Abstract
Multiple sclerosis (MS) is an inflammatory central nervous system (CNS) disorder characterized by T cell-mediated demyelination. In MS, prolonged T cell survival and increased T cell proliferation have been linked to disease relapse and progression. Recently, the autophagy-related gene 5 (Atg5) has been shown to modulate T cell survival. In this study, we examined the expression of Atg5 using both a mouse model of autoimmune demyelination as well as blood and brain tissues from MS cases. Quantitative real-time PCR analysis of RNA isolated from blood samples of experimental autoimmune encephalomyelitis (EAE) mice revealed a strong correlation between Atg5 expression and clinical disability. Analysis of protein extracted from these cells confirmed both upregulation and post-translational modification of Atg5, the latter of which was positively correlated with EAE severity. Analysis of RNA extracted from T cells isolated by negative selection indicated that Atg5 expression was significantly elevated in individuals with active relapsing-remitting MS compared to non-diseased controls. Brain tissue sections from relapsing-remitting MS cases examined by immunofluorescent histochemistry suggested that encephalitogenic T cells are a source of Atg5 expression in MS brain samples. Together these data suggest that increased T cell expression of Atg5 may contribute to inflammatory demyelination in MS.
Keywords: Atg5, autophagy, multiple sclerosis, T cell, autoimmune, neuroinflammation, encephalomyelitis, apoptosis, CNS, EAE
Introduction
Multiple sclerosis (MS) is a chronic demyelinating disease of the CNS. Many lines of experimental evidence indicate that MS is an autoimmune disorder characterized by the generation of T cells directed against myelin proteins that drive neuropathology and clinical development of this disease.1,2 Onset of MS typically occurs during early adulthood, making MS the most common neurological disease affecting people under the age of 30. The clinical presentation of MS is heterogeneous. In the majority of cases MS develops in an episodic fashion with phases of clinical disease followed by recovery. In this form of MS, called relapsing-remitting MS (RRMS), white matter lesions can progress toward permanent tissue injury associated with neural loss and clinical disability. Over time RRMS patients may develop chronic lesions that promote irreversible axonal injury resulting in conversion to secondary progressive MS (SPMS) characterized by minimal or no intermittent recovery of function.1
Current data support changes in the expansion and/or survival of autoreactive T cells as a primary event leading to inflammatory cascades mediating myelin loss and clinical relapse in MS. Accumulating evidence suggests that elevated expression of antiapoptotic factors in auto-reactive T cells prolongs their survival, thereby delaying the resolution of CNS inflammation.2–4 Recently, factors involved in the process of autophagy, known also as macroautophagy, have been implicated in a number of human disorders, including cancer, neurodegenerative diseases and infections.5–8 Autophagy is a cellular degradation process for the removal of damaged or surplus intracytosolic organelles through fusion with lysosomes.9,10 Of potential interest to autoimmunity, molecules involved in autophagy have also been found to profoundly affect T cell homeostasis.11–13 Specifically, autophagy-related gene-5 (Atg5)-deficient T lymphocytes exhibit multiple functional defects including reduced numbers in vivo, enhanced T cell apoptosis, and an inability to undergo T cell receptor-induced proliferation.13 Further studies on T cells showed that Atg5 post-translational cleavage can also induce apoptosis and imperil the viability of T cells.14
Given this newly identified role for autophagy in the regulation of T cells, we sought to determine whether the expression and post-translational modification of Atg5 were altered during a model of T cell-mediated experimental autoimmune encephalomyelitis (EAE) and in MS patients.
Results
Expression of Atg5 in peripheral blood correlates with clinical severity of EAE in mice
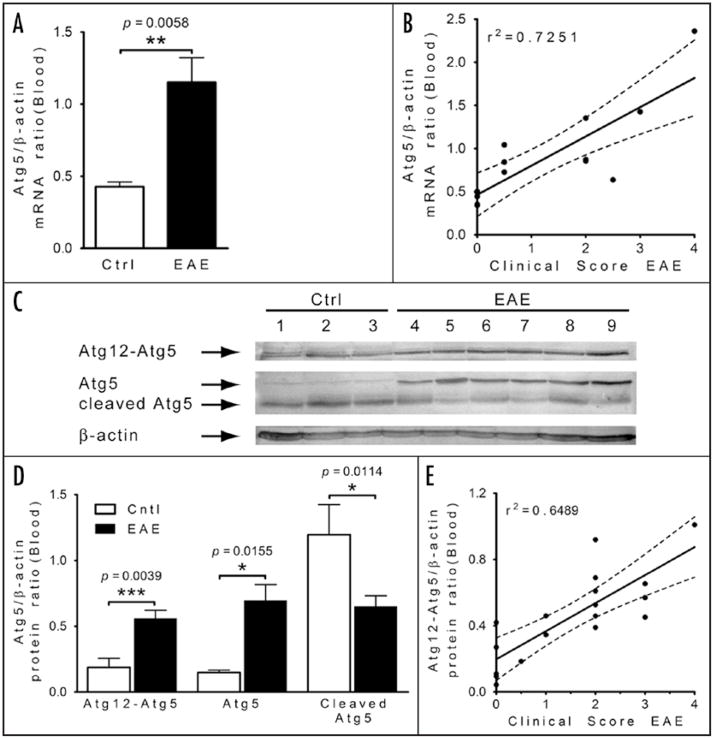
To determine whether expression of Atg5 in circulating immune cells is altered during inflammatory demyelination, we first examined blood samples from mice that had been immunized with MOG peptide and developed clinical signs of encephalomyelitis. Comparison of Atg5 mRNA expression by PCR between control (CFA treated) mice (n = 5) and EAE mice (n = 10) revealed a significant increase in the expression of Atg5 in blood cells associated with EAE (p < 0.01; Fig. 1A). Because we had collected samples from mice with varying degrees of clinical EAE, this allowed us to examine the expression of Atg5 mRNA from each subject sample relative to their clinical EAE score. This analysis revealed a strong positive correlation (r2 = 0.7251) between the expression of Atg5 in blood of EAE mice and the degree of their physical disability (Fig. 1B).
Figure 1.
Enhanced Atg5 expression in peripheral blood in mice correlates with clinical severity of EAE. (A) Comparative analysis of Atg5 mRNA expression by quantitative PCR of samples isolated from purified T cells of EAE (n = 10) and control (Ctrl) mice (n = 5). Student’s t-test used to derive the p value indicated on the graph. (B) Correlation study between the Atg5/β-actin ratio and clinical scores in EAE mice. Linear regression; r2 = 0.7251, p = 0.0001. (C) Western blot analysis of different forms of Atg5 (complex Atg12-Atg5, Atg5 and cleaved form of Atg5) isolated from T cells from Ctrl and EAE mice. Loading control was performed by re-probing the blot with an anti β-actin antibody. (D) Densitometry comparison of Atg12-Atg5 conjugates, Atg5 and cleaved form of Atg5 relative to β-actin as determined by western blot analysis (in C). Student’s t-test was used to derive the p value indicated on the graph, n = 5 for Ctrl and n = 13 for EAE mice. Student’s t-test used to derive the p value indicated on the graph. (E) Correlation analysis between the Atg12-Atg5/GAPDH ratio and clinical scores in EAE mice. Linear regression; r2 = 0.6489, p = 0.0001.
Next, we sought to determine the state of Atg5 protein in the blood samples by western blot analysis since the electrophoretic migration pattern of Atg5 can reflect differences in its post-translational state that can indicate a possible function of Atg5. Among blood samples from all cases, we resolved several dense Atg5-reactive bands; however, the pattern of these bands differed depending upon whether the animal had developed EAE with different clinical disability (Fig. 1C). Our first observation was that the intensity of the highest molecular weight band, representing an Atg12-Atg5 complex, was significantly increased among samples from EAE (n = 13) versus control mice (n = 5) (Fig. 1C and D). At lower molecular weights, differences in the apparent levels of free form of Atg5 and proteolytically cleaved Atg5 were also evident: Atg5 at protein expression level was increased among EAE mice and the proportion of the cleaved form of Atg5 was also reduced relative to controls (Fig. 1C and D). This increase in Atg5 protein expression, which is predicted to be in a complex with Atg12, also exhibited a correlation with clinical disability among EAE mice (r2 = 0.6489; Fig. 1E). These results indicate that transcriptional and translational upregulation of Atg5 occurs in the peripheral blood during inflammatory demyelination. These data also provide a significant post-translational distinction of Atg5 in EAE versus control mice. Moreover, this increase in Atg5 is also associated with reduced proapoptotic Atg5 cleavage, differences consistent with our current understanding of Atg5 function and altered expression among proliferative responses. Hence, significant differences in blood levels of Atg5 mRNA and protein were observed, and correlated with the clinical severity of EAE, suggesting a potential role in disease development.
Atg5 mRNA expression is elevated in T cells of RRMS patients
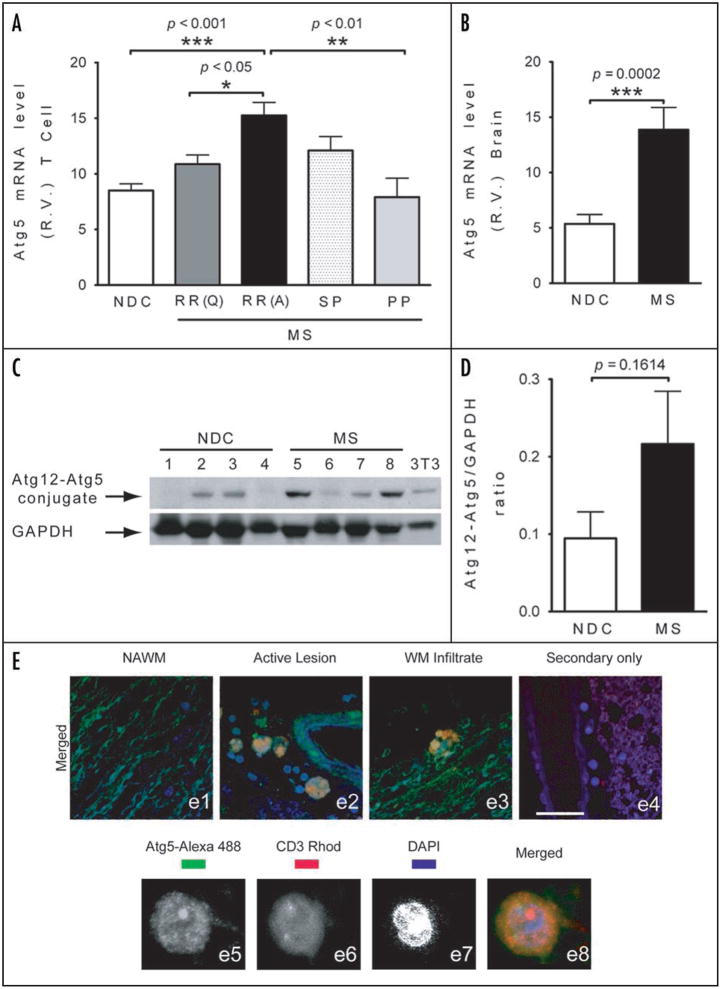
To ascertain whether changes in Atg5 expression observed in EAE mice were also present in humans with MS, we next performed qRT-PCR-based analyses on Atg5 mRNA using blood samples drawn from MS patients and nondiseased controls. Whole blood samples were collected from normal control individuals (n = 10) as well as from patients with a variety of clinically diagnosed categories of MS: SPMS (n = 14), RRMS (subdivided into active, n = 18, or quiescent predicated on whether patients had experienced a relapse during blood draw or in the year prior to blood draw, n = 19), and PPMS (n = 5). Initial screening of RNA extracted from whole blood did not reveal a significant difference in the expression of Atg5 between normal control specimens and those with MS among all clinical subtypes (data not shown). Based upon our findings in MOG-induced EAE in mice, a model of T cell mediated myelin injury, this finding was unexpected. However when we analyzed mRNA isolated from T cells purified from these blood samples, a significant difference in Atg5 expression was unveiled among active RRMS patients (Fig. 2A). The difference in Atg5 distinguished this cohort of active RRMS patients from those with quiescent RRMS, as well as normal controls and indicates that elevated Atg5 expression in peripheral T cells of MS patients is associated with an active clinical state of MS.
Figure 2.
Increased Atg5 expression in peripheral blood T cells and in the brain of MS subjects. (A) Comparative analysis in relative values (R.V.) of Atg5 mRNA expression by quantitative PCR of samples isolated from purified T cells of NDC and patients with different categories of MS. ANOVA revealed significant differences between the groups (p = 0.0003), and a post-hoc Tukey’s test revealed the differences denoted in the graph. (B) Comparative analysis of Atg5 mRNA expression by quantitative PCR of samples isolated from brain specimens of MS (n = 6) and controls (n = 14). Student’s t-test was used to derive the p value indicated on the graph (p = 0.0002). (C) Western blot analysis of Atg12-Atg5 complex isolated from NDC subjects (1–4) and MS (5–8) brain specimens, with 3T3 cells providing a positive control. Protein loading was assessed by re-probing the blot with an antiGAPDH antibody. (D) Densitometric comparison of Atg12-Atg5 conjugates relative to GAPDH as determined by western blot analysis from MS samples (n = 4) and controls (n = 4) (in C). Unpaired student’s t-tests were performed to obtain the p values indicated on the graph. (E) Immunohistofluorescent detection of Atg5 protein in MS brain. (e1–e4) Merged images of brain sections from MS patients stained with antibodies against Atg5 (green), CD3 expression (red) and DAPI (blue). (e1) Normal appearing white matter (NAWM) of a MS patient. (e2) Active perivascular lesion in the brain of the same MS patient. (e3) Image of T cell (CD3+) infiltrating the white matter (WM) of another MS patient. (e4) Negative control of MS brain, using only the secondary fluorescent antibodies and DAPI. Scale Bar, 50 μm. (e5–e8) magnified images of Atg5 expression (green, e5) in a CD3+ T cell (red, e6) located in a blood vessel close to an injured area in the brain of a MS subject, with nucleus reacted with DAPI (blue, e7) and the merged image (e8).
Atg5 expression in postmortem brain tissue from patients diagnosed with MS
Given that our analyses of EAE and human blood specimens identified a significant increase in Atg5 expression, we then sought to determine whether we could detect expression of Atg5 in postmortem brain specimens from MS cases. Consistent with analysis of peripheral blood samples, qRT-PCR analysis of mRNA from brain samples from SPMS patients identified a significant increase in transcript levels for Atg5 relative to age-matched cases from nondiseased control (NDC) patients (Fig. 2B). To determine whether this change in gene expression also resulted in an increase in Atg5 protein expression, we also performed western blot analyses on protein lysates from lesions localized in brain tissues from different subjects (Table 1, Fig. 2C and D). This also afforded us the opportunity to assess the presence of the Atg5 protein and its presumptive functional state based on migration and the formation of a complex with Atg12. An Atg12-Atg5 immunoreactive band was consistently observed, among MS cases, whereas only weak or no bands were seen in the NDC samples (Fig. 2C). Quantification of bands from western blot analysis showed increased level of Atg12-Atg5 formation in brain samples from MS cases, relative to age-matched control samples but the difference was not statistically significant (Fig. 2D). A larger sample size would increase the statistical power of the analysis. Moreover, both expression of free Atg5 and its cleaved form were not changed between the MS and NDC samples (data not shown). However, using anti-CD3 antisera (to identify T cells) and colabeling with Atg5, we performed immunohistochemical analysis of T cells present in brain tissues of MS cases to examine whether we could detect Atg5 in T cells in postmortem brain tissue (Fig. 2E). Since Atg5 is a ubiquitously expressed protein, we observed weak and diffuse labeling throughout the brain as seen by fluorescent microscopy (Fig. 2E; e1). However, intense Atg5 labeling was detected in cells that were also labeled with CD3 (Fig. 2E; e2 and e3). Of particular interest were areas of myelin loss, as determined by luxol fast blue staining in adjacent tissue sections, wherein Atg5+/CD3+ cells were noted at perivascular sites (Fig. 2E; e2). When examined by confocal microscopy these CD3+/Atg5+ cells exhibited a bright and punctate pattern of cytoplasmic labeling (Fig. 2E; e5–e8). Positive immunolabeling disappeared when primary antisera was omitted from the staining protocol (Fig. 2E; e4). These data show that Atg5 is expressed at higher levels in MS brain, and that Atg5 is detectable in T cells in MS brain tissue sections.
Table 1.
Human postmortem brain tissue samples
| Case | Age | Sex | PMI | Cause of death/diagnosis |
|---|---|---|---|---|
| Control cases | ||||
| 1 | 28 | M | 37.5 | Myocardial Infarction |
| 2 | 41 | M | 39 | Myocardial Infarction |
| 3 | 29 | M | 24 | Motor Cycle Accident |
| 4 | 89 | M | 9 | Heart Attack |
| MS cases | ||||
| 5 | 65 | F | 29.5 | Multiple Sclerosis |
| 6 | 43 | M | 28 | Multiple Sclerosis |
| 7 | 76 | M | 25 | Multiple Sclerosis |
| 8 | 61 | M | 10 | Multiple Sclerosis |
| 9 | 48 | F | 3.3 | Multiple Sclerosis |
| 10 | 64 | F | 17.3 | Multiple Sclerosis |
Characteristics of human brain tissue samples obtained from the Human Brain and Spinal Fluid Resource Center (VA West Los Angeles Healthcare Center, Los Angles, CA) that were used for real time-PCR and western blot analyses. Postmortem interval (PMI) is presented in hours between time of patient death and necropsy.
Discussion
Here we report for the first time an association between autophagy and immune- mediated demyelination. We found that both elevated mRNA and protein expression of the autophagy-related gene Atg5 in blood samples were correlated with the clinical severity of EAE, an animal model of MS. We also determined that circulating T cells in peripheral blood of RRMS patients express heightened levels of Atg5, and Atg5 was detected in T cells in the brain tissues of MS cases. In our analysis we determined that Atg5 expression increased significantly among patients diagnosed with RRMS experiencing an active relapse. This finding is consistent with clonal expansion of autoreactive T cells during active relapses of clinical disease.15,16 The lack of elevation in patients with quiescent RRMS is consistent with the absence of disease activity in these patients. The relative paucity of Atg5 expression in T cells among SPMS and PPMS patients compared to those with active RRMS is intriguing given that the effectiveness of immune therapies for MS is limited to RRMS.17 In addition, it is thought that the neuropathology of PPMS, a more aggressive from of this disease, is not associated with T cell-mediated myelin injury and may therefore be distinct from RRMS which involves resurgent T cell-mediated immune pathogenesis during clinical relapses.17,18 These data indicate a potential function for Atg5, probably in T cell physiology related to autoimmune processes and MS.
Development of autoimmunity has been proposed to be a consequence of inadequate homeostatic regulation of autoreactive T cells that contribute to disease. In the context of our study, previous work by others has shown that there exists crosstalk between autophagy and apoptosis signaling in cells.19 Under certain conditions, such as starvation stress, autophagy is considered a potent survival mechanism when apoptosis is prevented.19–21 For example, autophagy has been implicated in bystander T cell death of HIV-infected CD4+ T cells following exposure use CXCR4 (C-X-C chemokine Receptor 4) to trigger autophagy in uninfected T cells.22 These findings suggest that autophagy may play an important role in the regulation of homeostasis of T lymphocytes by promoting both survival and proliferation.12,13,23 Thus the process of autophagy may play an important role in homeostatic control of the immune system in human autoimmune disease but further studies need to be accomplished for getting functional effects of autophagy in MS. Additionally, studies showed the presence of cleaved form of Atg5, which in T cells has been shown by others to be associated with a switch to an apoptosis cell state from autophagic cell state.14 Thus, our findings suggest the presence of autophagy Atg12-Atg5 complex in blood cells or particularly in T cells from EAE and MS subjects consecutively, may play a role in MS.
What is the function of Atg5 in T cells and MS? Both Atg5 and Atg12 are key factors in regulating the formation of autophagic vacuoles (AV) within eukaryotic cells.24,25 Autophagy was originally described in yeast as a process activated by starvation stress. Additional reports examining cells lacking the Atg5 or Atg7 genes have demonstrated that autophagy plays pivotal roles in cell survival.26 Nevertheless, an important change in Atg5 in circulating cells, especially T cells, may relate to extending T cell survival and/or promoting T cell proliferation during active disease. Indeed, we observed a strong correlation between Atg5 protein complex formation in blood and clinical severity in EAE. We speculate that the changes in Atg5 expression measured in the blood of EAE mice and in T cells of RRMS patients are related to protracted survival of T cells and future studies may offer further insight to the potential relationship between Atg5 and the generation or propagation of T cells related to autoimmunity in MS. In a broader context, determining whether changes in autophagy are a common feature of autoimmune diseases in general and determining if there is a specific function for Atg5 within specific T cell sub-populations will require further studies.10,27,28 A T cell transgenic model of Atg5 will further help to see whether there is a potential development of EAE and how T cells survive versus wild-type mice. Recent evidence based on Atg5-deficient mice, generated by germline deletion, has revealed a central role for Atg5 in T cell function: T cell survival and expansion in response to antigen stimulation was compromised in Atg5KO mice.12,13 A previous report had also determined a potential role for autophagy in antigen processing.29 Additionally, deletion of Atg5 manifests intraneuronal inclusion formation accompanied by a spontaneous neurodegenerative phenotype.30 The strong immune and neurological phenotypes of Atg5-deficient mice make direct study of EAE in Atg5KO mice intractable, and thus it is necessary to use conditional knockout mice for Atg5 in T cells in order to examine the functional role of Atg5 in autoimmune disease.
In addition to our findings on T cells, recent data also suggest a potential role for Atg5 in B cells. In the context of autoimmunity and MS, B cells function as sensors and regulators of the immune response, which has strengthened the view that B cells and autoantibodies are fundamental for activating T cells and/or mediating tissue injury.31–35 It has been shown recently that Atg5 is required for the development and the maintenance of B cells. Although in our study we focused on T cells, it is tempting to speculate on a role for Atg5 in B cell development in the progression of EAE and MS.36 Therefore, future studies should analyze Atg5 and its post-translational forms in different subpopulation of blood cells in EAE and different subcategories of MS subjects.
In summary, we report here changes in expression of the autophagy-related gene Atg5, which are correlated with immune-mediated myelin injury in mice, and are associated with active relapse in patients with RRMS. In future studies it would be important to determine whether enhanced Atg5 expression is specific to MS or is perhaps a common feature of other autoimmune disorders, such as rheumatoid arthritis or systemic lupus erythematosus. Better understanding of mechanisms underlying autophagy and T cell function may provide additional therapeutic insights into MS and other auto-immune diseases.
Materials and Methods
Animal subjects and experimental autoimmune encephalomyelitis
Mice
Wild-type C57Bl/6 mice were immunized with 100 μg of myelin oligodendrocyte glycoprotein (MOG) peptide (amino acids 35–55) that was emulsified in complete Freund’s adjuvant (CFA) as described and employed in previous studies.37–39 Mice were evaluated daily for signs of EAE according to the following scale of clinical disability: 0—no signs of EAE; 1—flaccid tail; 2—hindlimb paresis; 3—unilateral hindlimb paralysis; 4—bilateral hindlimb paralysis; 5—moribund.
Human subjects and samples
Blood cells
Blood samples were collected from patients of the Dalhousie Multiple Sclerosis Research Unit (Halifax, Nova Scotia, Canada),40,41 as previously described.42 Blood for T cell isolation was drawn in an 8 ml sodium citrate CPT BD Ficoll gradient blood vacutainer while whole peripheral blood samples were collected into Paxgene RNA tubes. Highly purified T cells were isolated from whole blood by negative selection using the RosetteSepT (T cells) enrichment cocktail (Stem Cell Technologies, Vancouver, British Columbia).
Brain tissue
Postmortem tissues from six individuals with premortem clinical diagnosis of secondary progressive MS and neuropathological verification of MS and four without neurological symptoms or significant neuropathology on autopsy (Table 1), were acquired from The Human Brain and Spinal Fluid Resource Center (VA West Los Angeles Healthcare Center, Los Angeles, California).
RNA isolation
Animal samples
RNA was isolated from peripheral blood samples of EAE mice, which was obtained using cardiac puncture and a heparinized needle.37
Human samples
Isolation of RNA from T cell populations isolated from individual cases was performed using previously described protocols and procedures.42 For brain RNA, isolation and quantification of specific RNA was performed as described elsewhere.5
Reverse transcription and qRT-PCR
Atg5 primers were TTT GCA TCA CCT CTG CTT TC and TAG GCC AAA GGT TTC AGC TT, the double-labeled probe was CCA CTG CCA TCA TTA AAC CTC AGC TG; 18S RNA was used as the endogenous controls using the primers/probe described.5 β2-microglobulin (B2M) was the endogenous reference gene whose expression was assessed using TaqMan B2M Control Reagents Kit (Applied Biosystems). Mouse primers were GAC AAA GAT GTG CTT CGA GAT GTG (forward) and GTA GCT CAG ATG CTC GCT CAG (reverse).
All amplification was done in duplicate. Threshold cycle (Ct) scores were averaged for subsequent calculations of relative expression values. Data were generated using equipment and software from Stratagene (La Jolla, California) for the brain samples, and MJ Research Inc., (Waltham, Massachusetts) for blood and T cell samples. Data were exported into an Excel/SPSS spreadsheet for further statistical analysis. Quantification of gene expression was made relative to endogenous reference genes by calculating the differences in Ct (ΔCt) and relative values determined by 2(−ΔΔCt).
Western blot analysis
Brain tissues were homogenized in a mini bead beater, protein concentrations were measured using the acid bicinchoninic protein assay and samples were separated on NuPAGE 4%–12% Bis-Tris acrylamide gradient gels (Invitrogen) and transferred onto electrophoretically to HY-bond™ PVDF membranes (Invitrogen). Two different antibodies were used for Atg5 immunoblotting; rabbit polyclonal Atg5 (SO4),43 for human samples, clone FL-275 for mouse samples (Santa Cruz, California), followed by secondary antibody (1:10,000 HRP conjugated anti-rabbit; GE Healthcare, Little Chalfont, United Kingdom). Blots were developed with 1:1 solution of Super Signal West Pico Chemiluminescent Substrate and Luminol/Enhancer (Thermo Fisher Scientific, Rockford, Illinois). Blots for loading control were stripped subsequently using ReStore® western Blot stripping buffer (Thermo Fisher Scientific) then re-probed for GAPDH (Millipore, Billerica, Massachusetts) or β-actin (Sigma-Aldrich, St. Louis, Missouri). The optical density of bands was quantitated using the ImageJ v.1.38 software and the ratios to GAPDH or β-actin were analyzed and then expressed as values for the graphs.
Immunohistochemistry
Five-μm sections of formalin-fixed, paraffin embedded tissues were mounted on charged slides, then were deparaffinized and hydrated. The antigenic sites were unmasked with a 0.01 M citrate buffer (pH 6) in a steamer. Endogenous peroxidase was blocked by a 30-min treatment with 3% H2O2 in 10% methanol and nonspecific binding was blocked with 10% goat serum/0.1% Triton X-100 in PBS. Slides were incubated overnight with a chicken polyclonal anti-Atg5 (GenWay, San Diego, California) and rabbit polyclonal anti-CD3 (Biocare Medical, Concord, California) in PBS/10% goat serum block solution, under low agitation, at 4°C. Immunoreactivity was visualized using Alexa Fluor 488 (goat anti-chicken, green) and Rhodamine Red™ -X (goat antirabbit, red) conjugated secondary antisera. Sections from MS neurospecimens (n = 6) were counterstained with DAPI and images were acquired using a Rainbow Radiance 2100 Laser Scanning Confocal system attached to a Nikon TE2000-U inverted microscope (BioRad-Zeiss). Optical image slices (0.2 μm interval step slices) were acquired using Laser Sharp 2000 software and then imported and further analyzed with Image J (NIH Imaging; http://rsb.info.nih.gov/ij) and Image Pro Plus 3DS (Media Cybernetics, Silver Spring, Maryland). Negative controls were performed by omitting the primary antibodies.
Statistical analysis
Statistical analysis was carried out using GraphPad Prism (GraphPad Software, San Diego, California) software. Results are shown as the mean ± SEM. For two groups, data were compared by Student’s t-test while for multiple groups ANOVAs were performed, and if significant, post-hoc comparisons were performed using Tukey’s test to assess potential differences between patient groups. Significance was assessed at p < 0.05.
Acknowledgments
We are grateful for the generous gift of the Atg5 antibody (SO4) from Dr. N. Mizushima (Department of Physiology and Cell Biology; Tokyo Medical and Dental University, Japan). This work was supported by grants from the National Institutes of Health (AI-27028 to J.L.W; DA024467, MH072477 and MH062261 to H.S.F; and NRSA F32 NS048767 to M.A), Genome Canada (G.S.R), MS Society of Canada Post-Doctoral Fellowship (ALOH), MS Society of Canada Studentship (C.S.M), a Career Transition Fellowship from the National Multiple Sclerosis Society (NMSS:TA 3021A1/1) and startup funds from the University of Connecticut (both to S.J.C). We thank Michelle Zandonnati for technical assistance. We also thank the specimen repositories for brain specimens, obtained from the Human Brain and Spinal Fluid Resource Center, VA West Los Angeles Healthcare Center, which is sponsored by NINDS/NIMH, NMSS and the Department of Veterans Affairs. This is manuscript # 19308 from The Scripps Research Institute.
Abbreviations
- Atg
autophagy-related gene
- AV
autophagy vacuoles
- EAE
experimental autoimmune encephalomyelitis
- MS
multiple sclerosis
- RRMS
relapsing-remitting MS
- SPMS
secondary progressive MS
- PPMS
primary progressive MS
- qRT-PCR
quantitative real time-PCR
- B2M
β2-microglobulin
- NDC
non-diseased control
- NAWM
normal appearing white matter
- WM
white matter
References
- 1.Todaro M, Zeuner A, Stassi G. Role of apoptosis in autoimmunity. J Clin Immunol. 2004;24:1–11. doi: 10.1023/B:JOCI.0000018057.89066.c6. [DOI] [PubMed] [Google Scholar]
- 2.Trapp BD, Nave KA. Multiple sclerosis: an immune or neurodegenerative disorder? Annual review of neuroscience. 2008;31:247–69. doi: 10.1146/annurev.neuro.30.051606.094313. [DOI] [PubMed] [Google Scholar]
- 3.Hebb AL, Moore CS, Bhan V, Robertson GS. Targeting apoptosis to treat multiple sclerosis. Curr Drug Discov Technol. 2008;5:75–7. doi: 10.2174/157016308783769432. [DOI] [PubMed] [Google Scholar]
- 4.Zehntner SP, Bourbonniere L, Moore CS, Morris SJ, Methot D, St Jean M, Lacasse E, Hebb AL, Robertson GS, Durkin J, Gillard JW, Owens T. X-linked inhibitor of apoptosis regulates T cell effector function. J Immunol. 2007;179:7553–60. doi: 10.4049/jimmunol.179.11.7553. [DOI] [PubMed] [Google Scholar]
- 5.Alirezaei M, Kiosses WB, Flynn CT, Brady NR, Fox HS. Disruption of Neuronal Autophagy by Infected Microglia Results in Neurodegeneration. PLoS ONE. 2008;3:2906. doi: 10.1371/journal.pone.0002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alirezaei M, Kiosses WB, Fox HS. Decreased neuronal autophagy in HIV dementia: a mechanism of indirect neurotoxicity. Autophagy. 2008;4:963–6. doi: 10.4161/auto.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–7. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 8.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–55. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 9.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nature reviews. 2007;7:767–77. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, Jin S, Lu B. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol. 2006;177:5163–8. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- 12.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pua HH, He YW. Maintaining T lymphocyte homeostasis: another duty of autophagy. Autophagy. 2007;3:266–7. doi: 10.4161/auto.3908. [DOI] [PubMed] [Google Scholar]
- 14.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–32. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 15.Lublin FD, Baier M, Cutter G. Effect of relapses on development of residual deficit in multiple sclerosis. Neurology. 2003;61:1528–32. doi: 10.1212/01.wnl.0000096175.39831.21. [DOI] [PubMed] [Google Scholar]
- 16.Vollmer T. The natural history of relapses in multiple sclerosis. J Neurol Sci. 2007;2561:5–13. doi: 10.1016/j.jns.2007.01.065. [DOI] [PubMed] [Google Scholar]
- 17.Lin VW, Cardenas DD. Spinal cord medicine: principles and practice. New York: Demos; 2003. [Google Scholar]
- 18.Furlan R, Rovaris M, Martinelli Boneschi F, Khademi M, Bergami A, Gironi M, Deleidi M, Agosta F, Franciotta D, Scarpini E, Uccelli A, Zaffaroni M, Kurne A, Comi G, Olsson T, Filippi M, Martino G. Immunological patterns identifying disease course and evolution in multiple sclerosis patients. J Neuroimmunol. 2005;165:192–200. doi: 10.1016/j.jneuroim.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 19.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 20.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–12. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 21.Sarkar S, Rubinsztein DC. Huntington’s disease: degradation of mutant huntingtin by autophagy. The FEBS journal. 2008 doi: 10.1111/j.1742-4658.2008.06562.x. [DOI] [PubMed] [Google Scholar]
- 22.Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M, Codogno P, Biard-Piechaczyk M. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J Clin Invest. 2006;116:2161–72. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu B, Capan E, Li C. Autophagy induction and autophagic cell death in effector T cells. Autophagy. 2007;3:158–9. doi: 10.4161/auto.3637. [DOI] [PubMed] [Google Scholar]
- 24.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 25.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–6. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 26.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–48. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 27.Lleo A, Invernizzi P, Selmi C, Coppel RL, Alpini G, Podda M, Mackay IR, Gershwin ME. Autophagy: highlighting a novel player in the autoimmunity scenario. J Autoimmun. 2007;29:61–8. doi: 10.1016/j.jaut.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, Targan SR, Ippoliti AF, Bernard EJ, Mei L, Nicolae DL, Regueiro M, Schumm LP, Steinhart AH, Rotter JI, Duerr RH, Cho JH, Daly MJ, Brant SR. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Munz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–6. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 30.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 31.Browning JL. B cells move to centre stage: novel opportunities for autoimmune disease treatment. Nature reviews. 2006;5:564–76. doi: 10.1038/nrd2085. [DOI] [PubMed] [Google Scholar]
- 32.Dalakas MC. Invited article: inhibition of B cell functions: implications for neurology. Neurology. 2008;70:2252–60. doi: 10.1212/01.wnl.0000313840.27060.bf. [DOI] [PubMed] [Google Scholar]
- 33.Dalakas MC. B cells as therapeutic targets in autoimmune neurological disorders. Nature clinical practice. 2008;4:557–67. doi: 10.1038/ncpneuro0901. [DOI] [PubMed] [Google Scholar]
- 34.Hasler P, Zouali M. B lymphocytes as therapeutic targets in systemic lupus erythematosus. Expert opinion on therapeutic targets. 2006;10:803–15. doi: 10.1517/14728222.10.6.803. [DOI] [PubMed] [Google Scholar]
- 35.Shlomchik MJ, Craft JE, Mamula MJ. From T to B and back again: positive feedback in systemic autoimmune disease. Nature reviews. 2001;1:147–53. doi: 10.1038/35100573. [DOI] [PubMed] [Google Scholar]
- 36.Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, Mizushima NN, Iwasaki A, He YW, Swat W, Virgin HWt. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–14. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 37.Moore CS, Earl N, Frenette R, Styhler A, Mancini JA, Nicholson DW, Hebb AL, Owens T, Robertson GS. Peripheral phosphodiesterase 4 inhibition produced by 4-[2-(3,4-Bis-difluoromethoxyphenyl)-2-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)-phenyl]-ethyl]-3-methylpyridine-1-oxide (L-826,141) prevents experimental autoimmune encephalomyelitis. J Pharmacol Exp Ther. 2006;319:63–72. doi: 10.1124/jpet.106.106096. [DOI] [PubMed] [Google Scholar]
- 38.Moore CS, Hebb AL, Robertson GS. Inhibitor of apoptosis protein (IAP) profiling in experimental autoimmune encephalomyelitis (EAE) implicates increased XIAP in T lymphocytes. J Neuroimmunol. 2008;193:94–105. doi: 10.1016/j.jneuroim.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 39.Wheeler RD, Zehntner SP, Kelly LM, Bourbonniere L, Owens T. Elevated interferon gamma expression in the central nervous system of tumour necrosis factor receptor 1-deficient mice with experimental autoimmune encephalomyelitis. Immunology. 2006;118:527–38. doi: 10.1111/j.1365-2567.2006.02395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, McFarland HF, Paty DW, Polman CH, Reingold SC, Sandberg-Wollheim M, Sibley W, Thompson A, van den Noort S, Weinshenker BY, Wolinsky JS. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50:121–7. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 41.Polman CH, Reingold SC, Edan G, Filippi M, Hartung HP, Kappos L, Lublin FD, Metz LM, McFarland HF, O’Connor PW, Sandberg-Wollheim M, Thompson AJ, Weinshenker BG, Wolinsky JS. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann Neurol. 2005;58:840–6. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- 42.Hebb A, Moore C, Bhan V, Campbell T, Fisk J, Robertson H, Thorne M, Lacasse E, Holcik M, Gillard J, Crocker S, Robertson G. Expression of the inhibitor of apoptosis protein family in multiple sclerosis reveals a potential immunomodulatory role during autoimmune mediated demyelination. Mult Scler. 2008;14:577–94. doi: 10.1177/1352458507087468. [DOI] [PubMed] [Google Scholar]
- 43.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]