Abstract
Trans-translation is a process which switches the synthesis of a polypeptide chain encoded by a nonstop messenger RNA to the mRNA-like domain of a transfer-messenger RNA (tmRNA). It is used in bacterial cells for rescuing the ribosomes arrested during translation of damaged mRNA and directing this mRNA and the product polypeptide for degradation. The molecular basis of this process is not well understood. Earlier, we developed an approach that allowed isolation of tmRNA–ribosomal complexes arrested at a desired step of tmRNA passage through the ribosome. We have here exploited it to examine the tmRNA structure using chemical probing and cryo-electron microscopy tomography. Computer modeling has been used to develop a model for spatial organization of the tmRNA inside the ribosome at different stages of trans-translation.
Keywords: chemical probing, cryo-EM tomography, E.coli tmRNA, structure modeling, trans-translation
INTRODUCTION
Trans-translation is a complicated and well-organized process that switches the synthesis of a polypeptide chain encoded by a nonstop messenger RNA to the mRNA-like domain (MLD) of a transfer-messenger RNA (tmRNA) (Tu et al. 1995; Keiler et al. 1996). Bacteria cells use trans-translation for rescuing the ribosomes arrested during translation of damaged mRNA, thus directing this mRNA and the product polypeptide for degradation (Dulebohn et al. 2007). During its functioning, tmRNA interacts with several proteins and the ribosome. The interaction of tmRNA with alanyl-tRNA synthetase, elongation factor Tu, and small protein B (SmpB) has been studied in detail using biochemical techniques (Barends et al. 2000, 2001; Zvereva et al. 2001; Hanawa-Suetsugu et al. 2002; Sundermeier et al. 2005; Shimizu and Ueda 2006; Konno et al. 2007; Metzinger et al. 2008). The crystal structure of the SmpB complex with the tRNA-like domain (TLD) of tmRNA is also available (Gutmann et al. 2003; Bessho et al. 2007). Much less is known about the tmRNA interactions with the ribosome. Up to now, only the initiation step of trans-translation has been successfully subjected to structural analysis. In particular, the model of the ribosome interactions at this initiation stage with the tmRNA–protein complex was suggested on the basis of a three-dimensional cryo-electron microscopy (cryo-EM) study (Valle et al. 2003a; Kaur et al. 2006; Gillet et al. 2007). Recently, the hypothetical mechanism explaining how tmRNA distinguishes stalled ribosomes from the active ones in order to initiate trans-translation was described in detail (Moore and Sauer 2007). However, lack of sufficient information concerning the elongation and termination stages of trans-translation hampers a deeper understanding of the mechanism of this important process.
Previously, we developed an approach that allowed isolation of tmRNA–ribosomal complexes arrested at a desired step of tmRNA passage through the ribosome (Shpanchenko et al. 2005). In order to block the trans-translation process at the desired step, a translation termination signal was placed at a proper position within the MLD. The trans-translation process was blocked at the stop codon in the ribosomal A-site by inactivation of a thermosensitive termination factor 2 (RF2). The tmRNA-containing ribosomal complexes were separated from the other cellular ribosomes by affinity chromatography based on a streptavidin aptamer introduced into the tmRNA, instead of the pK3, together with the stop signal. We were able to isolate tmRNA–ribosomal complexes with an equimolar ratio of tmRNA to the ribosome and with a stop codon at the position of the second, fourth, fifth, and 11th codons in the MLD (Shpanchenko et al. 2005; Bugaeva et al. 2008). Here, we have exploited this approach to examine tmRNA structure using chemical probing and cryo-EM tomography. On the basis of computer modeling we have developed a model for spatial organization of the tmRNA inside the ribosome at different stages of trans-translation.
RESULTS
Chemical probing of tmRNA inside the ribosome at different steps of trans-translation
tmRNA–ribosomal complex block 2, 4, 5, and 11 codons of tmRNA ORF were prepared using an in vivo system as described (Shpanchenko et al. 2005). With the help of this approach, ribosomal complexes could be isolated only if a specific signal was placed into the tmRNA part. This signal (UGA stop codon with the context around) proved to stop translation of mRNA at the stop codon in the ribosomal A-site (Bjornsson and Isaksson 1996). In agreement with this, only tRNAPro was detected in the isolated complex. In order to get additional proof that the UGA stop codon is located at the ribosomal A-site in the isolated complexes they were analyzed by RelE RNase cleavage. RelE is known to introduce a single cut into an A-site codon in mRNA within the ribosomal complex with tRNA at the ribosomal P-site and empty A-site (Pedersen et al. 2003). An example of such analysis for tmRNA–ribosomal complexes 4 and 5 is shown in Supplemental Figure 1. Only one signal corresponding to the expected cleavage position at the stop codon in the A-site was observed in each case, proving that the major complex is indeed the tmRNA–ribosomal complex blocked at the stop codon at the ribosomal A-site.
Chemical probing is a powerful tool used to study the structure of RNA molecules and their interactions with other components of RNA–RNA and RNA–protein complexes. Four complexes with tmRNA-2, tmRNA-4, tmRNA-5, and tmRNA-11 (with a UGA stop codon in the indicated position, respectively), corresponding to different steps of translation of the tmRNA ORF, have been isolated and characterized, as has been described previuosly (Bugaeva et al. 2008). The complexes were treated with dimethyl sulfate (DMS), 1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide metho-p-toluene sulphonate (CMCT), or Kethoxal (KE), and the positions of the modifications in the tmRNA were determined by using the reverse transcription (RT) reaction. It is known that RT stops at the 3′ side of the nucleotide adjoining the modified base. The intensity of the band corresponding to the RT stop allows determination of the accessibility of every single nucleotide to the chemical modification, and thus enables speculation about its contacts with the other members of the complex, as well as the about the structure of the RNA. As controls, we have used the corresponding tmRNAs isolated from lyzed cells by affinity chromatography. Here, we have analyzed most of the tmRNA molecule in four complexes corresponding to different steps of the translation of the tmRNA ORF. The data are summarized in Figure 1. To quantify the probing results, the intensity of each band was normalized on the intensity of the whole lane, and the ratio of these values for complex and corresponding control was calculated. The results for the bands with the values that differ from 1 are shown in Supplemental Table 1.
FIGURE 1.
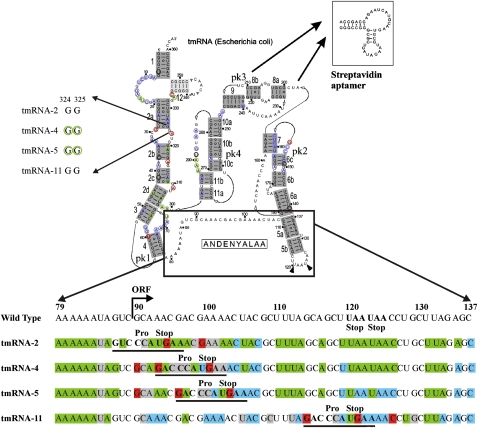
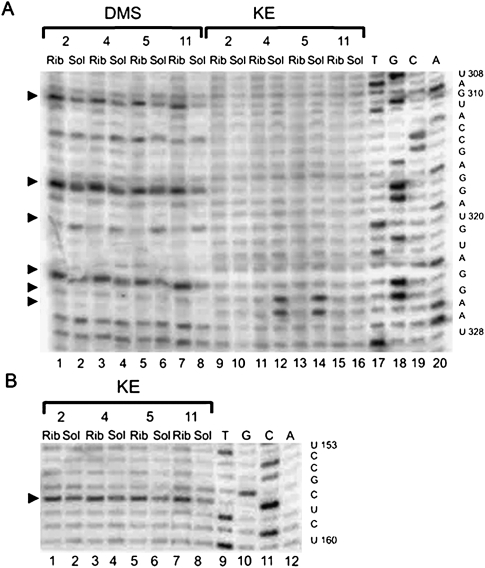
Protection pattern of tmRNA in ribosomal complexes. The secondary structure of tmRNA was adapted from data from rnp.uthct.edu/rnp/tmRDB/tmRDB.html. pk3 (nucleotides U212–A239) was substituted with an aptamer to streptavidin (inset). The effects that are similar for all complexes are shown at the secondary structure as a green circle for the protection, red for the exposure, blue for the nucleotides which are equally exposed for the modification both in the solution and in the complexes. Black circle indicates nucleotides that are inclined for degradation. The nucleotides 324GG325 which displayed different accessibility to the modifying reagents in different complexes are shown at the left side. The region A79–C137 is shown in details at the bottom. Protected nucleotides are colored in green, exposed in red, nucleotides which are equally exposed for the modification both in the solution and in the complexes are in blue, and nucleotides inclined for degradation are in gray.
Tag-encoding sequence of the tmRNA, the upstream region, and helix 5
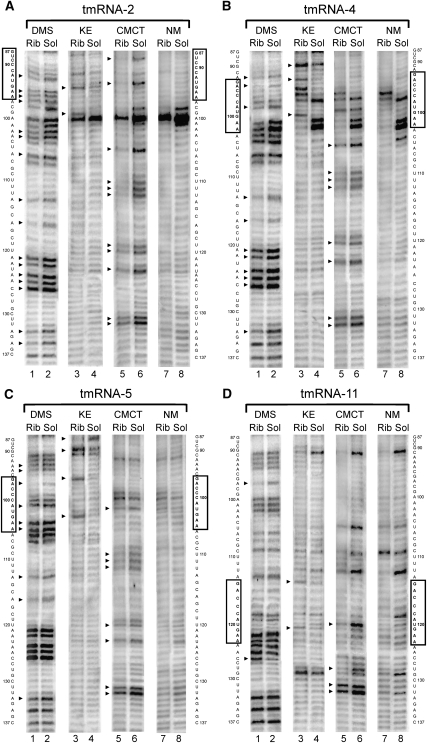
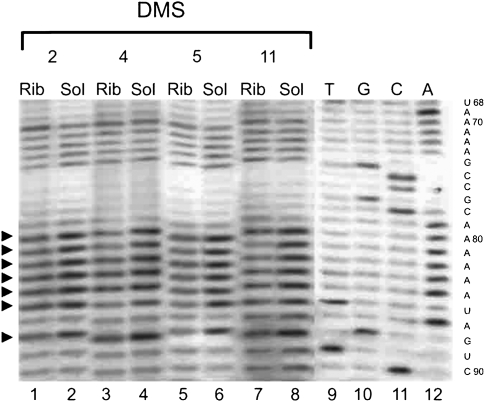
The accessibility of free tmRNA in solution and in a ribosomal complex to modifying agents was compared. Most of the differences in the modification patterns for different ribosomal complexes were observed in the MLD of the tmRNA and adjoined regions (Fig. 1, tmRNA, region 79–137; Figs. 2, 3). This region of tmRNA-2 in the complex seems to be the most protected (Figs. 1, 2A, 3). In total, 34 out of 59 nucleotides (nt) showed reduced accessibility to the different reagents in the tmRNA–ribosome complex. Nucleotides A79–84, A86, A92, A95–97, A102–103, A106, A113, A116, A121–122, A124–125, C126, A133, A135 and U88, U93, U105, U110–112, U119–120, U123, and U131–132, accessible to DMS and CMCT, respectively, in tmRNA-2 in solution (Fig. 2A, lanes 2,6; Fig. 3, lane 2) became protected from the modification when the tmRNA molecule is a constituent of the complex (Fig. 2A, lanes 1,5; Fig. 3, lane 1). At the same time, two nucleotides—G94 and G99—displayed higher accessibility to the modifying agent KE when tmRNA-2 entered the tmRNA–ribosome complex (Fig. 2A, cf. lanes 3 and 4).
FIGURE 2.
Chemical probing of the MLD and helix 5 tmRNA region in the ribosomal complexes (Rib) and in the solution (Sol) for tmRNA-2 (A), tmRNA-4 (B), tmRNA-5 (C), and tmRNA-11 (D) by DMS, KE, and CMCT. NM, nonmodified controls. The sequence of tmRNA is shown on the sides of the gels. The mutated sequences of the stop signal are boxed. Arrows indicate the position of nucleotides which displayed different accessibility to modifying reagents in the complexes and in solution.
FIGURE 3.
Chemical probing of the A79–A86 tmRNA region in the ribosomal complexes (Rib) and in the solution (Sol) for tmRNA-2 (2), tmRNA-4 (4), tmRNA-5 (5), and tmRNA-11 (11) by DMS. Dideoxy sequencing lanes are indicated by T, G, C, and A. The sequence of tmRNA is shown on the right side of the gels. Arrows indicate the position of nucleotides which displayed different accessibility to modifying reagents in the complexes and in solution.
The shift of the translation block to the fourth codon position at the ribosomal A-site reduced the number of the nucleotides protected in the complex to 27 (Figs. 1, 2B, 3). The protection pattern of the 3′-part of the region following the stop codon (A113, A116, A121–122, A124–125, C126, A133 and U105, U110–112, U120, U123, and U131–132) remains mostly the same as in the complex with the tmRNA-2 (Figs. 1, 2B, lanes 1,5), although the protection of A124–A125, C126, and U131–132 is weaker than for tmRNA-2 (Supplemental Table 1). However, in the 5′-part, where the tmRNA sequence has been changed, a pronounced difference in the protection pattern can be seen (Figs. 1, 2B). Nucleotide G87 became less available, while G90, G93, and G100 became more available for modification by KE (Fig. 2B, lanes 3,4), and A98 became less available for DMS (Fig. 2B, lanes 1,2). The nucleotide at position 94, which showed increased reactivity in the tmRNA-2 complex, became protected in the complex with tmRNA-4 (Fig. 2B, lanes 1,2). The reactivity of the nucleotide at position 99 in tmRNA-4 was the same in the complex and in the solution (Fig. 2B, lanes 5,6). This is in contrast to its increased reactivity in the complex of tmRNA-2 with the ribosome (Fig. 2A, lanes 3,4). Nucleotides 79–84 and 86 remained protected (Fig. 3, lanes 3,4).
Further translation block along the ORF (tmRNA-5) decreased the number of nucleotides protected by complex formation to 22 (Figs. 1, 2C, 3). The reactivity of A121–122, A124–125, C126, and U105, which was reduced in the complexes with tmRNA-2 and 4 became similar for tmRNA-5 in the complex and in the solution. The changes in the reactivity of A94, G87, and G90 in tmRNA-5 remained the same as in the complex with tmRNA-4 (Fig. 2C, lanes 1–4), as well as of nucleotides 79–84, 86 (Fig. 3, lanes 5,6). The nucleotide at position 93 became protected (Fig. 2C, lanes 1,2). The nucleotides at positions 96 and 103 became available for modification by KE (Fig. 2C, lanes 3,4).
Only 12 nt were protected from modification in the complex with tmRNA-11 (Figs. 1, 2D, 3). Protection of A79–84, A86, A97, U120, and U131–132 has been shown to be the same as for previously described complexes (Supplemental Table 1). The enhanced availability of G114, G121, and C126 and protection of U128 were specific for the complex with tmRNA-11 (Fig. 2D).
Some nucleotide residues are more prone to give degradation of different tmRNAs (U85, C91, C98, and A100–101 in tmRNA-2; U85, C95–97, A101–102, and C104 in tmRNA-4; U85, C91, A92, and C99–100 in tmRNA-5; and U85, C91, C109, U112, C118, and G129 in tmRNA-11). Due to this reason we could not examine the changes in their reactivity upon complex formation.
Helix 2 and pK1, pK4, and pK2
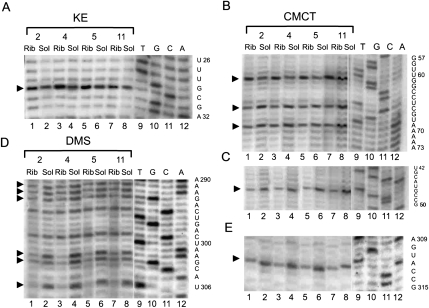
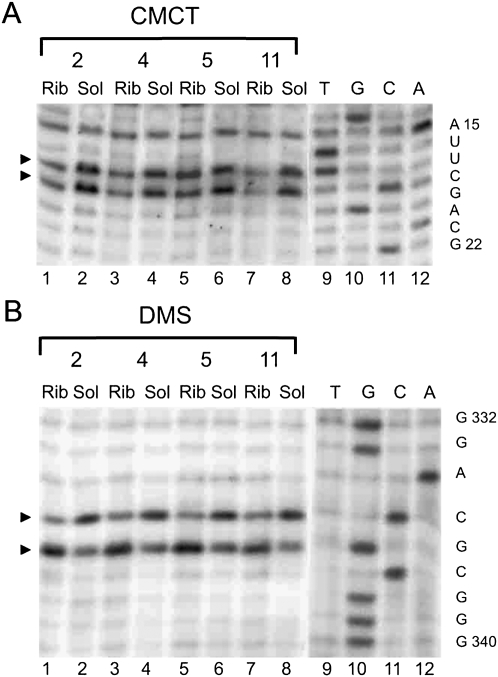
Trans-translation block at different positions has introduced a set of changes in the accessibility of different nucleotides in helix 2 and pK1 of tmRNA for modifying reagents (Figs. 1, 4, 5). Nucleotides G29 (Fig. 4A), U60 (Fig. 4B), A309, A316, and A323 (Fig. 5A) became more available for modification while nucleotides U46 (Fig. 4C), U65, U68 (Fig. 4B), A301, A302, A305 (Fig. 4D), U311 (Fig. 4E), and A319 (Fig. 5A) became protected from modification in all tmRNA–ribosome complexes (Supplemental Table 1).
FIGURE 4.
Chemical probing of the pK1, pK4, and helix 2 tmRNA region in the ribosomal complexes (Rib) and in the solution (Sol) for tmRNA-2 (2), tmRNA-4 (4), tmRNA-5 (5), and tmRNA-11 (11) by KE (A), CMCT (B,C,E), and DMS (D). Dideoxy sequencing lanes are indicated by T, G, C, and A. The sequence of tmRNA is shown on the right side of the gels. Arrows indicate the position of nucleotides which displayed different accessibility to modifying reagents in the complexes and in solution.
FIGURE 5.
Chemical probing of the helix 2 (A) and pK2 (B) of tmRNA in the ribosomal complexes (Rib) and in the solution (Sol) for tmRNA-2 (2), tmRNA-4 (4), tmRNA-5 (5), and tmRNA-11 (11) by DMS and KE. Dideoxy sequencing lanes are indicated by T, G, C, and A. The sequence of tmRNA is shown on the right side of the gels. Arrows indicate the position of nucleotides which displayed different accessibility to modifying reagents in the complexes and in solution.
Nucleotides G324 and G325 were accessible for modification by KE in tmRNA-4 and tmRNA-5 in solution (Fig. 5A, lanes 12,14) but were protected from modification in corresponding complexes with the ribosome (Fig. 5A, lanes 11,13).
Nucleotides A290, A291, and A292 in pK4 were protected from modification by DMS in all the studied complexes (Fig. 4D).
G156 in pK2 was more available for modification for all four complexes (Fig. 5B).
tRNA-like domain of tmRNA
The protection pattern for the TLD region of tmRNA was nearly the same in all of the complexes (Figs. 1, 6), (Supplemental Table 1). Nucleotides U16, U17 (Fig. 6A), and A334 (Fig. 6B) were found to be protected (although less pronounced in the case of the complex with tmRNA-5) while C335 became more accessible for modification (Fig. 6B).
FIGURE 6.
Chemical probing of the TLD of tmRNA in the ribosomal complexes (Rib) and in the solution (Sol) for tmRNA-2 (2), tmRNA-4 (4), tmRNA-5 (5), and tmRNA-11 (11) by CMCT (A) and DMS (B). Dideoxy sequencing lanes are indicated by T, G, C, and A. The sequence of tmRNA is shown on the right side of the gels. Arrows indicate the position of nucleotides which displayed the different accessibility to modifying reagents in the complexes and in solution.
Cryo-electron tomography of the tmRNA-4 complex
To visualize tmRNA in the tmRNA-4 ribosomal complex cryo-EM tomography was applied. This technology allows morphological investigation of individual particles in heterogeneous sample preparations. Structures of individual free 30S and 50S ribosomal subunits in solution reveal their high conformation flexibility (Zhao et al. 2004a,b). The method gives useful morphological information, even though the resolution is lower than can be achieved by single particle reconstruction (Frank 2006).
A general overview of the whole reconstruction area was made and particles suitable for detailed reconstruction and further analysis were manually selected. A gallery of particles with extra density corresponding to the tmRNA was obtained. Some representative examples of the reconstituted particles are shown in Supplemental Figure 2, where the extra density interpreted as being tmRNA represents an arch similar to the one visualized by cryo-EM in the pre-initiation complex (Valle et al. 2003a). The location of the arch on the 70S ribosome varies among the individual particles from a position on the shoulder to the top of the head (Supplemental Figure 2).
DISCUSSION
Chemical probing of the tmRNA structure in ribosomal complexes blocked at different stages of trans-translation allowed us to follow the structural changes starting from early steps of tmRNA passage, with the TLD in tmRNA at the ribosomal E-site, up to the termination step of trans-translation. Our data show that neither pseudoknots nor stable helices (except for helix 5) are unfolded in the process of trans-translation. Helix 5 is somehow stabilized in the complex with tmRNA-2 and 4. It is partly unwound in the complex with tmRNA-5 and is completely melted at the trans-translation termination stage in the complex with tmRNA-11 (Fig. 1).
The nucleotides in TLD with altered reactivity (Fig. 1) are the same for all steps of tmRNA passage through the ribosome. These data are in agreement with structural probing data (Barends et al. 2001; Ivanova et al. 2007) and the X-ray structure for SmpB interacting with TLD in solution (Gutmann et al. 2003; Bessho et al. 2007). Earlier, we found that one SmpB molecule is present in all studied complexes (Bugaeva et al. 2008). We propose that SmpB remains bound to the TLD of tmRNA starting from the second codon at the ribosomal A-site—when the TLD is at the ribosomal E-site—up to trans-translation termination.
In all of the complexes we observed an enhanced reactivity for the G base in the UGA stop codon located at the ribosomal A-site. For tmRNA-4, 5, and 11 (elongation and termination of trans-translation), we found conserved enhancement of the reactivity of the base located 6-nt residues upstream of the stop codon, indicating that the conformation of this mRNA part is similar for all later stages of trans-translation. Corresponding kinks in mRNA were found in an X-ray study (Yusupova et al. 2006). For all of the complexes, an A-rich single-stranded region (79–86) was protected. The reactivity of G90 (first base in the resume codon) was increased for tmRNA-4 and 5.
We suggest that the region 79–86 represents a fixed binding site on the ribosome, and the rest of the message can occupy any open ribosomal space. Such a fixed binding site would represent the interaction with the corresponding mRNA binding pocket on the 30S ribosomal subunit. This pocket is revealed by X-ray analysis of the region upstream of the P-site in an mRNA lacking a Shine–Dalgarno sequence (Yusupova et al. 2006).
The complex with tmRNA-2 is different from the others as judged by the protection pattern. This complex represents the initial step of trans-translation when TLD occupies the ribosomal E-site and the resume codon is at the ribosomal P-site. Since the single-stranded region A79–A86 also is protected in this complex, we suggest that these protections reflect the MLD entering the mRNA binding pocket.
A number of the bases whose reactivity was changed by complex formation are located in different single-stranded regions of tmRNA in all complexes, indicating conformational changes in tmRNA upon complex formation. In the case of helix 2 and pK1, some bases appear to be more strongly protected in the complex than in the solution (Fig. 1), suggesting that these structural elements interact with ribosomal components.
In order to understand how tmRNA is arranged on the ribosome at different steps of trans-translation, we did modeling of the tmRNA structure in the ribosome. For modeling, we used the published ribosome X-ray structure with a defined mRNA path (Yusupova et al. 2006). We have assumed that the tmRNA ORF should occupy the mRNA binding region for canonical mRNA. For tmRNA-2, the stop codon is at the A-site and the preceding codon together with tRNA should occupy the P-site. The TLD of tmRNA at the ribosomal E-site region keeps the structure of the acceptor arm of tRNA. The position of the L1 stalk was taken from a previous study (Harms et al. 2001). Only the above-mentioned tmRNA elements were fixed. The program allowed the formation of the tmRNA structure with known secondary structure elements in the empty space in the ribosome or at the ribosome surface. The structure was optimized by energy minimization and manual corrections. The details of the models are available at http://dualopt1.cmm.msu.ru/bin/view/Projects/TmrnaEnglish.
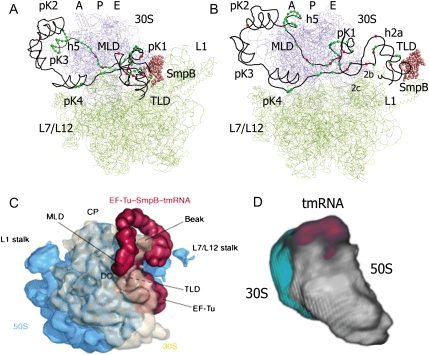
The structural model for tmRNA-2 inside the ribosome encompasses the footprinting data presented here as well as other available data (see Fig. 7A). The SmpB protein occupies the site on the TLD as known from its X-ray structure in complex with the tmRNA fragment (Gutmann et al. 2003). The TLD is located at the ribosomal E-site. The protected A79–A86 loop is located at the entrance of the mRNA binding pocket; pK1 is nearby at the E-site of tRNA exit channel. The L1 stalk is moved from its position in the original X-ray structure (Yusupova et al. 2006), as suggested by cryo-EM data for conformational changes in the ribosome caused by exiting deacylated tRNA (Harms et al. 2001; Valle et al. 2003b). Only in this case, there is enough space between the subunits at the L1 side of the ribosome to accommodate the TLD with SmpB and pK1 simultaneously. Helices 2a, 2b, and 2c support the TLD location at the E-site; helix 2d creates a link between pK1 and the arch consisting of pK4, pK2, and pK3. The arch is surrounding the head of the 30S subunit starting from the shoulder; helix 5 is located at the entrance to the mRNA channel and can be easily unwound during subsequent steps of ORF translation without influencing the arch structure. The resume codon is at the P-site of the ribosome and the second ORF codon is at the A-site. One can see that all protected bases in the single-stranded regions are indeed involved in the interactions with the ribosome, and the position of the bases with enhanced reactivity corresponds to possible distortions in the RNA chain.
FIGURE 7.
Structural models of tmRNA-2 (A) and tmRNA-4 (B) in complex with the ribosome (top views). 30S ribosomal subunit is colored in blue, 50S in green. tmRNA is shown as a black line connecting the phosphorus atoms. SmpB is presented as rose spheres of Cα atoms. Nucleotides of tmRNA protected by the ribosome from the chemical modification are shown as green spheres. Nucleotides that became more accessible to modification after the complex formation are shown as red spheres. Structural elements of tmRNA are marked. (C) Cryo-EM structure of the pre-initiation tmRNA–ribosomal complex adapted with permission (from Elsevier © 2004, Haebel et al. 2004). SmpB–tmRNA–EF-Tu (red) emerging from the intersubunit space between the 50S subunit (blue) and 30S subunit (cream). (D) Cryo-electron tomography structure of elongating tmRNA–ribosomal complex. 30S ribosomal subunit is colored in light gray, 50S in light blue, and tmRNA in magenta.
The same approach was applied to create a model for tmRNA-4 in the ribosome (Fig. 7B). Again, there is good agreement between the chemical probing data, cryo-EM tomography data, and the proposed model. At this step of trans-translation the TLD–SmpB complex is moved out from the ribosome and is located at the platform side of the 30S subunit. It is not tightly fixed on the ribosome and can occupy any position such that it does not interfere with the deacylated tRNA that leaves the ribosome through the exit site. Helices 2a, 2b, and 2c link the TLD to pK4 (part of the arch) on one side and pK1 and the A79–A86 loop on the other side supported by helix 2d. This helix could be partially unwound, but hidden in the ribosome. It is thus protected from chemical modification even more strongly than in tmRNA being in solution. The protected A79–A86 loop is tightly bound in the mRNA binding pocket (Yusupova et al. 2006) that causes the conformational change of the first nucleotide of the resume codon, which becomes more exposed for modification. The arch that can be visualized by cryo-EM tomography consists of three pseudoknots. This arch surrounds the head of the 30S subunit starting from the shoulder, as in the case of tmRNA-2. The model allows movement of the arch around the head of the 30S subunit in agreement with cryo-EM tomography data (Supplemental Fig. 2). Helix 5 is located at the entrance to the mRNA channel. The third and fourth codons of the tmRNA ORF are located at the P- and A-site positions, as determined by X-ray analysis (Yusupova et al. 2006), in the decoding center on the 30S subunit. Again, all protected bases in the single-stranded regions are indeed involved in the interactions with the ribosome, and the position of the bases with enhanced reactivity corresponds to possible distortions in the RNA chain.
Our model considers one SmpB molecule at the stages when tmRNA has already entered the ribosome, in agreement with Shpanchenko et al. (2005), Sundermeier and Karzai (2007), and Bugaeva et al. (2008). However, two molecules of SmpB could bind the ribosome at the pre-initiation step of trans-translation (Hallier et al. 2004; Kaur et al. 2006), stimulating recognition of the stalled ribosome by tmRNA.
MATERIALS AND METHODS
tmRNA–ribosome complexes were isolated from Escherichia coli SKZ-1 cells carrying the plasmid pGEM-stra-2, pGEM-stra-4, pGEM-stra-5, or pGEM-stra-11 (coding for tmRNA-2, tmRNA-4, tmRNA-5, and tmRNA-11, respectively), with mutated termination signals (stop codons in position 2, 4, 5, or 11 of the tag sequence), and purified on Streptavidin Sepharose (GE Healthcare) as described previously (Shpanchenko et al. 2005). The RNA content and protein content of the complex were analyzed as described before (Bugaeva et al. 2008).
Preparation and purification of mutant tmRNAs
For isolation of mutant tmRNA, a cell lysate was prepared as described before (Shpanchenko et al. 2005).
Structure probing
Structure probing of tmRNA–ribosome complexes and mutant tmRNAs was done using a previously published procedure (Moazed et al. 1986). The radioactivity on a gel was monitored by a Bio-Image Analyzer BAS3000 (Fuji Film). The intensities of the bands were determined using the ImageQuant program 5.0.Ink.
Cryo-EM tomography
Electron microscope grids, coated with holey carbon film, were glow discharged. Five microliters of the solution of 10 nm colloidal gold particles coated with BSA (British Biocell International) were mixed with 10 μL of the ribosome–tmRNA complex. Subsequently, 2 μL of a 5-nm Streptavidin-gold solution (Sigma) were added, having the effect of concentrating the particles in the grid hole. Both colloidal gold solutions contained 10 mM MgSO4.
A 4 μL droplet of the suspension was deposited onto a grid, blotted with filter paper, and plunged into liquid ethane (Adrian et al. 1984). The plunging was performed in a climate chamber at a humidity of 80%. The specimens were transferred at −180°C into a FEG 200keV transmission electron microscope (Philips CM200), and kept cool with liquid nitrogen. Micrographs were recorded on a CCD detector with a pixel size of 7.838E at magnification 20,000. The specimen was tilted from −60° to 60°. The images were recorded either every one or every second degree giving a set of either 121 or 61 images. The accumulated total dose was <20 electrons/E2. Post-images showed no visible specimen damage during data collection.
The 10-nm colloidal gold particles were used to align the micrographs to one another, with the alignment error being <8E. A filtered back-projection algorithm was used to reconstruct the three-dimensional densities followed by refinement using COMET software for 24 cycles at regularization parameters of 0.7 in the span of 0.0–1.0, where 1.0 means entropy regularization, or total trust in the data (Skoglund et al. 1996; Gherardi et al. 2006; Rullgerd et al. 2007).
The electron tomograms were visualized using iBOB (GNU General Public License). A general overview of the whole reconstruction area was made, and particles suitable for detailed reconstructions and further analysis were manually selected. Separate particles, showing two ribosomal subunits together were selected. The reconstructed particles were visually analyzed and compared with known X-ray and cryo-electron microscopy structures of 70S bacterial ribosomes.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dr. A. Myasnikov for electron microscope grids, coated with holey carbon film, and Dr. M. Ehrenberg for providing us with RelE. This work was supported by grants from the Howard Hughes Medical Institute, INTAS, FEBS, the Russian Foundation for Basic Research and Leading Scientific Schools, from the Swedish Research Council, and the Swedish Institute (Visby program) to L.A.I., and from Knut and Alice Wallenbergs Stiftelse, the European Union “3D-EM” Network of Excellence to U.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1584209.
REFERENCES
- Adrian M, Dubochet J, Lepault J, McDowall AW. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- Barends S, Wower J, Kraal B. Kinetic parameters for tmRNA binding to alanyl-tRNA synthetase and elongation factor Tu from Escherichia coli. Biochemistry. 2000;39:2652–2658. doi: 10.1021/bi992439d. [DOI] [PubMed] [Google Scholar]
- Barends S, Karzai AW, Sauer RT, Wower J, Kraal B. Simultaneous and functional binding of SmpB and EF-Tu•GTP to the alanyl acceptor arm of tmRNA. J Mol Biol. 2001;314:9–21. doi: 10.1006/jmbi.2001.5114. [DOI] [PubMed] [Google Scholar]
- Bessho Y, Shibata R, Sekine S, Murayama K, Higashijima K, Hori-Takemoto C, Shirouzu M, Kuramitsu S, Yokoyama S. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc Natl Acad Sci. 2007;104:8293–8298. doi: 10.1073/pnas.0700402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson A, Isaksson LA. Accumulation of a mRNA intermediate by ribosomal pausing at a stop codon. Nucleic Acids Res. 1996;24:1753–1757. doi: 10.1093/nar/24.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaeva EY, Shpanchenko OV, Felden B, Isaksson L, Dontsova OA. One SmpB molecule accompanies tmRNA during its passage through the ribosomes. FEBS Lett. 2008;582:1532–1536. doi: 10.1016/j.febslet.2008.03.049. [DOI] [PubMed] [Google Scholar]
- Dulebohn D, Choy J, Sundermeier T, Okan N, Karzai AW. Trans-translation: The tmRNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry. 2007;46:4681–4693. doi: 10.1021/bi6026055. [DOI] [PubMed] [Google Scholar]
- Frank J. Electron tomography: Methods for three-dimensional visualization of structures in the cell. Springer Science; New York: 2006. [Google Scholar]
- Gherardi E, Sandin S, Petoukhov MV, Finch J, Youles ME, Ofverstedt LG, Miguel RN, Blundell TL, Vande Woude GF, Skoglund U, et al. Structural basis of hepatocyte growth factor/scatter factor and MET signalling. Proc Natl Acad Sci. 2006;103:4046–4051. doi: 10.1073/pnas.0509040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet R, Kaur S, Li W, Hallier M, Felden B, Frank J. Scaffolding as an organizing principle in trans-translation. The roles of small protein B and ribosomal protein S1. J Biol Chem. 2007;282:6356–6363. doi: 10.1074/jbc.M609658200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N. Crystal structure of transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature. 2003;424:699–703. doi: 10.1038/nature01831. [DOI] [PubMed] [Google Scholar]
- Haebel PW, Gutmann S, Ban N. Dial tm for rescue: tmRNA engages ribosomes stalled on defective mRNAs. Curr Opin Struct Biol. 2004;14:58–65. doi: 10.1016/j.sbi.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Hallier M, Ivanova N, Rametti A, Ehrenberg M, Felden B. Pre-binding of small protein B to stalled ribosome triggers trans-translation. J Biol Chem. 2004;279:25978–25985. doi: 10.1074/jbc.M314086200. [DOI] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K, Takagi M, Inokuchi H, Himeno H, Muto A. SmpB functions in various steps of trans-translation. Nucleic Acids Res. 2002;30:1620–1629. doi: 10.1093/nar/30.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Lindell M, Pavlov M, Holmberg Schiavone L, Wagner EG, Ehrenberg M. Structure probing of tmRNA in distinct stages of trans-translation. RNA. 2007;13:713–722. doi: 10.1261/rna.451507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Gillet R, Li W, Gursky R, Frank J. Cryo-EM visualization of transfer messenger RNA with two SmpBs in a stalled ribosome. Proc Natl Acad Sci. 2006;103:16484–16489. doi: 10.1073/pnas.0607438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Konno T, Kurita D, Takada K, Muto A, Himeno H. A functional interaction of SmpB with tmRNA for determination of the resuming point of trans-translation. RNA. 2007;13:1723–1731. doi: 10.1261/rna.604907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzinger L, Hallier M, Felden B. The highest affinity binding site of small protein B on transfer messenger RNA is outside the tRNA domain. RNA. 2008;14:1761–1772. doi: 10.1261/rna.1185808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Stern S, Noller HF. Rapid chemical probing of conformation in 16s ribosomal RNA and 30s ribosomal subunits using primer extension. J Mol Biol. 1986;787:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNA in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- Rullgerd H, Oktem O, Skoglund U. A component-vise iterated relative entropy regularization method with updated prior and regularization parameter. Inverse Probl. 2007;23:2121–2139. [Google Scholar]
- Shimizu Y, Ueda T. SmpB triggers GTP hydrolysis of elongation factor Tu on ribosomes by compensating for the lack of codon-anticodon interaction during trans-translation. J Biol Chem. 2006;281:15987–15996. doi: 10.1074/jbc.M512165200. [DOI] [PubMed] [Google Scholar]
- Shpanchenko OV, Zvereva MI, Ivanov PV, Bugaeva EY, Rozov AS, Bogdanov AA, Kalkum M, Isaksson LA, Nierhaus KH, Dontsova OA. Stepping transfer messenger RNA through the ribosome. J Biol Chem. 2005;280:18368–18374. doi: 10.1074/jbc.M409094200. [DOI] [PubMed] [Google Scholar]
- Skoglund U, Ofverstedt LG, Burnett RM, Bricogne G. Maximum-entropy three-dimensional reconstruction with deconvolution of the contrast transfer function: A test application with adenovirus. J Struct Biol. 1996;117:173–188. doi: 10.1006/jsbi.1996.0081. [DOI] [PubMed] [Google Scholar]
- Sundermeier TR, Karzai AW. Functional SmpB-ribosome interactions require tmRNA. J Biol Chem. 2007;282:34779–34786. doi: 10.1074/jbc.M707256200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundermeier TR, Dulebohn DP, Cho HJ, Karzai AW. A previously uncharacterized role for small protein B (SmpB) in transfer messenger RNA-mediated trans-translation. Proc Natl Acad Sci. 2005;102:2316–2321. doi: 10.1073/pnas.0409694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu GF, Reid GE, Zhang JG, Moritz RL, Simpson RJ. C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J Biol Chem. 1995;270:9322–9326. doi: 10.1074/jbc.270.16.9322. [DOI] [PubMed] [Google Scholar]
- Valle M, Gillet R, Kaur S, Henne A, Ramakrishnan V, Frank J. Visualizing tmRNA entry into a stalled ribosome. Science. 2003a;300:127–130. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003b;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Ofverstedt LG, Skoglund U, Isaksson LA. Morphological variation of individual Escherichia coli 50S ribosomal subunits in situ, as revealed by cryo-electron tomography. Exp Cell Res. 2004a;300:190–201. doi: 10.1016/j.yexcr.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Ofverstedt LG, Skoglund U, Isaksson LA. Morphological variation of individual Escherichia coli 30S ribosomal subunits in vitro and in situ, as revealed by cryo-electron tomography. Exp Cell Res. 2004b;297:495–507. doi: 10.1016/j.yexcr.2004.03.049. [DOI] [PubMed] [Google Scholar]
- Zvereva MI, Ivanov PV, Topilina NI, Dontsova OA, Bogdanov AA, Kalkum M, Teraoka Y, Nierhaus KH, Shpanchenko OV. Complex of tmRNA and EF-Tu: Unexpected modes of interaction. J Biol Chem. 2001;276:47702–47708. doi: 10.1074/jbc.M106786200. [DOI] [PubMed] [Google Scholar]