Abstract
Deep sequencing technologies such as Illumina, SOLiD, and 454 platforms have become very powerful tools in discovering and quantifying small RNAs in diverse organisms. Sequencing small RNA fractions always identifies RNAs derived from abundant RNA species such as rRNAs, tRNAs, snRNA, and snoRNA, and they are widely considered to be random degradation products. We carried out bioinformatic analysis of deep sequenced HeLa RNA and after quality filtering, identified highly abundant small RNA fragments, derived from mature tRNAs that are likely produced by specific processing rather than from random degradation. Moreover, we showed that the processing of small RNAs derived from tRNAGln is dependent on Dicer in vivo and that Dicer cleaves the tRNA in vitro.
Keywords: small RNA, miRNA, deep sequencing, Dicer, tRNA
INTRODUCTION
The recent development of high-throughput sequencing technology has accelerated the identification of small regulatory RNAs. Deep sequencing of small RNA fractions from cell culture and animal tissues in which key small RNA processing factors have been impaired, or by cataloging small RNAs bound to different Argonaute proteins, has started to provide valuable data showing how different species of small RNAs are processed and sorted into different regulatory complexes (Calabrese et al. 2007; Azuma-Mukai et al. 2008; Babiarz et al. 2008; Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008b; Tam et al. 2008; Watanabe et al. 2008b; for review, see Okamura and Lai 2008).
Animal cells encode a wide arsenal of small RNAs that regulate the expression of protein coding genes and control selfish genetic elements. miRNAs are 21–23-nucleotides (nt) long, fast evolving regulatory RNAs that are mainly coded by short hairpin structures in intronic sequences. They are processed cotranscriptionally into pre-miRNAs by the microprocessor that contains an RNase III enzyme called Drosha, and an RNA binding protein, DGCR8 (Lee et al. 2003; Han et al. 2004). However, mirtrons, miRNAs located in small introns, bypass this process and are generated by splicing (Berezikov et al. 2007; Okamura et al. 2007; Ruby et al. 2007). Pre-miRNAs are transported into the cytoplasm where they are further processed into miRNAs by a complex containing Dicer (Grishok et al. 2001; Hutvagner et al. 2001; Yi et al. 2003). Finally, mature miRNAs are loaded into an Argonaute complex that regulates gene expression by altering the protein synthesis of mRNAs or interfering with transcription (for review, see Hock and Meister 2008; Hutvagner and Simard 2008). siRNAs are another source of small regulatory RNAs that are processed from diverse naturally formed double-stranded RNA precursors. The majority of endo-siRNAs are derived from transposable and repetitive elements, and they are believed to regulate the expression of transposons (Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008). Hairpin-derived endogenous siRNAs were shown to be processed to their mature form in Drosophila and mouse ES cells (Babiarz et al. 2008; Okamura et al. 2008b). Cis- and trans-derived endogenous siRNA loci have also been discovered in flies and mammalian cells (Okamura et al. 2008a; Tam et al. 2008; Watanabe et al. 2008a,b). The common feature of these siRNAs is that their processing is not dependent on Drosha and DGCR8 (Babiarz et al. 2008; Ghildiyal et al. 2008). Some classes of endo-siRNAs were shown or predicted to target mRNAs (Okamura et al. 2008b; Tam et al. 2008; Watanabe et al. 2008b). Piwi-associated small RNAs (piRNAs) are longer (27–31-nt long) small RNAs that exclusively regulate the expression of transposable elements in the germline. The maturation of these types of regulatory RNAs are Drosha and Dicer independent, and they are generated by consecutive cleavage activity of different Piwi proteins (Brennecke et al. 2007; Gunawardane et al. 2007).
Cloning and sequencing of small RNA libraries always generates fragments that are derived from abundant noncoding RNAs such as rRNAs, tRNAs, snoRNAs, and snRNAs. These RNAs are generally considered to be random degradation products, and in some cases it has been shown that their abundance does not change in a mutant background that impairs canonical regulatory small RNA production (Calabrese et al. 2007). Furthermore, in yeast, a mechanism has been discovered that prevents rRNA and tRNA species from entering the small RNA pathway (Buhler et al. 2008). However; recent data suggest that abundant noncoding RNAs can produce stable small siRNA-sized RNA fragments that are unlikely to be random degradation products, and one tRNA has been shown to produce a Dicer-dependent small RNA that is expressed with low abundance in mouse ES cells (Babiarz et al. 2008; Kawaji et al. 2008). A similar tRNA fragment was also detected by Northern hybridization in immortalized human cell lines; however, its processing properties were not investigated (Kawaji et al. 2008). In addition, it has been recently demonstrated that snoRNAs can produce small RNAs that are processed in a Drosha-independent and Dicer-dependent manner and incorporate into functional Argonaute complexes (Ender et al. 2008). In this paper we show that quality filtering of deep sequencing data reveals a wide range of small RNAs that are likely products of processing events. Interestingly, we have found that an abundant small RNA species in HeLa cells is derived from the 5′ ends of a series of mature tRNAs, and have shown that the production of small RNAs derived from tRNAGln is Dicer dependent.
RESULTS
Purification of small RNAs
Since miRNAs (Politz et al. 2006) and small RNAs processed from snoRNAs (AI Lamond, pers comm.) could be detected in the nucleolus, we initiated deep sequencing to identify the small RNA populations of this cellular compartment. We isolated total RNA from purchased purified HeLa nucleoli (Supplemental Fig. 1A) and deep sequenced the small RNA fraction by the Solexa method following a published protocol (Lu et al. 2007). However, post-sequencing testing of the nucleoli and other fractions derived from the purification steps have revealed that our original sample was contaminated with nucleoplasmic and, to a lesser extent, cytoplasmic materials (Supplemental Fig. 1B).
Small RNA sequence analysis
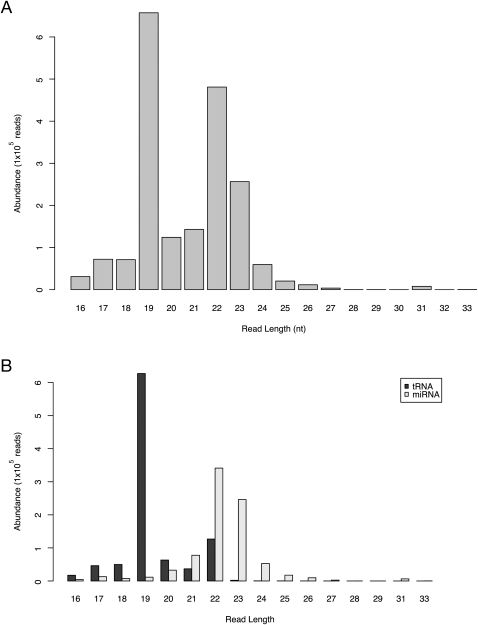
A total of 2.3 million sequence reads were obtained from the sequencing experiment and following filtering and collation (see Materials and Methods), 22,386 unique reads remained. The sequences were size-selected small RNAs, with a distribution of lengths up to 33 nt, as shown in Figure 1A. The length distributions show peaks at around 19 and 22 nt, which are discussed below.
FIGURE 1.
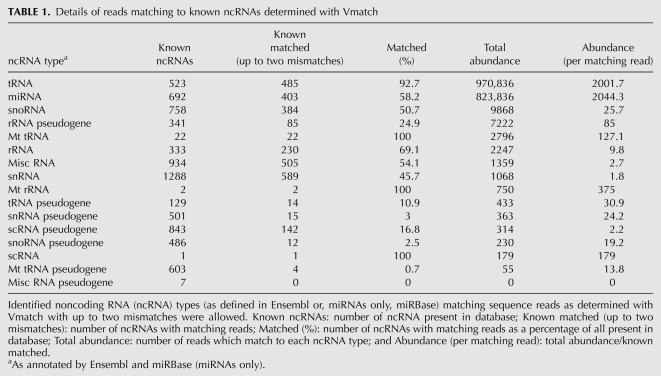
Length distribution of all sequence reads. (A) Plot shows the length distribution by abundance of each of the 22,836 unique sequences obtained from deep sequencing of HeLa cell extracts. (B) Length distribution of tRNA and miRNA matching reads. The length distribution of the sequence reads of classified by matching to either tRNA (dark gray bars) or miRNA (light gray bars).
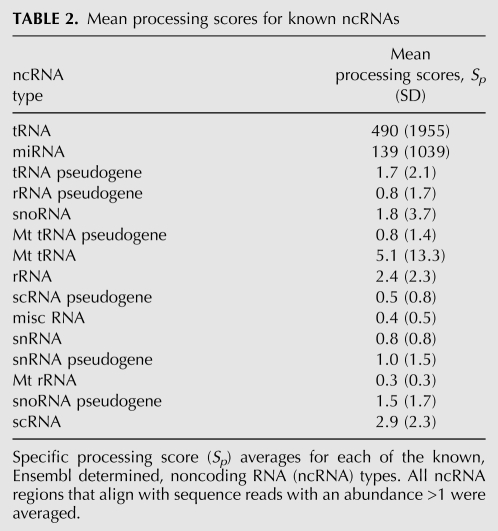
The sequence reads were compared by Vmatch (Abouelhoda et al. 2004) to the miRNA database miRBase (Griffiths-Jones 2004) and the human complement of noncoding RNA (ncRNA) sequences available from Ensembl (Birney et al. 2004). Table 1 details matches to known ncRNAs and Supplemental Table 1 contains all noncoding matches ranked by abundance. The vast majority of matches found were to known tRNA and miRNA sequences, but other RNA species were also represented. For example, the recently published (Ender et al. 2008) snoRNA (ACA45, ENST00000390972) was found in our data set with an abundance of 22 reads and the matching region overlaps that found by Ender and coworkers. Figure 1B details the length distribution of the tRNA and miRNA read sequences. Comparing Figure 1, A and B, it is clear that the two peaks in the overall length distribution correspond to the tRNA (Fig. 1B, dark gray bars) and the miRNA (Fig. 1B, light gray bars) reads, revealing that the tRNA sequences have a preferred length of 19 nt and the miRNAs have a preferred length of 22 nt. This is consistent with the understood maturation of miRNAs following processing by the Dicer enzyme. The reads match to 190 known human miRNA mature sequences as detailed in Supplemental Table 2.
TABLE 1.
Details of reads matching to known ncRNAs determined with Vmatch
Specific processing of miRNAs and tRNAs
The tRNA fragments observed might be the result of random degradation of tRNA, or some specific processing event that leads to 19-mer reads. Visualization of the aligned tRNA sequence reads and comparing to other noncoding RNAs (ncRNAs) shows two types of matching. Ladder-type matching, as illustrated in Supplemental Figure 2A, suggests the reads are the result of random degradation of RNA. In contrast, reads that are “stacked” as shown in Supplemental Figure 2B, with the majority of reads matching to one region of the RNA suggest that the RNA sequences observed are the result of specific processing by cleavage enzymes, such as Dicer in the case of miRNAs.
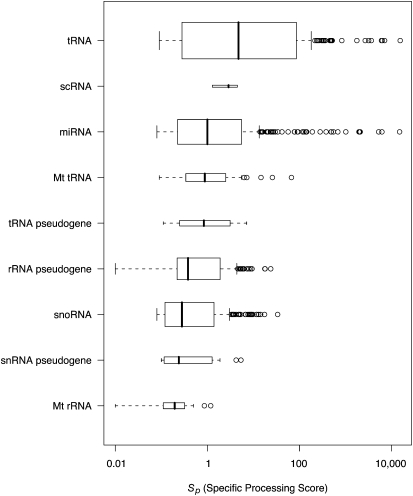
In order to quantify the amount of specific processing observed for the ncRNA matches, a simple metric Sp was defined as the number of reads matching to an RNA region divided by the length of the matching region. A larger Sp indicates a higher propensity for specific processing. Values of Sp for the different ncRNA matches are detailed in Table 2 and box plots shown in Figure 2. miRNAs were found to have a mean Sp of 139, but interestingly, tRNA sequences showed a higher Sp than the miRNAs with a mean Sp of 490 and includes very high scoring regions with Sp of up to 15,333. This is significantly different to the other ncRNAs (P ≤ 6.2 × 10−4 except for the tRNA pseudogenes, scRNA and snoRNA pseudogenes) whose highest Sp values are in the region of 9.3 (e.g., rRNAs) (Fig. 2). Overall, the tRNAs appear to be more precisely processed than the miRNA population as measured by Sp (Table 2), with the exception of miR-21, which had an Sp of 14,893.
TABLE 2.
Mean processing scores for known ncRNAs
FIGURE 2.
Box plot showing the distribution of specific processing scores, Sp. Only noncoding RNA types with an abundance per matching read of >20 (see Table 1) are plotted. The left and right of the boxes represent the first and third quartiles of the data (the interquartile range), and the bold line within the box is the median. The dotted lines represent 1.5 times the interquartile range and data points outside deemed to be outliers (open circles). The height of the boxes represent the amount of data there is with taller boxes having more data.
miRNAs have a well-characterized processing pathway, involving the enzyme Dicer, which cleaves the pre-miRNA hairpin to the mature miRNA at a length of ∼21 nt. The high Sp for the matched miRNAs reflects this processing. Similarly, the high Sp for the matched tRNAs suggests the presence of specific processing for these species.
A number of matches to pseudogenes were also found (Table 2). In particular, matches to 85 rRNA pseudogenes were found with a total abundance of 7222, the fourth most abundant ncRNA in the data set and also more abundant than “real” rRNAs (Table 2). It should be noted that ncRNAs found in Ensembl are annotated by sequence matching to known RNAs of similar function (Griffiths-Jones et al. 2005) and by prediction algorithms such as tRNAscan-SE (Lowe and Eddy 1997). Thus, the matched pseudogene ncRNAs may not be genuine ncRNAs or, as the RNA extracts are from HeLa cells, it is possible that they are a product of uncontrolled transcription known to happen in cancerous cells and may not have a biological function.
miRNA
miRNAs undergo several processing steps before becoming “mature” transcripts. The final step is the cleavage by Dicer of the stem–loop from the hairpin structure formed by the single-stranded RNA molecule. The cleavage produces a short (∼21 nt in length) section of double-stranded RNA where one strand is retained as the mature miRNA molecule and the other strand is degraded. The mature miRNA in association with the Argonaute complex then silences its target messenger RNA as previously described. However, sometimes the complementary strand of the stem is detected, but at much lower levels than the main product, and is referred to as the “star” sequence. Supplemental Figure 4 shows the mature and star sequence reads aligning to the human let-7a-1 hairpin sequence. Reads aligning to the major product can be seen at position 5 on the hairpin sequence with a total abundance of 58,490 reads, whereas the star sequence aligns to position 56 with a total abundance of four reads. A total of 22 miRNAs matching both the major and star regions of the hairpin sequence were found and are detailed in Supplemental Table 3.
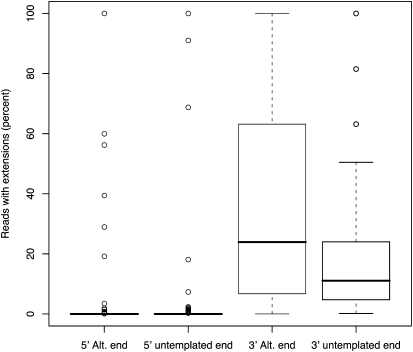
It has been previously noted that in vitro miRNA processing of the 5′ and 3′ ends is not perfect (Azuma-Mukai et al. 2008). In particular, at the 3′ end, reads were found to have additional bases compared with the mature miRNA sequence annotated in miRBase. Figure 3 details the 5′ and 3′ end processing for miRNAs found in vivo in this experiment and shows that the 5′ end of reads are only rarely extended, whereas the 3′ ends show a much greater prevalence of extension (P ≤ 3.8 × 10−10). Extensions can be either “alternative” (i.e., identical to the native hairpin sequence) or “untemplated” (i.e., differing from the native hairpin sequence). As shown in Figure 3 the 3′ ends of miRNA matching reads show a preference for “alternative” reads with an average of 37.6% reads containing this extension (P < 0.05, when compared with 3′ “untemplated” ends), confirming previously published in vitro data (Azuma-Mukai et al. 2008).
FIGURE 3.
5′ and 3′ processing variability of miRNA matching reads. The box plots detail the distributions of the percentage of reads that show extensions at the 5′ and 3′ ends for each of the miRBase annotated mature sequences. To avoid matching to degradation products, extensions at the 5′ end were limited to a maximum of two bases. Two types of extensions are determined for each end; when at least one of the extension bases match the hairpin miRNA sequence (Alt. end), and instances when one or more of the extended bases do not match the hairpin miRNA sequence (untemplated end).
tRNA
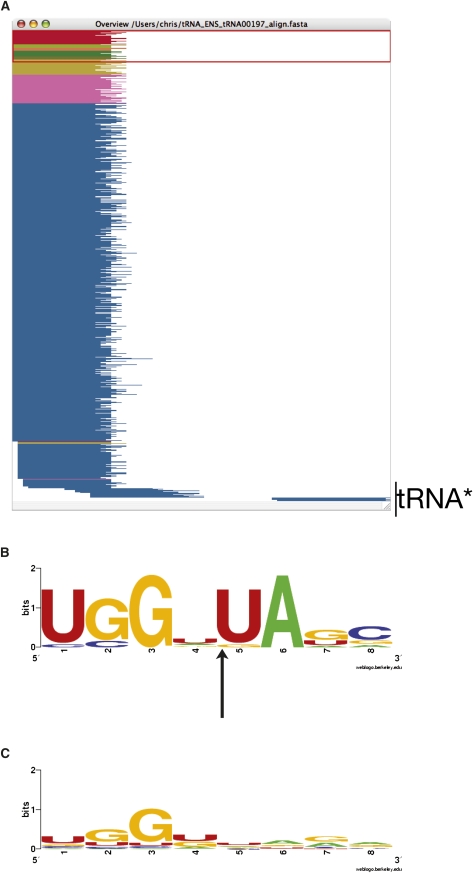
The tRNA sequence matches also have low abundance reads at the 3′ end that are complementary to the high abundance 5′ end. This suggests that they may be being cleaved from a hairpin intermediate via an equivalent process to the miRNAs which produces a mature and a star (*) strand. Figure 4A shows an example tRNA with the major product aligning to position 1 of the whole tRNA sequence with a total abundance of 567,275 reads and a minor product (the star sequence) aligning to position 50 with an abundance of 18. There is some evidence of degradation products from the tRNA near the bottom of the alignment, but overall the alignment is dominated by reads aligning to the 5′ end of the tRNA.
FIGURE 4.
(A) Overview window from Jalview (Clamp et al. 2004; Waterhouse et al. 2009) of sequence reads aligning to a known tRNA sequence as determined by Vmatch. Sequence reads are color coded by abundance with red >500 reads and blue <10 reads. The majority of the reads align to the 5′ end of the tRNA. A small number of complementary sequences can be seen aligning to the 3′ end: tRNA*. (B) Sequence motif logo for tRNA 8mer sequence around the 19–20 position of selective cleavage. Nonredundant set of tRNAs with matching read abundances of at least 2000 (B), and a nonredundant set of all human tRNAs (C). Arrow indicates the tRNA cleavage site. Images were created with WebLogo (Crooks et al. 2004).
The 3′ end of the tRNA matching 19mer reads is located in the D-loop of the tRNA structure. The D-loop forms a stabilizing contact with the T-loop at the conserved nucleotides G18G19 and in Drosophila has been shown to be a required contact for RNase P and 3′-tRNase processing (Levinger et al. 1998). In humans, the GG motif is found at bases 17 and 18. Interestingly, the processing observed here occurs at the base immediately following this motif. Analyzing the sequence motif four bases either side of the tRNA cleavage position shows that the bases 3′ to the cleavage site have a distinct preference for nucleotides U20A21 (Fig. 4B) when compared with a nonredundant set defined from all tRNA sequences (Fig. 4C). The nonredundant set includes the unique 8mer sequences centered around position 19 for all the known human tRNA sequences. Dicer shows a preference for cleavage at positions 5′ to uridine (Aravin et al. 2003) and, thus, is a possible candidate for the selective processing of the tRNAs.
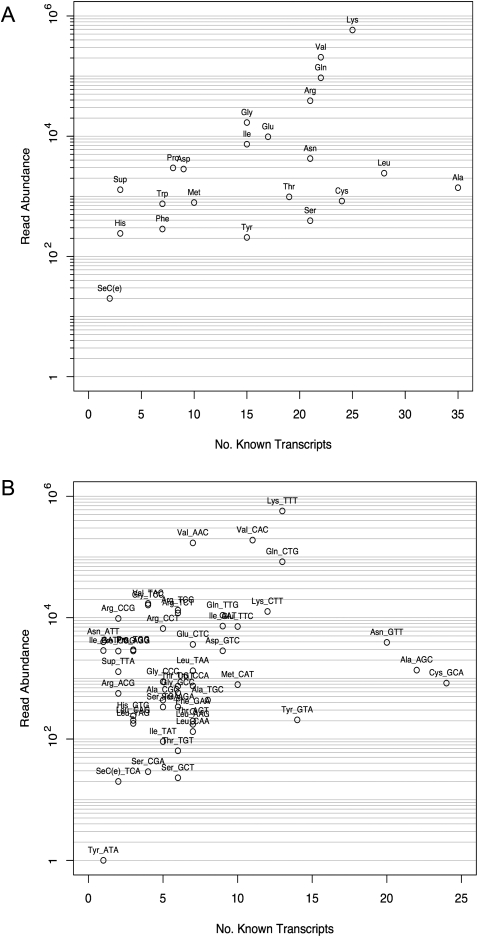
The abundance of tRNA genes is nonuniform across the genome, with the number of tRNA genes varying between 3 (Selenocysteine) and 43 (Alanine), giving 523 tRNA genes in total. Many of the genes are identical at the transcript level, and removing identical representatives leaves 353 unique examples. Figure 5A shows a plot of the number of reads for each tRNA type against the number of unique transcripts and reveals that four types dominate in terms of abundance: lysine, valine, glutamine, and arginine. There does appear to be a general trend where the more abundant tRNAs are those with the most transcripts, but leucine and alanine seem to be underrepresented in abundance. It could mean also, that the tRNAs exist at a basal or steady-state level of ∼10,000 reads or fewer and the high abundance tRNAs show an overexpression over the basal levels.
FIGURE 5.
tRNA abundance by amino acid type and anticodon usage. Scatterplots showing the relationship between tRNA read abundance and the number of unique transcripts for each amino acid type (A) and anticodon usage (B). The number of known transcripts is for all sequence unique transcripts.
Owing to the degenerate nature of mRNA codons required during translation there are several different codons possible for each amino acid. Differences in the abundance of each tRNA anticodon could reveal selection for specific anticodons within tRNA types. Of the high abundance tRNAs mentioned previously, tRNALys and tRNAGln have two anticodons, tRNAVal has three anticodons and tRNAArg has five. Figure 5B shows that four anticodons are present at significantly higher abundance than the others: tRNAVal AAC and CAC, tRNALys TTT, and tRNAGln CTG, each having around 100,000 reads or more, where the remainder have around 10,000 or fewer. The tRNAArg abundance appears to be spread across all five anticodons with none dominating over the majority of the tRNAs. The underrepresented anticodons tRNAAla AGC, tRNAAsn GTT, and tRNACys GCA show a low abundance despite their high number of transcripts.
This suggests that the high abundance tRNAs found in HeLa cells are found at higher levels than expected based on the number of known tRNA genes in the genome, and is even more marked when looking at anticodon abundance. HeLa cells are fast growing cells and have a high requirement for processes involved in translation, such as the availability of tRNAs. The specific requirement for high abundance of tRNAGln, tRNALys, and tRNAVal is not clear in a search for proteins with large requirements for these amino acids (data not shown), but may still be of biological relevance.
Detection of small RNA derived from a tRNA
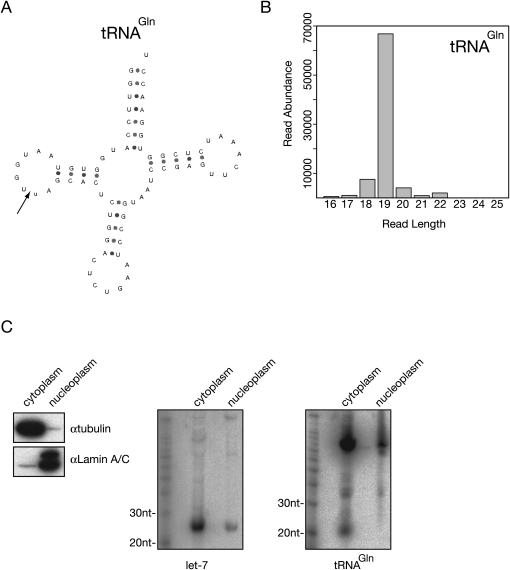
A tRNA that showed high specific processing was chosen from the sequence data analysis for further investigation. tRNAGln (Fig. 6A, total abundance of 82,739) shows a preference for 19mer fragments (Fig. 6B) and has an Sp of 2,669. The abundance and processing score are comparable with miRNA let-7a. An RNA probe, complementary to the tRNAGln 5′ fragment, was used to detect a discrete band at around 20 nt (Fig. 6C) via Northern blotting. This fragment showed exclusive cytoplasmic localization in contrast to the miRNA let-7 that was also detected in the nuclear fraction (Fig. 6C).
FIGURE 6.
Small RNA derived from tRNAGln. (A) Putative folding of tRNAGln, arrow shows the predicted cleavage site. (B) Size distribution of small RNAs derived from tRNAGln. (C) Northern hybridization shows that a ∼20-nt-sized fragment derived from tRNAGln exclusively localized in the cytoplasm (right panel) while the let-7 miRNA also could be detected in the nucleoplasmic fraction (middle panel). The efficiency of cell fractionations was checked with Western blotting for cell fraction specific proteins (left panel; tubulin for cytoplasm, Lamin A/C for nucleoplasm).
Processing of small RNA derived from tRNAGln is Dicer dependent
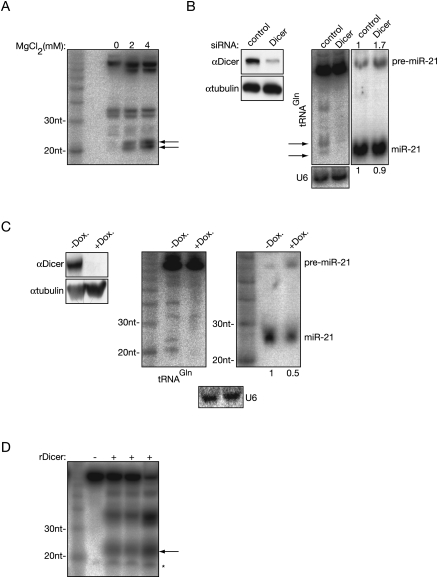
Dicer activity is Mg2+ dependent (Zhang et al. 2002), thus incubating cell extract with varying amounts of MgCl2 may give evidence of Dicer-dependent processing of tRNAGln. Figure 7A shows the effect of incubating HeLa S100 extract with 0, 2, and 4 mM concentration of MgCl2 where the amount of tRNAGln cleavage product at ∼20 nt increases with MgCl2 concentration.
FIGURE 7.
The processing of small RNAs derived from tRNAGln is Dicer dependent. (A) The processing of endogenous tRNAGln into small RNA could be stimulated with MgCl2. S100 extract was supplemented with increasing concentration of MgCl2 and incubated for 1 h at 37°C. The tRNA fragments were detected with Northern hybridization. (B–D) Dicer is required for the generation of the small RNA derived from tRNAGln. (B) Dicer was knocked down with siRNA in HeLa cells. The efficiency of the knock down was tested with Western blotting and tubulin was used as a loading control (left panel). Northern blots show the marked decrease of the tRNA-derived small fragment and the accumulation of pre-miRNA in the Dicer knocked down cells (right panel). U6 was used as a loading control. The relative abundance of pre-miR-21 and mature miR-21 to the U6 loading control was normalized to the RNA level of the control siRNA transfected cells. The result of the quantification is indicated on the top and the bottom of the Northern blot (right panel). (C) Similar experiment described in B, only Dicer was knocked down in 293 cells with doxycyline (Dox.) inducible shRNA. (D) Recombinant Dicer was incubated with uniformly labeled tRNAGln, and the products were separated in 15% denaturing gel. “–,” RNA incubated with reaction buffer only; “+,” independent processing experiments; “arrows,” tRNA-derived small RNAs; and “*,” a nonspecific fragment present in the mock Dicer processing assay.
Suppression of Dicer expression with siRNA as shown in Figure 7B reveals a marked decrease in the small RNA fraction derived from tRNAGln, compared with the nontargeting siRNA, further suggesting that Dicer is involved in the generation of this small RNA species. Since this approach resulted in a very modest effect on the steady state level of miR-21 the experiment was repeated in HEK 293 cells that encode doxycycline inducible siRNA against Dicer (Schmitter et al. 2006). A more efficient knock down of Dicer was achieved that resulted in a significant decrease of endogenous miR-21, and again, it showed that the abrogation of Dicer causes a strong decrease of the steady-state level of tRNAGln small RNAs (Fig. 7C).
Having shown that the production of small tRNA fragments was dependent on having active Dicer present, an in vitro experiment was performed to determine any direct effect of Dicer. Recombinant Dicer was incubated with uniformly labeled tRNAGln precursor and as Figure 7D demonstrates, small fragments were produced from Dicer in triplicate experiments. The pattern of tRNAGln processing in vitro is very similar to the pattern observed in vivo in HeLa S100 extract (Fig. 7A).
Small RNA derived from tRNAGln is poorly associated with Argonaute complexes
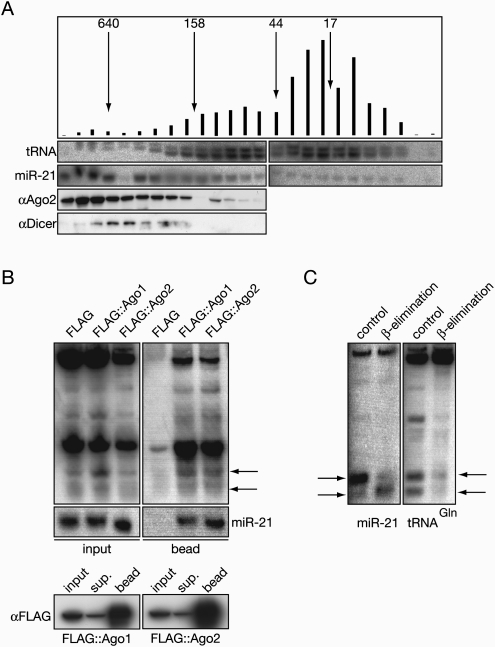
In both Dicer siRNA knock-down experiments, the effect of the decrease of Dicer level on the steady-state level of the small RNA fragment derived from tRNA was more prominent than its effect on the mature miRNA level. This suggests that this type of small RNA is either not incorporated into effector complexes as efficiently as bone fide miRNAs or the turnover rate of the complexes formed by tRNA-derived small RNAs is faster. In order to determine whether small tRNAs could associate with human Ago2, Hela S100 extract was size-fractionated on a Superdex-200 column and the fractions were assayed for the presence of Ago2, Dicer, miR-21, and small tRNA fragments, as shown in Figure 8A. Both small tRNAs and Ago2 were detected in the eluates that correspond to the minimal RISC (Martinez et al. 2002), but the majority of the processed small tRNAs were present in small molecular weight fractions lacking Ago2. Figure 8B shows the immunoprecipitation of human Argonautes with small tRNA fragments. When Flag-tagged hAgo1 and hAgo2 were overexpressed in Hela cells and immunoprecipitated with FLAG, the bound fraction contained tRNAGln small RNAs. However, they were less enriched in the beads than miR-21 (Fig. 8B). Interestingly, larger than 19–21-nt-long tRNA fragments and full-length tRNAs also associate with the immunoprecipitates.
FIGURE 8.
Small RNAs derived from tRNAGln poorly associate with Argonaute proteins. (A) small tRNAGln RNAs partially cofractionate with Ago2. HeLA S100 extract was fractionated through a Superdex-200 column. Every second fraction was tested for Ago2 and Dicer with Western blotting, for miR-21 and small tRNAGln (labeled as tRNA) species with Northern hybridizations. Fractions containing the corresponding size markers (kDa) are indicated with arrows. Vertical bars are representing the level of small tRNA in the corresponding fractions in arbitrary units. (B) RNAs derived from tRNAGln incorporate into Ago1 and Ago2 complexes. FLAG immunoprecipitations were carried out from cells that were transfected with empty FLAG, FLAG∷Ago1, and FLAG∷Ago2 fusion plasmids. The total lysate and the bound fractions were assayed with Northern hybridization using complementary RNA to the sequenced small tRNAGln fragment as a probe. Black arrows indicate the small RNAs derived from the tRNA. (C) small RNAs derived from tRNAGln are resistant to β-elimination. Northern hybridizations show the results of the effect of β-elimination on miR-21 and small RNA processed from tRNA. Black arrows indicate miRNA and small tRNA fragments.
The modification of the 2′ and 3′ OH of the last nucleotide of a miRNA and siRNA could be characteristic of their association with specific Argonaute proteins (Forstemann et al. 2007; Tomari et al. 2007) and the presence of the modification in tRNA-derived small RNA would suggest it may have miRNA or siRNA function. Performing β-elimination reactions on miR-21 and the tRNAGln fragment reveals that tRNAGln is resistant to the treatment in contrast to miR-21 where mobility is increased (Fig. 8C). This suggests that the 3′ terminal ribose of tRNA-derived small RNA is modified. This finding could potentially explain why tRNA fragments are not efficiently incorporated into Ago complexes since human miRNAs are unmodified on the 3′terminal ribose.
DISCUSSION
Deep sequencing of mixed HeLa cell extracts revealed that miRNAs and small RNAs derived from tRNAs are the two major sources of small RNAs in this cell type. The four most abundant tRNAs (tRNALys, tRNAVal, tRNAGln, and tRNAArg) in HeLa cells produce small RNA fragments with an abundance of >20,000 reads each, comparable to the five most abundant miRNAs (miR-21, let-7f-1, let-7a-1, let-7c, and let-7f-2). The tRNAs are almost exclusively processed from the 5′ end with cleavage by Dicer at the D-loop that results in the generation of ∼19-nt-long RNA fragments. We have shown evidence that the tRNA fragments we identified are not random degradation products. First, their pattern shows a very precise processing and the Sp value of these tRNA-derived small RNAs exceed even the Sp of the specifically processed miRNAs. Second, we demonstrated through tRNAGln that the small RNA produced from tRNAs requires the endonuclease Dicer.
Although these small tRNA fragments are generated using Dicer, the same enzyme as miRNAs, they differ from miRNAs in several ways. First, they are more sensitive to the ablation of Dicer than miRNAs. miRNAs are efficiently incorporated into Argonautes, forming a stable complex; therefore, Dicer knock down with siRNAs for a short period of time sometimes does not result in a significant change in the miRNA steady-state level. In contrast, we observed that the tRNA fragment disappeared after a short siRNA treatment against Dicer. Our fractionation and immunoprecipitation experiments suggest that the tRNAGln-derived small RNAs did not efficiently associate with human Argonautes, and the majority of these small RNAs are seen in fractions smaller than the minimal RISC on a size-exclusion column. Therefore, it is likely that small RNAs from tRNAs either do not associate with the proteins at all, or that these small RNAs incorporate into non-Argonaute complexes that have a faster turnover rate.
A second feature that separates tRNA-derived small RNAs from normal mammalian miRNAs is that they are modified at the terminal 3′ ribose as demonstrated by resistance to β-elimination. Plant miRNAs, piRNAs, and siRNAs and fly siRNAs are all methylated at the terminal ribose and miRNAs in these organisms that are sorted into Ago2 complexes are also 3′ modified (Matranga and Zamore 2007). Mammalian miRNAs are free from terminal ribose modifications. We do not know whether the small RNA processed from tRNAGln is modified after Dicer processing or if it is modified prior to processing. The latter would be unsurprising since tRNA bases are heavily modified and ribose 2′-O-methylation is one very common modification.
Specific cleavage of tRNAs has been reported in various organisms. Ago associated tRNA fragments were identified in Schizosaccharomyces pombe in a mutant in which the TRAMP complex, a complex that is responsible for preventing abundant RNAs such as rRNAs and tRNAs to enter the siRNA pathways, was disrupted. This small tRNA fragment is also generated from the 5′ end of the tRNAs; however, it is 23-nt long and its processing is Dicer independent (Buhler et al. 2008). A Dicer-dependent small tRNA fragment processed from the 3′ terminus has been annotated in mouse ES cells from a tRNA, which could alternatively fold into miRNA-like secondary structure (Babiarz et al. 2008). Short, 22-nt-long tRNA fragments corresponding to the 3′ end of certain tRNAs have been sequenced, and in the case of tRNAGlu this fragment was detected with Northern hybridization in several human cells; however, the enzyme that generates such fragments had not been identified (Kawaji et al. 2008). In addition to these short RNA molecules, tRNA cleavage to produce stable half-tRNAs has been reported in Aspergillus fumigatus, where a developmentally regulated endonuclease activity was detected that cleaves tRNAs at the anticodon (Jochl et al. 2008). Similar activity has been recently demonstrated in yeast, plants, and mammalian cells to be a conserved response to oxidative stress (Thompson et al. 2008).
The discovery of Dicer-dependent small tRNA fragments in human cells automatically raises the question of their biological function. It cannot be ignored that a minor fraction of tRNA-derived small RNAs cofractionate and coimmunoprecipitate with Argonautes; therefore, it is plausible that these tRNA-programmed RISCs have messenger RNA targets. However, another possibility—supported by the relatively high turnover rate of these RNA molecules —is that the act of tRNA cleavage itself has a biological effect. It has been shown that improper folding of tRNAs in E. coli produces a hairpin structure that could conceivably cause the tRNA to be a Dicer target (Madore et al. 1999). Why does tRNA become a substrate of Dicer in HeLa cells, while other deep sequenced human cells do not show the accumulation of this species of small RNA? If we exclude the possibility that Dicer-dependent tRNA-derived small RNAs were overlooked in other cells so far (we could not identify similar species of small RNAs in deep sequenced primary neurons) (Remenyi J, Hunter JC, Cole C, Impey S, Mouk EC, Martin K, Barton GJ, Hutvagner G, and Arthur JS, unpubl.) then one explanation is that the accumulation of these RNAs represents an aberrant state of the cells.
One can envision that tRNAs, as an abundant RNA species, compete with pre-miRNAs for Dicer resulting in impaired miRNA homeostasis. Changes in Dicer activity and miRNA level have been proposed multiple times to be a significant factor in cancer development. tRNAs may be accessible by Dicer due to a lack of RNA quality control in HeLa cells, analogous to the mechanism to one that has been described in S. pombe (Buhler et al. 2008). Alternatively, increased tRNA levels might be responsible for the Dicer-dependent processing of tRNAs by forcing Dicer to accept tRNA as a substrate. It has recently been shown that overexpression of tRNA can induce cell transformation, although the exact mechanism is not known yet (Marshall et al. 2008). It is tempting to speculate that there is a connection between the accumulation of the Dicer dependent small tRNAs and the aberrantly elevated tRNA expression in transformed cells and the transformation is caused by the competition between tRNAs and pre-miRNAs for Dicer accessibility, resulting in impaired miRNA homeostasis, particularly as only certain tRNA anticodon types are expressed at high levels and processed. Alternatively, the cytoplasmic miRNA processing machinery may be actively processing misfolded inactive tRNAs as a defense response to tRNA overexpression.
MATERIALS AND METHODS
Antibodies and oligonucleotides
Antibodies and dilutions used for Western blotting were as follows: monoclonal mouse anti-α-tubulin (Sigma), 1:5000; monoclonal mouse antilamin A/C (Santa Cruz), 1:1000; mouse antidicer (Abcam), 1:250; and HRP-conjugated goat–antimouse IgG (Jackson Immunoresearch), 1:10,000. For Northern blot hybridization, the following RNA oligonucleotides were synthesized (Eurofins MWG):
let-7 (5′-UAUACAACCUACUACCUCAUU-3′);
mir-21 (5′-UCAACAUCAGUCUGAUAAGCUA-3′); and
tRNAGln (5′-ACCAUUACACCAUGGAACC-3′).
For U6 hybridization the following DNA oligonucleotides were used:
U6-fwd (5′-GGAACGATACAGAGAAGATTAGCATGGCCCCTGCGCAAGG-3′); and
U6-rev (5′-CCTTGCGCAG-3′).
Oligonucleotides used for siRNA knockdown of Dicer-1 were RNA duplexes (Dharmacon) with the following sense sequences:
5′-UAAAGUAGCUGGAAUGAUGUU-3′;
5′-GAAUAUGGUUGUUUGAAGAUU-3′;
5′-ACACAGCAGUUGUCUUAAAUU-3′; and
5′-GAAUAUCGAUCCUAUGUUCUU-3′.
The nontargeting duplex (MWG) had the following sense sequence:
5′-AGGUAGUGUAAUCGCCUUGdTdT-3′.
For making GlnCTG tRNA, the following DNA oligonucleotides were used:
Transcript: 5′-AGGTCCCAGTGAGACTTGAACTCTGATCACTGTATTCAGAGTCCAAAGTGCTCACCATTACACCATGGAACCCTATAGTGAGTCGTATTACC-3′;
Forward primer: 5′-AGGTCCCAGTGAGACTTGAAC-3′; and
Reverse primer/T7 promoter: 5′-GGTAATACGACTCACTATAGG-3′.
Subcellular fractionation
Cytoplasm and nuclei prepared as per Dignam et al. (1983) were purchased from Biovest International. Nuclei were resuspended in 250 mM sucrose, 10 mM MgCl2, and centrifuged through a sucrose cushion (880 mM sucrose, 0.5 mM MgCl2). Protein for Western blotting was obtained by resuspending equal cell equivalents of material in Laemmli's sample buffer and boiling (5 min, 95°C). For RNA isolation, equal cell equivalents of material were resuspended in 2× proteinase K buffer (200 mM Tris-Cl at pH 7.5; 300 mM NaCl; 25 mM EDTA; 2% (w/v) SDS), and 200 μg proteinase K (VWR) were added. The samples were incubated at 65°C for 1 h and RNA isolated by extraction with phenol:chloroform:isoamyl alcohol (24:25:1) (pH 6.6). RNA was precipitated with 3 vol EtOH (abs) in the presence of 35 μg glycogen as a carrier.
In order to produce cytoplasmic S-100 extract, cytoplasm was obtained as above and mixed with 0.11 vol buffer B (300 mM HEPES at pH 7.9, 1.4 M KCl, 30 mM MgCl2) and then centrifuged at 28,400 rpm (1 h, 4°C, SW41 Ti Rotor). The supernatant was dialyzed against an excess of buffer D (20 mM HEPES at pH 7.9, 20% [v/v] glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT), aliquots flash frozen in liquid nitrogen, and stored at −80°C until required.
Gel fractionation
Twenty milligrams (1 mL) of S-100 extract were injected onto a Superdex-200 XK16/60 gel filtration column (GE Healthcare) and eluted with 0.65 mL/min buffer D. A total of 1.5 mL fractions were collected, of which RNA was precipitated from 500 μL.
Northern blotting
Northern blots used the enhanced sensitivity method described by Pall et al. (2007), and were probed with the RNA oligonucleotides described above after 5′ end labeling with polynucleotide kinase (NEB). Hybridization was done at 37°C overnight and the blots were washed twice for at least 1 h at 37°C in 2× SSC/0.1% (w/v) SDS.
For U6, a DNA probe was synthesized from a single-stranded template using Klenow reagent (Stratagene) on the oligonucleotides described above in the presence of labeled dATP. After synthesis, the duplex was denatured at 95°C and hybridization and washing were performed at 55°C.
Cell culture, transfection, and siRNA knockdown
Four siRNA duplexes targeting the Dicer-1 ORF (sequences as above) were mixed in a 1:1:1:1 ratio to make a final siRNA concentration of 20 μM. A nontargeting duplex was transfected in parallel into control cells.
HeLa cells were transfected in a six-well plate format using oligofectamine (Invitrogen) following the manufacturer's instructions. Cells were harvested 48 h post-transfection, and RNA isolated using Trizol reagent (Invitrogen).
To knock down Dicer-1 in HEK 293 cells, we used HEK 293 cells stably expressing a short hairpin targeting Dicer-1 as described in Schmitter et al. (2006). Briefly, cells were cultured in the presence or absence of 1 μg/mL doxycycline for 6 d with the media and doxycycline replaced on 2, 3, and 4 d post-induction. At 6 d post-induction, RNA was isolated using Trizol reagent (Invitrogen).
S-100 processing assay
HeLa RNA corresponding to 70–80 nt was gel extracted and incubated in a 10 μL final reaction volume with cytoplasmic S-100 extract (40 μg total protein) in buffer D supplemented with 1 mM ATP and varying amounts of MgCl2 at 37°C for 2 h. The cleavage products were run on a gel that was subsequently Northern blotted and probed for tRNAGln.
Dicer assay
A DNA template oligo containing the tRNAGln sequence and T7 RNA polymerase promoter was amplified and made double stranded in a standard PCR reaction (15 cycles). The PCR product was gel purified and used for T7 in vitro transcription in the presence of 32P-radiolabeled UTP. The RNA product was gel purified.
This RNA was incubated in the presence or absence of 1 U recombinant Dicer (Genlantis) in a 10 μL reaction containing 4 μL 2.5× reaction buffer, 1 mM ATP, and 2.5 mM MgCl2 at 37°C for 2 h.
β-Elimination
A total of 10 μg HeLa RNA (2.5 μg/μL), isolated using Trizol reagent (Invitrogen), was resuspended in borate buffer (pH 8.6) (60 mM borax, 60 mM boric acid) and 25 mM NaIO4. The reaction was set up in duplicate. After incubation at 22°C (10 min), the reaction was quenched by addition of 2 μL glycerol. After a further incubation at 22°C (10 min), the RNA pellet was dried in a SpeedVac (45 min, 30°C).
One pellet was resuspended in 50 μL H2O and immediately precipitated as below. The other was subjected to β-elimination by resuspension in borate buffer (pH 9.5, adjusted by addition of NaOH) and incubation (90 min, 45°C). After β-elimination, 150 μL EtOH (abs) and 35 μg glycogen were added and RNA precipitated at −80°C for 1 h. The pellet was recovered by centrifugation, washed in 70% (v/v) EtOH, and resuspended in 15 μL H2O.
Small RNA isolation and sequencing
RNA was isolated with Trizol reagent (Invitrogen) according to the manufacturer's instructions. Small RNA libraries and sequencing was carried out as it was described (Lu et al. 2007)
Database and genome matching
A total of 2,253,266 raw sequence reads were obtained from the instrument. The native RNA sequences were retrieved by removing the 5′ “AG” and 3′ “TCGTATGCCGTCTTCTGCTTGT” adaptor tags. The 3′ adaptor tags were removed by exact matching to the 5′ end of the adaptor sequence with a minimum matching length of four bases. Sequences containing poly-A regions tend to match very frequently to reference genomes, so the 10 reads containing (A)13 were also removed from the data set. In addition, since errors tend to occur toward the 3′ end of reads, the raw reads were clipped in order to retain only the bases which satisfied the following quality protocol. Starting from the 5′ end, a minimum mean quality score of 20 was required over a sliding window of four bases with the reads clipped at the first position where the mean score was below this minimum. The clipped reads were then detagged and collated to include only reads longer than 16nt and read abundance of >1, giving 22,386 unique reads.
The collated reads were matched to annotated noncoding RNAs (ncRNAs) (Ensembl release 50) (Birney et al. 2004) and annotated mature/hairpin miRNA sequences (miRBase release 12) (Griffiths-Jones 2004). tRNA sequences are not present in the ncRNA data set available from Ensembl; thus, they were retrieved as “Simple Features” with a perl script accessing the Ensembl API. Any miRNA annotations found in the Ensembl data were removed in preference for the miRBase annotations on the basis that the miRBase data were more recent. Any reads without matches to either the miRNA or ncRNA datasets were searched against the Human genome and annotations retrieved via the Ensembl perl API. Matching of reads to the RNA sets and the Human genome was performed with the Vmatch algorithm (Abouelhoda et al. 2004), allowing for up to two mismatches per match in all cases.
Specific processing sites
In order to differentiate between read sequences arising from random degradation versus specific processing the following metric was applied to the ncRNA aligned data.
where Sp is the specific processing score, l is the maximal length of the aligned region, and n is the total abundance of sequence reads matching to the aligned region. A higher score indicates a higher presence of regions which appear to be the result of specific processing.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
The work has been funded by the Wellcome Trust and European Framework 6 SIROCCO consortium fund (G.H.), and grant nos. 548569 and 0445638 from the National Science Foundation (P.J.G.). C.C. is funded by the Scottish Funding Council under the Scottish Bioinformatics Research Network (SBRN) grant. G.H. is a Wellcome Trust CD fellow. A.S. is funded by a BBSRC Doctoral Training Grant. S.R.T. was supported in part by National Institutes of Health Training Grant T32 GM-08550. We acknowledge Dr. Thomas Walsh for the technical support of the College of Life Sciences High Performance Computing service, and Dr. David Martin for constructive discussions. We are also very grateful to Dr. Witold Filipowicz for providing the inducible Dicer knock down cells.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1738409.
REFERENCES
- Abouelhoda MI, Kurtz S, Ohlebusch E. Replacing suffix trees with enhanced suffix trees. J Discrete Algorithms. 2004;2:53–86. [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes & Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Andrews TD, Bevan P, Caccamo M, Chen Y, Clarke L, Coates G, Cuff J, Curwen V, Cutts T, et al. An overview of Ensembl. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Buhler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol. 2008;15:1015–1023. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: Annotating noncoding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha–DGCR8 complex in primary microRNA processing. Genes & Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Jochl C, Rederstorff M, Hertel J, Stadler PF, Hofacker IL, Schrettl M, Haas H, Huttenhofer A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008;36:2677–2689. doi: 10.1093/nar/gkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-12164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Levinger L, Bourne R, Kolla S, Cylin E, Russell K, Wang X, Mohan A. Matrices of paired substitutions show the effects of tRNA D/T loop sequence on Drosophila RNase P and 3′-tRNase processing. J Biol Chem. 1998;273:1015–1025. doi: 10.1074/jbc.273.2.1015. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Meyers BC, Green PJ. Construction of small RNA cDNA libraries for deep sequencing. Methods. 2007;43:110–117. doi: 10.1016/j.ymeth.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Madore E, Florentz C, Giege R, Lapointe J. Magnesium-dependent alternative foldings of active and inactive Escherichia coli tRNAGlu revealed by chemical probing. Nucleic Acids Res. 1999;27:3583–3588. doi: 10.1093/nar/27.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L, Kenneth NS, White RJ. Elevated tRNAiMet synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Matranga C, Zamore PD. Small silencing RNAs. Curr Biol. 2007;17:R789–R793. doi: 10.1016/j.cub.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol. 2008a;15:998. doi: 10.1038/nsmb0908-998c. [DOI] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008b;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by Northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Zhang F, Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc Natl Acad Sci. 2006;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34:4801–4815. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Imai H, Minami N. Identification and expression analysis of small RNAs during development. Methods Mol Biol. 2008a;442:173–185. doi: 10.1007/978-1-59745-191-8_13. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008b;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]