Abstract
7,8-dihydro-8-oxoguanine (8-oxoG) is the major oxidative product of guanine and the most prevalent base lesion observed in DNA molecules. Because 8-oxoG has the capability to form a Hoogsteen pair with adenine (8-oxoG:A) in addition to a normal Watson-Crick pair with cytosine (8-oxoG:C), this lesion can lead to a G:C → T:A transversion after replication. However, 8-oxoG is recognized and excised by the 8-oxoguanine DNA glycosylase (Ogg) of the base excision repair pathway. Members of the Ogg1 family usually display a strong preference for a C opposite the lesion. In contrast, the atypical Ogg1 from Clostridium actetobutylicum (CacOgg) can excise 8-oxoG when paired with either one of the four bases, albeit with a preference for C and A. Here we describe the first high resolution crystal structures of CacOgg in complex with duplex DNA containing the lesion 8-oxoG paired to cytosine and to adenine. A structural comparison with human OGG1 provides a rationale for the lack of opposite base specificity displayed by the bacterial Ogg.
Keywords: 8-oxoguanine glycosylase, crystal structure, DNA repair, 8-oxoguanine, base excision repair, opposite base specificity
1. Introduction
DNA is particularly sensitive to ionizing radiation and to oxidative stress from the cell’s own environment as well as from exogenous sources. [1] Because of its unsaturated N7-C8 bond, guanine is prone to oxidative damage and its oxidative product, 7,8-dihydro-8-oxoguanine (8-oxoG) [2], constitutes the most frequent base lesion observed in DNA. [3] 8-Oxoguanine is particularly pernicious among DNA lesions because of its miscoding properties. [3, 4] 8-oxoG can form a normal Watson-Crick base pair with cytosine (8-oxoG:C); however, it has also been shown to form a stable Hoogsteen pair with adenine (8-oxoG:A) [4, 5], which can lead to a G:C→T:A transversion after replication [5, 6]. In contrast to many replicative polymerases, DNA polymerase β, a base excision repair polymerase, preferentially inserts a C opposite 8-oxoG rather than A [7] and thus the integrity of DNA is maintained.
Formamidopyrimidine-DNA glycosylase (Fpg) and 8-oxoguanine DNA glycosylase (Ogg) [8, 9], two enzymes which belong to two different families of BER enzymes, recognize and cleave the 8-oxoG lesion from the DNA duplex. Both Fpg and Ogg are bifunctional glycosylases: they catalyze the excision of the oxidized base by cleaving the N-glycosylic bond between the base and the deoxyribose moiety (glycosylase activity) and subsequently cleave the DNA backbone (lyase activity). Fpg enzymes are mostly found in bacteria whereas Ogg members are more widespread in eukaryotes and archaea. However, some bacterial species like Clostridium acetobutylicum use Ogg rather than Fpg for the removal of 8-oxoG [10, 11]. In this case, the gain of Ogg function seems to be associated with a loss of the Fpg function.
The Ogg DNA glycosylases belong to three different families: Ogg1, which includes the well characterized human OGG1 (hOGG1) and the bacterial Clostridium acetobutylicum Ogg (CacOgg) [11–19], Ogg2 which was the last Ogg family to be structurally characterized and comprises mostly archaeal enzymes [20, 21] and finally, AGOG (Archaeal GO Glycosylase) [22] represented by Pyrobaculum aerophilum AGOG (Pa-AGOG). [23, 24] Overall CacOgg shares a similar tertiary fold with hOGG1. Our previously published crystal structure of CacOgg in complex with the nucleoside 8-oxo-2′-deoxyguanosine (8-oxodG) revealed that CacOgg binds the damaged base similarly to hOGG1. However, in contrast to hOGG1 which displays a strong preference for C opposite 8-oxoG with no recognition of 8-oxoG opposite A, CacOgg can cleave the lesion regardless of the opposite base, recognizing 8-oxoG opposite A as well as with C. [11, 19] This relaxed opposite base specificity makes CacOgg quite unusual among the Ogg1 enzymes. Interestingly, two of the four amino acids making interactions with the cytosine in hOGG1 are not conserved in CacOgg, i.e., residues Arg154 and Tyr203 in hOGG1 correspond to Met132 and Phe179 in CacOgg. Previous structural and functional studies revealed that the hOGG1-Arg154 variant exhibits a decreased opposite base specificity and an impaired glycosylase function. [25, 26]
Here we describe two high-resolution crystal structures of CacOgg in complex with a 13-mer DNA molecule containing 8-oxoG:C or 8-oxoG:A. The CacOgg/8-oxoG:A model is the first structure of an Ogg enzyme in complex with an adenine opposite the lesion. These structures provide a unique opportunity to identify the molecular determinants responsible for the specific recognition of the base opposite the lesion. We observe a structural reorganization in CacOgg upon binding DNA similar to what was reported for hOGG1. Furthermore, the structure unveiled a major role for CacOgg residues Asn127, Met132, Phe179 and Arg180 in the recognition of the opposite base and provides a rationale as to why this enzyme is less specific for the base opposite the lesion compared to its human counterpart. Our structural data concur with previously published biochemical studies of the CacOgg Met132 and Phe179 variants.
2. Material and methods
2.1 Recombinant CacOgg expression and purification
Recombinant CacOggK222Q was expressed and purified essentially as described. [11] Briefly, the CacOggK222Q variant was expressed in ER2566 fpg- E. coli co-transfected with a pLysS RIR vector. [27] After overnight expression with IPTG at 16°C, bacterial pellets were sonicated in lysis buffer (50 mM Tris pH 8.0, 500 mM NaCl, 1 mM EDTA, 1 mM PMSF). The variant protein was purified using the same purification protocol as for the wild type enzyme: The centrifuged lysate was loaded on a chitin column and eluted using 50 mM DTT. Pooled fractions of CacOgg were loaded on a Q column (GE Healthcare) and eluted with a NaCl gradient. The purified protein was dialyzed in crystallization buffer (20 mM Tris-HCl pH 8.5, 100 mM NaCl, 1 mM DTT and 10 % (v/v) glycerol) then concentrated to 30 mg/ml and flash frozen in liquid nitrogen.
2.2 Crystallization of the recombinant CacOgg in complex with 13mer duplex DNA containing 8-oxoG:C/A
Crystals of CacOggK222Q in complex with DNA were obtained by hanging-drop vapor diffusion at 12°C. DNA oligonucleotides (13-mer) were ordered from Midland Certified Reagent Co. (Midland, TX) and purified on an acrylamide gel. The sequences were as follows: 5′-ATC-CAX-GTC-TAC-C-3′ and 5′-TGG-TAG-ACY-TGG-A-3′ where X is 8-oxoG and Y is either C or A. Protein and duplex DNA were mixed in a 1:1 ratio. Crystals grew in 2 μL drops containing a 1:1 ratio of protein/DNA mix and well solution (16% (w/v) PEG-4000, 0.1 M Na Acetate pH 4.8 and 0 to 0.1 M MgCl2). Typical crystals grew to dimensions suitable for X-ray diffraction experiments (200 × 70 × 70 μm3) in about a week.
2.3 X-ray analysis and structure determination of CacOggK222Q in complex with DNA
All X-ray diffraction experiments were done at 100K after crystals were allowed to equilibrate for 1–2 minutes in a cryoprotectant solution consisting of the crystallization buffer supplemented with 20% (v/v) ethylene glycol. X-ray diffraction images for CacOgg/13mer-8-oxoG:C were recorded on our laboratory MAR345 detector (MAR Research) mounted on a Rigaku RU-200 rotating anode-generator equipped with Xenocs focusing mirrors while data for the CacOgg/8-oxoG:A complex were collected at a wavelength of 0.9765 Å at the ALS synchrotron by Reciprocal Space Consulting, LLC (beamline 5.0.3).
The diffraction images were integrated using XDS [28] and merged and scaled with XSCALE. The structures of CacOggK222Q in complex with DNA were solved by molecular replacement with MOLREP software from the CCP4 suite [29] using the coordinates of apo-CacOgg (PDB ID code 3F0Z) [19] as a model. For both models, a very clear Fo-Fc electron density map corresponding to DNA (including 8-oxoG and opposite base) was observed immediately after molecular replacement. The initial models were submitted to rigid body refinement and one cycle of simulated annealing at 3000 K followed by energy minimization and B-factor refinement cycle. Afterwards, the model was refined with CNS [30] by simple energy minimization followed by isotropic B-factors refinement (restrained and individual) and corrected by manual rebuilding using O. [31] Missing parts of the model, nucleotides, ion and water molecules were progressively added during the refinement procedure. Finally, the quality of the model was verified with PROCHECK. [32] There are no residues in the disallowed region of the Ramachandran plot for either model. A Na+ ion bound to the helix-hairpin-helix motif was refined in both structures. The identity of the metal ion was deduced from coordination geometry (octahedral) and bond distances (average = 2.43 Å for 8-oxoG:C and 2.39 Å for 8-oxoG:A complex). [33] In the CacOgg 8-OxoG:A complex several of the solvent exposed glutamates and aspartates appear to have undergone decarboxylation, a structural damage caused by synchrotron radiation. [34]
2.4 Protein Data Bank accession codes
Atomic coordinates and structure factor amplitudes have been deposited with the Protein Data Bank and are available under the following accession codes: 3I0W for CacOggK222Q in complex with DNA containing 8-oxoG:C and 3I0X for CacOggK222Q in complex with DNA containing 8-oxoG:A.
3. Results
3.1 Crystallization and structure determination of CacOggK222Q in complex with DNA containing 8-oxoG:C or 8-oxoG:A
A single crystal of CacOggK222Q in complex with a 13mer DNA oligonucleotide containing 8-oxoG:C was used to collect a 1.73 Å data set on our laboratory X-ray equipment (See Table 1 for diffraction statistics). Crystals of the CacOggK222Q/8-oxoG:C complex belong to the hexagonal space group P6522 with unit-cell dimensions a=b=92.4 c=191.05Å. A molecular replacement solution (correlation factor of 0.70) was found using the atomic coordinates of apo-CacOgg as a search model. The resulting electron density map was well defined for the unique molecule per asymmetric unit. The CacOggK222Q/8-oxoG:C complex model was rebuilt and refined to a crystallographic R-factor of 0.195 (Rfree=0.218). The final model comprises 290 residues lacking only the last two C-terminal residues. Residues His36, Ile46, Arg83, Met217 and Gln222 appear to adopt alternate conformations. A clear Fo-Fc electronic density (at 3) corresponding to the DNA molecule including the 8-oxoG base and the estranged C was obtained after molecular replacement. (see figure 1a)
Table 1.
| CacOggK222Q/8-oxoG:C | CacOggK222Q/8-oxoG:A | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 1.5418 | 0.9765 |
| Resolution (Å)a | 20–1.73 (1.8–1.73) | 20–1.8 (1.9–1.8) |
| Space group | P6522 | P6522 |
| Unit-cell parameters | ||
| a,b,c (Å) | 92.4, 92.4, 191.05 | 92.13, 92.13, 190.73 |
| Total reflection | 688339 (49662) | 899273 (64868) |
| Unique reflection | 48376 (4455) | 41903 (2881) |
| Redundancy | 14.2 (11.1) | 21.5 (22.5) |
| Completeness (%) | 94.7 (79.2) | 92.8 (82.7) |
| I/σ(I) | 32.2 (4.9) | 19.6 (2.9) |
| Rmerge (%) | 5.6 (32.5) | 8.5 (49.0) |
| Refinement | ||
| Rcryst (%) | 19.5 | 18.7 |
| Rfree (%)b | 21.8 | 21.0 |
| Rmsd from ideal bond length (Å)/angles (°) | 0.007/1.2 | 0.005/1.1 |
| Non-hydrogen atoms | ||
| All atoms | 3315 | 3240 |
| Protein | 2454 | 2486 |
| Water | 319 | 242 |
| DNA | 508 | 508 |
| Na+ | 1 | 1 |
| Average B factors (Å2) | 27.3 | 30.5 |
| Ramachandran plot (%) | ||
| Most favored regions | 90.2 | 89.1 |
| Allowed regions | 9.8 | 10.9 |
| Disallowed regions | 0 | 0 |
High-resolution shell is shown in parentheses.
Rfree was calculated with 5% of the reflections not used in refinement
Figure 1. Overall fold of CacOggK222Q in complex with DNA containing A) 8-oxoG:C and B) 8-oxoG:A.
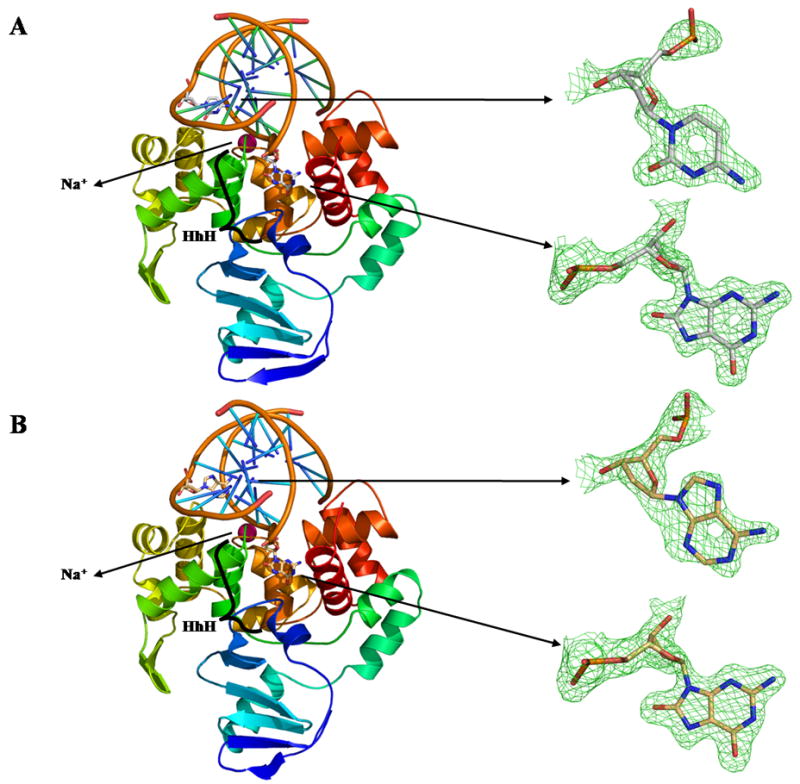
Ribbon diagrams of CacOggK222Q in complex with DNA containing A) 8-oxoG:C and B) 8-oxoG:A Proteins are colored according to the amino acid sequence going from cold blue to warm red from N- to C-terminal. A simulated annealing omit map (green) contoured at 3σ is shown for each of the estranged bases and 8-oxoG. The sodium atom is colored in pink in both panels and the HhH motif is labeled. All figures were prepared using PYMOL [38].
A 1.8 Å data set was acquired from a single crystal of CacOggK222Q in complex with a 13mer DNA-8-oxoG:A at beamline 5.0.3 of the Advanced Light Source (λ =0.9765Å) (See Table 1 for diffraction statistics). Crystals of CacOggK222Q/8-oxoG:A complex belong to the same space group as the 8-oxoG:C complex but with slightly different unit-cell dimensions: a=b=92.13 c=190.73Å. The CacOggK222Q/8-oxoG:A complex model was refined to a crystallographic R-factor of 0.187 (Rfree=0.21). The final model lacks only the last C-terminal residue. Residues Met5, His36, Ile46, Val52, Arg83, Asp189, Met217 and Gln222 appear to adopt alternate conformations. An Fo-Fc omit map (contoured at 3) corresponding to the DNA molecule and including the 8-oxoG base and the estranged A was clearly observable after molecular replacement. (see figure 1b)
3.2 Overall structure description
The overall fold of both CacOggK222Q/8-oxoG:C and/8-oxoG:A is nearly identical to the previously published apo-CacOgg model [19] (RMSD of 1.04Å for either CacOgg/DNA complex). Briefly, the protein is composed of three domains (A, B and C) around the central HhH motif (αK-L, residues 206-232). The N-terminal domain (domain A) comprises a twisted antiparallel β-sheet made of six β-strands (β1; βA-βE), and two α-helices (αA and αB). Domain B (αE–αJ) is composed of six α-helices and two antiparallel β-strands (βF and βG) whereas domain C (αC–D and αM–O) comprises five α-helices. Both domains B and C are well conserved among the Ogg1 family. The central element of the Ogg1 proteins, the well conserved HhH motif (αK-L) is considered the fingerprint of DNA repair glycosylases of this superfamily [10] and contains the conserved catalytic lysine (Lys222). The second strictly conserved catalytic residue (Asp241) belongs to αM, the helix immediately following the HhH motif. It is noteworthy that three residues of the middle loop of the HhH motif, namely Lys214, Phe216 and Val219, establish interactions with one of the DNA phosphates through a well-coordinated sodium ion.
The high isomorphism between the two CacOgg/DNA complexes (overall cross-R of 0.17 calculated on amplitudes) made it possible to calculate an Fo8-oxoG:A-Fo8-oxoG:C isomorphous difference map to pinpoint the difference between the two models. [35] An isomorphous difference map centered on the estranged base and contoured at 4σ is shown in Supplemental Figure 1. The only significant observable difference in the unbiased map is found at the site of the estranged base. The presence of an adenine at the estranged position instead of a cytosine induces a minor displacement in the position of the neighboring base.
3.3 Binding site for 8-oxoG and structural reorganization after substrate binding
The 8-oxoG binding site is located at the junction of the three domains in a deep cavity delineated by polar residues. The oxidized guanine engages in several hydrophilic interactions with the enzyme. However, as previously shown in the CacOgg complex with 8-oxo-2′-deoxyguanosine [19], and similarly to human OGG1 [26], the 8-oxygen atom of 8-oxoG is devoid of any hydrophilic interaction with the protein. Figure 2 shows the interactions made by 8-oxoG with the protein. The 8-oxoG base forms a H-bond with the side chains of Gln222, Asp241, Gln278 and Gln279 and main chain atoms of Gly30 and Pro239 while Phe282 stacks with the purine ring. The H-bond between the carboxyl group of Gly30 and the N7-H atom of 8-oxoG is of particular interest because this interaction has been shown to be critical for the distinction of 8-oxoG/G by hOGG1. [26]. The phosphate group of 8-oxoG is well stabilized by several H-bonds involving the main chain of Ile130 and the side chains of Trp243 and Arg286. Such interactions were not observable in the CacOgg/8-oxodG [19] complex because of the absence of the phosphate group; in that complex, Arg286 forms an H-bond with the O5′ atom of the nucleoside.
Figure 2. Close-up view of CacOgg residues interacting with 8-oxoG.
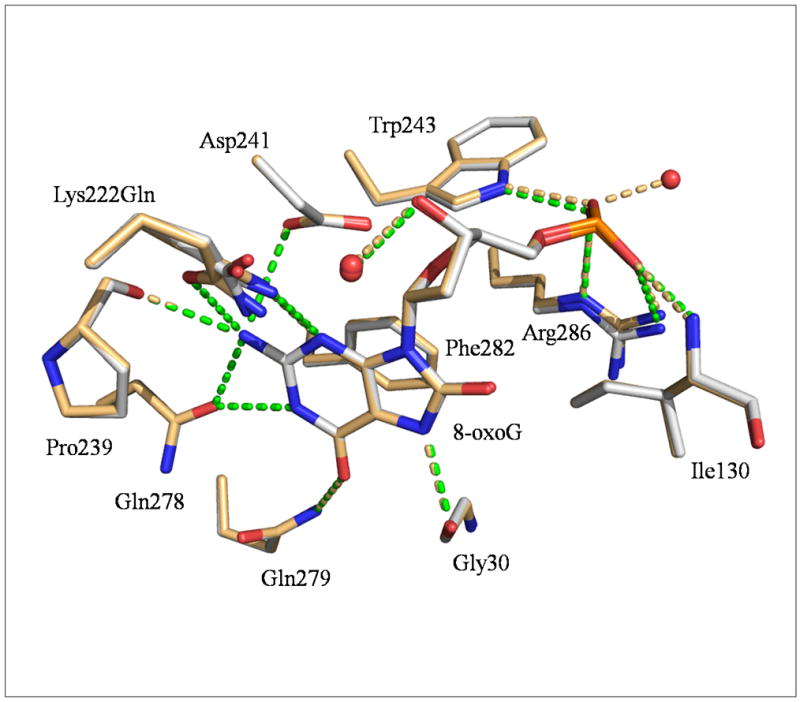
CacOggK222Q/8-oxoG:C and/8-oxoG:A are superimposed. Only residues involved in H-bonds and stacking interactions with 8-oxoG are depicted. The CacOggK222Q/8-oxoG:C carbon atoms are shown in light grey and those of the 8-oxoG:A complex in light orange. Gln222 appears to adopt alternate conformations in both complexes. H-bonds are represented by green dashed lines for 8-oxoG:C complex and light orange dash line for 8-oxoG:A complex. The H-bond between 8-oxoG N7 H atom and the main chain carbonyl of Gly30 has been shown to be essential for the recognition of 8-oxoG in hOGG1[26, 39].
A surface representation of CacOgg reveals a long groove from the HhH motif to the N-terminal domain in which DNA binds (Supplementary figure 2). The binding of DNA in this groove and/or the binding of 8-oxoG in its binding pocket induce a structural reorganization of three helices (αD, αM and αO) of the C-terminal domain, which moves closer to the 8-oxoG binding site, allowing the formation of several interactions with the ligand. A similar reorganization of the C-terminal domain has been described for CacOgg in complex with 8-oxo-2′-deoxyguanosine [19] and hOGG1 bound to DNA containing 8-oxoG. [36]
3.4 Interactions of CacOgg with DNA and estranged base
Once bound to the protein, the 8-oxoG-containing DNA molecule induces a reorganization of the C-terminal domain which allows the formation of several hydrophilic interactions between the two partners. Most of these interactions involve 8-oxoG and residues lining its binding pocket. However, the remaining hydrophilic contacts are made with the DNA backbone of the strand containing the lesion as well as with the estranged base. The protein-DNA interactions are summarized in figure 3. It is noteworthy that there are no interactions between the enzyme and the DNA backbone of the strand containing the estranged base. In addition, all H-bonds between CacOgg and DNA are located in the distorted and locally widened minor groove.
Figure 3. Schematic representation of Ogg1-DNA interactions.
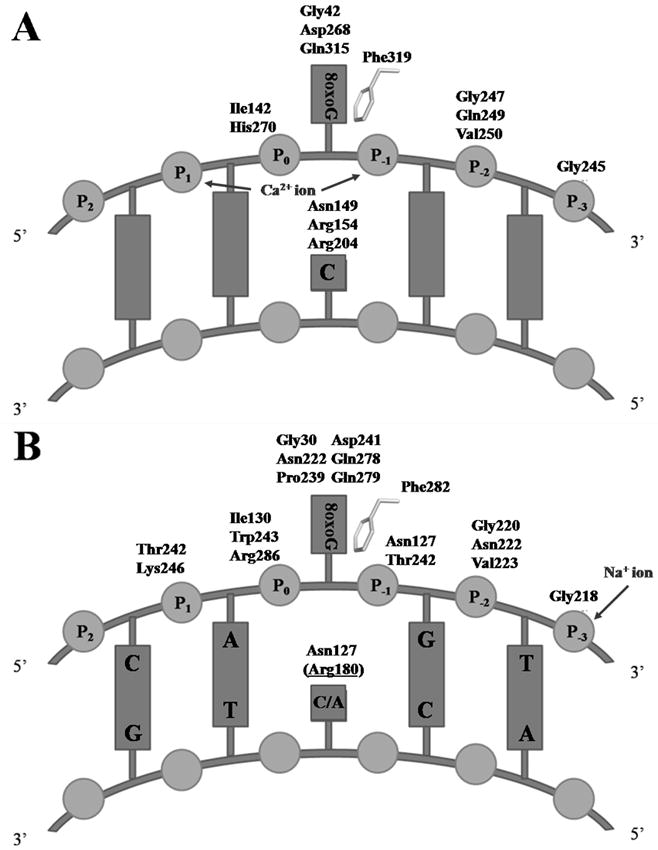
Schematic representation of A) DNA interactions with hOGG1 (PDB ID code 1EBM [26]) and B) interactions of DNA with CacOggK222Q/8-oxoG:C/A complexes. hOGG1-Phe319 and CacOgg-Phe282 stack against the 6-membered ring of 8-oxoG. Arg180 (in parenthesis and underlined) in panel B makes a H-bond only when the estranged base is a cytosine. Metal ion interactions are depicted.
The estranged base is involved in few H-bonds with the protein. These interactions are slightly different depending on the nature of the base. Figure 4a shows the estranged cytosine from the CacOgg/8-oxoG:C complex involved in H-bonds with both main chain and side chain atoms of Asn127 and the side chain of Arg180. In addition, the cytosine ring stacks on the edge of Phe179 aromatic ring. In this position, the cytosine is very well stabilized by a strong network of interactions. In contrast, the estranged adenine of the CacOgg/8-oxoG:A complex makes fewer contacts with the protein, making a H-bond only with the side chain of Asn127. However, the hydrophobic contacts with Phe179 are conserved when A is present.
Figure 4. CacOgg interactions with the estranged nucleoside.
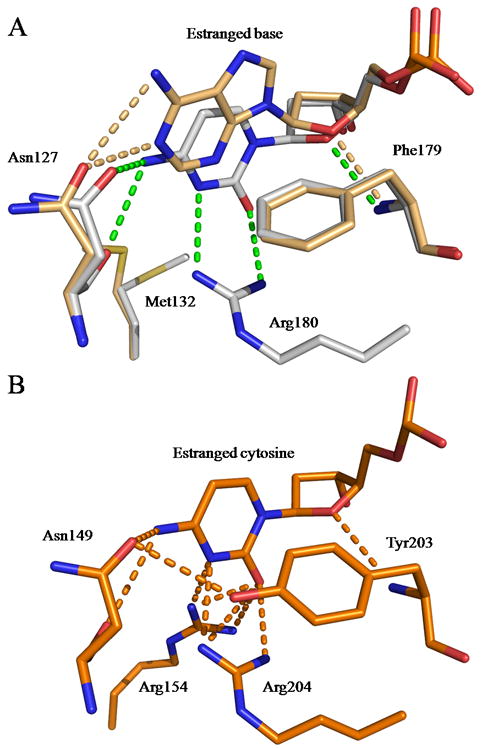
A) CacOggK222Q/8-oxoG:C and/8-oxoG:A are superimposed. Only residues involved in H-bonds and stacking interactions with the estranged base are depicted. CacOggK222Q/8-oxoG:C carbon atoms are shown in light grey and those in the 8-oxoG:A complex are colored in light orange. Phe179 is involved in both hydrophobic and hydrophilic interactions. B) Interactions made by hOGG1 (PDB ID code 1EBM) [26] with estranged cytosine. Tyr203 is involved in both hydrophobic and hydrophilic interactions and helps stabilize the side chain of Asn149. H-bonds are represented by green dashed lines for CacOgg/8-oxoG:C or by the color corresponding to their structure.
4. Discussion
4.1 Structural comparison of CacOgg/8-oxoG:C/A with apo-CacOgg and CacOgg/8-oxodG models
The major consequence of DNA binding to CacOgg is the structural rearrangement of the C-terminal domain which moves closer to the B-domain, narrowing the DNA binding groove and engaging in several interactions with both DNA backbone and bases. Binding of the ligand by the enzyme stabilizes the C-terminal helix (αO), which contains five additional ordered residues in the CacOGG/DNA model compared to the apo-enzyme. This is not surprising because several residues in this helix (Gln278, Gln279, Phe282 and Arg 286) form a H-bond with DNA or 8-oxoG itself
As observed previously in the complex with 8-oxodeoxyguanosine [19], Phe282 undergoes a shift of ~5Å going from a distal position in the apo-CacOgg to a proximal position in the CacOgg/DNA complexes, where it stacks with 8-oxoG (supplemental figure 3). Asp241, and more importantly Trp243, display a similar reorganization upon binding 8-oxoG, allowing the formation of a H-bond with the ligand stabilizing it in an optimal position for enzymatic reaction. There is, however, a notable difference between the CacOgg/DNA and CacOgg/8oxodG complexes. The presence of the DNA backbone generates new interactions that were not observed when the deoxynucleoside was bound to the enzyme. Arg286 was observed to form a H-bond with the 5′-OH group of 8-oxodG while in the CacOgg/DNA complexes the side chain of this residue forms a H-bond with the phosphate group of the oxidized guanine. Another very important observation can be drawn by comparing the CacOgg/DNA complexes to CacOgg/8-oxodG. In the CacOgg/8-oxodG complex, no hydrophilic interactions were observed between 8-oxodG and domain B. In contrast, the main chain N atom of Ile130 makes a H-bond with the phosphate group of 8-oxoG in the CacOGG/DNA complexes (see figure 2). Also, Arg129 undergoes a ~180° rotation in the CacOGG/DNA complexes compared to both apo- and 8-oxodG complexes to form a H-bond with a DNA base (Cytosine 17). All these interactions contribute to stabilize the DNA molecule and 8-oxoG and facilitate the enzymatic reaction. In addition, a well defined and coordinated sodium ion also contributes to the stabilization of one of the DNA phosphate groups by the HhH motif. It is known that metal ion binding by the HhH depends on the presence of a DNA substrate, explaining why the metal ion was not observed in the apo-enzyme model or CacOgg/8-oxodG complex. [37]
4.2 Structural comparison of CacOgg and hOGG1
Human OGG1 and CacOgg share a very similar architecture [19] despite the fact their sizes differ significantly (345 residues in hOGG1 vs. 292 for CacOgg). hOGG1 has longer connecting loops and some helices are longer than in CacOgg. However, the ligand binding residues appear to be very well conserved and superimpose well onto the corresponding amino acids in CacOgg. Furthermore hOGG1, with the exception of amino acids interacting with the estranged base (see below), shares similar hydrophilic interactions with DNA to those observed for CacOgg. Very few differences can be found in the manner in which hOGG1 and CacOgg bind their substrate. One of these differences involves the binding mode of the observed metal ion (see figure 3). As described previously, the CacOgg sodium ion connects the hairpin loop of the HhH motif and P−3 of DNA. Such metal binding by the HhH appears to be DNA dependent and is consistent with earlier characterization of the HhH motif. [37] Surprisingly, there is no metal ion observed linking HhH to the DNA in the hOGG1/8-oxoG:C complex (PDB ID code 1EBM; [26]). However, in the hOGG1/8-oxoG:C complex, a calcium ion was observed away from the HhH making direct and water-mediated interactions to both the P1 and P−1 groups of the DNA molecule. The presence of this calcium ion is in all likelihood due to the addition of CaCl2 in the crystallization buffer (as suggested by the authors) and is unlikely to be physiological. [26]
4.3 Structural basis for the estranged base specificity in Ogg1 enzymes
Both hOGG1 and CacOgg can recognize and cleave 8-oxoG and 8-oxoA, but unlike CacOgg, hOGG1 can cleave the damaged base only when paired to a cytosine. [11, 26] CacOgg is less specific for the opposite base, cleaving the damaged guanine when paired to either of the four bases, with a preference for C and A. [11] Until now, it was unclear how such different nucleosides (purine vs. pyrimidine) could be recognized by the same enzyme. The availability of atomic structures of both hOGG1/8-oxoG:C and CacOgg bound to both 8-oxoG:C and 8-oxoG:A presents a unique opportunity to identify the molecular determinants responsible for the binding and the specific recognition of the estranged base.
A structural superposition between the two CacOgg/DNA complexes revealed that the adenine is far less stabilized by protein interactions than cytosine (figure 4a). The adenine is H-bonded by only one residue (Asn127) whereas the cytosine base makes a total of four H-bonds with two residues (Asn127 and Arg180). CacOgg binds the estranged cytosine with a H-bond on only one side of the base and the pyrimidine ring is sandwiched between the edge of the Phe179 aromatic ring and the adjacent 3′-base. In contrast to cytosine, the adenine makes few hydrophilic interactions with the protein. However, with its larger purine ring, hydrophobic interactions might be more important: the adenine is wedged between the adjacent 3′-base on one side and Phe179 and the neighboring 5′-base on the other side.
hOGG1 displays a more extensive interaction network with the estranged cytosine than CacOgg. As shown on figure 4b, the estranged cytosine is tightly bound by a total of seven H-bonds originating from three residues (Asn149, Arg154 and Arg204) located on each side of the base. Similarly to CacOgg, the cytosine is sandwiched between Tyr203 and the adjacent 3′-base. However, in contrast to CacOgg-Phe179, hOGG1-Tyr203 is involved in a hydrophilic interaction with the side chain carbonyl of Asn149, certainly contributing to stabilize this residue in a suitable position to form a H-bond with the estranged cytosine N4 atom. Of note is that cytosine is the only natural DNA base that contains two adjacent H-bond acceptors. This combination of acceptors is perfectly complemented by NH1 and NH2 (both H-bond donors) of arginine residues. In hOGG1 N2 and O3 of the estranged cytosine are involved in hydrophilic interactions with the Arg154 and Arg204 side chains. [26]
The hOGG1 arginine pair (Arg154/204) and Asn149 create a very selective binding site for the estranged base where only a cytosine can match all the potential H-bond acceptors and donors from the protein. In contrast in CacOgg, only one of the arginine residues is conserved (Arg180) while the other is replaced by a methionine (Met132) (see figure 4a), which contributes to a weaker binding and lessened selectivity for cytosine. Mutating Met132 to an arginine (Met132Arg), as in hOGG1, increases the glycosylase efficiency for 8-oxoG:C by more than 4-fold compared to wild-type CacOgg and a 3.6 fold decrease in the glycosylase efficiency for the 8-oxoG:A pair. [11] This result indicates that restoration of the missing arginine increases the opposite base specificity substantially.
The asparagine corresponding to hOGG1-Asn149 (Asn127 in CacOgg) participates in fewer interactions than its human counterpart. The shift of Asn127 in CacOgg/8-oxoG:A compared to the CacOgg/8-oxoG:C complex is enough to accommodate the bulkier adenine base (Figure 5a). Such movement might be more difficult in hOGG1 because the corresponding Asn149 is H-bonded to Tyr209, which might impede the displacement of Asn149. This observation is supported by enzyme kinetics analysis of the wild-type and CacOgg-Phe179Tyr variant. [11] There is no significant glycosylase efficiency change between the wild-type and the Phe179Tyr variant for binding the estranged cytosine. We observed, however, a 14-fold activity decrease when adenine is found across 8-oxoG probably due to steric interactions. In addition, the double mutant (Met132Arg/Phe179Tyr) shows a 50-fold increase in specificity for C versus A compared to wild type CacOgg.
Figure 5. Putative interactions of guanine and thymine with CacOgg at the estranged base binding site.
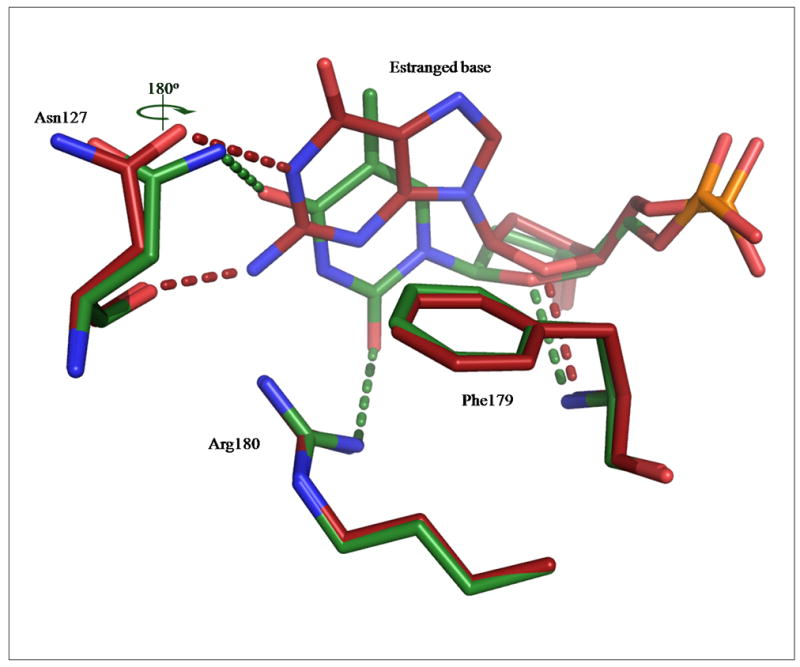
The figure shows the putative interactions that CacOgg may make with G (red) or T (green) as the estranged base. The putative H-bonds are depicted in the same color as their corresponding model. Asn127 most likely performs a 180° rotation around its Cγ to interact with the O4 acceptor of thymine. In both models, the estranged base stacks against of Phe179. The hydrophilic interactions are extrapolated from the CacOgg/8-oxoG:C and/8-oxoG:A complexes.
As mentioned earlier CacOgg differs greatly from its human counterpart by its ability to bind any of the four bases opposite the lesion. [11] The availability of the CacOgg models in complex with 8-oxoG:C and 8-oxoG:A allows us to extrapolate how the enzyme may bind the two other bases. Figure 5 shows the putative interactions made by estranged G and T with the enzyme. The overall number of hydrophilic interactions is comparable to what was observed with C or A. As shown in figure 5, a guanine might interact with both side- and main chain atoms of Asn127, where the two donors (N1 and N2 atoms) might H-bond with the acceptor atoms (Oδ1 and O) of Asn127. The guanine O6 atom appears to be devoid of any hydrophilic interactions with the protein. The thymine may be H-bonded to the Arg180 donor groups via its O2 keto group. Asn127 might undergo a 180° rotation around its Cγ (see figure 5) in order to accommodate the acceptor O4 atom of thymine. The lack of the hOGG1-Arg154 equivalent in CacOgg creates a more flexible binding site allowing the binding of diverse bases with different pattern of acceptors and donors.
4.4 Concluding remarks
Structural data from this study along with previously published structures of CacOgg [19] and hOGG1/8-oxoG:C [26] show that Ogg1 enzymes undergo a structural rearrangement to accommodate the binding of DNA containing 8-oxoG. We also confirmed by direct observation and comparison with hOGG1 the major role of Met132, Asn149, Phe179 and Arg180 in the binding specificity of the estranged base. Our structural data concur with the conclusions drawn by biochemical experiments previously published for CacOgg. [11] The ability of CacOGG to cleave 8-oxoG opposite each of the four bases might be puzzling at first glance because removal of 8-oxoG from a 8-oxoG:A mispair, for example, can lead to deleterious mutations. However, Clostridium acetobutylicum is an obligate anaerobic bacterium and thus its exposure to oxygen is likely to be sporadic. Moreover, its growth rate under aerobic conditions is presumably considerably slower and insertion of an A by a polymerase less likely. Most anaerobic bacteria can tolerate brief exposures to oxygen and CacOgg may be needed only in this rare event. There is therefore little selective pressure to maintain an opposite base specificity similar to that of eukaryotic 8-oxoG glycosylases.
Supplementary Material
Supplemental figure 1: Unbiased isomorphous difference map at the estranged base site of CacOggK222Q/8-oxoG:C and/8-oxoG:A complexes.
Close-up view of the superposition of the CacOggK222Q/8-oxoG:C and/8-oxoG:A complexes. The cytosine carbon atoms are colored in light grey while the adenine is colored in light orange. The Fo8-oxoG:A-Fo8-oxoG:C map is contoured at 4σ (positive density is shown in green, negative density in red). The presence of the larger base at the estranged position causes a shift in the position of the adjacent base.
Supplemental figure 2: Surface representation of CacOgg/DNA complex.
Surface representation of CacOgg in complex with DNA showing the binding groove (in red) from the N-terminal domain to the helix-hairpin-helix (HhH) motif. The sodium ion bound to the hairpin of the HhH is depicted in pink.
Supplemental figure 3: Close-up view of CacOgg residues undergoing a conformational change upon ligand binding
Superposition of CacOgg/8-oxoG:C (standard CPK colors), apo-CacOgg (red; PDB ID code 3F0Z) [19] and the CacOgg/8-oxodG complex (blue; PDB ID code 3F10) [19] illustrating residues undergoing a conformational change upon ligand binding. Residues Asp241 and Trp243 move closer to 8-oxoG/8-oxodG. Trp243 and Phe282 move from a distal position to a proximal position. In the proximal position, Phe282 stacks against the six-membered ring of 8-oxoG/8-oxodG. Arg286 is disordered in the apo-CacOgg model. CacOgg/8-oxoG:C Ile130 interacts with the phosphate group of 8-oxoG while Arg129 undergoes a 180° rotation to interact with cytosine 17 and the deoxyribose of adenine 18. DNA is not shown for clarity.
Acknowledgments
We thank April Averill and Lauren M. Harvey for help with protein purification and Dr. Susan Robey-Bond for critical reading of the manuscript. This research was supported by National Institutes of Health grants R01CA33657 and P01CA098993 awarded by the National Cancer Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fraga CG, Shigenaga MK, Park JW, Degan P, Ames BN. Oxidative damage to DNA during aging: 8-hydroxy-2′-deoxyguanosine in rat organ DNA and urine. Proc Natl Acad Sci U S A. 1990;87:4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasai H, Crain PF, Kuchino Y, Nishimura S, Ootsuyama A, Tanooka H. Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis. 1986;7:1849–1851. doi: 10.1093/carcin/7.11.1849. [DOI] [PubMed] [Google Scholar]
- 3.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 4.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 5.Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Miura K, Ohtsuka E, Nishimura S. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature. 1987;327:77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- 6.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 7.Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH. Structure of DNA polymerase beta with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure. 2003;11:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
- 8.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 9.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denver DR, Swenson SL, Lynch M. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol Biol Evol. 2003;20:1603–1611. doi: 10.1093/molbev/msg177. [DOI] [PubMed] [Google Scholar]
- 11.Robey-Bond SM, Barrantes-Reynolds R, Bond JP, Wallace SS, Bandaru V. Clostridium acetobutylicum 8-oxoguanine DNA glycosylase (Ogg) differs from eukaryotic Oggs with respect to opposite base discrimination. Biochemistry. 2008;47:7626–7636. doi: 10.1021/bi800162e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- 13.Arai K, Morishita K, Shinmura K, Kohno T, Kim SR, Nohmi T, Taniwaki M, Ohwada S, Yokota J. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- 14.Bjørås M, Luna L, Johnsen B, Hoff E, Haug T, Rognes T, Seeberg E. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 1997;16:6314–6322. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagashima M, Sasaki A, Morishita K, Takenoshita S, Nagamachi Y, Kasai H, Yokota J. Presence of human cellular protein(s) that specifically binds and cleaves 8-hydroxyguanine containing DNA. Mutat Res. 1997;383:49–59. doi: 10.1016/s0921-8777(96)00045-6. [DOI] [PubMed] [Google Scholar]
- 16.Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roldan-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenquist TA, Zharkov DO, Grollman AP. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faucher F, Robey-Bond SM, Wallace SS, Doublie S. Structural characterization of Clostridium acetobutylicum 8-oxoguanine DNA glycosylase in its apo form and in complex with 8-oxodeoxyguanosine. J Mol Biol. 2009;387:669–679. doi: 10.1016/j.jmb.2009.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gogos A, Clarke ND. Characterization of an 8-oxoguanine DNA glycosylase from Methanococcus jannaschii. J Biol Chem. 1999;274:30447–30450. doi: 10.1074/jbc.274.43.30447. [DOI] [PubMed] [Google Scholar]
- 21.Faucher F, Duclos S, Viswanath B, Wallace SS, Doublie S. Crystal structure of two archeal 8-oxoguanine DNA glycosylases provide structural insight into guanine/8-oxoguanine distinction. Structure. 2009;17:703–712. doi: 10.1016/j.str.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sartori AA, Lingaraju GM, Hunziker P, Winkler FK, Jiricny J. Pa-AGOG, the founding member of a new family of archaeal 8-oxoguanine DNA-glycosylases. Nucleic acids research. 2004;32:6531–6539. doi: 10.1093/nar/gkh995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingaraju GM, Sartori AA, Kostrewa D, Prota AE, Jiricny J, Winkler FK. A DNA glycosylase from Pyrobaculum aerophilum with an 8-oxoguanine binding mode and a noncanonical helix-hairpin-helix structure. Structure. 2005;13:87–98. doi: 10.1016/j.str.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Lingaraju GM, Prota AE, Winkler FK. Mutational studies of Pa-AGOG DNA glycosylase from the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. DNA Repair (Amst) 2009 doi: 10.1016/j.dnarep.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Audebert M, Radicella JP, Dizdaroglu M. Effect of single mutations in the OGG1 gene found in human tumors on the substrate specificity of the Ogg1 protein. Nucleic Acids Res. 2000;28:2672–2678. doi: 10.1093/nar/28.14.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 27.Bandaru V, Blaisdell JO, Wallace SS. Oxidative DNA glycosylases: recipes from cloning to characterization. Methods Enzymol. 2006;408:15–33. doi: 10.1016/S0076-6879(06)08002-5. [DOI] [PubMed] [Google Scholar]
- 28.Kabsch W. Automatic Processing of Rotation Diffraction Data from Crystals of Initially Unknown Symmetry and Cell Constants. Journal of Applied Crystallography. 1993;26:795–800. [Google Scholar]
- 29.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallographica Section D-Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron-Density Maps and the Location of Errors in These Models. Acta Crystallographica Section A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 33.Harding MM. Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr D Biol Crystallogr. 2006;62:678–682. doi: 10.1107/S0907444906014594. [DOI] [PubMed] [Google Scholar]
- 34.Ravelli RB, Garman EF. Radiation damage in macromolecular cryocrystallography. Curr Opin Struct Biol. 2006;16:624–629. doi: 10.1016/j.sbi.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Rould MA. The same but different: isomorphous methods for phasing and high-throughput ligand screening. Methods Mol Biol. 2007;364:159–182. doi: 10.1385/1-59745-266-1:159. [DOI] [PubMed] [Google Scholar]
- 36.Bjørås M, Seeberg E, Luna L, Pearl LH, Barrett TE. Reciprocal “flipping” underlies substrate recognition and catalytic activation by the human 8-oxo-guanine DNA glycosylase. J Mol Biol. 2002;317:171–177. doi: 10.1006/jmbi.2002.5400. [DOI] [PubMed] [Google Scholar]
- 37.Pelletier H, Sawaya MR. Characterization of the metal ion binding helix-hairpin-helix motifs in human DNA polymerase beta by X-ray structural analysis. Biochemistry. 1996;35:12778–12787. doi: 10.1021/bi960790i. [DOI] [PubMed] [Google Scholar]
- 38.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: 2008. http://www.pymol.org. [Google Scholar]
- 39.Radom CT, Banerjee A, Verdine GL. Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations. J Biol Chem. 2007;282:9182–9194. doi: 10.1074/jbc.M608989200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure 1: Unbiased isomorphous difference map at the estranged base site of CacOggK222Q/8-oxoG:C and/8-oxoG:A complexes.
Close-up view of the superposition of the CacOggK222Q/8-oxoG:C and/8-oxoG:A complexes. The cytosine carbon atoms are colored in light grey while the adenine is colored in light orange. The Fo8-oxoG:A-Fo8-oxoG:C map is contoured at 4σ (positive density is shown in green, negative density in red). The presence of the larger base at the estranged position causes a shift in the position of the adjacent base.
Supplemental figure 2: Surface representation of CacOgg/DNA complex.
Surface representation of CacOgg in complex with DNA showing the binding groove (in red) from the N-terminal domain to the helix-hairpin-helix (HhH) motif. The sodium ion bound to the hairpin of the HhH is depicted in pink.
Supplemental figure 3: Close-up view of CacOgg residues undergoing a conformational change upon ligand binding
Superposition of CacOgg/8-oxoG:C (standard CPK colors), apo-CacOgg (red; PDB ID code 3F0Z) [19] and the CacOgg/8-oxodG complex (blue; PDB ID code 3F10) [19] illustrating residues undergoing a conformational change upon ligand binding. Residues Asp241 and Trp243 move closer to 8-oxoG/8-oxodG. Trp243 and Phe282 move from a distal position to a proximal position. In the proximal position, Phe282 stacks against the six-membered ring of 8-oxoG/8-oxodG. Arg286 is disordered in the apo-CacOgg model. CacOgg/8-oxoG:C Ile130 interacts with the phosphate group of 8-oxoG while Arg129 undergoes a 180° rotation to interact with cytosine 17 and the deoxyribose of adenine 18. DNA is not shown for clarity.


