Abstract
Objective
Uric acid is the primary end product of purine metabolism. Increased serum uric acid has been associated with gouty arthritis as well as with a variety of cardiovascular related phenotypes.
Methods
A 500,000 SNP genome wide association study of serum uric acid levels was performed in a cohort of Old Order Amish from Lancaster County, Pennsylvania.
Results
The scan confirmed a previously identified region on chromosome 4 to be strongly associated with uric acid levels (p = 4.2×10−11 for rs10489070). Follow-up genotyping revealed a non-synonymous coding SNP (Val253Ile; rs16890979) in GLUT9 that was most strongly associated with uric acid levels, with each copy of the minor allele associated with 0.47 mg/dl less uric acid (95% confidence interval: 0.31 − 0.63; p = 1.43 × 10−11). The effect of this variant tended to be stronger in women than in men (p = 0.16 for sex × genotype interaction). The genotypic effect was not modified by the inclusion of several cardiovascular risk factors suggesting that GLUT9 is directly related to uric acid homeostasis. The same SNP (rs10489070) identified in the Amish genome wide scan was significantly associated with gout in the Framingham Heart Study (p = 0.004).
Conclusions
We conclude that GLUT9, which is expressed in the kidney may be a novel regulator of uric acid elimination and a common non-synonymous variant in this gene contributes to abnormalities in uric acid homeostasis and gout.
INTRODUCTION
Uric acid is the primary end product of purine metabolism by xanthine oxidase. An elevated serum uric acid level is associated with gouty arthritis and kidney stones due to deposition of uric acid crystals in the joints and collecting ducts of the kidney, respectively. Serum uric acid is also an independent predictor of several cardiovascular and metabolic syndrome phenotypes in both healthy and at risk populations (1-3). While the direct causal mechanisms linking uric acid metabolism to these endpoints have not been unequivocally determined, clinical and experimental evidence supporting such an effect is mounting (4;5), and it has been suggested that decreasing uric acid levels may attenuate cardiovascular disease risk (6). Identifying genetic factors influencing variation in serum uric acid levels may contribute to the understanding of uric acid homeostasis and facilitate the identification of new targets for intervention. To this end, we performed a genome wide association study of serum uric acid levels in a cohort of 868 Old Order Amish from Lancaster County, Pennsylvania. Our findings confirm previous associations identified on chromosome 4 in Caucasian cohorts (7;8). We extend these findings to the identification of a common non-synonymous variant, Val253Ile in GLUT9 that is likely functional. Finally, we demonstrate that a SNP in high linkage disequilibrium with this variant is associated with uric acid levels and gout in the Framingham Heart Study, thus linking the gene with this common and often disabling disease.
METHODS
Research subjects
The Heredity and Phenotype Intervention (HAPI) Heart Study began in 2003 with the goal of identifying genes that interact with environmental exposure to alter risk of cardiovascular disease (9). Old Order Amish individuals aged 20 years and older who were relatively healthy were recruited into the study. Exclusion criteria included severe hypertension (BP > 180/105 mm Hg), malignancy, and kidney, liver or untreated thyroid disease. The study protocol was approved by the Institutional Review Board at the University of Maryland, School of Medicine and informed consent was obtained from each study participant.
Physical examinations were conducted at the Amish Research Clinic in Strasburg, PA in the early morning following an overnight fast and a blood sample was taken. Uric acid levels drawn non-fasting at the screening exam were assayed by Quest Diagnostics (Baltimore, MD) and measured to the nearest 0.1 mg/dL.
Genotype analysis
Participants of the HAPI Heart Study were genotyped with the use of the Affymetrix GeneChip® Human Mapping 500K Array set. The GeneChip Genotyping Analysis Software (GTYPE 4.0) was used for automated genotype calling as part of the GeneChip Operating Software (GCOS) platform. The GTYPE-generated chip files were re-analyzed using the BRLMM genotype calling algorithm which provided improved call accuracy compared with the DM algorithm. Only samples with call rates > 93% on both microassays (the NspI and StlyI digestions) were used for analysis. The resulting mean call rates of the 861 resulting samples was 97.5%. Marker call rate was then assessed across the acceptable samples and markers with call rates of > 90% across samples and had minor allele frequency > 5% were considered for analysis (n = 361,034).
Data analysis
Genome wide association was performed using the measured genotype approach that modeled variation in uric acid as a function of measured environmental covariates (age, age2, and sex), measured genotype and a polygenic component to account for phenotypic correlation due to relatedness. The polygenic component was modeled using the relationship matrix derived from the complete pedigree structure since all subjects are related. Specifically, the covariance between each pair of individuals within the pedigree is estimated as a function of their degree of relationship, the trait heritability, and the phenotypic variance of the trait. The model is thus defined as:
where Y is a vector of uric acid values, and X is a design matrix accommodating an intercept and a vector of covariates and individual genotype values coded as 0, 1 or 2. β is a vector containing the estimates of the fixed effects. The g term is the polygenic component that is distributed multivariate normally with a mean of zero and a covariance equal to two times the kinship matrix times the expected variance due to the additive effect of genes. The e term is a normally distributed error component with mean zero. Generalized least squares estimates of the parameters of interest are given by:
where V is the variance-covariance matrix and is a function of residual trait heritability and the relationships implied by the pedigree structure. A 1-degree of freedom likelihood ratio test is used to assess significance of the measured genotype under the additive model. The genome-wide analysis was carried out using software developed in our group.
RESULTS
A genome wide association scan of uric acid levels was performed in 868 Amish participants of the Heredity and Phenotype Intervention (HAPI) Heart Study. The study sample included slightly more men (n=460) than women (n=408). Uric acid levels were higher in men than in women (4.54 ± 1.0 vs. 3.71 ± 0.9 mg/dl, p < 0.0001), see Table 1.
Table 1.
Characteristics [mean (standard deviation)] by sex of HAPI Heart Study participants.
| Trait | Women (n=408) | Men (n=460) | p |
|---|---|---|---|
| Age (yrs) | 45.4 (14.2) | 42.2 (13.6) | 0.0007 |
| Body Mass Index (kg/m2) | 27.8 (5.5) | 25.6 (3.2) | <0.0001 |
| Percent Body Fat (%) | 34.4 (6.4) | 18.4 (6.5) | <0.0001 |
| Uric Acid (mg/dl) | 3.71 (0.9) | 4.54 (1.0) | <0.0001 |
| SBP (mmHg) | 121.4 (16.9) | 121.5 (12.6) | 0.9775 |
| DBP (mmHg) | 75.8 (8.4) | 77.6 (8.8) | 0.0019 |
| Triglycerides (mg/dl) | 73.8 (45.4) | 63.9 (37.3) | 0.0005 |
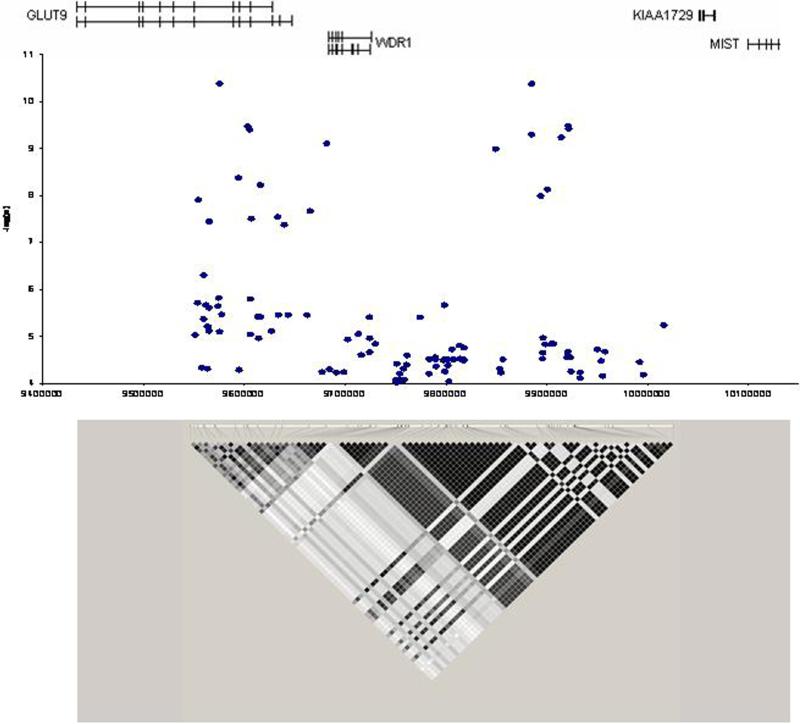
A total of 361,034 SNPs passed quality control measures with a minor allele frequency greater than 5% and comprised the genome wide scan. The results of the association tests for those SNPs with very strong evidence for association (n = 246 SNPs with p < 0.0001) are given in Supplemental Table S.1. The strongest association signal was on chromosome 4, in the same region as reported previously (7;8) (Figure 1). The most strongly associated SNP was rs10489070 (p = 4.2 × 10−11) with a cluster of 20 SNPs in linkage disequilibrium with rs10489070 that all provided strong evidence (p < 10−7) for association with uric acid levels. These SNPs encompass an approximately 367 KB region that include GLUT9 and WDR1.
Figure 1.
Region on chromosome 4 strongly associated with serum uric acid levels. Two clusters of SNPs approximately 300 kb apart are in linkage disequilibrium and showed strong association with uric acid level. The region contains GLUT9 and WDR1.
GLUT9 is a class II member of the facilitated hexose transporter family (SLC2A). Substrate specificity is varied with some able to translocate both glucose and fructose (10). GLUT9, which codes for a 540 amino acid protein, is expressed primarily in liver, kidney and placenta and to some extent in chondrocytes, brain, lung and leukocytes (11). GLUT9 also has a demonstrated splice variant, GLUT9ΔN which codes for a 512 amino acids protein expressed only in kidney and placenta (11). GLUT9ΔN was shown to be located in the apical membrane of human kidney proximal tubule epithelial cells, the primary site for renal uric acid regulation (12). The WDR1 gene appears to affect actin disassembly and help regulate cell morphologic changes during mitosis (13). No potential functional correlation between WDR1 and uric acid are known, and we therefore choose GLUT9 as our target for further study.
GLUT9 contains 12 exons spanning 195 Kb and is described to have four non-synonymous coding SNPs, Ala17Thr (rs6820230), Val253Ile (rs16890979), Arg265His (rs3733591) and Pro321Leu (rs2280205) (dbSNP, build 128). We genotyped all four non-synonymous coding SNPs in our HAPI Heart sample; Val253Ile GLUT9, was in linkage disequilibrium with rs10489070 (D’ = 0.92, r2 = 0.71) and showed the strongest association with uric acid in an additive fashion, p = 1.43 × 10−11 (Table 2). The Val253Ile substitution is in exon 8 of GLUT9 and is located in the region between transmembrane domains 6 and 7. Valine at this position is highly conserved among the orthologs of GLUT9 and is found in all known primate, rodent, and even tetraodon GLUT9 proteins, Table 3. This variant was the only significant association among the 4 coding SNPs when all were included in a single model providing evidence that it is associated with uric acid levels independently of other coding variants in the gene and thus is the most likely functional variant.
Table 2.
Non-synonymous coding SNPs in GLUT9, linkage disequilibrium with strongest signal in genome wide scan and each other, effect size controlling for age, sex and family structure (simple model) and age, sex, family structure and other SNPs (full model).
| Allele | Linkage Disequilibrium (r2) | Simple Model | Full Model | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | rsNumber | Frequency | Rs10489070 | Val253Ile | Arg265His | Pro321Leu | Effect Size (SE); mg/dl | p | Effect Size (SE); mg/dl | p |
| Ala17Thr | rs6820230 | 0.16 | 0.01 | 0.00 | 0.02 | 0.14 | 0.01 (0.07) | 0.92 | 0.01 (0.08) | 0.87 |
| Val253Ile | rs16890979 | 0.17 | 0.71 | 0.02 | 0.09 | 0.44 (0.06) | 1.43 × 10−11 | 0.43 (0.07) | 7.89 × 10−9 | |
| Arg265His | rs3733591 | 0.12 | 0.02 | 0.00 | 0.10 (0.07) | 0.16 | 0.03 (0.07) | 0.65 | ||
| Pro321Leu | rs2280205 | 0.46 | 0.18 | 0.12 (0.05) | 0.01 | 0.00 (0.06) | 0.94 | |||
Table 3.
Amino acid sequences flanking the uric acid-associated non-synonymous SNP Val253Ile (rs16890979) in several species.
| Species | Ref. Protein No. | Amino Acid Sequence (Val253Ile in bold) |
|---|---|---|
| Human | NP_064425 | LLLEKHNEARAVKAFQTFLGKADVSQEVEEVLAESRVQRSIRLVSVLELL |
| Chimpanzee | XP_520688.2 | LLLEKHNEARAVKAFQTFLGKADVSQEVEEVLAESRVQRSIRLVSVLELL |
| Orangutan | Q5RB09 | LLLEKRNEARAVKAFQTFLGKADVSREVEEV-AESRVQRSIRLVSVLELL |
| Mouse | NP_001012363.2 | LLFEKHDEAGAMKAFQTFLGKADVSQELEEALAESRVQRNLRLVSVLELL |
| Dog | XP_536240.2 | LLFEKHDQAGAEKAFQTFLGKEDVSREVEEVLAESRVQRNIQLVSVLELL |
| Rat | XP_577349.2 | LLFEKHDEAGATKAFQTFLGKADVSQELEEALAESRVQRNLRLVSVFELL |
| Chicken | XP_420789.2 | LLLEKHNTSKAEKAFQTFLGKDDVSQEVEEVLAESRVQRNTKLVSVLQLL |
| Platypus | XP_001512025.1 | LLFEKHDEAAATKAFQTFLGKDDVSQEIEDILAESRAQRNLRLESVPQLL |
| Opossum | XP_001371233.1 | LLFEKHDEDGAEKAFQTFLGKMDVSQEMEEALEESRVQRNIRLVSVWELL |
| Pufferfish | CAG02006.1 | LLMERRDEEGAKRAFQKFLGKDDVSEELEEVHAEARAQETLQTASVLQLM |
| Conserved | LL-E---------AFQ-FLGK-DVS-E-----AE-R-Q------SV--L- |
Since women have significantly lower uric acid levels than men, we examined the effect of Val253Ile in sex-stratified analysis. After adjusting for age, each copy of the Ile allele was associated with 0.47 mg /dl lower uric acid (95% confidence interval 0.31 − 0.63) among women and 0.27 mg/dl lower uric acid (95% confidence interval 0.10 − 0.45) among men (sex by genotype interaction p-value = 0.16). Additional analyses were carried out in women. These results were consistent with a potential modifying effect of estrogen on genotype - uric acid association. Among the 153 women reporting that they had reached menopause, the effect of the Ile allele was more similar to that observed in men, 0.35 mg/dl lower uric acid (95% confidence interval 0.05 − 0.64), while the greatest effect was observed among the 227 women who reported not yet reaching menopause, 0.53 mg/dl less uric acid per Ile allele (95% confidence interval 0.35 − 0.72).
Serum uric acid has been shown to be associated with a number of cardiovascular inflammation, and metabolic traits (14;15). We similarly found strong associations between uric acid levels and a panel of cardiovascular risk factors, including percent body fat, triglycerides, HDL, LDL, glucose, insulin, and estimated glomerular filtration rate (eGFR) calculated by the MDRD equation (16) (Table 4). However, no consistent significant associations were identified between the Val253Ile GLUT9 variant and these cardiovascular and metabolic traits (Table 5). Similarly, inclusion of each risk factor into the model did not affect the relationship between Val253Ile GLUT9 and uric acid. This result suggests that Val253Ile may affect serum uric acid levels independent of eGFR and known cardiovascular risk factors.
Table 4.
Association between uric acid and other quantitative traits in the HAPI Heart Study. Point estimates are effect on trait with each increase of 1 mg/dl of uric acid.
| Men | Women | |||
|---|---|---|---|---|
| Trait | Point Estimate (95% CI) | p | Point Estimate (95% CI) | p |
| Triglycerides (mg/dl) | 10.12 (13.36, 6.88) | <0.0001 | 19.42 (23.76, 15.09) | <0.0001 |
| Fasting HDL Cholesterol (mg/dl) | −4.14 (−2.99, −5.28) | <0.0001 | −5.97 (−4.35, −7.59) | <0.0001 |
| Cholesterol HDL Ratio | 0.28 (0.38, 0.17) | <0.0001 | 0.43 (0.57, 0.3) | <0.0001 |
| eGFR (mL/min/1.73m2) | −5.03 (−3.5, −6.55) | <0.0001 | −4.27 (−2.56, −5.97) | 0.0001 |
| Creatinine (mg/dL) | 0.04 (0.05, 0.03) | <0.0001 | 0.03 (0.05, 0.02) | <0.0001 |
| Glucose ln(mg/dl) | 0.01 (0.03, 0) | 0.01 | 0.04 (0.05, 0.02) | 0.0002 |
| Insulin ln(mU/ml) | 0.11 (0.15, 0.07) | <0.0001 | 0.16 (0.22, 0.1) | <0.0001 |
| Adiponectin ln(mg/ml) | −0.09 (−0.05, −0.12) | 0.0007 | −0.15 (−0.1, −0.2) | <0.0001 |
| Leptin ln(pg/ml) | 0.61 (0.86, 0.36) | 0.0003 | 0.42 (0.57, 0.27) | <0.0001 |
| Percent Body Fat (%) | 2.01 (2.83, 1.19) | 0.0003 | 2.1 (2.91, 1.3) | <0.0001 |
| Body Mass Index (kg/m2) | 1.23 (1.5, 0.95) | <0.0001 | 2.27 (2.77, 1.76) | <0.0001 |
| Whole Body Fat Mass ln(g) | 0.16 (0.23, 0.1) | <0.0001 | 0.14 (0.18, 0.09) | <0.0001 |
| Whole Body Lean Mass ln(g) | 0.02 (0.04, 0.01) | 0.0008 | 0.04 (0.06, 0.02) | <0.0001 |
| Hemoglobin (g/dL) | 0.19 (0.27, 0.11) | 0.0002 | 0.22 (0.31, 0.12) | <0.0001 |
| C Reactive Protein ln(mg/L) | 0.2 (0.31, 0.1) | 0.0002 | 0.28 (0.38, 0.18) | <0.0001 |
| Hematocrit (%) | 0.49 (0.71, 0.26) | <0.0001 | 0.65 (0.92, 0.38) | 0.0003 |
| ALT (U/L) | 1.28 (1.98, 0.57) | 0.0004 | 1.95 (2.68, 1.22) | <0.0001 |
| Red Blood Cell Count (mill/mcl) | 0.06 (0.08, 0.03) | <0.0001 | 0.08 (0.12, 0.05) | <0.0001 |
| White Blood Cell Count (thous/mcl) | 0.1 (0.22, −0.02) | 0.09 | 0.27 (0.39, 0.15) | <0.0001 |
| DBP (mmHg) | 1.72 (2.52, 0.92) | <0.0001 | 0.67 (1.54, −0.21) | 0.13 |
| SBP (mmHg) | 2.03 (3.12, 0.94) | 0.0003 | 0.86 (2.42, −0.7) | 0.28 |
Table 5.
Association between Val253Ile (rs16890979) and other quantitative traits in the HAPI Heart Study.
| Men | Women | |||
|---|---|---|---|---|
| Trait | Point Estimate (95% CI) | p | Point Estimate (95% CI) | p |
| Triglycerides (mg/dl) | −2.43 (−9.11, 4.24) | 0.48 | −2.45 (−10.56, 5.67) | 0.55 |
| Fasting HDL Cholesterol (mg/dl) | 1.15 (−1.17, 3.46) | 0.33 | 3.04 (0.1, 5.98) | 0.04 |
| Cholesterol HDL Ratio | 0.08 (−0.13, 0.3) | 0.44 | −0.24 (−0.48, −0.01) | 0.05 |
| eGFR (mL/min/1.73m2) | −0.74 (−3.88, 2.4) | 0.65 | −0.13 (−3.15, 2.9) | 0.93 |
| Creatinine (mg/dL) | 0.01 (−0.02, 0.03) | 0.57 | 0 (−0.03, 0.02) | 0.75 |
| Glucose ln(mg/dl) | 0 (−0.02, 0.02) | 0.92 | −0.01 (−0.04, 0.01) | 0.30 |
| Insulin ln(mU/ml) | 0.04 (−0.03, 0.12) | 0.27 | 0.03 (−0.07, 0.13) | 0.58 |
| Adiponectin ln(mg/ml) | −0.04 (−0.12, 0.03) | 0.24 | 0.04 (−0.04, 0.13) | 0.32 |
| Leptin ln(pg/ml) | 0.02 (−0.46, 0.5) | 0.94 | −0.08 (−0.35, 0.19) | 0.56 |
| Percent Body Fat (%) | 0.8 (−0.91, 2.51) | 0.36 | 0.3 (−1.26, 1.86) | 0.71 |
| Body Mass Index (kg/m2) | 0.04 (−0.55, 0.62) | 0.90 | −0.39 (−1.35, 0.57) | 0.42 |
| Whole Body Fat Mass ln(g) | 0.08 (−0.05, 0.21) | 0.25 | 0.02 (−0.08, 0.11) | 0.74 |
| Whole Body Lean Mass ln(g) | 0.02 (−0.01, 0.04) | 0.28 | 0 (−0.03, 0.03) | 0.91 |
| Hemoglobin (g/dL) | 0.01 (−0.14, 0.16) | 0.89 | −0.02 (−0.19, 0.15) | 0.80 |
| C Reactive Protein ln(mg/L) | 0.02 (−0.19, 0.23) | 0.85 | 0.01 (−0.17, 0.19) | 0.94 |
| Hematocrit (%) | 0.25 (−0.19, 0.69) | 0.27 | 0.12 (−0.37, 0.6) | 0.64 |
| ALT (U/L) | −0.66 (−2.07, 0.75) | 0.36 | −1.48 (−2.78, −0.18) | 0.03 |
| Red Blood Cell Count (mill/mcl) | 0.02 (−0.03, 0.07) | 0.42 | 0.01 (−0.04, 0.07) | 0.65 |
| White Blood Cell Count (thous/mcl) | 0.16 (−0.07, 0.39) | 0.17 | −0.16 (−0.38, 0.06) | 0.15 |
| DBP (mmHg) | 0.28 (−1.33, 1.88) | 0.74 | −1.42 (−2.89, 0.04) | 0.06 |
| SBP (mmHg) | −0.36 (−2.5, 1.79) | 0.75 | −1.02 (−3.68, 1.65) | 0.46 |
Subjects of the HAPI Heart Study were relatively healthy and gout phenotypes were not available. We thus sought to examine association between this clinically significant consequence of elevated uric acid levels and GLUT9 genotype in subjects from the Framingham Heart Study (FHS). The Val253Ile GLUT9 variant was not genotyped in the 100K GWAS that is publicly available (17), however, rs10489070 was on both the 100K FHS GWAS and the 500K Amish GWAS. This SNP is in linkage disequilibrium with Val253Ile GLUT9 in the HapMap CEU samples (D’ = 0.68, r2 = 0.42) and was associated with uric acid levels in FHS (ex1 GEE p = 0.0001, ex2 GEE p = 0.002). The allele associated with increased uric acid levels was also strongly associated with gout in the FHS (GEE ß (SE) = −0.03 (0.009); p = 0.004). This demonstrates that common variation in GLUT9 in addition to being associated with serum levels of uric acid has direct clinical relevance.
DISCUSSION
We performed a genome wide association study of serum uric acid levels and found strong association of multiple SNPs on chromosome 4 that exceeded that of genome-wide significance and replicates a previously identified association in the region of GLUT9. We further narrowed the most likely causative variant to a non-synonymous coding SNP in exon 8, rs16890979, which codes for a highly conserved Val to Ile amino acid change at position 253. The effect size associated with this non-synonymous SNP is large and resulted in our most significant association. The age adjusted difference between Ile homozygotes and the Val homozygotes in our sample was nearly 1 mg/dl. A model of age, sex, genotype and BMI revealed that the Val253Ile variant explains 33% of the variation of uric acid in our sample.
As in other studies (14;15), our study indicates that uric acid is associated with a number of metabolic, inflammatory and CVD risk factors in the Amish. Uric acid was strongly associated with triglycerides, HDL cholesterol, creatinine and whole body fat mass, among others CVD risk factor traits Although genotype at the Val253Ile locus was strongly associated with uric acid, it was not consistently associated with these CVD-related markers; nor did adjustment for these CVD-related markers or eGFR alter the association of uric acid with genotype. The latter observation suggests that genotype may be in the direct causal pathway for uric acid homeostasis and not secondary to other associated factors. The lack of association between genotype and metabolic/CVD markers could either be due to a lack of causality between uric acid and increased metabolic/CVD risk or due to insufficient power since our sample is healthy with a low prevalence of elevated uric acid levels and clinically significant CVD. However, association of GLUT9 variation with gout in FHS strongly supports the role of this gene in uric acid homeostasis that is clinically significant.
Serum urate level reflects the balance between production and excretion. The production is dependent on dietary protein intake and endogenous production and breakdown of purine by xanthine oxidase. The excretion is dependent primarily on renal elimination, which accounts for about 70% of urate excretion, the remaining being dependent on intestinal excretion (12). Interestingly, women have lower serum uric acid levels than their male counterparts, which has been shown to relate, at least in part, to increase renal excretion of uric acid in response to estrogen (18). Of potential relevance, in the Amish population as well as the Sardinia and Chianti populations, GLUT9 genotype effect tends to be more pronounced in pre-menopausal women compared to post-menopausal women and males. This raises the possibility that GLUT9 activity could be modulated by estrogen. Evidence for a gender interaction with GLUT9 genotype was not identified in a study of British hypertensive subjects (8), but their sample had a mean age at time of phenotyping of 64 years (19) and it is possible that a sufficient number of pre-menopausal women were not available to detect an interaction. Sufficiently powered studies will be needed to formally test the hypothesis of a gene – sex interaction.
The mechanism by which GLUT9 may affect uric acid levels is not known. However, there are at least two plausible mechanisms. The first relates to GLUT9's potential role in fructose homeostasis in kidney and liver. Increased fructose is known to increase uric acid levels secondary to increase production (20-23), and has been implicated as a potential cause of gout (24), kidney stones (25) and the metabolic syndrome (26;27). In accordance, hereditary fructosemia, which is caused be aldolase deficiency in the liver, is associated with hypoglycemia, jaundice, and hyperuricemia (28). GLUT9 has also shown to be significantly up regulated in liver and kidney of diabetic rats (29), creating a potential link between the metabolic syndrome and hyperuricemia. A second plausible mechanism relates to uric acid excretion. The GLUT9ΔN splice variant is not only exclusively expressed in kidney and placenta but is located in kidney proximal tubules epithelial cells, the primary site of renal uric acid regulation and clearance. Future studies of GLUT9's role in uric acid homeostasis will be required to effectively test the proposed hypotheses.
Both valine and isoleucine are hydrophobic amino acids and thus the Val253Ile substitution may be regarded as conservative. However, in some proteins such substitutions at key positions leads to altered structure and function (30;31). Clinically relevant phenotypes involving valine to isoleucine substitutions have been implicated in disorders such as rheumatoid arthritis and Alzheimer's disease (32-34). The mechanism by which the substitution alters function of the GLUT9 protein will require further investigation.
In summary, we identified GLUT9 as an important genetic determinant of serum uric acid levels. This highly significant replication of the previous reports, including very similar effect size estimates, indicates that the association represents a true signal. The robustness of the association to adjustment of uric acid related covariates, association with gout, and identification of a highly conserved non-synonymous SNP, provide context for future mechanistic studies related to GLUT9, uric acid homeostasis and gout.
Supplementary Material
ACKNOWLDEGEMENTS
We thank the Amish Research Clinic Staff for their excellent work conducting the HAPI Heart Study. This study would not have been possible without the outstanding cooperation and support of the Amish community. We also thank Dr. Robert Bloch for valuable discussion on protein function and investigators of the Framingham Heart Study for making their genome-wide association data available to the public.
FUNDING
This work supported by the NIH grants U01 HL72515, U01HL084756, the University of Maryland General Clinical Research Center (GCRC)(M01 RR 16500), the Johns Hopkins University GCRC (M01 RR 000052), National Center for Research Resources, and the Clinical Nutrition Research Unit of Maryland (P30 DK072488) and the National Research Initiative Grant no. 2007-35205-17883 from the USDA Cooperative State Research, Education, and Extension Service Animal Genome Program.
Footnotes
COMPETING INTERESTS
The authors have declared that no competing interests exist.
Reference List
- 1.Alderman M, Aiyer KJ. Uric acid: role in cardiovascular disease and effects of losartan. Curr Med Res Opin. 2004;20(3):369–79. doi: 10.1185/030079904125002982. [DOI] [PubMed] [Google Scholar]
- 2.Strazzullo P, Puig JG. Uric acid and oxidative stress: relative impact on cardiovascular risk? Nutr Metab Cardiovasc Dis. 2007;17(6):409–14. doi: 10.1016/j.numecd.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Baker JF, Krishnan E, Chen L, Schumacher HR. Serum uric acid and cardiovascular disease: recent developments, and where do they leave us? Am J Med. 2005;118(8):816–26. doi: 10.1016/j.amjmed.2005.03.043. [DOI] [PubMed] [Google Scholar]
- 4.Johnson RJ, Kang DH, Feig D, Kivlighn S, Kanellis J, Watanabe S, et al. Is there a pathogenetic role for uric acid in hypertension and cardiovascular and renal disease? Hypertension. 2003;41(6):1183–90. doi: 10.1161/01.HYP.0000069700.62727.C5. [DOI] [PubMed] [Google Scholar]
- 5.Johnson RJ, Segal MS, Srinivas T, Ejaz A, Mu W, Roncal C, et al. Essential hypertension, progressive renal disease, and uric acid: a pathogenetic link? J Am Soc Nephrol. 2005;16(7):1909–19. doi: 10.1681/ASN.2005010063. [DOI] [PubMed] [Google Scholar]
- 6.Dawson J, Quinn T, Walters M. Uric acid reduction: a new paradigm in the management of cardiovascular risk? Curr Med Chem. 2007;14(17):1879–86. doi: 10.2174/092986707781058797. [DOI] [PubMed] [Google Scholar]
- 7.Li S, Sanna S, Maschio A, Busonero F, Usala G, Mulas A, et al. The GLUT9 Gene Is Associated with Serum Uric Acid Levels in Sardinia and Chianti Cohorts. PLoS Genet. 2007;3(11):e194. doi: 10.1371/journal.pgen.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82(1):139–49. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell BD, McArdle PF, Shen H, Rampersaud E, Pollin TI, Peyser PA, et al. The Genetic Response to Short-term Interventions Affecting Cardiovascular Function: Rationale and Design of the HAPI Heart Study. Am Heart Journal. 2008 doi: 10.1016/j.ahj.2008.01.019. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manolescu AR, Augustin R, Moley K, Cheeseman C. A highly conserved hydrophobic motif in the exofacial vestibule of fructose transporting SLC2A proteins acts as a critical determinant of their substrate selectivity. Mol Membr Biol. 2007;24(5−6):455–63. doi: 10.1080/09687680701298143. [DOI] [PubMed] [Google Scholar]
- 11.Augustin R, Carayannopoulos MO, Dowd LO, Phay JE, Moley JF, Moley KH. Identification and characterization of human glucose transporter-like protein-9 (GLUT9): alternative splicing alters trafficking. J Biol Chem. 2004;279(16):16229–36. doi: 10.1074/jbc.M312226200. [DOI] [PubMed] [Google Scholar]
- 12.Anzai N, Kanai Y, Endou H. New insights into renal transport of urate. Curr Opin Rheumatol. 2007;19(2):151–7. doi: 10.1097/BOR.0b013e328032781a. [DOI] [PubMed] [Google Scholar]
- 13.Fujibuchi T, Abe Y, Takeuchi T, Imai Y, Kamei Y, Murase R, et al. AIP1/WDR1 supports mitotic cell rounding. Biochem Biophys Res Commun. 2005;327(1):268–75. doi: 10.1016/j.bbrc.2004.11.156. [DOI] [PubMed] [Google Scholar]
- 14.Cirillo P, Sato W, Reungjui S, Heinig M, Gersch M, Sautin Y, et al. Uric Acid, the metabolic syndrome, and renal disease. J Am Soc Nephrol. 2006;17(12 Suppl 3):S165–S168. doi: 10.1681/ASN.2006080909. [DOI] [PubMed] [Google Scholar]
- 15.Feig DI, Mazzali M, Kang DH, Nakagawa T, Price K, Kannelis J, et al. Serum uric acid: a risk factor and a target for treatment? J Am Soc Nephrol. 2006;17(4 Suppl 2):S69–S73. doi: 10.1681/ASN.2005121331. [DOI] [PubMed] [Google Scholar]
- 16.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med. 1999;130(6):461–70. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 17.Framingham Heart Study 100K Analyses. 2008 http://www.ncbi.nlm.nih.gov/projects/gap/cgibin/study.cgi?study_id=phs000007.v1.p1. Ref Type: Unpublished Work.
- 18.Adamopoulos D, Vlassopoulos C, Seitanides B, Contoyiannis P, Vassilopoulos P. The relationship of sex steroids to uric acid levels in plasma and urine. Acta Endocrinol (Copenh) 1977;85(1):198–208. doi: 10.1530/acta.0.0850198. [DOI] [PubMed] [Google Scholar]
- 19.Caulfield M, Munroe P, Pembroke J, Samani N, Dominiczak A, Brown M, et al. Genome-wide mapping of human loci for essential hypertension. Lancet. 2003;361(9375):2118–23. doi: 10.1016/S0140-6736(03)13722-1. [DOI] [PubMed] [Google Scholar]
- 20.Choi JW, Ford ES, Gao X, Choi HK. Sugar-sweetened soft drinks, diet soft drinks, and serum uric acid level: the Third National Health andNutrition Examination Survey. Arthritis Rheum. 2008;59(1):109–16. doi: 10.1002/art.23245. [DOI] [PubMed] [Google Scholar]
- 21.Emmerson BT. Effect of oral fructose on urate production. Ann Rheum Dis. 1974;33(3):276–80. doi: 10.1136/ard.33.3.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao X, Qi L, Qiao N, Choi HK, Curhan G, Tucker KL, et al. Intake of added sugar and sugar-sweetened drink and serum uric acid concentration in US men and women. Hypertension. 2007;50(2):306–12. doi: 10.1161/HYPERTENSIONAHA.107.091041. [DOI] [PubMed] [Google Scholar]
- 23.Perheentupa J, Raivio K. Fructose-induced hyperuricaemia. Lancet. 1967;2(7515):528–31. doi: 10.1016/s0140-6736(67)90494-1. [DOI] [PubMed] [Google Scholar]
- 24.Choi HK, Curhan G. Soft drinks, fructose consumption, and the risk of gout in men: prospective cohort study. BMJ. 2008;336(7639):309–12. doi: 10.1136/bmj.39449.819271.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor EN, Curhan GC. Fructose consumption and the risk of kidney stones. Kidney Int. 2008;73(2):207–12. doi: 10.1038/sj.ki.5002588. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa T, Tuttle KR, Short RA, Johnson RJ. Hypothesis: fructose-induced hyperuricemia as a causal mechanism for the epidemic of the metabolic syndrome. Nat Clin Pract Nephrol. 2005;1(2):80–6. doi: 10.1038/ncpneph0019. [DOI] [PubMed] [Google Scholar]
- 27.Nakagawa T, Kang DH, Feig D, Sanchez-Lozada LG, Srinivas TR, Sautin Y, et al. Unearthing uric acid: an ancient factor with recently found significance in renal and cardiovascular disease. Kidney Int. 2006;69(10):1722–5. doi: 10.1038/sj.ki.5000391. [DOI] [PubMed] [Google Scholar]
- 28.Wong D. Hereditary fructose intolerance. Mol Genet Metab. 2005;85(3):165–7. doi: 10.1016/j.ymgme.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Keembiyehetty C, Augustin R, Carayannopoulos MO, Steer S, Manolescu A, Cheeseman CI, et al. Mouse glucose transporter 9 splice variants are expressed in adult liver and kidney and are up-regulated in diabetes. Mol Endocrinol. 2006;20(3):686–97. doi: 10.1210/me.2005-0010. [DOI] [PubMed] [Google Scholar]
- 30.Iwasaki M, Darden TA, Pedersen LG, Negishi M. Altering the regiospecificity of androstenedione hydroxylase activity in P450s 2a-4/5 by a mutation of the residue at position 481. Biochemistry. 1995;34(15):5054–9. doi: 10.1021/bi00015a016. [DOI] [PubMed] [Google Scholar]
- 31.Monplaisir N, Merault G, Poyart C, Rhoda MD, Craescu C, Vidaud M, et al. Hemoglobin S Antilles: a variant with lower solubility than hemoglobin S and producing sickle cell disease in heterozygotes. Proc Natl Acad Sci U S A. 1986;83(24):9363–7. doi: 10.1073/pnas.83.24.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349(6311):704–6. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 33.Graessler J, Verlohren M, Graessler A, Zeissig A, Kuhlisch E, Kopprasch S, et al. Association of chondromodulin-II Val58Ile polymorphism with radiographic joint destruction in rheumatoid arthritis. J Rheumatol. 2005;32(9):1654–61. [PubMed] [Google Scholar]
- 34.Valdes AM, Wolfe ML, O'Brien EJ, Spurr NK, Gefter W, Rut A, et al. Val64Ile polymorphism in the C-C chemokine receptor 2 is associated with reduced coronary artery calcification. Arterioscler Thromb Vasc Biol. 2002;22(11):1924–8. doi: 10.1161/01.atv.0000038486.48400.e7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.