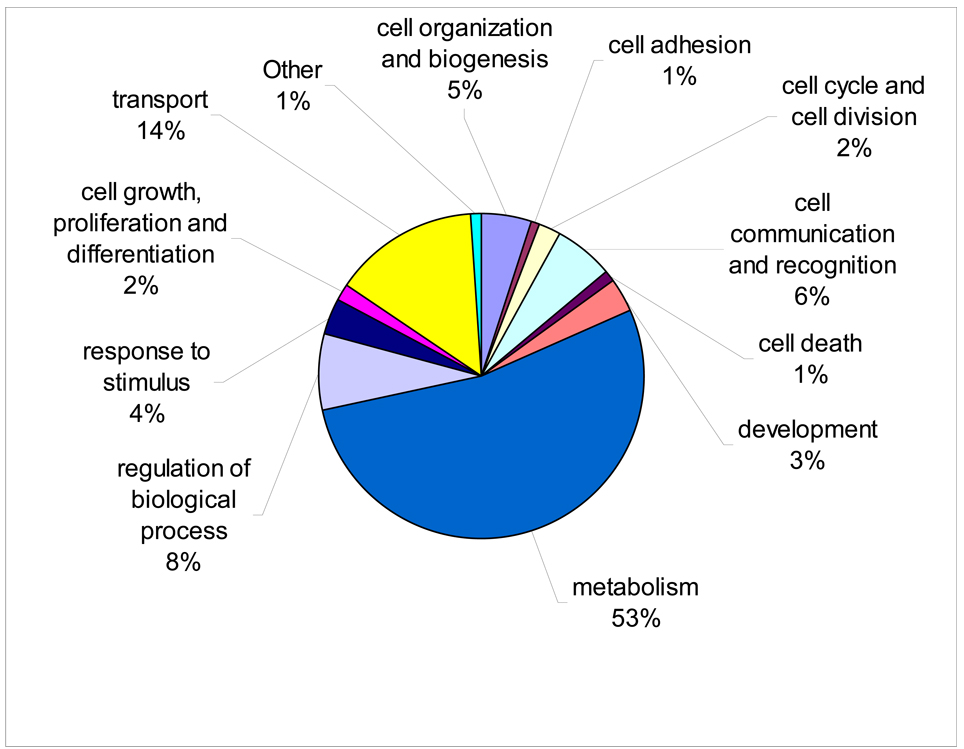
The distribution of all genes on the array within biological process categories is shown in Fig. 6 and is listed more finely in Supplemental Table S3. To identify biological processes altered by œstradiol treatment, we examined genes that were significantly altered (P < 0.01) which could be assigned to gene ontology (GO) orthologs and for which pathways were enriched in our list as determined by Fisher’s exact test. In the liver, 21 biological process categories were up-regulated and 33 categories were down-regulated (Table 2). In the gonad, 17 categories were significantly up-regulated and none were significantly down-regulated (Table 3). The pathways most significantly altered include development, signal transduction, morphogenesis, cell communication, and response to chemical stimulus.
Figure 6.
Distribution of genes on the array for GO terms belonging to the biological process category.
Table 2.
| A. GO biological processes that are up-regulated in the liver. | ||||||
|---|---|---|---|---|---|---|
| GO ID | GO Name | # of Genes Selected | # of Genes on Array | Fisher p Value | FDR | |
| 1 | GO:0030163 | protein catabolism | 23 | 87 | 3.93E-10 | 7.31E-08 |
| 2 | GO:0006511 | ubiquitin-depend protein catabolism | 16 | 68 | 8.17E-07 | 3.80E-05 |
| 3 | GO :0019941 | modification-depend protein catabolism | 16 | 68 | 8.17E-07 | 3.04E-05 |
| 4 | GO:0051603 | proteolysis -- cell protein catabolism | 16 | 71 | 1.34E-06 | 3.12E-05 |
| 5 | GO:0009056 | catabolism | 23 | 189 | 7.74E-05 | 1.60E-03 |
| 6 | GO:0006508 | proteolysis | 22 | 197 | 3.43E-04 | 6.39E-03 |
| 7 | GO:0006512 | ubiquitin cycle | 21 | 201 | 1.04E-03 | 1.62E-02 |
| 8 | GO:0019538 | protein metabolism | 67 | 1022 | 3.30E-03 | 4.73E-02 |
| 9 | GO:0044260 | cellular macromolecule metabolism | 64 | 1001 | 7.49E-03 | 8.71E-02 |
| 10 | GO:0006465 | signal peptide processing | 2 | 2 | 1.17E-02 | 1.28E-01 |
| 11 | GO:0006518 | peptide metabolism | 2 | 2 | 1.17E-02 | 1.21E-01 |
| 12 | GO:0006354 | RNA elongation | 3 | 9 | 1.52E-02 | 1.49E-01 |
| 13 | GO:0006412 | protein biosynthesis | 25 | 330 | 1.83E-02 | 1.70E-01 |
| 14 | GO:0000004 | biological process unknown | 70 | 1161 | 1.83E-02 | 1.62E-01 |
| 15 | GO:0043170 | macromolecule metabolism | 86 | 1492 | 2.35E-02 | 1.98E-01 |
| 16 | GO:0043283 | biopolymer metabolism | 52 | 833 | 2.59E-02 | 2.09E-01 |
| 17 | GO:0006626 | protein targeting to mitochondrion | 3 | 13 | 3.39E-02 | 2.63E-01 |
| 18 | GO:0008285 | neg regulation of cell proliferation | 3 | 15 | 4.62E-02 | 3.44E-01 |
| 19 | GO:0006413 | translational initiation | 5 | 39 | 4.85E-02 | 3.47E-01 |
| 20 | GO:0043412 | biopolymer modification | 30 | 455 | 4.98E-02 | 3.43E-01 |
| 21 | GO:0006464 | protein modification | 29 | 437 | 4.99E-02 | 3.32E-01 |
| B. GO biological processes that are down-regulated in the liver. | ||||||
|---|---|---|---|---|---|---|
| GO ID | GO Name | # of Genes Selected | # of Genes on Array | Fisher p Value | FDR | |
| 1 | GO:0008406 | gonad development | 3 | 5 | 1.29E-03 | 2.63E-01 |
| 2 | GO:0045137 | develop of primary sex characters | 3 | 5 | 1.29E-03 | 1.31E-01 |
| 3 | GO:0040007 | growth | 6 | 43 | 3.03E-03 | 2.05E-01 |
| 4 | GO:0008105 | asymmetric protein localization | 3 | 9 | 4.66E-03 | 2.36E-01 |
| 5 | GO:0007548 | sex differentiation | 3 | 10 | 5.92E-03 | 2.40E-01 |
| 6 | GO:0000902 | cellular morphogenesis | 5 | 35 | 6.17E-03 | 2.09E-01 |
| 7 | GO:0008361 | regulation of cell size | 4 | 22 | 6.74E-03 | 1.95E-01 |
| 8 | GO:0016049 | cell growth | 4 | 22 | 6.74E-03 | 1.71E-01 |
| 9 | GO:0007275 | development | 17 | 284 | 7.33E-03 | 1.65E-01 |
| 10 | GO:0045167 | Asymmetric protein -- cell fate commitment | 2 | 3 | 8.29E-03 | 1.68E-01 |
| 11 | GO:0006629 | lipid metabolism | 7 | 73 | 9.21E-03 | 1.70E-01 |
| 12 | GO:0009653 | morphogenesis | 8 | 94 | 1.04E-02 | 1.76E-01 |
| 13 | GO:0006869 | lipid transport | 3 | 13 | 1.09E-02 | 1.70E-01 |
| 14 | GO:0045165 | cell fate commitment | 3 | 14 | 1.29E-02 | 1.87E-01 |
| 15 | GO:0006006 | glucose metabolism | 5 | 46 | 1.70E-02 | 2.30E-01 |
| 16 | GO:0001649 | osteoblast differentiation | 3 | 16 | 1.76E-02 | 2.24E-01 |
| 17 | GO:0009888 | tissue development | 4 | 31 | 1.92E-02 | 2.29E-01 |
| 18 | GO:0006957 | complement activation, alternative pathway | 2 | 6 | 2.19E-02 | 2.47E-01 |
| 19 | GO:0009401 | PEP-sugar phosphotransferase system | 2 | 6 | 2.19E-02 | 2.34E-01 |
| 20 | GO:0045087 | innate immune response | 2 | 7 | 2.76E-02 | 2.80E-01 |
| 21 | GO:0006096 | glycolysis | 4 | 36 | 2.99E-02 | 2.89E-01 |
| 22 | GO:0016197 | endosome transport | 2 | 8 | 3.38E-02 | 3.12E-01 |
| 23 | GO:0005996 | monosaccharide metabolism | 5 | 56 | 3.40E-02 | 3.00E-01 |
| 24 | GO:0019318 | hexose metabolism | 5 | 56 | 3.40E-02 | 2.88E-01 |
| 25 | GO:0006956 | complement activation | 2 | 9 | 4.06E-02 | 3.29E-01 |
| 26 | GO:0008643 | carbohydrate transport | 2 | 9 | 4.06E-02 | 3.17E-01 |
| 27 | GO:0044255 | cellular lipid metabolism | 6 | 81 | 4.39E-02 | 2.97E-01 |
| 28 | GO:0001503 | ossification | 3 | 24 | 4.48E-02 | 2.93E-01 |
| 29 | GO:0031214 | biomineral formation | 3 | 24 | 4.48E-02 | 2.84E-01 |
| 30 | GO:0006066 | alcohol metabolism | 6 | 82 | 4.60E-02 | 2.83E-01 |
| 31 | GO:0046849 | bone remodeling | 3 | 25 | 4.91E-02 | 2.93E-01 |
| 32 | GO:0048771 | tissue remodeling | 3 | 25 | 4.91E-02 | 2.85E-01 |
| 33 | GO:0046164 | alcohol catabolism | 4 | 43 | 4.99E-02 | 2.74E-01 |
Table 3.
GO biological processes that are up-regulated in the Gonad.
| GO ID | GO Name | # of Genes Selected | # of Genes on Array | Fisher p Value | FDR | |
|---|---|---|---|---|---|---|
| 1 | GO:0007275 | development | 12 | 284 | 5.93E-05 | 0.003 |
| 2 | GO:0007165 | signal transduction | 12 | 368 | 6.33E-04 | 0.016 |
| 3 | GO:0009653 | morphogenesis | 6 | 94 | 7.12E-04 | 0.012 |
| 4 | GO:0000902 | cellular morphogenesis | 4 | 35 | 8.08E-04 | 0.010 |
| 5 | GO:0007154 | cell communication | 12 | 397 | 1.22E-03 | 0.012 |
| 6 | GO:0042221 | response to chemical stimulus | 5 | 70 | 1.28E-03 | 0.010 |
| 7 | GO:0009628 | response to abiotic stimulus | 5 | 75 | 1.71E-03 | 0.012 |
| 8 | GO:0016043 | cellular organization and biogenesis | 10 | 373 | 7.74E-03 | 0.047 |
| 9 | GO:0009987 | cellular process | 41 | 2980 | 8.72E-03 | 0.047 |
| 10 | GO:0048513 | organ development | 5 | 112 | 8.78E-03 | 0.043 |
| 11 | GO:0008283 | cell proliferation | 4 | 83 | 1.49E-02 | 0.066 |
| 12 | GO:0044248 | cellular catabolism | 5 | 162 | 3.55E-02 | 0.145 |
| 13 | GO:0050874 | organismal physiologic process | 4 | 121 | 4.78E-02 | 0.180 |
| 14 | GO:0007242 | intracellular signaling cascade | 5 | 182 | 5.34E-02 | 0.187 |
| 15 | GO:0009056 | catabolism | 5 | 189 | 6.08E-02 | 0.198 |
| 16 | GO:0050896 | response to stimulus | 6 | 264 | 7.42E-02 | 0.227 |
| 17 | GO:0006091 | Generation of precursor metabolites | 6 | 278 | 8.98E-02 | 0.259 |
DISCUSSION
MICROARRAY CONSTRUCTION
Because genome sequencing for ecologically relevant fish species is still in the early stages, compared to mammalian species use of gene expression analysis has been hampered. There are only a few fish species with completed sequenced genomes, including the spotted puffer fish (Tetraodon nigroviridis), the Japanese puffer fish (Takifugu rubripes) and the zebrafish (Danio rerio). Of these, there are commercial arrays available only for zebrafish from Affymetrix and Agilent.
LMB are present throughout the United States and have been impacted by environmental chemicals (Denslow and Sepúlveda, 2007), making it a useful species for ecotoxicological testing purposes. Previously, the Denslow lab made great efforts to develop a useful LMB microarray, mostly using SSH, providing roughly 2,616 different reads, corresponding to 758 genes. This number of unique sequences is small compared to the 32,882 sequences generated by two runs on the GS20. Additionally, the normalized cDNA library we present here resulted in a highly normalized transcript pool in considerably less time (approximately 3 days) than is required for traditional SSH. The cDNA normalization is an important component of this process, as it minimizes the sequencing of redundant sequences, allowing an efficient gene-discovery process.
Pyrosequencing data, however, is not perfect for construction of arrays due to short reads which can make it difficult to identify directionality and perform annotation. By using probes against the sense and antisense strands for genes where directionality is not known, one can overcome this disadvantage. This stategy, however, reduces the number of transcripts that can be examined on a single array. Newer sequencing technology (e.g. the 454 GS-FLX) produces longer reads (average 200–300 bp) and should reduce the problems associated with this data. It is likely that sequencing costs will also continue to decrease.
E2 EXPOSURE RESULTS
Using the custom microarray, many genes in the liver that were affected by œstrogen exposure in LMB were similar to those reported in other fish species. In addition to Vtg and ERα, treatment with œstrogens has been reported to up-regulate mitochondrial import inner membrane translocase subunit TIM8 A, proteasome delta, pyruvate kinase, and aspartic protease; while down-regulating coagulation factor X, CYP1A1 (cytochrome P450 1A), GAPDH, cyclin G2, alcohol dehydrogenase 5 and transferrin in fathead minnow, European flounder, zebrafish, rainbow trout, and carp (Hoffmann et al., 2006; Filby et al., 2007; Hook et al., 2007; Moens et al., 2007b; Williams et al., 2007). Similar changes were observed in LMB liver using the custom microarray. In LMB gonad, StAR was down-regulated and ERα, ERβb, growth hormone, and insulin-like growth factor 1 (IGF-1) were unchanged, which is consistent with previous reports (Hook et al., 2006; Filby et al., 2007).
Some responses observed in LMB treated with œstradiol did not agree with previous reports. For instance, ribosomal protein L13 and cyclin G1 were found up-regulated in flounder liver (Williams et al., 2007) but were unchanged in the LMB results. Also, ERβb and aromatase (CYP19) were down-regulated in the gonad of fathead minnows (Filby et al., 2007), but were unchanged in LMB. It is likely that these discrepancies are due to differences between species and exposure paradigms. It is very interesting that some of the changes are consistent among the different species and exposures and these common genes are ideal targets for creating a broader gene expression profile in response to œstrogenic exposure.
PATHWAY ANALYSIS OF ARRAY RESULTS WITH PATHWAY STUDIO®
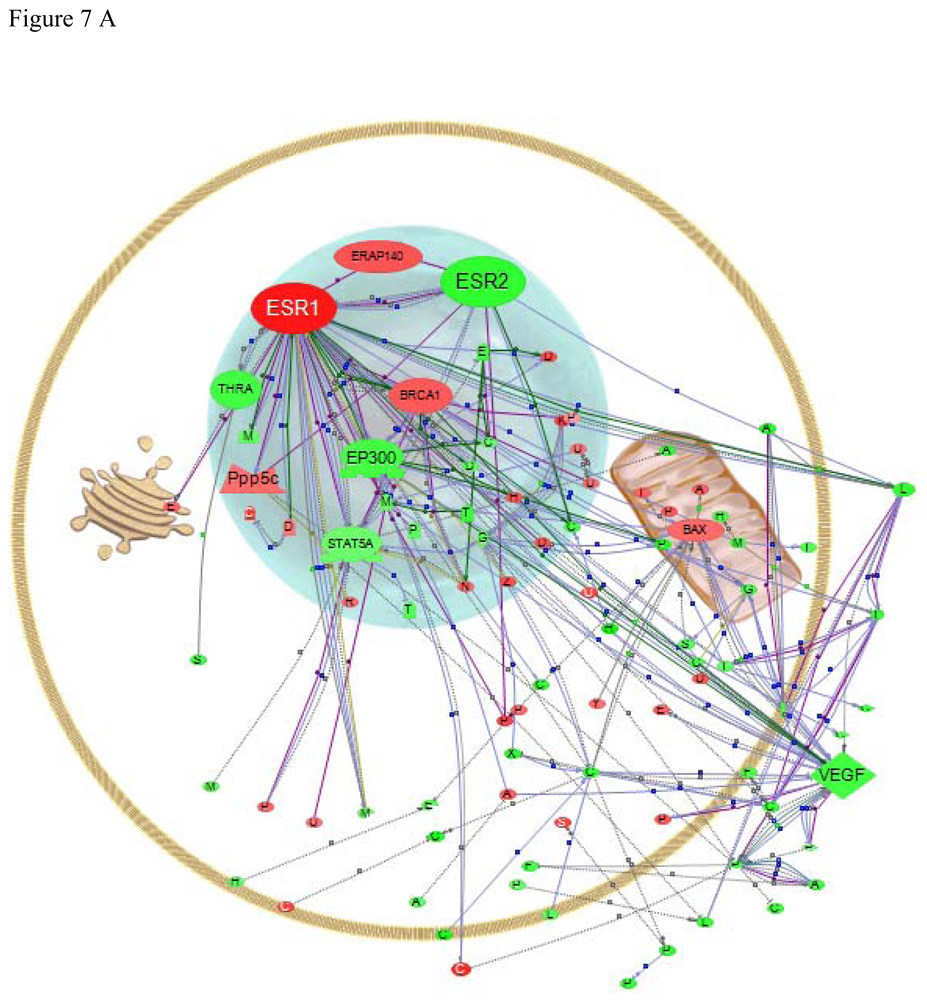
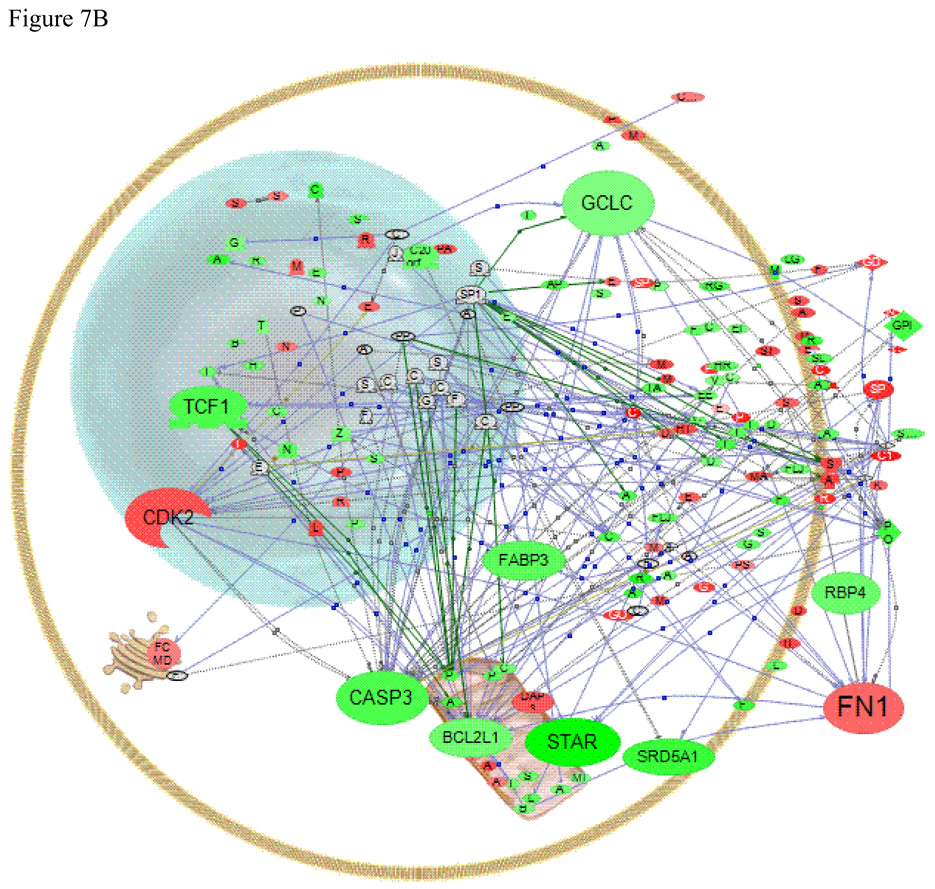
Data analysis is also a problem for non-mammalian species as databases of gene function and pathways are sparse. One option to overcome this is to map genes to mammalian homologs. Here, we used the program Pathway Studio® (Nikitin et al., 2003) to map interactions between human homologs of LMB genes identified by the custom microarray as being significantly affected by E2 exposure (Fig. 7). These results are useful to help place the observed changes into context by comparing them to the much larger database of human protein interactions, although only genes for which we could identify human homologs are presented in the figures. For example, since there are no human homologs for Vtg and ERβa, they are not shown. In addition, we are making the assumption that the genes are involved in similar pathways in fish as they are in humans. While both are vertebrates, the gene duplications that have occurred in teleost fish produces multiple genes with high sequence similarity but sequence homology is not necessarily sufficient to infer similarity of function. With those caveats, several genes with identifiable human homologues can be used to highlight the possible utility of Pathway Studio® for identifying gene or protein interactions of interest in non-model fish species and for establishing hypotheses about what might be occurring after exposure to an œstrogen. We used the list of genes regulated at the P < 0.05 for this analysis, which revealed that many of the affected genes were related within established pathways.
Figure 7.
Pathway Studio® analysis of common regulators that are changed in (A) liver or (B) gonad with exposure of male fish to E2. Red indicates up regulation, green indicates down regulation. The type of regulator molecule is depicted by the shape of the symbol and includes  transcription factors,
transcription factors,  nuclear receptors,
nuclear receptors,  kinases,
kinases,  phosphatases,
phosphatases,  membrane receptors and
membrane receptors and  ligands.
ligands.
Effects of E2 exposure on gene expression in the liver
Exposure to high doses of E2 produces dramatic gene expression changes in male LMB liver. We searched our expression data for direct interactions among the proteins that were differentially expressed. Figure 7A shows the results of this analysis. We accentuated only the direct interactions among our differentially expressed genes and two œstrogen receptors (ERα is ESR1 and ERβ is ESR2, in these figures). The lines that emanate from the two ERs show the interactions that have been mapped by previous research in mammalian systems. The multiple red and green dots represent the regulated genes from the current study, which are up- or down-regulated in the liver in response to E2. A full list of the altered genes, for which we could find human homologs, appears in supplemental Table S1.
Among the hundreds of genes that are altered by E2 in the liver, there are several important regulatory genes directly connected to ER signalling, either by ERα or ERβ. There are few studies of E2 regulation of expression in mammalian liver, but some interactions observed in mammalian uterus and breast tissues among E2 regulated genes also appear in our LMB liver dataset. Among those that interact with ERα (ESR1) in mammals are two transcription factors EP300 (E1A binding protein p300), a coactivator for ERα (Kim et al., 2001) and STAT5A (signal transducer and activator of transcription 5A) (Stoecklin et al., 1999; Frasor et al., 2001; Wang and Cheng, 2004), both of which are down-regulated in our experiment. EP300 and STAT5A in turn control expression of a large number of genes, including BRCA1 (breast cancer 1) which is up-regulated in our expression experiment. BRCA1 has been shown to directly interact with ESR1 in mammals (Jeffy et al., 2005) as well as with both EP300 and STAT5A, and to potentiate a number of down stream interactions. LMB BAX (Bcl2-associated × protein) was up-regulated by the E2 exposure. This protein is correlated with an increase in apoptosis (Knudson and Brown, 2008). In MCF7 cells, however, E2 was shown to increase bcl-2 but not BAX (Wang and Phang, 1995) suggesting that E2 exposure may play a different role in apoptosis in LMB than in mammalian models. VEGF (vascular endothelial growth factor A) was down-regulated in LMB liver, but up-regulated by E2 in mammalian studies (Kawai et al., 2002), suggesting a key regulatory difference between fish and mammals in this response, although this may also be attributed to differences in tissue context.
Here, THRA (Thyroid hormone receptor alpha) was down-regulated in LMB liver, an effect that has also been observed in male fathead minnows (Filby et al., 2007). In addition, ERα (ESR1) and ERβ (ESR2) in LMB, as in mammalian models (Kietz et al., 2004), may modulate each other. We have seen direct confirmation of this interaction in transfection experiments with the LMB ERs (Sabo-Attwood et al., 2007). Figure 7a suggests that the two LMB ERs may be modulated by similar negative regulators such as Pppsc (protein phosphatase 5), which has been shown to dephosphorylate both receptors in mammalian systems (Ikeda et al., 2004), although this needs further study.
Effects of E2 exposure on gene expression in the gonad
The regulatory effects of E2 in the male LMB gonad are muted by comparison to the liver, but the interactions between nucleus, mitochondria, cellular metabolism and exported proteins are quite striking (Fig. 7b). While we have highlighted only a few of the genes in the figure, it is clear that many of the green and red spots that represent down- and up-regulated genes, are connected via pathways. A full list of the altered genes for which we could find human homologs appears in supplemental Table S2.
Of note, StAR (steroidogenic acute regulatory protein) is down-regulated by the treatment, as shown by the array data and validated by Q-PCR. This protein is critical for cholesterol delivery to the mitochondria for steroidogenesis (Clark et al., 1994). The LMB StAR promoter is highly complex with many nuclear factor binding sites (Kocerha, 2005) including one for ER. The gonadal protein SRD5A1 (steroid 5-alpha-reductase 1) which catalyzes the conversion of testosterone to dihydrotestosterone in mammalian systems is also down-regulated by E2 treatment. This enzyme is involved in metabolism of testosterone and progesterone in goldfish brain (Pasmanik and Callard, 1985), but there is scant information about it in other fish tissues.
Pathway analysis of the LMB data suggests that some important genes may be regulated differently in fish and mammals. Genes that responded opposite the way they respond in mammalian models included CASP3 (caspase 3) and BCL2L1 (Bcl2-like 1 protein) (Thompson et al., 2002; Nair and Shaha, 2003); and glutathione synthesis (GCLC, glutamate-cysteine ligase, catalytic subunit) (Urata et al., 2006) all of which were down-regulated in LMB gonad in response to E2. Peroxisome proliferator-activated receptor gamma (PPARgamma) (Huss et al., 2004) did not respond at the mRNA level in the LMB study. Several extracellular matrix proteins that are regulated by E2 including RBP4 (retinol binding protein 4), which is associated with the transport of plasma Vitamin D, and FN1 (fibronectin 1) also appear to be regulated by E2 differently in LMB and mammals (Li et al., 2004; Horii et al., 2006). Care must be exercised when comparing gene regulation among species and tissues and more work is needed to sort this out. This type of data illustrates how pathway analysis can facilitate this type of investigation.
The fact that some genes of interest for fish reproductive/endocrine research were able to be mapped to human homologues implies that ecotoxicological researchers can leverage the tremendous amount known about gene interactions in human and mammalian systems into understanding toxic responses in fish. By utilizing this information, it may be possible to develop novel hypotheses regarding gene/contaminant interactions. For a better understanding of mechanistic changes in aquatic organisms, it is crucial that fish-specific pathway analyses be developed and incorporated into programs such as Pathway Studio®.
Aquatic toxicologists have long struggled with the need for molecular biomarkers to assess contaminant – induced changes in gene expression for ecologically relevant species. While there are huge strides in making custom microarrays for these species, the lack of genomic sequences is still an impediment to using these technologies in ecotoxicology. The new pyrosequencing technology has provided a fast and highly efficient way to obtain EST sequences for these species, allowing a fast microarray development and a much better and deeper understanding of the species’ response to toxicants. Our results show that the use of a well normalized cDNA library as input to the 454 sequencer provides a much broader spectrum of sequences, avoiding the repetition of the highly abundant ones. The gene expression profile from the LMB exposure to E2 analyzed with the newly developed LMB microarrays was consistent with the expected results, confirming that the microarray development was highly successful.
Clearly, the decrease in time and effort, as well as the massive increase in sequence data generated, justify the increased cost of 454 sequencing. The quantum leap in high-quality sequence data generated by the process we have outlined here means a huge improvement in the effort of developing the LMB microarray, and offers the potential to revolutionize ecotoxicogenomics by allowing researchers to construct high-quality microarrays for ecologically relevant species, rather than focusing research efforts in model species that may not necessarily be the most appropriate in a given ecological setting.
Supplementary Material
Acknowledgements
These studies were funded by the Superfund Basic Research Program from the National Institute of Environmental Health Sciences (ND and DB), P42 ES 07375 and RO1 ES015449 and by a fellowship from the Spanish Ministry of Sciences and Technology (NG-R) (EX-2004-0986), cofunded by the European Union.
REFERENCES
- Benjamini Y, Hochberg Y. Controlling the false discovery rate, a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- Blum JL, Knoebl I, Larkin P, Kroll KJ, Denslow ND. Use of suppressive subtractive hybridization and cDNA arrays to discover patterns of altered gene expression in the liver of dihydrotestosterone and 11-ketotestosterone exposed adult male largemouth bass (Micropterus salmoides) Marine Environmental Research. 2004;58:565–569. doi: 10.1016/j.marenvres.2004.03.046. [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genetics. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Clark BJ, Wells J, King SR, Stocco DM. The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the steroidogenic acute regulatory protein (StAR) Journal of Biological Chemistry. 1994;269:28314–28322. [PubMed] [Google Scholar]
- Denslow ND, Bowman CJ, Ferguson RJ, Lee HS, Hemmer MJ, Folmar LC. Induction of gene expression in sheepshead minnows (Cyprinodon variegatus)treated with 17beta-estradiol, diethylstilbestrol, or ethinylestradiol: the use of mRNA fingerprints as an indicator of gene regulation. General and Comparative Endocrinology. 2001a;121:250–260. doi: 10.1006/gcen.2001.7605. [DOI] [PubMed] [Google Scholar]
- Denslow ND, Lee HS, Bowman CJ, Hemmer MJ, Folmar LC. Multiple responses in gene expression in fish treated with estrogen. Comparative Biochemistry and Physiology B Biochemistry and Molecular Biology. 2001b;129:277–282. doi: 10.1016/s1096-4959(01)00322-0. [DOI] [PubMed] [Google Scholar]
- Denslow ND, Garcia-Reyero N, Barber DS. Fish 'n' chips: the use of microarrays for aquatic toxicology. Molecular Biosystems. 2007;3:172–177. doi: 10.1039/b612802p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow ND, Sepúlveda MS. Ecotoxicological effects of endocrine disrupting compounds on fish reproduction. In: Babin PJ, Cerda J, Lubzens E, editors. The Fish Oocyte: From Basic Studies to Biotechnological Applications. The Netherlands: Springer Publishing Co, Dordrecht; 2007. pp. 255–322. [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proceedings of the National Academy of Sciences U S A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SE. Microarray studies of gene expression in fish. Omics. 2006;10:474–489. doi: 10.1089/omi.2006.10.474. [DOI] [PubMed] [Google Scholar]
- Farmerie WG, Hammer J, Liu L, Sahni A, Schneider M. Biological workflow with BlastQuest. Data & Knowledge Engineering. 2005;53:75–97. [Google Scholar]
- Filby AL, Thorpe KL, Maack G, Tyler CR. Gene expression profiles revealing the mechanisms of anti-androgen- and estrogen-induced feminization in fish. Aquatic Toxicology. 2007;81:219–231. doi: 10.1016/j.aquatox.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Frasor J, Park K, Byers M, Telleria C, Kitamura T, Yu-Lee LY, Djiane J, Park-Sarge OK, Gibori G. Differential roles for signal transducers and activators of transcription 5a and 5b in PRL stimulation of ERalpha and ERbeta transcription. Molecular Endocrinology. 2001;15:2172–2181. doi: 10.1210/mend.15.12.0745. [DOI] [PubMed] [Google Scholar]
- Garcia-Reyero N, Barber DS, Gross TS, Johnson KG, Sepulveda MS, Szabo NJ, Denslow ND. Dietary exposure of largemouth bass to OCPs changes expression of genes important for reproduction. Aquatic Toxicology. 2006;78:358–369. doi: 10.1016/j.aquatox.2006.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reyero N, Adelman I, Liu L, Denslow ND. Gene expression profiles of fathead minnows exposed to surface waters above and below a sewage treatment plant in Minnesota. Marine Environmental Research. 2008;66(1):134–136. doi: 10.1016/j.marenvres.2008.02.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffitt RJ, Greig TW, Chandler GT, Quattro JM. Serial analysis of gene expression reveals identifiable patterns in transcriptome profiles of palaemonetes pugio exposed to three common environmental stressors. Environmental Toxicology and Chemistry. 2007;26:2413–2419. doi: 10.1897/07-158R.1. [DOI] [PubMed] [Google Scholar]
- Hoffmann JL, Torontali SP, Thomason RG, Lee DM, Brill JL, Price BB, Carr GJ, Versteeg DJ. Hepatic gene expression profiling using Genechips in zebrafish exposed to 17alpha-ethynylestradiol. Aquatic Toxicology. 2006;79:233–246. doi: 10.1016/j.aquatox.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Hook SE, Skillman AD, Small JA, Schultz IR. Gene expression patterns in rainbow trout, Oncorhynchus mykiss, exposed to a suite of model toxicants. Aquatic Toxicology. 2006;77:372–385. doi: 10.1016/j.aquatox.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook SE, Skillman AD, Small JA, Schultz IR. Temporal changes in gene expression in rainbow trout exposed to ethynyl estradiol. Comparative Biochemistry and Physiology C Toxicology and Pharmacology. 2007;145:73–85. doi: 10.1016/j.cbpc.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii Y, Takei H, Koibuchi Y, Horiguchi J, Maemura M, Iino Y, Morishita Y. The regulatory effect of tamoxifen on fibronectin expression in estrogen-dependent MCF-7 breast carcinoma cells. Oncology Reports. 2006;15:1191–1195. [PubMed] [Google Scholar]
- Huss JM, Torra IP, Staels B, Giguere V, Kelly DP. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Molecular Cell Biology. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Ogawa S, Tsukui T, Horie-Inoue K, Ouchi Y, Kato S, Muramatsu M, Inoue S. Protein phosphatase 5 is a negative regulator of estrogen receptor-mediated transcription. Molecular Endocrinology. 2004;18:1131–1143. doi: 10.1210/me.2003-0308. [DOI] [PubMed] [Google Scholar]
- Iseli c, Jongeneel CV, Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proceedings of the 7th International Conference on Intelligent Systems for Molecular Biology. 1999:138–148. [PubMed] [Google Scholar]
- Jeffy BD, Hockings JK, Kemp MQ, Morgan SS, Hager JA, Beliakoff J, Whitesell LJ, Bowden GT, Romagnolo DF. An estrogen receptor-alpha/p300 complex activates the BRCA-1 promoter at an AP-1 site that binds Jun/Fos transcription factors: repressive effects of p53 on BRCA-1 transcription. Neoplasia. 2005;7:873–882. doi: 10.1593/neo.05256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Z, Wells MC, Walter RB. DNA microarray technology in toxicogenomics of aquatic models: methods and applications. Comparative Biochemistry and Physiology C Toxicology and Pharmacology. 2007;145:5–14. doi: 10.1016/j.cbpc.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Kawai H, Li H, Chun P, Avraham S, Avraham HK. Direct interaction between BRCA1 and the estrogen receptor regulates vascular endothelial growth factor (VEGF) transcription and secretion in breast cancer cells. Oncogene. 2002;21:7730–7739. doi: 10.1038/sj.onc.1205971. [DOI] [PubMed] [Google Scholar]
- Kietz S, Thomsen JS, Matthews J, Pettersson K, Strom A, Gustafsson JA. The Ah receptor inhibits estrogen-induced estrogen receptor beta in breast cancer cells. Biochemical Biophysical Research Communication. 2004;320:76–82. doi: 10.1016/j.bbrc.2004.05.132. [DOI] [PubMed] [Google Scholar]
- Kim MY, Hsiao SJ, Kraus WL. A role for coactivators and histone acetylation in estrogen receptor alpha-mediated transcription initiation. Embo J. 2001;20:6084–6094. doi: 10.1093/emboj/20.21.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson CM, Brown NM. Mitochondria potential, bax "activation," and programmed cell death. Methods in Molecular Biology. 2008;414:95–108. doi: 10.1007/978-1-59745-339-4_9. [DOI] [PubMed] [Google Scholar]
- Kocerha R. Biochemistry and Molecular Biology. Gainesville, Florida: University of Florida; 2005. Regulation of the steroidogenic acute regulatory protein by 3’-5’ dibutyryl cyclic AMP and transforming growth factor-beta dependent pathways. [Google Scholar]
- Larkin P, Sabo-Attwood T, Kelso J, Denslow ND. Analysis of gene expression profiles in largemouth bass exposed to 17-beta-estradiol and to anthropogenic contaminants that behave as estrogens. Ecotoxicology. 2003;12:463–468. doi: 10.1023/b:ectx.0000003031.05390.b5. [DOI] [PubMed] [Google Scholar]
- Larkin P, Villeneuve DL, Knoebl I, Miracle AL, Carter BJ, Liu L, Denslow ND, Ankley GT. Development and validation of a 2,000-gene microarray for the fathead minnow (Pimephales promelas) Environmental Toxicology and Chemistry. 2007;26:1497–1506. doi: 10.1897/06-501r.1. [DOI] [PubMed] [Google Scholar]
- Li XH, Kakkad B, Ong DE. Estrogen directly induces expression of retinoic acid biosynthetic enzymes, compartmentalized between the epithelium and underlying stromal cells in rat uterus. Endocrinology. 2004;145:4756–4762. doi: 10.1210/en.2004-0514. [DOI] [PubMed] [Google Scholar]
- Lorenz WW, Dean JF. SAGE profiling and demonstration of differential gene expression along the axial developmental gradient of lignifying xylem in loblolly pine (Pinus taeda) Tree Physiology. 2002;22:301–310. doi: 10.1093/treephys/22.5.301. [DOI] [PubMed] [Google Scholar]
- Marburger JE, Johnson WE, Gross TS, Douglas DR, Di J. Residual organochlorine pesticides in soils and fish from wetland restoration areas in central Florida, USA. Wetlands. 2002;22:705–711. [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyniuk CJ, Xiong H, Crump K, Chiu S, Sardana R, Nadler A, Gerrie ER, Xia X, Trudeau VL. Gene expression profiling in the neuroendocrine brain of male goldfish (Carassius auratus) exposed to 17alpha-ethinylestradiol. Physiological Genomics. 2006;27:328–336. doi: 10.1152/physiolgenomics.00090.2006. [DOI] [PubMed] [Google Scholar]
- Miracle AL, Toth GP, Lattier DL. The path from molecular indicators of exposure to describing dynamic biological systems in an aquatic organism: microarrays and the fathead minnow. Ecotoxicology. 2003;12:457–462. doi: 10.1023/b:ectx.0000003030.67752.04. [DOI] [PubMed] [Google Scholar]
- Moens LN, van der Ven K, Van Remortel P, Del-Favero J, De Coen WM. Expression profiling of endocrine-disrupting compounds using a customized Cyprinus carpio cDNA microarray. Toxicological Sciences. 2006;93:298–310. doi: 10.1093/toxsci/kfl057. [DOI] [PubMed] [Google Scholar]
- Moens LN, Smolders R, van der Ven K, van Remortel P, Del-Favero J, De Coen WM. Effluent impact assessment using microarray-based analysis in common carp: a systems toxicology approach. Chemosphere. 2007a;67:2293–2304. doi: 10.1016/j.chemosphere.2006.09.092. [DOI] [PubMed] [Google Scholar]
- Moens LN, van der Venog K, Van Remortel P, Del-Favero J, De Coen WM. Gene expression analysis of estrogenic compounds in the liver of common carp (Cyprinus carpio) using a custom cDNA microarray. Journal of Biochemistry and Molecular Toxicology. 2007b;21:299–311. doi: 10.1002/jbt.20190. [DOI] [PubMed] [Google Scholar]
- Nair R, Shaha C. Diethylstilbestrol induces rat spermatogenic cell apoptosis in vivo through increased expression of spermatogenic cell Fas/FasL system. Journal Biological Chemistry. 2003;278:6470–6481. doi: 10.1074/jbc.M209319200. [DOI] [PubMed] [Google Scholar]
- Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio--the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–2157. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- Pasmanik M, Callard GV. Aromatase and 5 alpha-reductase in the teleost brain, spinal cord, and pituitary gland. General and Comparative Endocrinol. 1985;60:244–251. doi: 10.1016/0016-6480(85)90320-x. [DOI] [PubMed] [Google Scholar]
- Perkins EJ, Garcia-Reyero N, Villeneuve DL, Martinovic D, Brasfield SM, Blake LS, Brodin JD, Denslow ND, Ankley GT. Perturbation of gene expression and steroidogenesis with in vitro exposure of fathead minnow ovaries to ketoconazole. Marine Environmental Research. 2008;66:113–115. doi: 10.1016/j.marenvres.2008.02.072. [DOI] [PubMed] [Google Scholar]
- Reinartz J, Bruyns E, Lin JZ, Burcham T, Brenner S, Bowen B, Kramer M, Woychik R. Massively parallel signature sequencing (MPSS) as a tool for in-depth quantitative gene expression profiling in all organisms. Briefings in Functional Genomics and Proteomics. 2002;1:95–104. doi: 10.1093/bfgp/1.1.95. [DOI] [PubMed] [Google Scholar]
- Sabo-Attwood T, Kroll KJ, Denslow ND. Differential expression of largemouth bass (Micropterus salmoides) estrogen receptor isotypes alpha, beta, and gamma by estradiol. Molecular and Cellular Endocrinology. 2004;218:107–118. doi: 10.1016/j.mce.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Santos EM, Paull GC, Van Look KJ, Workman VL, Holt WV, van Aerle R, Kille P, Tyler CR. Gonadal transcriptome responses and physiological consequences of exposure to oestrogen in breeding zebrafish (Danio rerio) Aquatic Toxicology. 2007;83:134–142. doi: 10.1016/j.aquatox.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Sheader DL, Williams TD, Lyons BP, Chipman JK. Oxidative stress response of European flounder (Platichthys flesus) to cadmium determined by a custom cDNA microarray. Marine Environmental Research. 2006;62:33–44. doi: 10.1016/j.marenvres.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Stoecklin E, Wissler M, Schaetzle D, Pfitzner E, Groner B. Interactions in the transcriptional regulation exerted by Stat5 and by members of the steroid hormone receptor family. Journal Steroid Biochemistry and Molecular Biology. 1999;69:195–204. doi: 10.1016/s0960-0760(99)00052-7. [DOI] [PubMed] [Google Scholar]
- Thompson KE, Sipes IG, Greenstein BD, Hoyer PB. 17beta-estradiol affords protection against 4-vinylcyclohexene diepoxide-induced ovarian follicle loss in Fischer-344 rats. Endocrinology. 2002;143:1058–1065. doi: 10.1210/endo.143.3.8665. [DOI] [PubMed] [Google Scholar]
- Urata Y, Ihara Y, Murata H, Goto S, Koji T, Yodoi J, Inoue S, Kondo T. 17Beta-estradiol protects against oxidative stress-induced cell death through the glutathione/glutaredoxin-dependent redox regulation of Akt in myocardiac H9c2 cells. Journal Biological Chemistry. 2006;281:13092–13102. doi: 10.1074/jbc.M601984200. [DOI] [PubMed] [Google Scholar]
- van der Meer DL, van den Thillart GE, Witte F, de Bakker MA, Besser J, Richardson MK, Spaink HP, Leito JT, Bagowski CP. Gene expression profiling of the long-term adaptive response to hypoxia in the gills of adult zebrafish. American Journal of Physiology- Regulatory, Integrative and Comparative Physiology. 2005;289:R1512–R1519. doi: 10.1152/ajpregu.00089.2005. [DOI] [PubMed] [Google Scholar]
- van der Ven K, Keil D, Moens LN, van Leemput K, van Remortel P, De Coen WM. Neuropharmaceuticals in the environment: mianserin-induced neuroendocrine disruption in zebrafish (Danio rerio) using cDNA microarrays. Environmental Toxicology and Chemistry. 2006;25:2645–2652. doi: 10.1897/05-495r.1. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Wang TT, Phang JM. Effects of estrogen on apoptotic pathways in human breast cancer cell line MCF-7. Cancer Research. 1995;55:2487–2489. [PubMed] [Google Scholar]
- Wang Y, Cheng CH. ERalpha and STAT5a cross-talk: interaction through C-terminal portions of the proteins decreases STAT5a phosphorylation, nuclear translocation and DNA-binding. FEBS Letters. 2004;572:238–244. doi: 10.1016/j.febslet.2004.06.098. [DOI] [PubMed] [Google Scholar]
- Williams TD, Gensberg K, Minchin SD, Chipman JK. A DNA expression array to detect toxic stress response in European flounder (Platichthys flesus) Aquatic Toxicology. 2003;65:141–157. doi: 10.1016/s0166-445x(03)00119-x. [DOI] [PubMed] [Google Scholar]
- Williams TD, Diab AM, George SG, Sabine V, Chipman JK. Gene expression responses of European flounder (Platichthys flesus) to 17-beta estradiol. Toxicology Letters. 2007;168:236–248. doi: 10.1016/j.toxlet.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Yang F, Xu HT, Dai ZM, Yang WJ. Molecular characterization and expression analysis of vitellogenin in the marine crab Portunus trituberculatus. Comparative Biochemistry and Physiology B Biochemistry and Molecular Biology. 2005;142:456–464. doi: 10.1016/j.cbpb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Zahurak M, Parmigiani G, Yu W, Scharpf RB, Berman D, Schaeffer E, Shabbeer S, Cope L. Pre-processing Agilent microarray data. Biomed Central Bioinformatics. 2007;8:142. doi: 10.1186/1471-2105-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.