Abstract
tRNA splicing is essential for the formation of tRNAs and therefore for gene expression. A circularly permuted sequence of an amber-suppressor pre-tRNA gene was inserted into the sequence encoding the mouse NEMO protein. We demonstrated that, in mouse cells, the hybrid pre-tRNA/pre-mRNAs can be spliced precisely at the sites of the pre-tRNA intron. This splicing reaction produces functional tRNAs that suppress amber codons as well as translatable mRNAs that sustain the NF-κB activation pathway. The RNA molecules extracted from mouse cells were amplified by RT-PCR, and their sequences were determined, confirming the identity of the splice junctions. We then applied the Archaea-express technology, in which an archaeal RNA endonuclease is expressed in mouse cells. We show that both the endogenous eukaryal endonuclease and the archaeal one cleave the hybrid pre-tRNA/pre-mRNAs in the same manner with an additive effect.
Keywords: pre-tRNA splicing, hybrid pre-tRNA/pre-mRNA, archaeal tRNA-endonuclease, NF-κB signaling
INTRODUCTION
Introns interrupt the continuity of many eukaryal genes, and therefore their removal by splicing is a crucial step in gene expression. Within Eukarya, mRNA splicing takes place in the nucleus on a complex and dynamic machine, the spliceosome (Moore et al. 1993), whereas tRNA splicing is catalyzed by three enzymes: the tRNA endonuclease, the tRNA ligase, and a phosphotransferase (Phizicky and Greer 1993; Abelson et al. 1998; Hopper and Phizicky 2003).
In yeast and vertebrates the tRNA-splicing reaction has a different subcellular localization: in Saccharomyces cerevisiae the tRNA-splicing endonuclease is associated with the outer mitochondrial membrane (Yoshihisa et al. 2003, 2007), while in vertebrates this enzyme is localized in the nucleus (Mattoccia et al. 1979; Melton et al. 1980; Paushkin et al. 2004).
We recently showed that in yeast a pre-tRNA sequence contained in a hybrid pre-mRNA/pre-tRNA can be correctly spliced by the tRNA-splicing machinery, producing a functional tRNA and a translatable mRNA (Di Segni et al. 2008). Here, we used the NF-κB signaling pathway present in mouse cells as a model system to determine whether the ability to process a hybrid pre-mRNA/pre-tRNA (hybrid precursor) is common to all eukaryal cells, despite the differences in subcellular distribution of splicing factors between yeast and vertebrates cells. This represents a powerful system to detect a weak signal, since a reporter gene at the end of the pathway is amplified hundreds of times when cells are stimulated by a multitude of extracellular signals. Activation of NF-κB requires the IκB kinase (IKK) complex, which contains two kinase subunits, named IKKα and IKKβ, and a critical regulatory subunit, named IKKγ/NEMO (NF-κB essential modulator). In the absence of NEMO, the IKK complex does not assemble and fails to transmit NF-κB activation signals via the classical pathway (Rothwarf et al. 1998; Yamaoka et al. 1998). Therefore NEMO-deficient cells are blocked in the NF-κB signaling pathway, and the NF-κB-dependent transcription of a reporter gene can be restored by the expression of a low amount of the NEMO protein (Fusco et al. 2004).
RESULTS AND DISCUSSION
Hybrid pre-mRNA/pre-tRNA genes
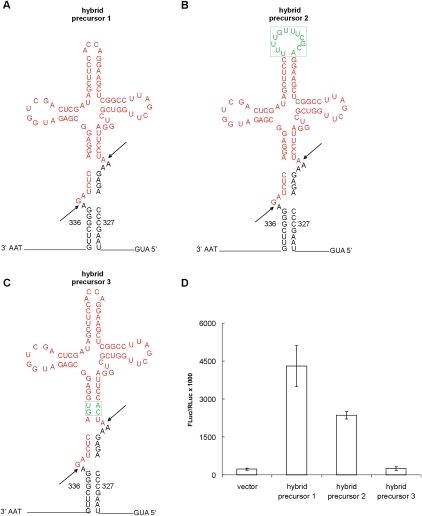
We inserted a circularly permuted sequence of suppressor tRNATyr(Su+) derived from Xenopus laevis tRNATyr (Laski et al. 1982) into the coding sequence of the mouse NEMO gene, under the control of the SV40 Pol II promoter. The native 5′ and 3′ ends of the tRNA were linked by a synthetic loop containing the CCA sequence, two new termini were generated at the tRNA 5′ and 3′ splice sites, and these were joined to the mRNA sequence. A similar permuted tRNA gene organization has been found in Cyanidioschyzon merolae (Soma et al. 2007), a red alga that is likely to represent one of the most ancestral forms of eukaryotes. The mRNA sequence of the hybrid precursor was modified to allow the formation of the bulge–helix–bulge structure (BHB) (Fig. 1A, hybrid precursor 1; Thompson et al. 1989). The BHB structure is typical of archaeal tRNA introns, and we have already demonstrated in vitro and in vivo that it is recognized and efficiently spliced by the eukaryal tRNA-splicing endonuclease (Fabbri et al. 1998; Fruscoloni et al. 2001; Di Segni et al. 2005). It is conceivable that the BHB structure facilitates the folding of the tRNA body. Correct cleavage of the hybrid precursor by the tRNA-splicing endonuclease, followed by RNA ligation and by RNase P cut (Carrara et al. 1995), should release the suppressor tRNA with the mature 5′-end and the 3′-terminal CCA. The other expected product should be the NEMO mRNA with the Leu at position 110 mutated to Pro and a Gly codon inserted at position 113.
FIGURE 1.
Amber suppression in NEMO-deficient cells expressing hybrid mRNA/tRNA precursors. (A) Hybrid pre-mRNA/pre-tRNA structure. Hybrid precursor 1 consists of the mouse NEMO pre-mRNA (in black) and the pre-tRNATyr(Su+) (in red) and contains the BHB motif at the pre-mRNA/pre-tRNA junction. In order to form the BHB structure, the mouse NEMO mRNA was modified from nucleotide 327 to nucleotide 336. We changed T328 into C, T329 into C, G330 into A, and we inserted the 5′-AGG sequence between nucleotide 335 and 336. In the spliced mRNA these changes result in the following modifications: the original Leu110 TTG codon is changed to a Pro CCA codon, the Lys111 AAG codon is changed to a Lys AAA codon, and a Gly GGG codon is inserted after the modified Lys111 AAA codon. Note that the 5′-side is shown on the right and the 3′-end on the left. Arrows indicate 5′ and 3′-splice sites. (B) In hybrid precursor 2, the sequence that contains the Pol III-transcription termination signal is in green and boxed. (C) In hybrid precursor 3, the two-base-pair insert in the anticodon stem of the pre-tRNA is in green and boxed. (D) NEMO-deficient MEFs were cotransfected with 200 ng of the reporter pGL3-Luc Amber, 20 ng of pRL-SV40 plasmid, and 400 ng of the plasmid containing the indicated hybrid precursor gene. Luciferase activity was determined as indicated in Materials and Methods. Data represent the mean value from two independent experiments in triplicate ±1 SD.
Analysis of amber suppression
In order to test whether a functional tRNATyr (Su+) was produced by correct splicing of hybrid precursor 1, NEMO-deficient mouse embryonic fibroblasts (MEFs) were transfected with a plasmid containing the hybrid precursor gene and a reporter plasmid coding for a defective firefly luciferase (FLuc) with a premature amber stop codon (Köhrer et al. 2004). The presence of mature tRNATyr(Su+) in the transfected cells should allow the complete translation of the luciferase gene. As shown in Figure 1D, FLuc activity was detected in extracts derived from these cells; processing of hybrid precursor 1 produces enough mature tRNATyr (Su+) to suppress the amber stop codon. In order to exclude the possibility that the suppressor tRNA is originated by splicing of a pre-tRNA transcribed under the control of the intragenic Pol III promoter, whose elements—the A box and the B box—even if circularly permuted, are indeed present, we substituted the synthetic loop that links the acceptor stem with the termination sequence of the X. laevis tRNATyr gene. The loop of this hybrid precursor, therefore, contains the signal to stop Pol III transcription (Fig. 1B, hybrid precursor 2). FLuc activity measured in the extract obtained from cells expressing this hybrid precursor is 50% of the activity measured in the case of hybrid precursor 1 (Fig. 1D): this activity is, therefore, presumably due to the processing of the hybrid precursor whose transcription is under the control of a Pol II promoter.
It was previously shown that, in vivo, an artificial pre-tRNA containing a BHB motif with a 2-base-pair (bp) insert in the anticodon stem is efficiently cleaved by the X. laevis tRNA endonuclease and that a mature transcript that is 4 nucleotides (nt) longer than mature wild-type tRNA is produced (Fabbri et al. 1998; Fruscoloni et al. 2001). This mode of action was called the mature-domain independent mode since the endonuclease fixes the sites of cleavage of this particular substrate not by reference to the mature domain, as in natural eukaryal pre-tRNA, but by recognition of local structures at the intron–exon boundaries. We thus set out to test whether in mouse cells the hybrid precursor could be cleaved in the mature-domain independent mode. Two base pairs were inserted in the anticodon stem of the hybrid precursor (Fig. 1C, hybrid precursor 3). Although no functional suppressor tRNA could be expected from this precursor, either if processed in the mature-domain dependent mode or in the independent one (Fig. 1D), the specificity of the cleavage could be verified by the analysis of the mRNA.
Analysis of spliced mRNA
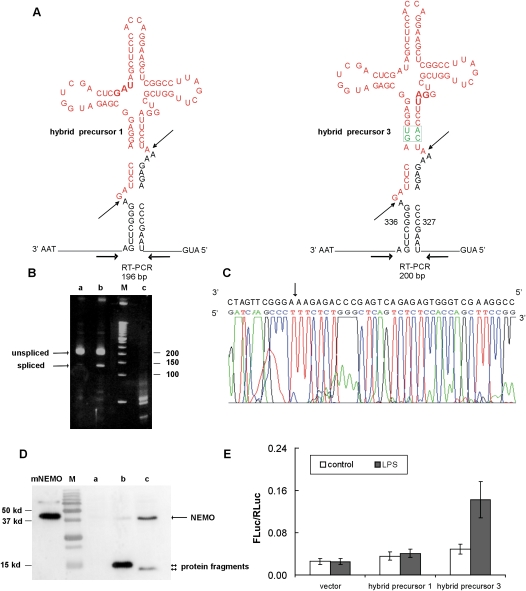
Total RNAs were obtained from NEMO-deficient MEFs transfected with plasmid encoding the indicated hybrid precursor 1 and 3 (Fig. 2A). The precursor and the putative spliced product were simultaneously amplified in a PCR reaction by using specific primers corresponding to sequences upstream of and downstream from the tRNA sequence inserted into the pre-mRNA (Fig. 2A). To increase the visibility of the putative spliced product, we exploited the fact that the X. laevis tRNATyr(Su+) gene bears a Bsu36I site. After Bsu36I digestion of the RT-PCR products derived from the unspliced precursor, the reaction was reamplified with the same pair of primers and loaded on a polyacrylamide gel. As shown in Figure 2B, lanes a and b, we detected in each lane a major band with the size expected for the unspliced precursor and, below it, a minor band whose size corresponds to the spliced product. From the intensity of the putative spliced product bands, hybrid precursor 3 (Fig. 2B, lane b) appears to be a better substrate than hybrid precursor 1 (Fig. 2B, lane a). We sequenced the DNA purified from the acrylamide gel and confirmed that the amplified products in both bands were indeed exactly as expected for a splicing reaction occurring at the sites typical of tRNA introns (Fig. 2C, Supplemental Fig. 1A). The observation that the spliced product derived from processing of hybrid precursor 3 has the same sequence as the product derived from hybrid precursor 1 (see the sequence comparison in Supplemental Fig. 1A) allowed us to conclude that the mouse tRNA-endonuclease cleaves hybrid precursor 3 in the mature-domain independent mode, recognizing local structure at the intron–exon boundaries. However, it is worth pointing out that the tRNA structure is important for the processing of these hybrid precursors since we could not detect splicing in mRNA precursors containing the BHB structural motif alone, i.e., without the mature tRNA domain. The presence of the mature domain could contribute to the stabilization of the BHB (data not shown).
FIGURE 2.
The spliced mRNA is properly ligated and translated. (A) Structures of hybrid precursors 1 and 3 with the positions of the RT-PCR primers used for the experiments shown in B and C. Stop codons are indicated in boldface type. (B) Analysis of mRNAs. NEMO-deficient MEFs were transfected with the plasmid containing the indicated hybrid precursor gene. Twenty-four hours after transfection, total RNA was isolated, reverse-transcribed, amplified, and analyzed on acrylamide gel. (Lane a) Transfection with hybrid precursor 1-SV40; (lane b) transfection with hybrid precursor 3-SV40; (lane M) 50-bp ladder (Invitrogen); (lane c) no transfection. Fragment sizes (in bp) are indicated. (C) Sequence analysis of the spliced product. Chromatogram of the sequence of the cDNA purified from the gel primed with P23-reverse primer. The arrow indicates the splice junction. (D) Analysis of the proteins: Splicing leads to synthesis of full-length NEMO protein. NEMO-deficient MEFs were transfected with an empty plasmid (lane a), a plasmid containing the hybrid precursor 1 gene (lane b), or a plasmid containing the hybrid precursor 3 gene (lane c). Total cell lysate (50 μg) was analyzed by Western blot using a horseradish peroxidase-conjugated 3F10 anti-HA antibody. Lane mNEMO is a positive control where 4 μg of total cell lysate of cells transfected with wild-type NEMO plasmid were loaded; (lane M) Bio-Rad Precision Protein Standards. Two-headed arrows indicate the short protein encoded by the unspliced hybrid precursors. (E) analysis of NF-κB signaling pathway. NEMO-deficient MEFs were cotransfected with vectors containing the gene for the indicated hybrid precursor together with NF-κB-Luc reporter and pRL–SV40 plasmid. After 20 h, cells were treated with LPS or not (control). The ratio of firefly luciferase activity to Renilla luciferase activity (FLuc/RLuc) in each sample served to normalize FLuc activity. Values are shown as the mean ±1 SD from four independent experiments in triplicate.
Analysis of proteins and fragments
To determine whether spliced mRNAs are indeed full length and capable of being translated, NEMO-deficient MEF cells were transfected with a plasmid with the coding sequence for the HA epitope added at the 5′ end of the hybrid precursors. The insertion of the tRNA sequence in the hybrid precursors causes a frameshift of the reading frame that results in the presence of several stop codons, the first of them being indicated in boldface type in Figure 2A. Efficient translation can occur only on correctly spliced mRNA where the open reading frame is restored. Western blotting analysis of lysates of cells transfected with the indicated two plasmids showed in both cases the presence of a band at the expected position for NEMO protein (∼45 kDa) (Fig. 2D). We observed a difference in the amount of NEMO protein produced from the two spliced precursors in agreement with the RT-PCR product amounts. The two-headed arrows in Figure 2D, lanes b and c, indicate the fragments produced from the unspliced hybrid precursors that contain the premature termination codons, in lane c shorter than in Figure 2D, lane b, as expected.
Functional analysis of proteins
NEMO was shown to be essential for NF-κB activation, and its deficiency results in a complete block of NF-κB activation by proinflammatory stimuli (Schmidt-Supprian et al. 2000). In unstimulated cells, NF-κB is present as a latent and inactive complex in the cytoplasm. When a cell receives any stimulatory signal, for example, the bacterial lipopolysaccharide (LPS), NF-κB rapidly enters the nucleus and activates gene expression. Almost all signals that lead to activation of NF-κB converge on the activation of a high molecular weight complex that contains at least three distinct subunits: IKKα, IKKβ, and NEMO. IKKα and IKKβ are related catalytic kinase subunits whereas NEMO is a regulatory subunit. We tested whether the proteins derived from the splicing of the NEMO-hybrid precursors complement the defect in NEMO-deficient MEFs and therefore support induction of the NF-κB signaling pathway by LPS. The plasmids encoding the hybrid precursors were cotransfected into NEMO-deficient MEF cells with a luciferase reporter construct. Transfected cells were treated with LPS to enhance NF-κB activity, and FLuc activity was measured 4 h later. The results in Figure 2E show that LPS induces the NF-κB-dependent transcription of the luciferase gene only in cells expressing hybrid precursor 3 (Fig. 2E). This experiment allows us to conclude that the product of splicing of precursor 3 is a functional protein. Hybrid precursor 1 does not give a significant response, as could be expected due to the fact that it is processed to a low extent (Fig. 2B) and produces only a modest amount of NEMO protein (Fig. 2D). It is possible to speculate that, given the reduced number of base-pairings, hybrid precursor 1 is less stable than hybrid precursor 3 and, therefore, it is poorly processed.
Effect of the application of the Archaea-express technology
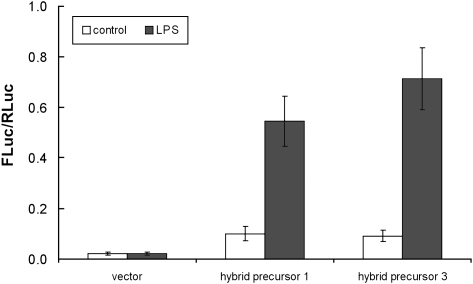
The pre-tRNA/pre-mRNA hybrid precursors were weakly processed by the endogenous splicing machinery, and the NF-κB signaling pathway was only slightly activated. Consequently, we asked if a more efficient NF-κB-dependent gene activation could be achieved in these cells by application of the Archaea-express technology. We have shown that the tRNA endonuclease from the archaebacterium Methanococcus jannaschii (MJ endonuclease), when expressed in mammalian cells, cleaves RNAs that contain the archaeal BHB structure and that the resulting cleavage products are subsequently joined together by an endogenous ligase (Deidda et al. 2003). The presence of the BHB motif should confer to the hybrid precursors the substrate characteristics that are recognized by the archaeal enzyme. As shown in Figure 3, cotransfection of a plasmid expressing the archaeal endonuclease results in a robust luciferase activity driven by NF-κB-dependent gene activation in cells expressing not only hybrid precursor 3 but also hybrid precursor 1. Total RNAs were isolated and subjected to RT-PCR analysis. We found that the precursors are correctly cleaved at the boundaries pre-tRNA/pre-mRNA (see the chromatograms in Supplemental Fig. 1B).
FIGURE 3.
Induction of the NF-κB pathway in cells expressing the M. jannaschii endonuclease. NEMO-deficient MEFs were cotransfected with pOPTI-MJ and the plasmid containing the indicated hybrid precursor gene. Twenty hours after transfection, cells were treated with LPS or not (control). Values are shown as in Fig. 2.
In cells expressing both endonucleases, processing of hybrid precursor 1 gives rise to a strong activation of the NF-κB pathway that was not observed at all in the absence of the archaeal enzyme. Precursor 3 promotes a more highly efficient activation when MJ endonuclease is present. These results point to an additive effect, implying that both the archaeal and endogenous eukaryal endonucleases cleave the pre-tRNA/pre-mRNA precursor in the same manner. The processing of both precursors results in the activation of the NF-κB pathway, even in the absence of LPS since in cultured cells there is a basal level of NF-κB activity (see, for instance, Fontan et al. 2007). It is worthwhile to notice that this activity requires the presence of the NEMO protein, since it does not occur in the control samples.
In this work we have shown that pre-mRNAs, in which pre-tRNA sequences have been inserted, are cleaved at the specific pre-tRNA splicing sites and ligated in mouse cells. The enzyme responsible for the cleavage reaction is presumably the tRNA-splicing endonuclease. In fact, the splice sites of the hybrid precursors are precisely those one would expect for a cleavage performed by this enzyme. The most likely candidate to ligate the cleaved pre-mRNA sequences is the tRNA ligase, producing both a mature functional tRNA and a correctly translatable mRNA. We can conclude that, despite the fact that the localization of the tRNA-splicing endonuclease is not conserved between vertebrates and yeast, mouse cells share with yeast cells the ability to recognize and process the tRNA splice sites contained in a hybrid pre-mRNA/pre-tRNA.
MATERIALS AND METHODS
Plasmids containing hybrid precursor genes
The 1245-bp DNA fragment encoding the mouse NEMO protein was amplified by PCR (primer pairs P1-P2) from pBlueNemo (a gift from M Pasparakis, University of Cologne) and inserted into the NcoI/XbaI sites of pGL3 Control (Promega) to generate pNEMO-SV40. To construct the DNA sequence of hybrid precursor 1, overlap extension PCR was performed (Ho et al. 1989). The hybrid gene was amplified using primer pairs P3-P2, P6-P2, P7-P2, P8-P2, P5-P4, and P5-P2, and pNEMO-SV40 as template. The last PCR product was digested with the restriction enzymes NcoI and PstI and cloned into the NcoI/PstI sites of pNEMO-SV40. To obtain hybrid precursor 2-SV40, the Pol III-termination sequence 5′-GCTTTGTTTT-3′ was introduced at the 3′-end of the acceptor stem in hybrid precursor 1, replacing the two 5′-CA-3′-nucleotides by PCR with primer pairs P6-P11, P1-P9, and P1-P10. The last two PCR fragments were digested and cloned into the NcoI/PstI sites of pNEMO-SV40. Hybrid precursor 3-SV40 was generated as hybrid precursor 1 with primer pairs P3-P2, P12-P2, P13-P2, P14-P2, P5-P4, and P5-P2.
Plasmids encoding the HA epitope at the N terminus of NEMO-derived proteins
In order to increase the level of expression of the hybrid precursor RNAs, their genes were placed under the control of the cytomegalovirus (CMV) promoter. The DNA sequences of NEMO, hybrid precursor 1, and hybrid precursor 3 were amplified by PCR from the respective SV40-plasmids with primers P15 and P16. The PCR products were digested with EcoRI and NotI and ligated to pEGFP-N1 (Clontech) restricted with XhoI-NotI, together with an EcoRI/XhoI linker consisting of the coding sequence for the HA-epitope (MYPYDVPDYA-) (oligonucleotides P17 and P18). These plasmids allow expression of proteins with an HA-tag at the N terminus.
Plasmid encoding the luciferase amber reporter
A single-base mutation from T to G at position 210 introduces a premature amber stop codon into the coding sequence of the firefly luciferase gene. The reporter pGl3-Luc Amber was constructed by PCR site-directed mutagenesis (Ho et al. 1989) with primer pairs P19-P20, P5-P21, and P5-P20, and pGL3 Control as template. The modified luciferase gene was reinserted into the NcoI and XbaI sites of pGL3 Control.
Transfection of NEMO-deficient MEFs
NEMO-deficient MEFs (E8 cells) were kindly provided by M Pasparakis (University of Cologne) (Schmidt-Supprian et al. 2000). E8 cells were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 2 mM glutamine, 100 U/mL of penicillin, and 100 μg/mL of streptomycin at 37°C in a 5% CO2 atmosphere. Twenty to 24 h before transfection, cells were subcultured into 24-well plates for the luciferase assay or in 6-well plates for RNA and protein isolation. Cells were transfected with plasmid DNA using JetPei (Polyplus transfection) according to the manufacturer's instructions.
Luciferase assay
To evaluate the NF-κB-dependent activation of the firefly luciferase reporter gene, plasmid with the hybrid precursor gene (400 ng/well) was cotransfected together with the reporter NF-κB-TA-Luc (Clontech) (200 ng/well). To normalize luciferase activity, the Renilla luciferase expression vector pRL-SV40 (Promega) was added (20 ng/well) to the transfection mixture as a control reporter. In the experiment of Figure 3, cells were cotransfected with 400 ng/well of pOPTIMJ (Deidda et al. 2003). Twenty hours post-transfection, cells were treated with LPS (Escherichia coli; Sigma) at 4 μg/mL for 4 h, harvested, and lysed in Passive lysis buffer (Promega). Firefly and Renilla luciferase activities were measured using the Dual luciferase reporter assay system (Promega) on a Lumat LB 9507 luminometer (EG&G Berthold). Luciferase activity is given as the ratio between firefly and Renilla activities (FLuc/RLuc). Error bars correspond to one standard deviation. The values shown represent the averages of at least three independent experiments.
To evaluate the suppression activity, cells were cotransfected with the pGl3-Luc Amber reporter (200 ng/well) together with the Renilla luciferase expression vector pRL-SV40 (20 ng/well). Twenty-four hours after transfection, cells were arrested and lysed, and firefly and Renilla luciferase activities were measured. The data are represented as the ratio of firefly to Renilla luciferase activity multiplied by 1000.
Total RNA isolation from cultured cells
E8 cells were transfected with 1.5 μg/well of plasmid DNA containing the hybrid precursor gene-SV40. Cells were harvested 24 h post-transfection. Total RNA was isolated using the Trizol reagent (Life Technologies) following the manufacturer's instructions. RNA (10 μg) was treated with 10 units of RQ1 DNase (RNase free; Promega) for 30 min at 37°C and extracted with phenol/chloroform. Single-stranded cDNA was obtained by poly(dT)-primed reverse transcription of 3 μg of total RNA with SuperScript III RT (Life Technologies). The cDNA was amplified by PCR using the primer pair P22 and P23 and 30 cycles with the Accuprime Taq DNA Polymerase System (Invitrogen). PCR products were digested with the restriction enzyme BSU36I and submitted to a new round of 30 cycles with the same pair of primers. PCR products were electrophoresed on 8% polyacrylamide gels.
Sequencing of RT-PCR products
Spliced and unspliced amplified cDNAs were eluted from the polyacrylamide gels and directly sequenced. Sequence reactions were done using the Bigdye Sequencing Kit (Perkin Elmer−Applied Biosystem), primed with reverse primer P23. Sequences were analyzed on an ABI Prism 310 (Applied Biosystem).
Western blot analysis
Cells were transfected with 3 μg/well of plasmid DNA containing the hybrid precursor gene–CMV. Thirty-six hours post-transfection, total cellular protein was isolated by freeze-and-thaw method. Protein concentration was measured by the dye-binding assay using protein assay reagent (Bradford Protein Assay, Bio-Rad) according to the manufacturer's instructions. A total of 50 μg of protein were loaded onto SDS/PAGE gels. After transfer, the membrane was probed with horseradish peroxidase-conjugated anti-HA (rat monoclonal anti-HA-Peroxidase, High affinity 3F10 Roche). The blots were developed using the ECL+ Western blotting detection system (Amersham Biosciences).
Oligodeoxyribonucleotides
The entire list of oligodeoxyribonucleotides can be found in Supplemental Table 1.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank the members of our laboratory for helpful discussions and G. Di Segni for help with the manuscript. We are grateful to RH Haselkorn for critical reading of the manuscript. We greatly thank G Di Franco for excellent technical assistance and A Ferrara and T Cuccurullo for secretarial work. This study was supported by Italian Ministry of Research grants (FIRB-G, Armenise Harvard Foundation 2005, FIRB-Idee Progettuali 2005, and PRIN-Cofin. 2007) and European Union Framework Programme 6 contracts (MUGEN, EURASNET, and EUMODIC).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1841609.
REFERENCES
- Abelson J, Trotta CR, Li H. tRNA splicing. J Biol Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- Carrara G, Calandra P, Fruscoloni P, Tocchini-Valentini GP. Two helices plus a linker: A small model substrate for eukaryotic RNase P. Proc Natl Acad Sci. 1995;92:2627–2631. doi: 10.1073/pnas.92.7.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deidda G, Rossi N, Tocchini-Valentini GP. An archaeal endoribonuclease catalyzes cis- and trans-nonspliceosomal splicing in mouse cells. Nat Biotechnol. 2003;21:1499–1504. doi: 10.1038/nbt908. [DOI] [PubMed] [Google Scholar]
- Di Segni G, Borghese L, Sebastiani S, Tocchini-Valentini GP. A pre-tRNA carrying intron features typical of Archaea is spliced in yeast. RNA. 2005;11:70–76. doi: 10.1261/rna.7138805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Segni G, Gastaldi S, Tocchini-Valentini GP. Cis- and trans-splicing of mRNAs mediated by tRNA sequences in eukaryotic cells. Proc Natl Acad Sci. 2008;105:6864–6869. doi: 10.1073/pnas.0800420105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri S, Fruscoloni P, Bufardeci E, Di Nicola Negri E, Baldi MI, Attardi DG, Mattoccia E, Tocchini-Valentini GP. Conservation of substrate recognition mechanisms by tRNA splicing endonucleases. Science. 1998;280:284–286. doi: 10.1126/science.280.5361.284. [DOI] [PubMed] [Google Scholar]
- Fontan E, Traincard F, Levy SG, Yamaoka S, Veron M, Agou F. NEMO oligomerization in the dynamic assembly of the IκB kinase core complex. FEBS J. 2007;274:2540–2551. doi: 10.1111/j.1742-4658.2007.05788.x. [DOI] [PubMed] [Google Scholar]
- Fruscoloni P, Baldi MI, Tocchini-Valentini GP. Cleavage of non-tRNA substrates by eukaryal tRNA splicing endonucleases. EMBO Rep. 2001;2:217–221. doi: 10.1093/embo-reports/kve040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco F, Bardaro T, Fimiani G, Mercadante V, Miano MG, Falco G, Israël A, Courtois G, D'Urso M, Ursini MV. Molecular analysis of the genetic defect in a large cohort of IP patients and identification of novel NEMO mutations interfering with NF-κB activation. Hum Mol Genet. 2004;13:1763–1773. doi: 10.1093/hmg/ddh192. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Phizicky EM. tRNA transfers to the limelight. Genes & Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- Köhrer C, Sullivan EL, RajBhandary UL. Complete set of orthogonal 21st aminoacyl-tRNA synthetase-amber, ochre and opal suppressor tRNA pairs: Concomitant suppression of three different termination codons in an mRNA in mammalian cells. Nucleic Acids Res. 2004;32:6200–6211. doi: 10.1093/nar/gkh959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laski FA, Belagaje R, RajBhandary UL, Sharp PA. An amber suppressor tRNA gene derived by site-specific mutagenesis: Cloning and function in mammalian cells. Proc Natl Acad Sci. 1982;79:5813–5817. doi: 10.1073/pnas.79.19.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoccia E, Baldi MI, Carrara G, Fruscoloni P, Benedetti P, Tocchini-Valentini GP. Separation of RNA transcription and processing activities from X. laevis germinal vesicles. Cell. 1979;18:643–648. doi: 10.1016/0092-8674(79)90119-3. [DOI] [PubMed] [Google Scholar]
- Melton DA, De Robertis EM, Cortese R. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature. 1980;284:143–148. doi: 10.1038/284143a0. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Query CC, Sharp PA. Splicing of precursors to mRNA by the spliceosome. In: Gesteland RF, Atkins JF, editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 303–357. [Google Scholar]
- Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell. 2004;117:311–321. doi: 10.1016/s0092-8674(04)00342-3. [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Greer CL. Pre-tRNA splicing: Variation on a theme or exception to the rule? Trends Biochem Sci. 1993;18:31–34. doi: 10.1016/0968-0004(93)90085-2. [DOI] [PubMed] [Google Scholar]
- Rothwarf DM, Zandi E, Natoli G, Karin M. IKK-γ is an essential regulatory subunit of the IκB kinase complex. Nature. 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- Schmidt-Supprian M, Bloch W, Courtois G, Addicks K, Israël A, Rajewsky K, Pasparakis M. NEMO/IKKγ-deficient mice model incontinentia pigmenti. Mol Cell. 2000;5:981–992. doi: 10.1016/s1097-2765(00)80263-4. [DOI] [PubMed] [Google Scholar]
- Soma A, Onodera A, Sugahara J, Kanai A, Yachie N, Tomita M, Kawamura F, Sekine Y. Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science. 2007;318:450–453. doi: 10.1126/science.1145718. [DOI] [PubMed] [Google Scholar]
- Thompson LD, Brandon LD, Nieuwlandt DT, Daniels CJ. Transfer RNA intron processing in the halophilic archaebacteria. Can J Microbiol. 1989;35:36–42. doi: 10.1139/m89-006. [DOI] [PubMed] [Google Scholar]
- Yamaoka S, Courtois G, Bessia C, Whiteside ST, Weil R, Agou F, Kirk HE, Kay RJ, Israël A. Complementation cloning of NEMO, a component of the IκB kinase complex essential for NF-κB activation. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T. Possibility of cytoplasmic pre-tRNA splicing: The yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell. 2003;14:3266–3279. doi: 10.1091/mbc.E02-11-0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Ohshima C, Yunoki-Esaki K, Endo T. Cytoplasmic splicing of tRNA in Saccharomyces cerevisiae. Genes Cells. 2007;12:285–297. doi: 10.1111/j.1365-2443.2007.01056.x. [DOI] [PubMed] [Google Scholar]