Abstract
Individuals with Down syndrome (DS) display a 40-fold greater risk of Hirschsprung disease (HSCR) than the general population of newborns implicating chromosome 21 in HSCR etiology. Here we demonstrate that the RET enhancer polymorphism RET+9.7 (rs2435357:C>T) at chromosome 10q11.2 is associated with HSCR in DS individuals both by transmission disequilibrium (P=0.0015) and case-control (P=0.0115) analysis of matched cases. Interestingly, the RET+9.7 T allele frequency is significantly different between individuals with DS alone (0.26±0.04), HSCR alone (0.61±0.04), and those with HSCR and DS (0.41±0.04), demonstrating an association and interaction between RET and chromosome 21 gene dosage. This is the first report of a genetic interaction between a common functional variant (rs2435357) and a not infrequent copy number error (chromosome 21 dosage) in two human developmental disorders.
Keywords: Down Syndrome, Hirschsprung Disease, RET, gene dosage, complex disorder
INTRODUCTION
Down syndrome (DS; MIM# 190685) arises from the genetic consequences of trisomy 21 and is a disorder of gene dosage. The incidence of DS is ~ 1/800 live births and arises each generation primarily from de novo meiosis I nondisjunction (NDJ) during oogenesis; a minority (6%) of cases arise from the segregation of a Robertsonian translocation in either parent [Epstein, 1986]. The central phenotype of DS is syndromic mental retardation, albeit quite variable, and is a consequence of the 50% increased dosage at many (but unknown) chromosome 21q genes. Additionally, individuals with DS have a gamut of associated developmental phenotypes that include, but are not limited to, early onset dementia, congenital heart defects, Hirschsprung disease (HSCR) and leukemia (in decreasing order of incidence). Cytogenetic mapping of rare patients with partial duplications of chromosome 21q and DS-associated phenotypes had led to the concept of a Down syndrome critical region (DSCR) at 21q22.2, minimal duplication of which is sufficient to lead to mental retardation and the DS facial gestalt [Delabar et al., 1993]. Indeed, specific genes (DSCR1, DYRK1A) within the DSCR in mouse models causing NFAT dysregulation lead to DS-like heart defects [Arron et al., 2006]. However, other features such as dementia map to the dosage effects of APP that is located outside the DSCR at band 21q21.3 [Wisniewski et al., 1985]. Thus, genes within and outside the DSCR can lead to DS-associated phenotypes. Nevertheless, the genetic etiologies of DS-associated disorders remain largely unknown, and, in particular, it is also likely that the etiologies of some of the DS associated phenotypes depend on genetic variants not mapping to chromosome 21.
HSCR (MIM# 142623), or congenital aganglionosis, is a multifactorial genetic defect of intestinal innervation with an incidence of ~1/5,000 live births; ~30% of HSCR patients have a recognized chromosomal abnormality, a recognized syndrome or additional congenital anomalies, the most frequent of which is DS [Chakravarti and Lyonnet, 2001]. DS is observed in HSCR patients with a frequency between 2–10% (average 5%) and is, consequently, 40-fold more common than in the general population of newborns; conversely, ~0.8% of individuals with DS have congenital aganglionosis. The existence of trisomy 21 increases the risk sharply but does not invariably lead to HSCR. Thus, the DS-HSCR association cannot be explained by increased dosage of chromosome 21 per se but requires variation in some additional ‘factor’, be it developmental noise, a chromosome 21 locus or even a non-chromosome 21 locus.
We hypothesize that this ‘factor’ is genetic and maps outside chromosome 21 since chromosome 21q is only 1% of the genome and contains few genes. Finally, non-21 genes are known to contribute to the DS phenotype: for example, somatic mutations at the X-linked GATA1 gene are etiologic for the DS-associated leukemias in trisomy 21 [Look, 2002]. Recently, de Pontual et al. [2007] have shown an elevation in the frequency of the RET (MIM# 164761; chromosome 10q11.2) enhancer polymorphism rs2435357 [Emison et al., 2005] in DS cases with HSCR; this association is not specific to DS and is also observed in other HSCR-associated syndromes. In this paper, we demonstrate that segregation of a common polymorphism at RET, residing on human chromosome 10q11.2, interacts with chromosome 21 and leads to the Hirschsprung disease association in Down syndrome patients.
MATERIALS AND METHODS
Samples
For nondisjunction (NDJ) analyses we used a set of 23 DS trios with HSCR matched with a set of 23 DS trios without HSCR (Table 1). Cases were matched with respect to maternal age at time of delivery (±4 years) and gender of proband; DS with HSCR individuals were matched to segment length of HSCR controls (7 long, 25 short, 4 total colonic aganglionosis and 26 unknown) in addition. These samples were also included in the analyses described in Table 2 for which we added 39 DS+HSCR trios, 7 DS-HSCR trios, 62 trios with HSCR only, and, 30 disease free control trios from the HapMap CEU. The additional DS+HSCR cases were ascertained by the Lyonnet, Borrego, Ceccherini, Hofstra, and Tam, the DS cases by the Sherman, and the additional HSCR cases by the Chakravarti laboratories.
Table 1.
Origin of non-disjunction (NDJ) and chromosome 21 gene-marker recombination analysis in Down syndrome (DS) individuals with or without Hirschsprung disease (HSCR).
| Number (%) of cases | ||
|---|---|---|
| DS with HSCR | DS without HSCR | |
| Sample size | 23 | 23 |
| Origin of NDJ | ||
| Maternal origin of NDJ | 21 (91) | 22 (96) |
| Meiosis I NDJ | 17 (74) | 18 (78) |
| Genes | Tetratype frequency | |
| SOD1 | 0.17 ±0.08 | 0.10 ±0.06 |
| IL10RB, IFNGR2, SON, CBR1 | 0.33 ±0.10 | 0.21 ±0.09 |
| TTC3 | 0.45 ±0.11 | 0.22 ±0.10 |
| TFF3 | 0.61± 0.10 | 0.96 ±0.04 |
| CSTB, PFKL | 0.64 ±0.10 | 1.00 ±0.00 |
The analyses are based on a set of 23 DS trios with HSCR matched with a set of 23 DS trios without HSCR.
Table 2.
Family-based (TDT) and case-control study of the RET+9.7 functional polymorphism rs2435357 on human chromosome 10q11.2.
| HSCR | + | + | − | − | − |
|---|---|---|---|---|---|
| DS | + | − | + | − | +/− |
| Number of probands | 62 | 62 | 30* | 30* | 60 |
| Transmission of allele T | 37 | 61 | 16 | 10 | 26 |
| Transmission of allele C | 13 | 8 | 15 | 20 | 35 |
| Transmission frequency (τ) | 0.74±0.06 | 0.88±0.04 | 0.52±0.09 | 0.33±0.09 | 0.43±0.06 |
| Proband genotype: TT | 12 | 30 | 2 | 1 | 3 |
| CT | 27 | 16 | 14 | 11 | 25 |
| CC | 23 | 16 | 14 | 18 | 32 |
| Frequency of T allele | 0.41±0.04 | 0.61±0.04 | 0.30±0.06 | 0.22±0.05 | 0.26±0.04 |
The individuals analyzed include those from Table 1 and additional samples as described in Methods.
HSCR-free controls (columns 4 and 5: total sample size of 60 matches the HSCR cases with and without DS in columns 2 and 3, respectively) do not demonstrate statistically significant differences in either transmission frequency (P= 0.15), genotype frequencies (P= 0.55) or allele frequency (P= 0.30) at rs2435357; consequently, the data were combined (column 6) for subsequent analysis.
Microsatellite genotyping
Assays for the following markers were purchased from Applied Biosystems: D21S1911, D21S1256, D21S1899, D21S1922, D21S1914, D21S263, D21S1252, D21S1255, and D21S266. Primers for the remaining markers were designed using sequences in the NCBI UniSTS database. Fluorescently labeled primers were as follows:
D21S1258: PET-CGTTTCAATATAGACCAGATAAAGG
D21S1898: 6-FAM-TGCAGGAACACTCAGTCTCTTCAG
D21S1893: 6-FAM TAACAAAATCCGCCACG
D21S1890: NED-AAAAACACTCTGAACGATTAAGG
D21S1912: PET-CCCTCATACAGATTTAAAACACAC
D21S1446: 6-FAM-ATGTACGATACGTAATACTTGACAA.
PCR products were pooled into two groups (Pool 1: D21S1256, D21S1899, D21S1914, D21S1258, D21S263, D21S1898, D21S1252; Pool 2: D21S1911, D21S1922, D21S1255, D21S1893, D21S266, D21S1890, D21S1912, D21S1446) and analyzed on the ABI Prism 3100 Genetic Analyzer. PCR amplifications and subsequent evaluations were performed according to the ABI Prism Linkage Mapping Sets v 2.5 protocol, with slight modifications as necessary to optimize PCR performance and equalize relative fluorescence intensities.
TaqMan Genotyping
All SNP assays were purchased from Applied Biosystems, run according to ABI protocol, and analyzed on the ABI Prism 7900HT Sequence Detection System.
Statistical Genetic Methods
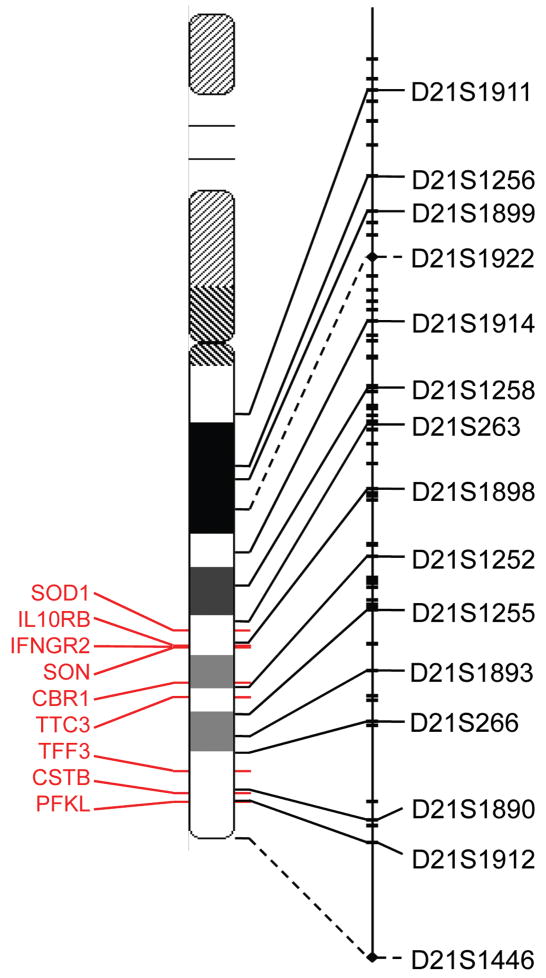
Parental origin and meiotic stage of NDJ was assessed using 15 chromosome 21 microsatellite markers (average heterozygosity: 76%) as listed in Figure 1. These were genotyped using methods described in Gabriel et al. [2002]; categorical NDJ assignment was performed as in Chakravarti and Slaugenhaupt [1987] by assuming no crossing-over between the centromere and the peri-centromeric marker D21S1911. This latter marker is located ~1,800kb from the centromere in a recombinationally suppressed region. The genetic map assumed was that combined from Kong et al. [2000] and McInnis et al. [1993] with common segments averaged.
Figure 1.
Chromosomal and genetic locations of microsatellite markers. The deCODE sex-equal genetic map of human chromosome 21 (male length 47.31cM, female length 76.40 cM, sex-equal length 61.86 cM14) is shown on the right, and microsatellite marker positions are extrapolated to their locations on the cytogenetic map in the center by solid or dashed lines. Only the microsatellite markers used in this study, spanning the entire long arm, are named: these and the remaining deCODE markers are represented by hashes along the length of the genetic map, and they are connected to the cytogenetic map by solid lines; two additional markers were genotyped and their positions, interpolated on the genetic map (based on the chromosome 21 DNA sequence), are represented by diamonds and connected to the cytogenetic map by dashed lines. Nine genes that are differentially expressed in mice with an HSCR phenotype9 are shown on the cytogenetic map (red text and lines)14,15.
The tetratype frequency (Table 1), or the frequency of recombination in the half-tetrad, is the frequency of reduction to homozygosity at a heterozygous marker in the parent in whom NDJ occurred and was estimated as described in Chakravarti and Slaugenhaupt [1987]. This was estimated from all informative parents either directly using SNPS within SOD1 (rs202445, rs4998557, and rs1041740) or TFF3 (rs225436, rs225360, and rs225362) or indirectly using all informative flanking microsatellite markers. Genes with infrequent meiotic recombination were treated as one locus. The genetic map assumed was that combined from Kong et al. [2000] and McInnis et al. [1993] with common segments averaged.
The transmission frequency (Table 2) was estimated as the transmission ratio of the enhancer deficiency allele T from all informative CT heterozygote parents.
RESULTS
To disentangle the roles of ‘21’ and ‘non-21’ genes, we ascertained four groups of subjects: 62 individuals with HSCR and DS, 62 individuals with HSCR alone and 60 controls (30 with DS alone and 30 disease-free). Where relevant, all probands were matched with respect to gender, segment-length for HSCR and maternal age at conception to within 4 years.
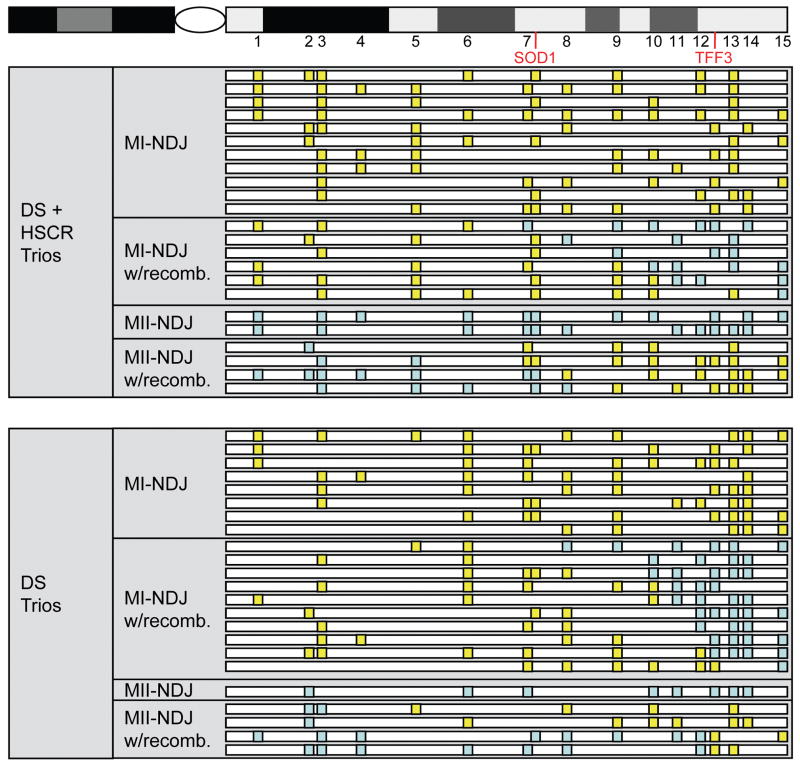
We first assessed the patterns of NDJ in a subset of 23 DS individuals with HSCR and 23 without HSCR using 15 microsatellite markers spanning the long arm of chromosome 21 (Figure 1). These results demonstrate no difference in the parental origin or meiotic stage of non-disjunction between the two groups of individuals with DS (Table 1). Our previous studies have convincingly shown that 9 genes in the syntenic mouse Dscr region are dysregulated in a mouse model of HSCR. Specifically, mice heterozygous for a Ret null mutation and homozygous for an Ednrb hypomorphic allele showed increased expression of Sod1 (MIM# 147450), Il10rb (MIM# 123889), Ifngr2 (MIM# 147569), Son (MIM# 182465), Cbr1 (MIM# 114830), Ttc3 (MIM# 602259), Cstb (MIM# 601145), Pfkl and absence of expression of Tff3 (MIM#600633) [McCallion et al., 2003]. The human orthologs of these genes are all on chromosome 21. These results imply that, in the human HSCR patient with RET or EDNRB mutations, genes on chromosome 21 may be dysregulated; this is exacerbated when DS is also present. We assessed whether these genes showed a different pattern of reduction during NDJ and thus altered genic homozygosity or heterozygosity on the NDJ chromosome. We found no statistically significant differences, but the proximal genes (SOD1 to TTC3) demonstrated a tendency to excess of reduction while the distal genes (TFF3 to PFKL) demonstrated a tendency to deficiency of reduction in probands with DS and HSCR as compared to DS individuals alone (Table 1, Figure 2, Supp. Table S1). Overall, individuals with DS, with and without HSCR, appear to be grossly identical with respect to their chromosome 21 NDJ and thus trisomic genotypes of common alleles. Admittedly, we have low statistical power to detect anything but very large differences. Consequently, larger sample sizes may yet show subtle chromosome 21 genotype differences along the lines we have indicated.
Figure 2.
Nondisjunction map for matched DS and DS+HSCR trios. The relative positions of microsatellite markers (numbered as in parentheses in Figure 1), SOD1 and TFF3 are indicated along the q arm of Chromosome 21 (top). Below, the analogous region of Chr 21 is represented for each proband, with definitive calls of reduction represented by yellow boxes and definitive calls of nonreduction represented by blue boxes for all relevant loci. Probands are further grouped according to the mode of inheritance (“with recombination” is abbreviated to w/recomb.) and phenotype.
A promising candidate for the DS-HSCR association is the common polymorphism rs2435357 at RET+9.7 [de Pontual et al., 2007]. This C→T variation arises from a hypomorphic allele within an intron 1 RET enhancer and is the mostcommon cause of short-segment HSCR with a 20-fold greater contribution to risk of HSCR than rare coding mutations [Emison et al., 2005, Grice et al., 2005]. Since the mutant allele (T) is quite common, with an average background frequency of 24% in Europe [Emison et al., 2005], it is expected to be present in many DS individuals and their parents.
Our analysis indicates that the effects of the RET enhancer variant rs2435357 are restricted to individuals with HSCR but with important differences between those with and those without DS. First, the variant shows no significant difference between disease free controls and those with DS only (P=0.30) with a background allele frequency of 0.26±0.04 (Table 2). Second, the T allele is increased both among individuals with only HSCR (0.61±0.04: P=2×10−8) and those with both DS and HSCR (0.41±0.04: P= 0.011). Lastly, and most intriguingly, the allelic (P=0.0015) and the genotypic (P=0.0028) distributions among individuals with HSCR with and without DS differ significantly. Since case control comparisons are sensitive to population stratification we assessed the rs2435357 effect using the transmission disequilibrium test (TDT) and found essentially the same results: 1) the transmission frequency was not different from expected for those with only DS (P= 0.86) or those who were disease free (P= 0.07) (Table 2); 2) both those with only HSCR (P = 1.9×10−10) and those with HSCR and DS (P = 6.9 ×10− 4) showed over-transmission of the T allele; 3) the transmission rates of the T allele between these two HSCR classes differed significantly (P = 0.042).
DISCUSSION
These results clearly demonstrate that a common RET enhancer polymorphism, not only has an etiologic role in the occurrence of HSCR but it does so in a specific manner with chromosome 21 dosage. Surprisingly, despite the increased 50-fold risk from the presence of an extra copy of chromosome 21, the RET genetic effect ‘appears’ to be attenuated since the mutant allele is less frequent (41 vs. 61%) and less frequently transmitted (74 vs. 88%). This apparent effect is due to the presence of an extra chromosome 21 increasing the risk of the C allele (relative to the T allele) or altering disease threshold. One explanation for this curious finding may be the specific negative regulation of RET protein by a chromosome 21-encoded protein. A cell culture study by Kato et al. [2000] suggests a mechanism for such an effect on RET dimerization mediated through the Cu-, Zn- superoxide dismutase (SOD1), an enzyme whose gene is on chromosome 21. This is intriguing since SOD1 is one of genes implicated as being dysregulated in a mouse model for HSCR [McCallion et al., 2003].
We conclude by raising two issues. First, although it has generally been assumed that all DS-associated phenotypes arise from genetic variation on chromosome 21, and thus the fine mapping of the DSCR [Delabar et al., 1993], the emerging data suggests that many dysregulated genes are on other chromosomes [Arron et al., 2006, Look, 2002, and this paper]. Second, genetic variation in DS has been implicitly assumed to be due to the variation between the 3 chromosome 21 alleles or haplotypes, but, it is also likely that polymorphic copy number variants on chromosome 21 may be responsible. Consequently, not all DS cases are trisomic everywhere along 21q but can segmentally harbor less than three copies and contribute to the phenotype. In other words, probing the genetic constitution of individuals with DS, in detail on chromosome 21 and the rest of the genome, may be necessary for understanding the features of Down syndrome.
Supplementary Material
Acknowledgments
We wish to thank the numerous patients and their families that have participated in our studies of Hirschsprung disease and Down syndrome over the past 20 years. We acknowledge the work of Jennifer Scott, Maura Kenton and Julie Muskett in family collection and the earlier genetic studies of Stacey (Bolk) Gabriel and Eileen Emison on DS and HSCR patients. This study was supported in part by grants from the US National Institutes of Health (HD28088 to A.C. and HD38979 to S.S.), the “Holes for Hirschsprung” fund-raiser, the Fondation Jérôme Lejeune and the Agence Nationale de la Recherche (HirGenet).
References
- Arron JR, Winslow MM, Polleri A, Chang CP, Wu H, Gao X, Neilson JR, Chen L, Heit JJ, Kim SK, Yamasaki N, Miyakawa T, Francke U, Graef IA, Crabtree GR. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- Chakravarti A, Slaugenhaupt SA. Methods for studying recombination on chromosomes that undergo nondisjunction. Genomics. 1987;1:35–42. doi: 10.1016/0888-7543(87)90102-9. [DOI] [PubMed] [Google Scholar]
- Chakravarti A, Lyonnet S. The Metabolic and Molecular Bases of Inherited Disease. 8. New York: McGraw-Hill; 2001. Hirschsprung Disease. Chapter 251. [Google Scholar]
- Delabar JM, Theophile D, Rahmani Z, Chettouh Z, Blouin JL, Prieur M, Noel B, Sinet PM. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur J Hum Genet. 1993;1:114–124. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- de Pontual L, Pelet A, Clement-Ziza M, Trochet D, Antonarakis SE, Attie-Bitach T, Beales PL, Blouin JL, Dastot-Le Moal F, Dollfus H, Goossens M, Katsanis N, Touraine R, Feinjold J, Munnich A, Lyonnet S, Amiel J. Epistatic interactions with a common hypomorphic RET allele in syndromic Hirschsprung disease. Hum Mut. 2007;28:790–796. doi: 10.1002/humu.20517. [DOI] [PubMed] [Google Scholar]
- Emison ES, McCallion AS, Kashuk CS, Bush RT, Grice E, Lin S, Portnoy ME, Cutler DJ, Green ED, Chakravarti A. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–863. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- Epstein CJ. The Consequences of Chromosome Imbalance: Principles, Mechanisms, and Models. Cambridge, UK: Cambridge Univ Press; 1986. [Google Scholar]
- Gabriel SB, Salomon R, Pelet A, Angrist M, Amiel J, Fornage M, Attie-Bitach T, Olson JM, Hofstra R, Buys C, Steffan J, Munnich A, Lyonnet S, Chakravarti A. Segregation at three loci explains familial and population risk in Hirschsprung disease. Nat Genet. 2002;31:89–93. doi: 10.1038/ng868. [DOI] [PubMed] [Google Scholar]
- Grice EA, Rochelle ES, Green ED, Chakravarti A, McCallion AS. Evaluation of the RET regulatory landscape reveals the biological relevance of a HSCR-implicated enhancer. Hum Mol Genet. 2005;14:3837–3845. doi: 10.1093/hmg/ddi408. [DOI] [PubMed] [Google Scholar]
- Kato M, Iwashita T, Takeda K, Akhand AA, Liu W, Yoshihara M, Asai N, Suzuki H, Takahashi M, Nakashima I. Ultraviolet light induces redox reaction-mediated dimerization and superactivation of oncogenic Ret tyrosine kinases. Mol Biol Cell. 2000;11:93–101. doi: 10.1091/mbc.11.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Look AT. A leukemogenic twist for GATA1. Nat Genet. 2002;32:83–84. doi: 10.1038/ng960. [DOI] [PubMed] [Google Scholar]
- McCallion AS, Emison ES, Kashuk CS, Bush RT, Kenton M, Carrasquillo MM, Jones KW, Kennedy GC, Portnoy ME, Green ED, Chakravarti A. Genomic varation in multigenic traits: Hirschsprung disease. Cold Spring Harb Symp Quant Biol. 2003;68:373–381. doi: 10.1101/sqb.2003.68.373. [DOI] [PubMed] [Google Scholar]
- McInnis MG, Chakravarti A, Blaschak J, Petersen MB, Sharma V, Avramopoulos D, Blouin JL, Konig U, Brahe C, Matise TC, Warren AC, Talbot CC, Jr, Van Broeckhoven C, Litt M, Antonarakis SE. A linkage map of human chromosome 21:43 PCR markers at average intervals of 2.5 cM. Genomics. 1993;16:562–571. doi: 10.1006/geno.1993.1231. [DOI] [PubMed] [Google Scholar]
- Wisniewski KE, Wisniewski HM, Wen GY. Occurrence of neuropathological changes and dementia of Alzheimer’s disease in Down’s syndrome. Ann Neurol. 1985;17:278–282. doi: 10.1002/ana.410170310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.