Abstract
Microdissection techniques have the potential to allow for transcriptome analyses in specific populations of cells that are isolated from heterogeneous tissues such as the nervous system and certain cancers. Problematically, RNA is not stable under the labeling conditions usually needed to identify the cells of interest for microdissection. We have developed an immunolabeling method that utilizes a high salt buffer to stabilize RNA during prolonged antibody incubations. We first assessed RNA integrity by three methods and found that tissue incubated in high salt buffer for at least 20 h yielded RNA of similar quality to that for RNA extracted from fresh-frozen tissue, which is considered highest quality. Notably, the integrity was superior to that for RNA extracted from tissue processed using rapid immunolabeling procedures (5 min total duration). We next established that high salt buffer was compatible with immunolabeling, as demonstrated by immunofluorescent detection of dopamine neurons in the brain. Finally, we laser microdissected dopamine neurons that were immunolabeled using high salt buffer and demonstrated that RNA integrity was preserved. Our described method yields high quality RNA from immunolabeled microdissected cells, an essential requirement for meaningful genomics investigations of normal and pathological cells isolated from complex tissues.
Keywords: RNA degradation, transcriptome preservation, quantitative PCR, microfluidics, capillary electrophoresis
INTRODUCTION
Evaluation of complete transcriptomes from various cells/tissues, at different developmental stages, with or without pathology, is an important goal for biologists and clinicians. It is now appreciated that >90% of the genome may be transcribed and that the transcriptome is far more complex than the traditional view of simply being a collection of protein-coding and structural RNAs (Birney et al. 2007; Carninci et al. 2008). Characterization of the various transcripts is a critical aspect of genomics, where accurate determination of the relative abundance of transcripts could be used for the identification of cell phenotype, the elucidation of cell function, and for the investigation of pathological processes. Indeed, tissue-specific transcriptomes are now being reported (Marioni et al. 2008; Mortazavi et al. 2008), the molecular taxonomy of the brain is underway (Sugino et al. 2006), and cancers can now be characterized and their prognoses predicted based on expression profiles (van't Veer and Bernards 2008). Transcript expression can be quantified either on a transcript-by-transcript basis using real-time PCR, by microarray analysis in which tens of thousands of transcripts are measured, or by next-generation RNA sequencing (RNA-seq) methods, which can assay millions of transcripts (Shendure and Ji 2008). With all these approaches, the accuracy and validity of the data are dependent on the quality of the sample RNA. Even minimal RNA degradation can lead to the loss of transcriptome complexity, compromising the ability to detect low abundance transcripts, such as the all important signaling molecules, for example. Therefore, certain RNA quality criteria must be met in order for transcriptome analyses to be meaningful. First, transcripts in the extracted sample need to reflect the in situ complexity, both in terms of the relative abundances and the dynamic range of the various transcripts. Second, if all forms of transcripts are to be detected, for example splice variant characterization by next-generation sequencing (Mortazavi et al. 2008), the full length of transcripts needs to be preserved.
To date, the majority of transcriptome studies have extracted RNA from homogenates of tissues. This practice seriously limits the potential of genomics, since important cell-specific transcript information is lost because of mixing and dilution effects. Characterizing transcriptomes in specific cells or cell populations is a problematic area of genomics, especially in highly heterogeneous tissue such as the brain. Techniques have been developed to enable collection of particular cells from mixed populations and generally involve either laser-assisted microdissection or fluorescence activated cell sorting (FACS) purification of dissociated cells (Espina et al. 2006; Lobo et al. 2006). In contrast to FACS, microdissection can be applied to most tissue types, including plant and bone. Target cells need to be identified and while it is possible in some cases to do this morphologically (e.g., Lobsiger et al. 2007), when this is not possible, a cell-specific label must be employed. This can be achieved either by immunolabeling or using cell-specific promoters to drive expression of fluorescent proteins (Grimm et al. 2004; Lobo et al. 2006). The latter approach obviates the need for an immunolabeling step but limits investigations to the availability of transgenic mice with appropriate cell-specific promoter–marker protein combinations. While the diversity of such transgenic lines is improving (Gong et al. 2003) there remain thousands of proteins of potential interest as well as the obvious interest in human and other nonmurine samples. Consequently, immunolabeling remains a vital tool for cell-specific genomics.
Unfortunately, the immunolabeling process itself results in rapid degradation of RNA and, therefore, loss of transcriptome (Fend et al. 1999; Gjerdrum et al. 2004). Once tissue is removed from fixative and placed into aqueous buffer, RNA degrades rapidly (Fend et al. 1999). Rapid immunolabeling procedures have been developed in an attempt to overcome this problem (Fend et al. 1999; Gjerdrum et al. 2004), but these have been inadequately validated with regard to RNA integrity. Usually the qualitative presence of PCR product or ribosomal RNA is reported as the only evidence of RNA preservation (Grimm et al. 2004). Furthermore, rapid procedures may not result in sufficient labeling for all antibodies, compromising the ability to reliably identify the cell of interest. Rapid immunolabeling has been successful in some instances (e.g., Greene et al. 2005), but its lack of routine use, even when the same target cell population is being studied (e.g., Yao et al. 2005; Lammel et al. 2008), likely indicates difficulties in obtaining adequate labeling or RNA integrity. Consistent with this view, close inspection of rRNA bands in the work of Grimm et al. (2004) reveals significant degradation after just 2 min in immunolabeling buffer containing RNase inhibitor (Grimm et al. 2004), and even relatively minor degradation during sample preparation can result in erroneous gene expression differences (Auer et al. 2003). Our primary goal was to develop a method that would allow extraction of the best quality RNA from relatively homogeneous cell populations, obtained by immunolabeling and microdissection. We report here that the addition of molar concentrations of NaCl to immunolabeling buffer results in superior protection of RNA, even for prolonged incubations, without compromising immunofluorescence detection of specific cells. This high salt buffer immunolabeling method will facilitate transcriptome analyses of microdissected cells.
RESULTS
Ribosomal RNA
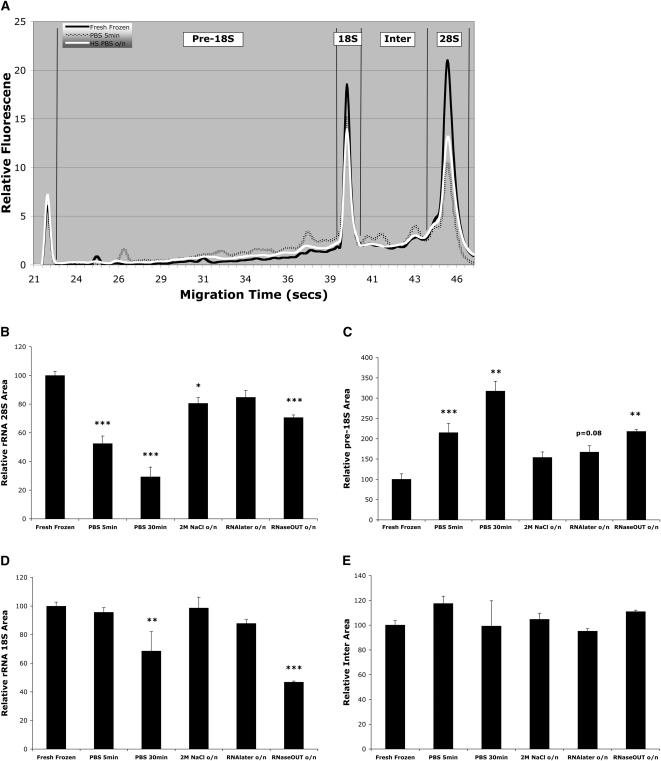
Ribosomal RNA (rRNA) intactness is the most commonly used indicator of RNA integrity. To accurately quantify the integrity of rRNA, we electrophoresed total RNA using microfluidic electrophoresis and compared four regions of the resulting electropherograms (Fig. 1A), similar to previous reports (Auer et al. 2003; Schroeder et al. 2006). RNA was extracted either from fresh-frozen (control) midbrain sections or sections that were ethanol fixed, and then incubated in PBS or PBS with various RNase inhibitors. As can be seen in Figure 1A–E, overnight incubation in high salt buffer (2M NaCl in PBS) was the most effective at maintaining rRNA integrity. There was no significant increase in pre-18S area and only a 20% decrease in 28S area with overnight high salt buffer (Fig. 1A–C). In contrast, rapid (5 min) PBS incubation resulted in a 120% increase in pre-18S area and a 50% decrease in 28S area (Fig. 1A–C). With the exception of prolonged (30 min) PBS and overnight RNaseOUT incubations, there were no significant differences between conditions for the 18S and Inter areas of the electropherograms (Fig. 1D,E).
FIGURE 1.
Effect of buffer conditions on the integrity of ribosomal RNA (rRNA). (A) Electropherogram showing rRNA integrity for fresh-frozen tissue (black line), which represents the best quality RNA, or for brain sections that were ethanol-fixed and then incubated in either PBS for 5 min (hatched line) or overnight (o/n) in 2M NaCl PBS (white line). To quantify the effect of buffer condition on rRNA, the electropherogram was divided into four regions for comparison: (B) the 28S region; (C) the pre-18S region; (D) the 18S region; (E) the Inter region (see Materials and Methods for details). 28S rRNA intactness is considered a sensitive indicator of RNA integrity in general. Note, even a PBS incubation of only 5 min duration resulted in a 50% reduction in 28S area and a 120% increase in the pre-18S area. In contrast, overnight incubation in 2M NaCl or RNA Later resulted in markedly less degradation. Data are the means (+SEM). * P < 0.05, ** P < 0.01, *** P < 0.001; n ≥ 5.
Transcript RNA
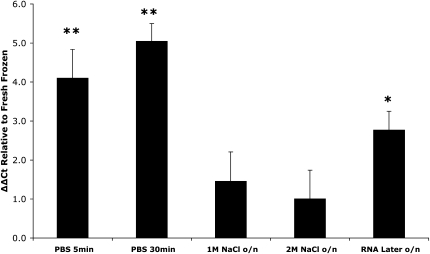
rRNA integrity may not accurately reflect the integrity of all types of transcripts. The relative abundance of messenger RNA (mRNA), the RNA species of interest in gene expression analyses, was used as an indicator of preservation of nonstructural RNAs. We determined the effect of buffer conditions on transcript levels for nine genes. 18S rRNA was chosen as the reference gene for normalization and was co-reversed transcribed with mRNA using a combination of 18S rRNA-specific primers and oligo(dT) (Zhu and Altmann 2005). Figure 2 and Table 1 show the effects of buffer condition on transcript level, as compared with that found in fresh-frozen tissue (ΔΔCt method). The pattern of stability was remarkably similar for all nine mRNAs, with overnight high salt buffer conferring the greatest stability for all genes, followed by RNA Later, and then the rapid PBS incubation. Indeed, the 2M NaCl condition was never significantly different to the fresh-frozen control condition and, averaged across all genes, there was ninefold less transcripts in samples from the rapid PBS incubation compared with those from the overnight 2M NaCl condition.
FIGURE 2.
Effect of buffer conditions on the level of mRNA transcripts relative to 18S rRNA, for the housekeeping gene Actb. The 18S rRNA was chosen as the reference gene as our electropherogram data indicated it was relatively stable under the various conditions used. Data are expressed as the means (+SEM) of the ΔΔ threshold cycle (Ct). First, the Ct for each gene (see Table 1 for the other eight genes) was normalized to the Ct for the 18S rRNA in that condition. Second, the ΔCt(Ctgene − Ct18S tRNA) for each experimental condition was normalized to the ΔCt(Ctgene − Ct18S tRNA) for the fresh-frozen condition (see Materials and Methods for details). Note that a similar pattern of transcript stability was found for each gene, with only the overnight (o/n) high salt conditions, in particular 2M NaCl, being consistently unchanged from the fresh-frozen condition (see Table 1). * P < 0.05, ** P < 0.01; n = 3.
TABLE 1.
Effect of buffer conditions on the relative levels of mRNA transcripts compared with controls
Transcript completeness
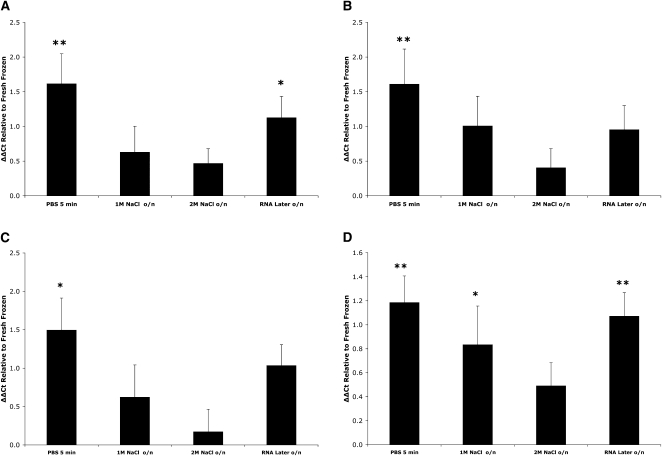
We next determined whether complete transcripts were being preserved. Representation of full-length transcripts is important in the analysis of splice variants and can be assessed using qPCR to compare the 5′ and 3′ levels of individual genes. qPCR primers were designed to amplify the 5′ and 3′ regions for β-actin (Actb), glyceraldehyde-3-phosphate dehydrogenase (Gapdh), tyrosine hydroxylase (Th), and the dopamine transporter (Slc6a3). The relative 5′ and 3′ levels for each gene, under the various incubation conditions, were compared with those obtained for fresh-frozen tissue. A similar degradation pattern resulted for each of the four genes, with overnight incubation in high salt buffer showing better preservation of full-length transcripts, when compared with rapid PBS incubation or RNA Later (Fig. 3A–D). Notably, the relative 5′–3′ levels for each of the four genes in the 2M NaCl overnight incubation were not significantly different from those found for fresh-frozen tissue, indicating complete preservation of full-length transcripts. In contrast, the relative 5′–3′ levels were 1.6–4.6 times lower in rapid PBS incubations compared with overnight high salt, indicating marked loss of the 5′ region of the transcripts.
FIGURE 3.
Effect of buffer conditions on the stability of full-length transcripts. The relative levels of the 5′ and 3′ ends of the mRNA for each of the genes, Actb (A), Gapdh (B), Th (C), and Slc6a3 (D), were determined. As in Figure 2, data are expressed as the means (+SEM) of the ΔΔCt, with the 5′–3′ ΔCt for each buffer condition normalized to the 5′–3′ ΔCt for the fresh-frozen condition. Again, only the overnight (o/n) high salt incubations, and the 2M NaCl buffer in particular, resulted in transcript completeness unchanged from that observed with the fresh-frozen condition. * P < 0.05, ** P < 0.01, n ≥ 5.
Transcript integrity following immunolabeling and laser capture microdissection
Our primary goal was to develop a method that would allow extraction of the best quality RNA from relatively homogeneous cell populations, obtained by immunolabeling and microdissection. Midbrain dopamine neurons were immunofluorescently labeled for Th, with all steps in the immunolabeling procedure carried out using high salt (2M NaCl) buffer. As is evident in Figure 4A, dopamine neurons are readily identifiable after high salt immunolabeling. Approximately 100–200 dopamine neurons were microdissected for each experiment (Fig. 4A). Since accurate rRNA quantification is difficult with extremely small samples and normal gene expression levels are not known for these neurons, we used the 5′–3′ assay to estimate the level of RNA degradation. The relative 5′–3′ levels for Gapdh, Actb, and Th were unchanged for RNA extracted from microdissected dopamine neurons compared with RNA extracted from fresh-frozen tissue. There was a small but significant decrease in the relative 5′–3′ levels for Slc6a3 (Fig. 4B).
FIGURE 4.
Effect of immunolabeling and laser microdissection on mRNA quality in microdissected dopamine neurons. (A) Laser-microdissected dopamine neurons. (Left) Tyrosine hydroxylase (TH) immunofluorescent neurons in ventral midbrain. Fresh-frozen midbrain was cryosectioned at 6–10 μm, the sections ethanol-fixed and immunolabeled for TH overnight in 2M NaCl PBS as described in detail in the Materials and Methods section. (Right) Image showing the midbrain section in the left panel after laser microdissection and catapulting a number of the immunofluorescently identified dopamine neurons. (B) The relative levels of the 5′ and 3′ ends of four genes were compared between laser-microdissected dopamine neurons (LCM) and fresh-frozen tissue (FF). Three of the four genes were not significantly different to FF. Data are expressed as the means (+SEM) of the 5′–3′ ΔCt. ns, not significantly different to FF; * P < 0.05; n = 6.
DISCUSSION
It is now clearly evident that if meaningful transcriptome analyses, whether at the single transcript or whole transcriptome level, are to be obtained, use of high quality RNA is critical (Auer et al. 2003; Imbeaud et al. 2005; Fleige and Pfaffl 2006; Lipska et al. 2006; Nolan et al. 2006; Copois et al. 2007; Thompson et al. 2007). Genomics approaches continue to be criticized because of inconsistent results (Miklos and Maleszka 2004; Draghici et al. 2006), and while some of this inconsistency arises from data processing (Shi et al. 2008), differences in sample RNA integrity markedly affect transcript quantification and likely are the cause of much of the inconsistency (Auer et al. 2003; Lipska et al. 2006; Atz et al. 2007; Thompson et al. 2007; Popova et al. 2008). Sample RNA of the highest quality is therefore imperative. To overcome RNA quality issues that arise when immunolabeling for microdissection, we have developed a method that allows prolonged antibody incubations without compromising RNA integrity.
The transcriptome is comprised of many RNA species that can be broadly divided into protein coding and noncoding RNAs. In order to evaluate the integrity of the transcriptome, we assessed the nonprotein coding, ribosomal RNA (rRNA) species and messenger RNA (mRNA) for the protein-coding component. Evaluating multiple RNA species is important, especially as a consensus for the best measure of RNA quality has not been reached (Imbeaud et al. 2005; Nolan et al. 2006). First, we assessed rRNA intactness, the most commonly used indicator of RNA integrity. The recent advent of microfluidic electrophoresis has made it possible to more accurately quantify rRNA degradation than traditional gel electrophoresis methods (Auer et al. 2003; Schroeder et al. 2006). In all cases we compared rRNA integrity to that obtained with fresh-frozen tissue, as this is considered to yield RNA of highest quality. Degradation of rRNA results in electrophoretic migration of smaller RNA fragments that appear in the pre-18S (“fast”) region and the region between the 28S and 18S bands, the Inter region, of electropherograms. We found RNA degradation effects were most pronounced and observed first with the 28S rRNA species, with degradation products appearing in the pre-18S region (see Fig. 1A–C; Schroeder et al. 2006). The pre-18S region and 28S peak are sensitive indicators of rRNA integrity, and while 18S/28S ratios have proved a poor predictor, comparison of pre-18S, 18S, and 28S regions is a reliable measure of RNA quality (Auer et al. 2003; Atz et al. 2007; Weis et al. 2007). We found relatively short (5 min) incubations in RNase-free aqueous buffer resulted in substantial rRNA degradation, with a 120% increase in the pre-18S area and 50% reduction in 28S. Consistent with these quantitative data, Grimm et al. (2004) show a reduction in rRNA after an even briefer (2 min) incubation (Grimm et al. 2004, see their Fig. 1C). These findings indicate that, when rRNA is accurately quantified, it is possible to detect degradation that might impact transcriptome analysis outcomes. Thus, rapid immunolabeling protocols may not adequately preserve RNA. Various salt solutions have long been used in RNA extraction protocols, and preliminary work in our laboratory indicated that addition of NaCl to immunolabeling buffers resulted in a concentration-dependent preservation of RNA, with the threshold at 0.5M and optimum at 2M (data not shown). Overnight incubation of cryosectioned, ethanol-fixed tissue in 2M NaCl buffer resulted in almost complete preservation of rRNA, with only a 20% drop in 28S area, an outcome clearly superior to that obtained with short duration PBS incubation. Also of note, 2M NaCl buffer was similar in effect to RNA Later (Ambion) and superior to RNaseOUT (Invitrogen), two commonly used commercial RNase inhibitors.
Analysis of the protein-coding component of the transcriptome requires intact mRNA. Typically, however, it is rRNA and not mRNA integrity that is assessed, primarily due to the relative abundance and ease of evaluation of rRNA. Of note though, rRNA integrity may not necessarily be a reliable indicator of the integrity of other transcripts (Weis et al. 2007). We therefore assessed whether the prolonged preservation of rRNA achieved with high salt buffer was also effective with mRNA. Different transcripts have varying stabilities; so quantitative PCR (qPCR) analyses were carried out for a variety of genes that encode membrane, cytoskeletal, neurotransmitter synthesizing, metabolic enzyme, vesicle, and transcription factor proteins. The choice of an appropriate reference gene for normalization is critical in gene expression comparisons and the geometric mean of multiple housekeeping genes has been proposed as a superior strategy compared with use of a single gene (Vandesompele et al. 2002; Huggett et al. 2005). However, in the present study, aqueous buffer incubation may affect the integrity of housekeeping genes, making them an unreliable internal reference. It has previously been shown that normalization genes are differentially affected by RNA degradation, meaning genes chosen from intact RNA samples may not be appropriate for normalization with degraded samples (Pérez-Novo et al. 2005). In contrast to a number of mRNAs, the 18S rRNA has been shown to be a relatively stable and reliable internal reference for qPCR normalization (Zhu and Altmann 2005). Moreover, our present studies show 18S rRNA to be very stable across conditions; only in prolonged PBS incubations did we see a reduction in 18S area (see Fig. 1D). Therefore, we chose 18S rRNA as our reference for normalization. The overall pattern of stability was relatively consistent; however, there were marked differences between genes in any one condition. For example, after 5 min in PBS there was a gene-dependent five- to 22-fold reduction in transcript level relative to fresh-frozen control (Fig. 2; Table 1). Differential effects of RNA degradation on gene transcripts have been previously reported and further underscore the importance of normalization choice (Pérez-Novo et al. 2005; Thompson et al. 2007). Surprisingly, 30-min PBS incubation did not result in a proportional reduction in transcripts relative to that found for 5-min PBS. This could be due to there being a highly labile population of transcripts that degrade rapidly. Overall these effects are consistent with our rRNA results and demonstrate that relatively short duration exposure to aqueous buffer results in substantial degradation of mRNA transcripts. In marked contrast, addition of either 1M or 2M NaCl for incubations lasting at least 20 h resulted in almost complete protection of transcripts. There were no significant reductions in transcript levels compared with fresh-frozen control, except for two genes with 1M NaCl (Table 1). Somewhat surprisingly, RNA Later, while better than PBS, did not afford the same degree of transcript protection as 2M NaCl, with four genes showing significant reduction in transcript levels compared with fresh-frozen control (Fig. 2; Table 1). Therefore, in terms of preservation of mRNA transcript abundance, high salt buffer is superior to short incubation in aqueous buffer and maintains transcripts for prolonged periods, potentially allowing for improved immunolabeling.
As the nature of genomes and their transcribed sequences become more fully characterized, the incredibly complex nature of the transcriptome is becoming increasingly evident (Birney et al. 2007). Consequently, transcriptome analyses that are limited to probing primarily the 3′ “end” of the transcriptome will no longer be sufficiently comprehensive. For example, if analyses of splice variants are the focus, ways need to be developed that ensure representation of complete transcripts. In order to assess whether the entire lengths of transcripts were being preserved, we used the 5′–3′ assay for two representative housekeeping and two cell-specific genes. This assay is considered the most reliable indicator of mRNA transcript integrity, as it is an independent measure (Nolan et al. 2006). Consistent with our rRNA and relative mRNA transcript abundance assays, we found that overnight incubation in 2M NaCl buffer resulted in the best preservation of full-length transcripts, with all four genes assayed being indistinguishable from fresh-frozen controls (Fig. 3A–D). That is, there were similar relative levels of the 5′ and 3′ ends. Again, short-duration PBS incubation resulted in substantial loss of 5′ levels for each gene relative to 3′. For example, for both Actb and Gapdh, there was on average threefold less 5′ relative to 3′. Therefore, in terms of ensuring preservation of full-length transcripts, which translates to more complete coverage of the transcriptome, high salt buffer is very effective.
Many thousands of microarray studies have been published, yet relatively few have used microdissected cells as source material. This is somewhat surprising given the importance of characterizing the molecular profiles of specific cells, especially when dealing with heterogeneous tissue such as brain or invasive cancers. Even fewer studies have combined immunolabeling and microdissection, which probably reflects the difficulty obtaining quality RNA and/or sufficient labeling with rapid labeling protocols. Our primary goal was to develop a method that would permit incubation durations typical of standard immunolabeling protocols for optimal labeling, yet at the same time preserve the transcriptome. The preceding paragraphs describe the effectiveness of high salt buffer for RNA preservation, so we then determined whether high salt buffer was compatible with immunolabeling and microdissection, using midbrain dopamine neurons as the cells of interest. One concern was whether high salt buffer would interfere with immunolabeling efficiency. As can be seen from Figure 4A, immunofluorescent dopamine neurons can be readily identified following immunolabeling in high salt buffer. In fact, we were able to immunolabel for TH with 3M NaCl buffer, but there was no improvement in RNA integrity and there was a slight decrease in labeling intensity (data not shown). It should be noted that RNA Later was completely incompatible with immunolabeling, we were never able to obtain TH immunofluorescence even with diluted RNA Later (data not shown). Furthermore, our high salt buffer immunolabeling did not compromise the ability to laser-microdissect the dopamine neurons, indicating that it does not interfere with the preparatory steps and the actual laser-assisted microdissection itself (Fig. 4A). The 5′–3′ assay, carried out on RNA extracted from microdissected dopamine neurons, indicated that, with the exception of Slc6a3, there was no significant difference in transcript completeness compared with fresh-frozen controls. The small loss in Slc6a3 5′ is indicative of variation in stability between transcripts, as alluded to above. Overall, high salt buffer is compatible with overnight antibody incubations, which are routine practice for immunolabeling, and with laser-assisted microdissection.
Midbrain dopamine neurons have been the focus of an enormous amount of research, primarily because of their role in a wide range of CNS functions. Dysfunction or degeneration of these neurons leads to pathologies such as Parkinson's disease and addiction. A number of microdissection studies investigating molecular events in dopamine neurons have been carried out (Backes and Hemby 2003; Grimm et al. 2004; Chung et al. 2005; Greene et al. 2005; Yao et al. 2005; Cantuti-Castelvetri et al. 2007; Stephenson et al. 2007; Gründemann et al. 2008; Lammel et al. 2008). However, not all have immunolabeled to identify dopamine neurons. Histological stains (Cantuti-Castelvetri et al. 2007; Stephenson et al. 2007) and presence of tracers transported back from known targets of dopamine neurons (Yao et al. 2005; Gründemann et al. 2008; Lammel et al. 2008) have been used as surrogate markers for the dopamine phenotype. While these approaches may be adequate for certain types of study, they are limited because histological stains are not cell-specific and tracers may be found in other, nondopaminergic neurons projecting to the same target region. Presumably, TH immunolabeling was not carried out in these studies because of the known RNA degradation issues with routine immunolabeling protocols. The most unequivocal way to identify dopamine neurons, or other cell types, is through immunolabeling for a cell-specific marker. Indeed, in the brain this approach could be combined with tracers to enable subpopulations, based on their targets, to be defined, resulting in an even more refined analysis (Margolis et al. 2006). A few groups have microdissected immunolabeled dopamine neurons (Backes and Hemby 2003; Grimm et al. 2004; Chung et al. 2005; Greene et al. 2005). However, RNA quality was either not assayed (e.g., Backes and Hemby 2003), or was done qualitatively based on the presence of a couple of genes (Chung et al. 2005; Greene et al. 2005) or the intactness of rRNA bands (Grimm et al. 2004). Thus, from these studies it would not be possible to accurately determine to what degree the transcriptome had been compromised and therefore to what degree the results accurately profile the cell. From the preceding examples it is clear there is a need for improved ways to stabilize the transcriptome while allowing immunolabeling for the purpose of identification of specific cell types. The method we have developed and demonstrated will resolve compatibility issues between RNA degradation and immunolabeling.
The immense complexity of the transcriptome is becoming increasingly appreciated, as is the need to study it comprehensively. To date, most genomics studies have focused on whole tissues, making it difficult to determine the precise cellular origin of transcriptomic changes. Microdissection of identified cells overcomes this difficulty and will greatly facilitate analysis of cell-specific transcriptomes. In the present study, we have demonstrated that, by the addition of molar concentrations of NaCl to immunolabeling buffer, RNA is preserved for relatively prolonged periods. Moreover, this preservation is compatible with standard immunolabeling and microdissection procedures, as demonstrated by our results with dopamine neurons. High salt buffer immunolabeling, in combination with modern microdissection techniques, will enable comprehensive genomics investigations on specific cell populations.
MATERIALS AND METHODS
Animals and tissue preparation
All animal housing and procedures were carried out in strict accordance with the University of Newcastle's Animal Care and Ethics Committee regulations, the NSW Animal Research Act and Regulations, and the Australian Code of Practice for the care and use of animals for scientific purposes. All tissue was obtained from 3 to 6 mo old, male Sprague Dawley rats. Animals were killed by pentobarbitone overdose and the brains rapidly removed and cooled in ice-cold PBS. Brains were then blocked into forebrain, midbrain, and hindbrain regions on an ice-cold stage and the blocks were then frozen in dry-ice chilled isopentane. Tissue was stored at −80°C until needed. Midbrain blocks were cryosectioned at 6–10 μm in the coronal plane and sections mounted onto RNase-free glass microscope slides. Slides were stored at −80°C until required.
Tissue incubation and RNA extraction
Frozen midbrain sections were rinsed in PBS (15 sec) and either immediately processed for RNA extraction (fresh-frozen controls) or fixed in 70% ethanol (5 min). Fixed sections were then briefly rinsed in PBS (5 sec) to remove excess ethanol and incubated in various buffers to mimic antibody incubations, as follows: standard PBS for 5 or 30 min; PBS with 1M or 2M NaCl overnight; PBS with 2.0 U/μL RNaseOUT (Invitrogen) overnight; RNA Later (Ambion) overnight. Note that RNA Later was used as a positive control for RNA preservation as it has previously been shown to be an effective RNase inhibitor. However, RNA Later is completely incompatible with immunolabeling procedures. All solutions were ice-cold and DEPC-treated where appropriate. All tissue incubations in the various buffer solutions were carried out at 4°C for the stated durations. Total RNA was extracted and contaminating genomic DNA (gDNA) removed by in-solution DNase-I digestion, followed by RNA Cleanup. RNA extraction, clean up, and DNA digestion were done using Qiagen's RNeasy mini kit and DNase reagents, according to the manufacturer's instructions. Concentration of pre- and post-DNase-treated RNA was determined by absorbance spectroscopy. It is recommended that column-based RNA purification methods be used with high salt-treated samples, as these incorporate multiple wash steps that remove excess salts that may potentially affect downstream processes such as cDNA synthesis.
RNA integrity assessment
The effects of the various incubation conditions on RNA integrity was determined at both the ribosomal RNA (rRNA) and messenger RNA (mRNA) levels.
rRNA
To accurately determine the effects on rRNA, total RNA was capillary electrophoresed on an Experion microfluidics station (Bio-Rad) and the resulting electropherograms compared. One microliter from each isolated total RNA sample was loaded onto a StdsSens chip and processed according to the manufacturer's instructions. Electropherograms were generated by the software, Experion Version 2.1, and were divided into pre-18S, 18S, Inter (18S–28S interval), and 28S regions. Partitioning the electropherogram in this manner was based on previous studies that addressed the issue of objective quantification of rRNA integrity (Auer et al. 2003; Schroeder et al. 2006). For each condition, the four designated areas under the electropherogram curve were quantified to determine the amount of RNA present in each region and then normalized to total area to control for variation in amount of RNA loaded on to the chip. Ninety five percent of total RNA in a cell is comprised of rRNA, the 18S, and 28S rRNA species. In nondegraded RNA samples, the vast majority of RNA will be represented by the 18S and 28S peaks in the electropherogram. In degraded samples, 18S and particularly the 28S peak decrease while there is a concomitant increase in degradation products in the pre-18S and Inter regions. Thus, in degraded samples, 18S and 28S areas decrease and pre-18S and Inter areas increase, relative to nondegraded samples. A minimum of five independent experiments were carried out.
mRNA
Reverse transcription
18S rRNA was chosen as the reference gene because our rRNA results indicated it was relatively stable. Furthermore, it cannot be determined, a priori, whether commonly used references genes, such as housekeeping genes, would be consistently and equally affected under buffer conditions used. A co-reverse transcription method (Zhu and Altmann 2005) was used to transcribe both the poly-A tail containing mRNAs and the poly-A tail lacking 18S-rRNA, in a single reaction using oligo(dT)20 primers and an 18S-specific primer, respectively. Reverse transcription was carried out using SuperScript III (Invitrogen), according to the manufacturer's instructions. Briefly, 100–500 ng of total RNA, 2.5 μM oligo(dT)20 primers, 0.5 μM 18S-rRNA specific primer (5′-GGAACTACGACGGTATCTGA-3′), 1 μL of 10 mM dNTP, and molecular biology grade water to 13 μL were mixed and heated for 5 min at 65°C, then chilled on ice for 1 min; 4 μL 5× First-Strand Buffer, 1 μL 0.1M DTT, 1 μL RNaseOUT (40 U/μL) and 1 μL SuperScript III RT (200 U/μL) were added and the mixture incubated for 60 min at 50°C, followed by 70°C for 15 min. Reverse transcription without reverse transcriptase was also carried out to assess gDNA contamination.
Real-time quantitative PCR (qPCR)
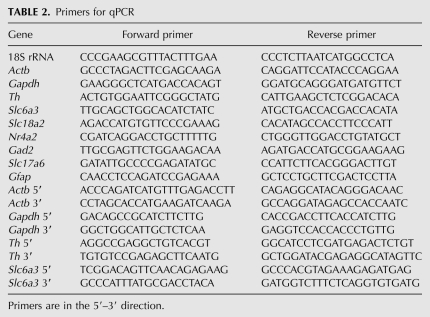
All qPCR primers (Table 2) were designed using standard qPCR primer design criteria with the web-based software, Primer 3. Reactions were carried out in 20 μL volumes containing: 10 μL 2× SensiMixPlus SYBR (Quantace); 200 nM each of forward and reverse primers, except for 18S RNA, where 1 μM was used; 2–20 ng cDNA; molecular biology grade water to 20 μL. After an initial 10-min, 95°C enzyme activation step, 40 cycles of 95°C for 30 sec (step 1) followed by 60°C for 30 sec (step 2) were completed. Melt curves were generated to confirm the presence of a single PCR product. Primers were deemed specific if a single amplified product was detected by both melt curve analysis and gel electrophoresis. Reactions were carried out on an ABI 7500 Real-Time PCR System (Applied Biosystems) and analyzed using the Applied Biosystems 7500 Sequence Detection Software (Version 1.4). For each gene, all conditions were run on the same plate in triplicate and each complete comparison was done three times. The effect of buffer condition on transcript levels was evaluated quantitatively for the housekeeping genes, β-actin (Actb) and glyceraldehyde-3-phosphate-dehydrogenase (Gapdh); the catecholaminergic genes, tyrosine hydroxylase (Th), the dopamine transporter (Slc6a3), the nuclear orphan receptor, NURR1 (Nr4a2), and the vesicular monoamine transporter 2 (Slc18a2); the glutamatergic gene, vesicular glutamate transporter 2 (Slc17a6); the GABAergic gene, glutamic acid decarboxylase 65 (Gad2); and the glial gene, glial fibrillary acidic protein (Gfap). Delta Ct (ΔCt, threshold cycle) was determined for each of the nine genes relative to 18S under each buffer condition. ΔCts were then normalized to the fresh-frozen condition (control), to enable a comparison with the best quality RNA possible (ΔΔCt method; Schmittgen and Livak 2008). To assess the effect of buffer condition on transcript completeness, the relative levels of the 5′ and 3′ ends for each of Actb, Gapdh, Th, and Slc6a3 were determined. In this case, the ΔCt was calculated by subtracting the 3′ Ct from the 5′ Ct for each of the four genes, under each condition, and then normalizing it to the 5′–3′ ΔCt found for control RNA from fresh-frozen tissue.
TABLE 2.
Primers for qPCR
Immuno-laser-microdissection of rat midbrain dopamine neurons
Immunolabeling and tissue preparation
Midbrain dopamine neurons were identified by TH immunofluoresence. Six to ten micron thick midbrain cryosections were rinsed in PBS (15 sec) and then fixed in 70% ethanol (5 min). Ethanol was removed by a rapid PBS rinse and the slide placed in 2M NaCl PBS. Sections were incubated overnight at 4°C with mouse anti-TH monoclonal antibody (MAB318, Chemicon, Millipore) diluted 1:100 in 2M NaCl PBS. Unbound primary antibody was removed by rinsing with 2M NaCl in PBS (5 min). Sections were then incubated with Alexa Fluor 488 conjugated donkey anti-mouse (Invitrogen), at 1:100 in 2M NaCl PBS, for 2 h at 4°C. Unbound secondary antibody was removed by a 2M NaCl PBS rinse. Immediately prior to microdissection, excess NaCl was removed by a PBS rinse (5 sec) and sections dehydrated in 90% and then 100% ethanol (3 min each). All solutions except the dehydration ethanols were ice-cold.
Laser microdissection and RNA extraction
Laser microdissection and pressure catapulting of dopamine neurons were done using a PALM MicroBeam system (Zeiss). TH immunofluorescent neurons, located in ventral midbrain, were visualized with the system's Axiovert 200M inverted fluorescent microscope. One to two hundred dopamine cell bodies were UVA laser-dissected and pressure-catapulted into 30 μL RLT lysis buffer (RNeasy, Qiagen). Following cell collection, lysis buffer volume was increased to 100 μL total, the sample homogenized by vortexing and then placed at −80°C for later use. Total RNA was extracted using the RNeasy micro kit (Qiagen) according to the manufacturer's instructions, including addition of poly-A carrier RNA to the lysate and an on-column DNase-I treatment.
Reverse transcription and qPCR of laser-microdissected sample
RNA extracted from microdissected dopamine neurons was reverse-transcribed as described above for whole midbrain sections. Half the total RNA from 100 to 200 cells was used in a 20 μL cDNA synthesis reaction, with the remaining half being used in the RT-minus reaction. RNA integrity of microdissected dopamine neurons was assessed using the 5′–3′ assay described above for whole section, for each of Actb, Gapdh, Th, and Slc6a3. One microliter of cDNA product was added to each qPCR reaction, with all other reagents and volumes as described above for this assay. The 5′–3′ ΔCt for each of the four genes for the microdissected samples were compared with the ΔCts for fresh-frozen control tissue. Laser microdissection of dopamine neurons and the 5′–3′ assay were carried out six independent times.
Statistical analysis
Effects of buffer condition on whole section rRNA, mRNA relative to 18S, and relative 5′ and 3′ levels, in comparison to RNA from fresh-frozen, control tissue, were analyzed using ANOVA with Dunnett's post-hoc tests. Comparison of relative 5′ and 3′ levels in microdissected samples to fresh-frozen control samples was done with Student's t-test for each of the four genes. Significance was set at the P < 0.05 level.
ACKNOWLEDGMENTS
We thank Drs. Henrik Steinberg, Severine Roselli, and Christopher Dayas for their comments on the manuscript, and Maggie Soliman for her expert technical assistance. This work was supported by grants from the Hunter Medical Research Institute, the Center for Brain and Mental Health Research at the University of Newcastle, the Clive and Vera Ramaciotti Foundation, and the National Health and Medical Research Council of Australia.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1733509.
REFERENCES
- Atz M, Walsh D, Cartagena P, Li J, Evans S, Choudary P, Overman K, Stein R, Tomita H, Potkin S, et al. Methodological considerations for gene expression profiling of human brain. J Neurosci Methods. 2007;163:295–309. doi: 10.1016/j.jneumeth.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer H, Lyianarachchi S, Newsom D, Klisovic MI, Marcucci G, Kornacker K. Chipping away at the chip bias: RNA degradation in microarray analysis. Nat Genet. 2003;35:292–293. doi: 10.1038/ng1203-292. [DOI] [PubMed] [Google Scholar]
- Backes E, Hemby SE. Discrete cell gene profiling of ventral tegmental dopamine neurons after acute and chronic cocaine self-administration. J Pharmacol Exp Ther. 2003;307:450–459. doi: 10.1124/jpet.103.054965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri I, Keller-McGandy C, Bouzou B, Asteris G, Clark TW, Frosch MP, Standaert DG. Effects of gender on nigral gene expression and parkinson disease. Neurobiol Dis. 2007;26:606–614. doi: 10.1016/j.nbd.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Yasuda J, Hayashizaki Y. Multifaceted mammalian transcriptome. Curr Opin Cell Biol. 2008;20:274–280. doi: 10.1016/j.ceb.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Chung CY, Seo H, Sonntag KC, Brooks A, Lin L, Isacson O. Cell type-specific gene expression of midbrain dopaminergic neurons reveals molecules involved in their vulnerability and protection. Hum Mol Genet. 2005;14:1709–1725. doi: 10.1093/hmg/ddi178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copois V, Bibeau F, Bascoul-Mollevi C, Salvetat N, Chalbos P, Bareil C, Candeil L, Fraslon C, Conseiller E, Granci V, et al. Impact of RNA degradation on gene expression profiles: Assessment of different methods to reliably determine RNA quality. J Biotechnol. 2007;127:549–559. doi: 10.1016/j.jbiotec.2006.07.032. [DOI] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Eklund AC, Szallasi Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006;22:101–109. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espina V, Wulfkuhle JD, Calvert VS, VanMeter A, Zhou W, Coukos G, Geho DH, Petricoin EF, III, Liotta LA. Laser-capture microdissection. Nat Protoc. 2006;1:586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- Fend F, Emmert-Buck M, Chuaqui R, Cole K, Lee J, Liotta L, Raffeld M. Immuno-LCM: Laser capture microdissection of immunostained frozen sections for mRNA analysis. Am J Pathol. 1999;154:61–66. doi: 10.1016/S0002-9440(10)65251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Gjerdrum LM, Abrahamsen HN, Villegas B, Sorensen BS, Schmidt H, Hamilton-Dutoit SJ. The influence of immunohistochemistry on mRNA recovery from microdissected frozen and formalin-fixed, paraffin-embedded sections. Diagn Mol Pathol. 2004;13:224–233. doi: 10.1097/01.pdm.0000134779.45353.d6. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Greene JG, Dingledine R, Greenamyre JT. Gene expression profiling of rat midbrain dopamine neurons: Implications for selective vulnerability in parkinsonism. Neurobiol Dis. 2005;18:19–31. doi: 10.1016/j.nbd.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Grimm J, Mueller A, Hefti F, Rosenthal A. Molecular basis for catecholaminergic neuron diversity. Proc Natl Acad Sci. 2004;101:13891–13896. doi: 10.1073/pnas.0405340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründemann J, Schlaudraff F, Haeckel O, Liss B. Elevated α-synuclein mRNA levels in individual UV-laser-microdissected dopaminergic substantia nigra neurons in idiopathic Parkinson's disease. Nucleic Acids Res. 2008;36:e38. doi: 10.1093/nar/gkn084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalization; strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, Mueller O, Schroeder A, Auffray C. Toward standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005;33:e56. doi: 10.1093/nar/gni054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Hetzel A, Hackel O, Jones I, Liss B, Roeper J. Unique properties of mesoprefrontal neurons within a dual mesocorticolimbic dopamine system. Neuron. 2008;57:760–773. doi: 10.1016/j.neuron.2008.01.022. [DOI] [PubMed] [Google Scholar]
- Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM, Kleinman JE. Critical factors in gene expression in postmortem human brain: Focus on studies in schizophrenia. Biol Psychiatry. 2006;60:650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Lobsiger CS, Boillee S, Cleveland DW. Toxicity from different SOD1 mutants dysregulates the complement system and the neuronal regenerative response in ALS motor neurons. Proc Natl Acad Sci. 2007;104:7319–7326. doi: 10.1073/pnas.0702230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis EB, Lock H, Chefer VI, Shippenberg TS, Hjelmstad GO, Fields HL. κ Opioids selectively control dopaminergic neurons projecting to the prefrontal cortex. Proc Natl Acad Sci. 2006;103:2938–2942. doi: 10.1073/pnas.0511159103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos GLG, Maleszka R. Microarray reality checks in the context of a complex disease. Nat Biotechnol. 2004;22:615–621. doi: 10.1038/nbt965. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Pérez-Novo CA, Claeys C, Speleman F, Van Cauwenberge P, Bachert C, Vandesompele J. Impact of RNA quality on reference gene expression stability. Biotechniques. 2005;39:52, 54–56. doi: 10.2144/05391BM05. [DOI] [PubMed] [Google Scholar]
- Popova T, Mennerich D, Weith A, Quast K. Effect of RNA quality on transcript intensity levels in microarray analysis of human post-mortem brain tissues. BMC Genomics. 2008;9:91. doi: 10.1186/1471-2164-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;6:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Shi L, Jones WD, Jensen RV, Harris SC, Perkins RG, Goodsaid FM, Guo L, Croner LJ, Boysen C, Fang H, et al. The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies. BMC Bioinformatics. 2008;9(Suppl 9):S10. doi: 10.1186/1471-2105-9-S9-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson D, Ramirez A, Long J, Barrezueta N, Hajos-Korcsok E, Matherne C, Gallagher D, Ryan A, Ochoa R, Menniti F, et al. Quantification of MPTP-induced dopaminergic neurodegeneration in the mouse substantia nigra by laser capture microdissection. J Neurosci Methods. 2007;159:291–299. doi: 10.1016/j.jneumeth.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Thompson KL, Pine PS, Rosenzweig BA, Turpaz Y, Retief J. Characterization of the effect of sample quality on high density oligonucleotide microarray data using progressively degraded rat liver RNA. BMC Biotechnol. 2007;7:57. doi: 10.1186/1472-6750-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–570. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034–0034.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis S, Llenos IC, Dulay JR, Elashoff M, Martinez-Murillo F, Miller CL. Quality control for microarray analysis of human brain samples: The impact of postmortem factors, RNA characteristics, and histopathology. J Neurosci Methods. 2007;165:198–209. doi: 10.1016/j.jneumeth.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Yao F, Yu F, Gong L, Taube D, Rao DD, MacKenzie RG. Microarray analysis of fluoro-gold labeled rat dopamine neurons harvested by laser capture microdissection. J Neurosci Methods. 2005;143:95–106. doi: 10.1016/j.jneumeth.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Zhu LJ, Altmann SW. mRNA and 18S-RNA coapplication-reverse transcription for quantitative gene expression analysis. Anal Biochem. 2005;345:102–109. doi: 10.1016/j.ab.2005.07.028. [DOI] [PubMed] [Google Scholar]