Abstract
In the prostate gland of adult mammals, most epithelial cells are in a state of proliferative quiescence. Androgens regulate this effect by inducing cell cycle arrest in the G0/G1 phase. Potential mediators of this androgen-induced proliferative shutoff were identified by means of subtracted cDNA libraries. The expression pattern of one of these sequences, AS3, strongly correlated with the expression of the androgen-induced proliferative shutoff both temporally and dosewise. The AS3 gene is located on chromosome 13 q12.3, in close proximity to the BRCA2 gene. The loss of chromosomal regions where AS3 alleles are located correlates with various human cancers, including prostate. The biological effect of AS3 was tested in two stable cell lines, one expressing sense and another expressing antisense AS3 constructs, both under tetracycline regulation. S9 cells were obtained by retroviral infection with virions containing a tetracycline-regulated sense AS3 construct. In these cells, sense AS3 was negatively regulated by tetracycline. Tetracycline withdrawal increased the expression of AS3 mRNA and protein. The expression of tetracycline-regulated AS3 resulted in inhibition of cell proliferation. A4 cells were obtained by retroviral infection with virions containing a tetracycline-regulated antisense AS3 construct. Vector-driven expression of antisense-AS3 blocked the induction of androgen-induced endogenous AS3 mRNA and blocked the inhibitory effect of androgens on cell proliferation. Tetracycline-regulated expression of the empty vector control had no effect on cell proliferation. These experiments strongly suggest that AS3 is a mediator of the androgen-induced proliferative shutoff.
Androgens regulate cell number in the prostate gland. They are involved in the control of three processes, namely inhibition of apoptosis, induction of cell proliferation, and inhibition of cell proliferation (proliferative shutoff) (1, 2). Androgen control of apoptosis and induction of cell proliferation have been studied extensively. In contrast, research on the androgen-induced shutoff has been hindered by the difficulty in isolating this effect from the other two when using animal models. Thus, research on proliferative shutoff has been performed mainly by using the human prostate cell line LNCaP-FGC and variants derived from it (1, 3). LNCaP-FGC cells are inhibited from proliferating in medium supplemented with charcoal dextran-stripped serum (1). The addition of androgens to this medium results in a biphasic proliferative response. At low physiological androgen doses, proliferation rates are significantly increased, while at high physiological doses a transient proliferative response is followed by a proliferative shutoff (1, 4). The LNCaP-TAC variant proliferates maximally in medium supplemented with androgens and lacks a shutoff response. The LNCaP-LNO variant instead responds only to androgens by expressing proliferative shutoff. Thus, the proliferative and inhibitory effects of androgens have become unambiguously separated in these variants (4, 5).
Two new LNCaP variants with a phenotype similar to that of LNCaP-LNO were recently reported (6). Like the LNCaP-LNO variant, one of these cell lines (LNCaP 104-S) grows in castrated athymic mice while tumor growth is repressed by androgens (7). These results suggest that this inhibitory mechanism may be present in prostate cancer, and hence it is a potential target for intervention in the management of this disease.
LNCaP variants provided the means to produce subtracted libraries to search for mRNAs that are expressed during proliferative shutoff and allowed the screening of the candidate cDNA sequences responsible for this effect (8, 9). The following criteria were used to identify putative shutoff mediators: (i) their expression was induced exclusively at the androgen concentrations that triggered proliferative shutoff; (ii) they were not induced by androgens in variants that do not express proliferative shutoff; (iii) the time course of induction preceded the expression of cell cycle arrest; (iv) they were not expressed in the rat prostate after castration and during androgen-mediated regeneration; and (v) they were expressed when the prostate ceased to proliferate (like in normal adult animals and after androgen-induced complete regeneration in castrated animals). Among the candidates isolated from the subtracted libraries, we found that the mRNA of the sequence identified as AS3 (Androgen Shutoff 3) (GenBank accession no. U95825) meets the above criteria (9).
LNCaP cells have a point mutation in the androgen receptor (AR) steroid-binding domain, which increases the binding affinity to progesterone and estradiol (10, 11). A stable wild-type AR transfectant, MCF7-AR1, was used to overcome concerns that results obtained with LNCaP cells may be the result of such mutation. MCF7-AR1 cells express androgen-induced proliferative shutoff (12), indicating that proliferative shutoff is not an effect caused by a mutated AR. A retrovirus-transduced tetracycline (tet)-regulated system was developed from MCF7-AR1 cells. Here we report the biological effects of the expression of tet-regulated sense and antisense AS3 constructs.
Materials and Methods
Cell Lines and Culture Conditions.
The LNCaP-FGC cell line was a generous gift from J. Horoszewicz (3). The MCF7-AR1 cell line, an AR-transfected MCF7 line, was established in this laboratory (12). Both cell lines express the androgen-induced proliferative shutoff (1, 4, 12). All cell lines used for these experiments were routinely maintained in 5% FBS in DMEM.
Detection of the AS3 Polypeptide.
An antigenic epitope was identified in positions 1369–1387 on the AS3 polypeptide by using the gcg protein analysis program (Genetics Computer Group, Madison, WI). The C terminus oligopeptide ‘N’–CTPQKGRGRPSKTPSPSQP–‘C’ was synthesized, conjugated to keyhole limpet hemocyanin, and injected into rabbits. The affinity-purified antibody (HiTrap NHS-activated column, Pharmacia) was used for immunoblot detection of the AS3 polypeptide. Briefly, LNCaP-FGC cells were kept in charcoal-dextran (CD) stripped FBS medium for 24 h to bring them to quiescence; then, the synthetic androgen R1881 was added at a concentration of 1 nM to induce proliferative shutoff (1). Control cells were treated with vehicle (0.01% ethanol in DMEM). Cells were harvested 48 h later. MCF7-AR1 cells were placed in 5% CD/FBS medium. Twenty-four hours later, they were treated with 100 pM of estradiol (control) or 100 pM of estradiol and 1 nM R1881 (shutoff). Cells were harvested 48 h later. The cells were then lysed by using RIPA (0.15 M NaCl/1% NP40/1% sodium deoxycholate/0.1% SDS/10 mM sodium phosphate, pH 7.2/2 mM EDTA) lysis buffer containing 0.2 mM sodium vanadate, 50 mM sodium fluoride, and protease inhibitors, following standard procedures (13). Proteins were separated on 4–20% polyacrylamide/SDS gradient gels (Ready Gel, Bio-Rad) and transferred to nitrocellulose membranes (Protoblot, Schleicher & Schuell) overnight. The membranes were blocked in 10% milk solution in PBS for 5 h at room temperature and washed 5 times over 30 min in Tris-buffered saline (TBS) containing 0.5 M NaCl. Then the membranes were incubated overnight at 4°C in anti-AS3 antibody diluted 1:800 in 2% milk solution in PBS. The membranes were washed in TBS containing 0.5 M NaCl and 0.05% Tween 20 (TBS-T) for 30 min at room temperature and incubated in peroxidase-conjugated goat-anti-rabbit antibody diluted 1:10,000 in 2% milk solution in PBS for 1 h at room temperature. The membranes were washed in TBS-T for 30 min before chemiluminescent reaction was performed with the Renaissance Chemiluminescence kit (NEN). Antibody specificity was assessed by competition with 50-fold molar excess of either the specific C terminus antigen or the unrelated N terminus oligopeptide with preincubation for 2 h at 4°C. The optical densities of the visualized bands were determined by using the Eagle Eye scanner (Stratagene).
Establishment of the Tet-Regulated Host Cell Line.
To create a model for the stable expression of the tet transactivator (tTA), the tTA construct (pTet-tTAk, Life Technologies, Gaithersburg, MD) was cotransfected into MCF7-AR1 cells with a puromycin-resistance gene (pPUR, CLONTECH) by using Lipofectamine (Life Technologies). After selection with 5 μg/ml puromycin, the clones were screened for tTA expression by reverse transcription–PCR (RT-PCR). Characterization by using transient expression of luciferase identified a clone, STFX1, with low baseline expression at 1 μg/ml tet and high-level induction by tet withdrawal. Stable transfection of this cell line with the green fluorescent protein (GFP) by using the pBI-EGFP plasmid (CLONTECH) also confirmed tet-inducible expression. The cells were grown on coverslips, fixed by using 2% paraformaldehyde in PBS, and visualized with an Axioscope fluorescence microscope (Zeiss). These cells were used for the expression of tet-regulated constructs delivered as retroviral particles.
Retroviral Cloning and Expression.
The MMTV promoter in the retrovirus vector (pRevTRE, CLONTECH) drives the hygromycin-resistance gene, and the tet-sensitive promoter regulates genes in the multiple cloning site. The AS3 cDNA was amplified by using terminal primers with restriction enzyme sites to clone in sense (HpaI and ClaI) and antisense (BamHI and HindIII) orientations into the pRevTRE vector. The constructs were confirmed by sequencing and then transfected into the packaging cell line PT67 (CLONTECH). Hygromycin-resistant clones (100 μg/ml) were tested for viral transcript expression. The virions in the supernatants were used to infect the STFX1 cell line with the empty vector control, the sense AS3, and the antisense AS3 constructs by using Polybrene (Sigma) (10 μg/ml) following the manufacturer's instructions. The STFX1-derived clones were selected by hygromycin and were tested for tet induction and analyzed by RT-PCR. Three clones were chosen: RT5, which contains the empty vector, A4, which expresses the antisense AS3, and S9, which expresses the sense AS3.
Analysis of the Antisense AS3 Effect on Proliferative Shutoff.
A4 (antisense) and RT5 (vector control) cells were seeded into six-well plates; each well contained two coverslips and 1 μg/ml of tet in 1.5 ml of DMEM with 5% FBS. Once the cells were attached, the medium was changed to 5% CD/FBS with 100 pM estradiol to maintain maximal cell proliferation rates, while the further addition of 1 μg/ml tet was administered to suppress gene expression. Tet was withdrawn for 36 h before androgen treatment in the antisense induction experiments. Each experimental condition was assayed in duplicate wells.
To assess the effect of antisense AS3 on the expression of the androgen-induced proliferative shutoff, the following conditions were used: (i) proliferation control (no R1881 added, tet added), (ii) androgen-induced shutoff (R1881 added, tet added), (iii) antisense induction (no R1881 added, tet withdrawn), and (iv) androgen-induced shutoff with antisense induction (R1881 added, tet withdrawn).
Thirty-six hours later, 10 μM BrdUrd was added for 6 h. Cells were fixed with 50% methanol/50% acetone. BrdUrd incorporation was detected by immunocytochemistry by using the 5-bromo-2′-deoxyuridine Labeling and Detection Kit (Boehringer–Mannheim Roche); cell nuclei were counterstained with Hoechst 33258. Images were captured with a SPOT RT color digital camera (Diagnostic Instruments, Sterling Heights, MI) attached to an Axioscope fluorescence microscope. The cells attached to the wells were used to assess the expression of the vector, the antisense AS3 transcript, and the endogenous AS3 mRNA by RT-PCR.
Analysis of the Sense AS3 Effect on Proliferation.
The S9 cultures were kept in CD/FBS medium (with tet) for 3 days to bring them to quiescence. The cultures undergoing AS3 induction by means of tet withdrawal were washed twice with PBS, placed in tet-free medium for 6 h, and then the induction was carried out for an additional period of 36 h. The control cultures were exposed to tet for the same period. Proliferation was induced with 100 pM estradiol, and the cells were kept for 52 h under the following conditions: (i) maximal proliferation control (tet added), (ii) androgen-induced shutoff (R1881 added, tet added), (iii) AS3 induction from the tet-regulated construct (no R1881 added, tet withdrawn), and (iv) androgen-induced shutoff together with AS3 induction from the tet-regulated construct (R1881 added, tet withdrawn). After incubation, the cells were treated with 10 μg/ml BrdUrd for 1 h. Immunocytochemistry and analysis were performed as described for testing antisense AS3. The cells attached to the wells were used to assess the expression of the retroviral tet-induced AS3 transcript by RT-PCR. For immunoblot analysis of AS3 protein induction, the cells were treated with the same combination of hormones and tet, following the above protocol, and then harvested for protein extraction as described above.
Expression Analysis by RT-PCR.
RT-PCR reactions were done by using oligo(dT) primed cDNA from total RNA (Qiagen, Chatsworth, CA) harvested after 36 h of induction (tet withdrawn) or suppression (tet added) of the AS3 transcript. PCR amplifications were performed with the Platinum Supermix PCR reagent (Life Technologies). For detection of the control vector transcript, a tet-promoter specific primer (5′-GTTTTGACCTCCATAGAAGACAC) and a 3′LTR-specific primer (5′-GATCTGAACTTCTCTATTCTCAG) were used. For the detection of antisense expression, an AS3 gene specific primer (5′-TAAGGTTCTGTATAGCTGGGTG) and the 3′LTR primer were used. For retroviral sense transcript detection, the same AS3 specific primer and the tet-promoter primer were used. For detection of the endogenous AS3 transcript, a 3′ untranslated area was amplified between positions 3969–4392. This area is not present in the retroviral antisense transcript (upstream primer: 5′-CCAAAAGAAGAGCCAACAAT; downstream primer: 5′-GTTCATCGCCGTTCCCTTTTAG).
Data Analysis.
Quantitative image analysis was performed by using the optimas software, Ver. 6.5 (Media Cybernetics, Silver Spring, MD). Cell colonies were mapped and counted in each coverslip. Fifty percent of the colonies in each coverslip were analyzed. Each colony contained between 50 and 200 cells. Data were expressed as the percent of BrdUrd-labeled cells in each colony relative to cells stained with Hoechst 33258. Data were analyzed by ANOVA and Sheffe's post hoc test. Data that did not satisfy the assumptions of an ANOVA were analyzed by the nonparametric Mann–Whitney test (14).
Results
Expression of the AS3 Polypeptide Is Up-Regulated in the Androgen-Induced Proliferative Shutoff.
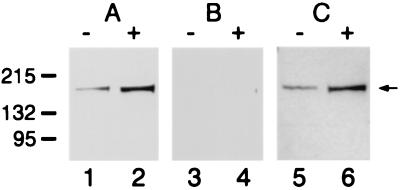
The molecular mass of AS3 was calculated from its amino acid sequence (159 kDa). The affinity-purified polyclonal antibody raised against the AS3 oligopeptide detected a single band in immunoblot analysis of protein extracts. The band detected by the anti-AS3 antibody from androgen-treated and vehicle-treated LNCaP-FGC cells is in the range of 165–170 kDa (Fig. 1A). Competition with 50-fold molar excess of the specific oligopeptide antigen (C terminus) resulted in the total disappearance of the AS3 band (Fig. 1B). Competition with an unrelated oligopeptide (N terminus) had no effect on the binding of the antibody (Fig. 1C). The AS3 protein is approximately 5-fold up-regulated in the androgen-induced proliferative arrest in the LNCaP-FGC cell line.
Figure 1.
Immunoblot analysis of AS3 protein induction in cellular extracts. Detection of AS3 in the LNCaP-FGC cell extract was carried out by using a rabbit polyclonal anti-AS3 antibody. The lysates used were from cells treated with 1 nM R1881 for 48 h (+) and cells treated for 48 h with vehicle only (−). The protein load was 35 μg/lane. (A) Uncompeted antibody; (B) peptide competition was performed with 50-fold excess of the specific C terminus peptide and (C) with a nonrelated N terminus peptide. The numbers (Left) indicate the molecular markers in kilodaltons; the arrow (Right) points to the AS3 band.
STFX1, the Host Cell Line for Tet-Regulated Gene Expression.
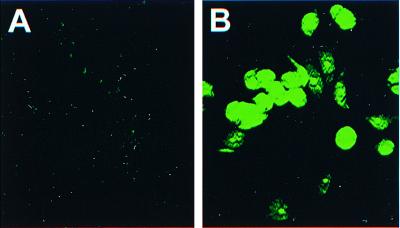
Cotransfection of MCF7-AR1 cells with the tet-transactivator plasmid (pTet-tTak) and the gene for puromycin-resistance (pPUR) resulted in the establishment of a cell line, STFX1, with a stable integration of the tTA transactivator. tTA expression was confirmed by RT-PCR (not shown). The stable expression of a tet-regulated GFP construct (pBI-EGFP) showed that tTA expression is functional. GFP expression was suppressed to a minimal baseline on exposure to 1 μg/ml tet (Fig. 2A), while tet withdrawal induced high-level expression (Fig. 2B). Quantitative analysis of the transient expression of a tet-regulated luciferase construct showed induction across three orders of magnitude (data not shown).
Figure 2.
Tet-regulated reporter gene expression in the host cell line STFX1. Tet regulation was tested in GFP stable transfectans of STFX1 cells. (A) Cells treated with 1 μg/ml tet; (B) tet withdrawal for 36 h. The photomicrographs were taken at ×400 by using standard FITC filters.
STFX1-Derived Cell Lines with Tet-Regulated Sense and Antisense AS3 Expression.
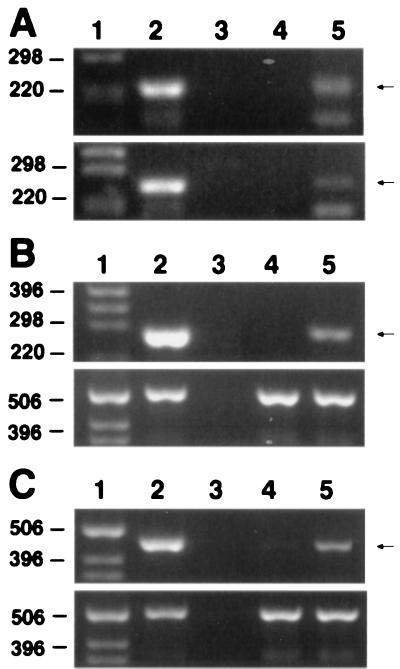
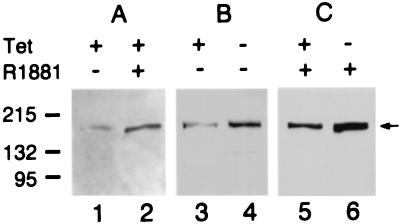
Screening by genomic PCR identified clones with integrated retroviral constructs (Fig. 3A). RT-PCR transcription analyses were performed on total RNA from tet-suppressed cells and from tet-withdrawn cells after 36 h of induction. Tet withdrawal resulted in induction of the vector control in RT5 cells (Fig. 3B), the antisense AS3 in A4 cells (Fig. 3C), and the sense AS3 in S9 cells (Fig. 4A). Antisense expression reduced the levels of the androgen-induced endogenous AS3 mRNA by 9-fold (Fig. 4B). Immunoblot analysis of AS3 protein expression in the host MCF7-AR1 cell line detected a 165-kDa band, which was up-regulated approximately 5-fold in androgen-induced proliferative arrest (Fig. 5A). Tet withdrawal induced the expression of the retroviral AS3 polypeptide approximately 4-fold in the sense-AS3-transfected S9 cell line (Fig. 5B). In androgen-arrested cells a 2-fold increase was detected on tet withdrawal, in addition to the AS3 androgen-induced control (Fig. 5C). These data confirm that in the selected clones, sense and antisense AS3 transcripts are functional, and their expression is under tet regulation.
Figure 3.
PCR and RT-PCR analysis of the integration and induction of the antisense transcripts. In A–C, lane 1 represents the size markers, lane 2 the positive PCR control (a plasmid containing the sequence of interest), and lane 3 the negative control (water). The expected PCR products are indicated (Right). (A) PCR of genomic DNA for detection of the integrated retrovirus constructs. Upper shows the integrated antisense vector; Lower shows the integrated control vector. Lane 4, uninfected cell DNA; lane 5, DNA from retrovirus infected cells. (B and C Lower) GAPDH RT-PCR normalization controls. (B) Expression of the control vector transcript was detected by RT-PCR of total RNA from RT5 cells. Lane 4, tet added (gene suppression); lane 5, tet withdrawal (gene induction). (C) Expression of antisense RNA was detected by RT-PCR of total RNA from A4 cells. Lane 4, tet added (gene suppression); lane 5, tet withdrawal (gene induction).
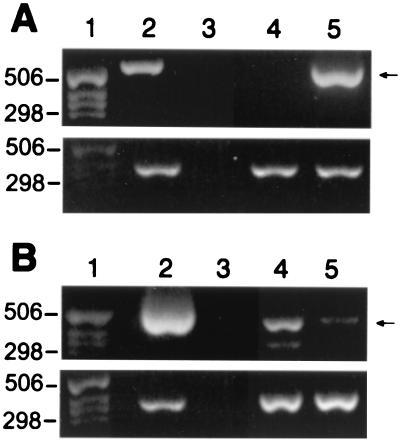
Figure 4.
RT-PCR analysis of the sense transcripts. (A and B Lower) GAPDH RT-PCR normalization controls. Lanes 1–3 indicate the size marker, the positive and negative PCR controls, respectively. The expected PCR products are indicated (Right). (A) RT-PCR of total RNA from S9 cells to detect the induction of the retroviral sense AS3 transcript. Lane 4, tet added to suppress AS3 expression; lane 5, induction by tet withdrawal of the retroviral sense AS3 transcript. (B) Suppression of the endogenous AS3 mRNA by the induction of the antisense transcript in the A4 cells. Lane 4, detection of the endogenous AS3 mRNA in conditions where the antisense transcript was suppressed by tet; lane 5, induction of the antisense RNA by tet withdrawal inhibited the expression of the endogenous AS3 mRNA.
Figure 5.
Expression of AS3 protein in MCF7-AR1, and S9 cellular extracts. (A) Androgen induction of AS3 in MCF7-AR1 cells. Lane 1, cells treated with 100 pM estradiol for 48 h; lane 2, cells expressing the androgen-induced proliferative shutoff after 48 h exposure to 100 pM estradiol and 1 nM R1881. (B) Tet induction of AS3 in S9 cells. Lane 3, cells were grown in the presence of 100 pM estradiol and 1 μg/ml tet for 4 days; lane 4, cells were grown in the presence of 100 pM estradiol and with 1 μg/ml tet for the first 2 days and then without tet for an additional 2 days. (C) Tet induction of AS3 in S9 cells treated with R1881. Lanes 5 and 6, cells received the same treatment as in lanes 3 and 4, but they were also exposed to 10 nM R1881 for 4 days. The protein load was 35 μg/lane. The numbers (Left) indicate the molecular markers in kDa; the arrow (Right) points to the AS3 band.
Antisense AS3 Expression Inhibits Androgen-Induced Proliferative Shutoff.
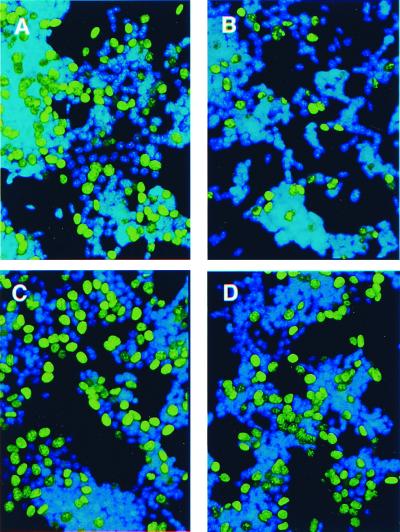
Expression of the empty vector did not affect RT5 cell proliferation in any of the conditions tested (Table 1). A4 cells proliferated maximally in the presence of tet (Table 1 and Fig. 6A). Administration of R1881 resulted in a significant (P < 0.001) decrease of A4 cells in the S phase when the antisense AS3 expression was repressed by tet (Table 1 and Fig. 6B). Cell proliferation was not affected when the antisense AS3 was expressed in the absence of androgen (Table 1 and Fig. 6C). R1881 no longer induced proliferative shutoff when the antisense AS3 was expressed in the absence of tet (Table 1 and Fig. 6D). Blocking the androgen-induced shutoff by antisense AS3 expression strongly suggests that AS3 mediates the androgen-induced proliferative shutoff.
Table 1.
Analysis of the effect of tet and androgen on the percentage of BrdUrd-labeled nuclei of STFX1 cells containing the empty vector (RT5 cells) and vector plus antisense AS3 construct (A4 cells)
| Treatment | % of BrdUrd-labeled cells |
|---|---|
| Vector, tet+, A− | 35.0 ± 1.7* |
| Vector, tet+, A+ | 9.4 ± 1.7† |
| Vector, tet−, A− | 36.6 ± 2.7* |
| Vector, tet−, A+ | 10.7 ± 0.9† |
| Antisense, tet+, A− | 34.7 ± 1.0* |
| Antisense, tet+, A+ | 9.7 ± 0.6† |
| Antisense, tet−, A− | 32.2 ± 1.5* |
| Antisense, tet−, A+ | 29.1 ± 1.6* |
“A” denotes 10 nM R1881. Each data point represents the mean ± SEM of 20 colonies. Means were considered significantly different at P < 0.001. The mean values with a * superscript are significantly different from those with a † superscript.
Figure 6.
Analysis of the antisense effect on proliferative shutoff. A4 cells were exposed to100 pM estradiol to allow for maximal proliferation in all of the experimental groups. Tet and R1881 were added as described below. Cells were stained for BrdUrd (green fluorescence) and Hoechst 33258 DNA stain (blue). (A) Maximal cell proliferation control (no R1881 added, tet added). (B) Androgen-induced shutoff without antisense AS3 induction (R1881 added, tet added). (C) Antisense AS3 induction (no R1881 added, tet withdrawn). (D) The effect of antisense induction on the androgen-induced shutoff (R1881 added, tet withdrawn). The photomicrographs were taken at ×200 by using standard FITC filters for BrdUrd detection and UV filters for Hoechst 33258 detection.
The Tet-Off-Regulated AS3 Expression Induces Proliferative Shutoff in S9 Cells.
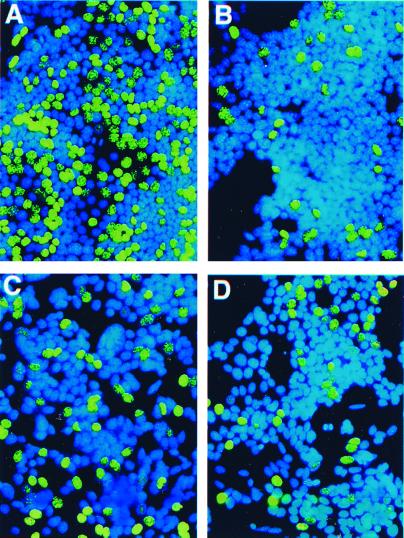
S9 cells showed maximal cell proliferation in the presence of tet, a condition in which AS3 was not expressed (Table 2 and Fig. 7A). Addition of R1881 treatment induced proliferative shutoff (Table 2 and Fig. 7B). Tet withdrawal induced the retroviral AS3 mRNA and protein in S9 cells and induced a significant (P < 0.0005) decrease of the number of cells that incorporated BrdUrd (Table 2 and Fig. 7C). There were no statistically significant differences among the cells expressing a proliferative shutoff induced by R1881 (Table 2 and Fig. 7B), tet withdrawal (Table 2 and Fig. 7C), or both (Table 2 and Fig. 7D). These findings strongly suggest that AS3 mediates proliferative shutoff.
Table 2.
Analysis of the effect of tet and androgen on the percent of BrdUrd-labeled nuclei of STFX1 cells containing the AS3 construct (S9 cells)
| Treatment | % of BrdUrd-labeled cells |
|---|---|
| tet+, A− | 34.6 ± 1.4* |
| tet+, A+ | 10.2 ± 0.8† |
| tet−, A− | 17.2 ± 0.7† |
| tet−, A+ | 13.5 ± 1.1† |
“A” denotes 10 nM R1881. The percent of BrdUrd positive nuclei was measured in each preparation. Each data point represents the mean ± SEM of 20–26 colonies. Means were considered significantly different at P < 0.0005. The mean values with a * superscript are significantly different from those with a † superscript.
Figure 7.
Analysis of the effect of sense AS3 expression on proliferation. S9 cells were treated with tet and hormones as described below and stained for BrdUrd (green fluorescence) and Hoechst 33258 nuclear stain (blue). (A) Maximal cell proliferation control (no R1881 added, tet added). (B) Androgen-induced shutoff control without AS3 induction (R1881 added, tet added). (C) AS3 induction by tet withdrawal (no R1881 added, tet withdrawn). (D) Androgen-induced shutoff and AS3 induction by tet withdrawal (R1881 added, tet withdrawn). The photomicrographs were taken ×200 by using standard FITC filters for BrdUrd detection and UV filters for Hoechst 33258 detection.
Discussion
In the adult prostate, most epithelial cells are cell-cycle arrested when plasma androgen levels are high. Castration induces cell death, and androgen treatment results in increased cell proliferation until the adult cell number is restored. At this point, no further proliferation occurs even if androgen treatment continues (2).
There is evidence that the shutoff mechanism remains unaltered in certain prostate cancers. Prostate carcinomas usually respond to androgen withdrawal. However, later on, they relapse. It is assumed that because they no longer respond to androgen ablation, they have become “autonomous.” The presence of the proliferative shutoff effect in some established prostate carcinoma cell lines argues against this notion (3). If this phenotype is present, treatment with antiandrogens or 5α-reductase inhibitors may result in the unintended proliferation of cells that otherwise would have become inhibited from proliferating when exposed to androgens (7). In fact, a small number of case reports indicate that subjective and objective remission responses occurred in men with advanced prostatic carcinoma who were treated with testosterone propionate (15–18). Hence, it would be desirable to understand the mechanism by which androgens inhibit cell proliferation and to identify markers of this phenotype to guide hormone therapy in prostate cancer patients.
The AS3 gene was identified as a putative candidate because of the androgen-induced expression pattern of the AS3 transcript in shutoff-positive MCF7-AR1 cells, in the LNCaP shutoff-positive variants, and in the rat prostate (9). We show here that the AS3 protein is also expressed in cells undergoing proliferative shutoff after androgen treatment. AS3 was not expressed in shutoff-negative variants (9). The AS3 gene is located on chromosome 13q12.3, in close proximity to the BRCA2 gene (19). Microsatellite marker data indicate that the loss of the genomic region coding for AS3 (with or without the loss of BRCA2) correlates with various human cancers (19–23), including prostate.
Sequence analysis data show that AS3 belongs to a novel gene family. It is related to fungal proteins involved in proliferation arrest, like the Aspergillus bimD protein (24), and in chromosome packaging, like the Sodaria Spo76p protein (25). AS3-related human sequences such as KIAA0648 (26) and KIAA0979 (27) were also found in brain cDNA libraries.
In the present study, a retroviral transduction tet-regulated model was developed to test the role of AS3. Antisense AS3-transformed A4 cells behaved as their parental cells in the presence of tet and expressed antisense AS3 in the absence of tet. The expression of antisense AS3 significantly decreased androgen-induced expression of endogenous AS3 mRNA and blocked the expression of androgen-induced Go/G1 arrest. In contrast, when A4 cells were grown in the presence of tet, a condition that suppresses the expression of antisense AS3, androgen treatment resulted in proliferation arrest, and BrdUrd incorporation was low. Expression of the empty vector did not interfere with the effect of androgen (Fig. 3 and Table 1).
Tet-regulated retroviral sense AS3 expression experiments revealed that AS3 induced the shutoff effect in the absence of androgens. Tet withdrawal increased the expression of AS3 mRNA from the retroviral construct. Expression of sense AS3 resulted in induction of proliferative shutoff in conditions that otherwise supported maximal cell proliferation. These data strongly suggest that AS3 mediates androgen-induced proliferative shutoff.
Kokontis et al. have shown that R1881 persistently increased the cyclin-dependent-kinase inhibitor p27kip1 level in shutoff-positive LNCaP 104-R2 cells (28). Transfection of these cells with p27kip1 resulted in cell cycle arrest in Go/G1. Androgens did not increase the p27kip1 mRNA level. This finding suggests that p27kip1 levels are regulated by androgens at the posttranscriptional level.
Sequence analysis revealed that AS3 has putative transactivating features, protein–protein interaction motifs (coiled–coil and a leucine zipper), and DNA-binding domains (19). This suggests that AS3 is a transcription factor. AS3 also has a protein–kinase motif (19); this suggests that it may act by phosphorylating a target protein, which in turn will be either activated or inactivated.
To integrate the evidence presented in this paper on the role of AS3 as inducer of the shutoff effect with the findings by Kokontis et al., we propose two hypotheses. The first posits that androgens induce expression of AS3, which acts as a transcription factor. AS3 would then bind to the promoter region of a gene(s) and control its expression. This gene may, in turn, interact with the cell cycle machinery to induce arrest in Go/G1. The second hypothesis posits that the AS3 protein interacts directly with the cell cycle machinery. Two types of interactions are predicted by the AS3 sequence, namely direct binding to a protein and/or activation of target proteins by phosphorylation.
In summary, we have identified a gene, AS3, which is up-regulated during androgen-induced proliferative shutoff and induces cell proliferation arrest when expressed in a retrovirus transduced model. The data collected argue that AS3 is one of a few gene products that inhibit the proliferation of prostate cells under androgen control. It is expected that testing the hypotheses outlined above may provide a better understanding of the mechanisms underlying the control of cell proliferation in normalcy and cancer.
Acknowledgments
The technical contributions of Lisa Stagon, Nancy Prechtl, and Cheryl Michaelson are appreciated. We are thankful to Drs. Charles Powell and Caroline Markey for their helpful suggestions in the preparation of the manuscript. This work was supported by National Institutes of Health Grants NIH-CA-55574, NCI-CA-13410, and NIH-AG-13807. M.V.M. is a recipient of a postdoctoral fellowship supported in part from the World Bank Program (Grant 815, Universidad Nacional del Litoral, Santa Fe, Argentina).
Abbreviations
- AR
androgen receptor
- tet
tetracycline
- RT-PCR
reverse transcription–PCR
- tTA
tet transactivator
- GFP
green fluorescent protein
References
- 1.Sonnenschein C, Olea N, Pasanen M E, Soto A M. Cancer Res. 1989;49:3474–3481. [PubMed] [Google Scholar]
- 2.Bruchovsky N, Lesser B, Van Doorn E, Craven S. Vitam Horm (San Francisco) 1975;33:61–102. doi: 10.1016/s0083-6729(08)60951-6. [DOI] [PubMed] [Google Scholar]
- 3.Horoszewicz J S, Leong S S, Kawinski E, Karr J P, Rosenthal H, Chu T M, Mirand E A, Murphy G P. Cancer Res. 1983;43:1809–1818. [PubMed] [Google Scholar]
- 4.Soto A M, Lin T M, Sakabe K, Olea N, Damassa D A, Sonnenschein C. Oncol Res. 1995;7:545–558. [PubMed] [Google Scholar]
- 5.Lin T-M. Ph.D. thesis. Boston, MA: Tufts University; 1993. [Google Scholar]
- 6.Kokontis J, Takakura K, Hay N, Liao S. Cancer Res. 1994;54:1566–1573. [PubMed] [Google Scholar]
- 7.Umekita Y, Hiipakka R A, Kokontis J M, Liao S. Proc Natl Acad Sci USA. 1996;93:11802–11807. doi: 10.1073/pnas.93.21.11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Brown D D. Proc Natl Acad Sci USA. 1991;88:11505–11509. doi: 10.1073/pnas.88.24.11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geck P, Szelei J, Jimenez J, Lin T M, Sonnenschein C, Soto A M. J Steroid Biochem Mol Biol. 1997;63:211–218. doi: 10.1016/s0960-0760(97)00122-2. [DOI] [PubMed] [Google Scholar]
- 10.Veldscholte J, Ris-Stalpers C, Kuiper G G, Jenster G, Berrevoets C, Claassen E, van Rooij H C, Trapman J, Brinkmann A O, Mulder E. Biochem Biophys Res Commun. 1990;173:534–540. doi: 10.1016/s0006-291x(05)80067-1. [DOI] [PubMed] [Google Scholar]
- 11.Brinkmann A O, Kuiper G G, Ris-Stalpers C, van Rooij H C, Romalo G, Trifiro M, Mulder E, Pinsky L, Schweikert H U, Trapman J. J Steroid Biochem Mol Biol. 1991;40:349–352. doi: 10.1016/0960-0760(91)90201-f. [DOI] [PubMed] [Google Scholar]
- 12.Szelei J, Jimenez J, Soto A M, Luizzi M F, Sonnenschein C. Endocrinology. 1997;138:1406–1412. doi: 10.1210/endo.138.4.5047. [DOI] [PubMed] [Google Scholar]
- 13.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Greene and Wiley Interscience; 1990. [Google Scholar]
- 14.Shott S. Statistics for Health Professionals. Philadelphia: Saunders; 1954. [Google Scholar]
- 15.Brendler H, Chase W E, Scott W W. Arch Surg. 1950;61:433–440. [PubMed] [Google Scholar]
- 16.Pearson O H. Cancer Res. 1957;17:473–479. [PubMed] [Google Scholar]
- 17.Prout G R, Brewer W R. Cancer. 1967;20:1871–1878. doi: 10.1002/1097-0142(196711)20:11<1871::aid-cncr2820201112>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 18.Fowler J E, Jr, Whitmore W F., Jr J Urol. 1981;126:372–375. doi: 10.1016/s0022-5347(17)54531-0. [DOI] [PubMed] [Google Scholar]
- 19.Geck P, Szelei J, Jimenez J, Sonnenschein C, Soto A M. J Steroid Biochem Mol Biol. 1999;68:41–50. doi: 10.1016/s0960-0760(98)00165-4. [DOI] [PubMed] [Google Scholar]
- 20.Gudmundsson J, Johannesdottir G, Bergthorsson J T, Arason A, Ingvarsson S, Egilsson V, Barkardottir R B. Cancer Res. 1995;55:4830–4832. [PubMed] [Google Scholar]
- 21.Cleton-Jansen A-M, Collins C, Lakhani S R, Weissenbach J, Devilee P, Cornelisse C J, Stratton M R. Br J Cancer. 1995;72:1241–1244. doi: 10.1038/bjc.1995.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caldas C, McGregor A, Wiedemann L, Catovsky D, Ashworth A, Garcia-Marco J. Proc Am Assoc Cancer Res. 1997;38:191. [Google Scholar]
- 23.Garcia-Marco J A, Caldas C, Price C M, Wiedemann L M, Ashworth A, Catovsky D. Blood. 1996;88:1568–1575. [PubMed] [Google Scholar]
- 24.Denison S H, Kafer E, May G S. Genetics. 1992;134:1085–1096. doi: 10.1093/genetics/134.4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Heemst D, James F, Poggeler S, Berteaux-Lecellier V, Zickler D. Cell. 1999;98:261–271. doi: 10.1016/s0092-8674(00)81020-x. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa K, Nagase T, Suyama M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. DNA Res. 1998;5:169–176. doi: 10.1093/dnares/5.3.169. [DOI] [PubMed] [Google Scholar]
- 27.Nagase T, Ishikawa K, Suyama M, Kikuno R, Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. DNA Res. 1999;6:63–70. doi: 10.1093/dnares/6.1.63. [DOI] [PubMed] [Google Scholar]
- 28.Kokontis J M, Hay N, Liao S. Mol Endocrinol. 1998;12:941–953. doi: 10.1210/mend.12.7.0136. [DOI] [PubMed] [Google Scholar]