Abstract
We show that microRNA-427 (miR-427) mediates the rapid deadenylation of maternal mRNAs after the midblastula transition (MBT) of Xenopus laevis embryogenesis. By MBT, the stage when the embryonic cell cycle is remodeled and zygotic transcription of mRNAs is initiated, each embryo has accumulated ∼109 molecules of miR-427 processed from multimeric pri-miR-427 transcripts synthesized after fertilization. We demonstrate that the maternal mRNAs for cyclins A1 and B2 each contain a single miR-427 target sequence, spanning less than 30 nucleotides, that is both necessary and sufficient for deadenylation, and that inactivation of miR-427 leads to stabilization of the mRNAs. Although this deadenylation normally takes place after MBT, exogenous miRNAs produced prematurely in vivo can promote deadenylation prior to MBT, indicating that turnover of the maternal mRNAs is limited by the amount of accumulated miR-427. Injected transcripts comprised solely of the cyclin mRNA 3′ untranslated regions or bearing a 5′ ApppG cap undergo deadenylation, showing that translation of the targeted RNA is not required. miR-427 is not unique in promoting deadenylation, as an unrelated miRNA, let-7, can substitute for miR-427 if the reporter RNA contains an appropriate let-7 target site. We propose that miR-427, like the orthologous miR-430 of zebrafish, functions to down-regulate expression of maternal mRNAs early in development.
Keywords: deadenylation, microRNA-427 function, miRNA biogenesis, Xenopus embryo-specific miRNA, maternal mRNA
INTRODUCTION
MicroRNAs (miRs) were discovered as gene products needed for the orderly transition between stages of larval development in C. elegans (Lee et al. 1993). Subsequent studies in many laboratories have shown that these 22–24-nucleotide (nt)-long RNAs function as guides that direct ribonucleoprotein particles—RNA-induced silencing complexes (RISCs)—to partially complementary sequences in the 3′ untranslated regions (UTRs) of specific messenger RNAs (mRNAs). Six-to-eight nucleotides in the 5′ proximal region of the miRNA (the “seed sequence”) are of primary importance in this recognition (Bartel 2004). Binding of RISCs and associated proteins results in translational repression (Nilsen 2007; Filipowicz et al. 2008) and can lead to deadenylation and degradation of the targeted mRNAs (Behm-Ansmant et al. 2006; Giraldez et al. 2006; Wu et al. 2006; Eulalio et al. 2007). Because they affect the activities of specific mRNAs, miRNAs that accumulate only at particular developmental stages are thought to affect stage-specific switches in gene expression.
Xenopus laevis miR-427 is an abundant miRNA that accumulates specifically in the early embryo (Watanabe et al. 2005; this report), reaching a maximum level after the midblastula transition (MBT), the time when zygotic mRNA synthesis starts and the cell cycle undergoes remodeling (Newport and Kirschner 1982). During the initial 12 rapid cell divisions prior to MBT, nuclear mRNA synthesis is not detectable, and translation in the cleavage embryo depends on the expression of maternal mRNAs that were made and stored during oogenesis (Davidson 1986). Several of these mRNAs encode cyclin proteins that must be continuously replenished to sustain the rapid cell divisions (Minshull et al. 1989; Hartley et al. 1996).
Shortly after MBT (∼7–8 h postfertilization [p.f.]), maternal cyclin A1 and B2 mRNAs undergo rapid deadenylation and subsequent decay, in a process that requires zygotic transcription (Audic et al. 2001, 2002). Hartley and coworkers identified regions in the 3′ UTRs of these two cyclin mRNAs that were both necessary and sufficient for the deadenylation (Audic et al. 2001, 2002; R Hartley, unpubl.).
In zebrafish and flies the clearance of many maternal mRNAs during the transition from exclusively maternal to maternal plus zygotic gene expression is promoted by microRNAs (Giraldez et al. 2006; Bushati et al. 2008). In particular, the zebrafish miR-430 that accumulates upon activation of zygotic transcription (Giraldez et al. 2005) is responsible for reducing the levels of numerous maternal mRNAs, and it mediates deadenylation of several of these (Giraldez et al. 2006; Mishima et al. 2006). Xenopus miR-427 is the ortholog of miR-430 (Chen et al. 2005), raising the possibility that it participates in the turnover of maternal mRNAs during early embryogenesis.
Here, we show that the rapid accumulation of high amounts of miR-427 by MBT is a consequence of transcription of a large multigene family that starts prior to the onset of zygotic mRNA synthesis. We demonstrate that miR-427 directs the efficient deadenylation and destabilization of cyclin A1 and B2 mRNAs after MBT, in a process that is independent of translation. Each of the targeted mRNAs possesses a single miR-427 recognition element (MRE427) that is both necessary and sufficient for this process, but other miRNA/MRE pairs can substitute for miR-427/MRE427 in directing deadenylation of reporter RNAs.
RESULTS
miR-427 is expressed prior to MBT
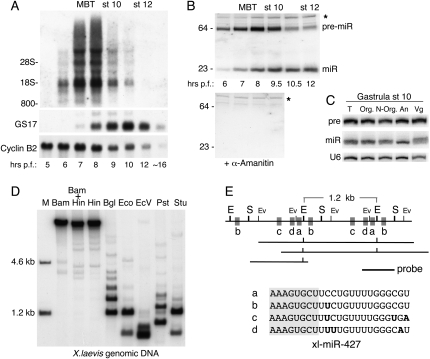
The onset of synthesis of miR-427 transcripts was determined by Northern blot analysis of RNAs isolated from embryos at various times after fertilization. Neither miR-427 nor its precursors, the 64-nt pre-miR-427 and the >1-kb primary transcripts (pri-miR-427) (analyzed using the probe indicated in Fig. 1E), were detectable at very early times (Figs. 1A, top; 2D, left; Watanabe et al. 2005), showing that miR-427 is produced by zygotic transcription rather than by processing of stored maternal RNAs. Synthesis of pri-miR-427 and pre-miR-427 was first detectable at ∼5–6 h p.f., which is several hours earlier than activation of general zygotic mRNA transcription, as monitored by accumulation of GS17 mRNA (Fig. 1A, middle; Krieg and Melton 1985).
FIGURE 1.
Biogenesis of miR-427 during early stages of Xenopus laevis embryogenesis. (A) Kinetics of accumulation and turnover of pri-miR-427, GS17 mRNA (an early indicator of zygotic transcription), and maternal cyclin B2 mRNA. The midblastula transition (MBT) and different stages of gastrulation (st 10, st 12) and hours post fertilization (p.f.) are shown above and below the autoradiograms of Northern blots containing one embryo-equivalent of total RNA per lane; the gel mobilities of ribosomal RNA and other size markers are indicated. (B) Northern blot analyses (1.5 embryo-equivalents of total RNA per lane) showing the precursor–product relationship of pre-miR-427 and mature miR-427 (top) and the absence of pre-miR-427 and miR-427 from embryos treated with α-amanitin (bottom). (*) Cross-hybridizing RNA that serves as loading control. (C) Northern blot analysis of pre-miR-427 and miR-427 in total RNAs from dissected pieces of stage 10.25 gastrula embryos. U6 RNA was also probed, for normalization. (T) Total; (Org) organizer half; (N-Org) nonorganizer half; (An) animal pole; (Vg) vegetal pole. (D) Southern blot of Xenopus genomic DNA (3.2 μg per lane) digested with the restriction enzymes indicated (BamH1, HindIII, BglII, EcoRI, EcoRV, PstI, StuI), showing the existence of a large number of identical miR-427-encoding regions that are devoid of BamH1 and HindIII cleavage sites. (E) Maps of representative EST clones (see Materials and Methods) used to deduce the structure of the 1.2-kb repeat unit in X. laevis genomic DNA (top) that encodes four (a–d) sequence variants of xla- miR-427 (bottom); (E) EcoRI cleavage sites; (S) StuI cleavage sites; (EV) EcoRV cleavage sites. The hybridization probe used in D is indicated.
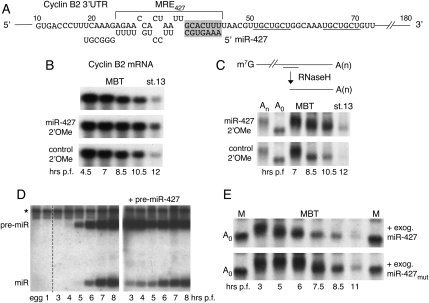
FIGURE 2.
miR-427-dependent destabilization and deadenylation of cyclin B2 mRNA. (A) Nucleotide sequences in the cyclin B2 3′ UTR showing the predicted seed-match (shaded box) for miR-427 within a potential MRE427, and potential miR-16 seed-matches (underlined). (B) Stabilization of cyclin B2 mRNA upon inactivation of miR-427. Northern blot analyses of endogenous cyclin B2 mRNA in early embryos that were untreated (top) or injected at the one-cell stage with 2′-OMe-antisense oligonucleotides against miR-427 (middle) or a let-7 control (bottom). (C) Impaired deadenylation of endogenous cyclin B2 mRNA upon inactivation of miR-427. (Top) Schematic of RNase H assay for determination of changes in poly(A) tail length, using a deoxyoligonucleotide that targets sequences ∼500 nt from the 3′ end of the mRNA. (Lower panels) Northern blot analyses of the 3′-terminal fragments of cyclin B2 mRNA generated by digestion with RNase H, showing the kinetics of deadenylation in the absence (top) or presence (bottom) of functional miR-427. Lanes An and A0 show size markers generated by digestion of cyclin B2 mRNA of pre-MBT embryos with RNase H plus the deoxy-oligonucleotide, in the absence (An) or presence (A0) of oligo-dT15. (D) Northern blot analyses of pre-miR-427 and miR-427 in normal embryos (left) and in embryos injected with exogenous pre-miR-427 (right); a similar pattern of miRNA accumulation was seen when pre-miR-427mut was injected. Analyses of RNAs from unfertilized eggs and one-cell (1 h p.f.) embryos (left panel) show the lack of pre-miR- and miR-427 in those cells. (E) Accelerated deadenylation of endogenous cyclin B2 mRNA upon premature expression of miR-427, but not miR-427mut. Exogenous pre-miR-427 or pre-miR-427mut was injected into one-cell embryos, and RNase H digested RNAs were analyzed as in C.
Production of miR-427 was sensitive to α-amanitin (Fig. 1B), indicating that it required RNA polymerase II (Pol II) function, but the factor(s) responsible for the unusually early activation of Pol II transcription remains to be identified. Accumulation of both pri-miR-427 and pre-miR-427 ceased ∼9 h p.f., indicating that transcription of miR-427 stops during gastrulation. At that time, each embryo contained ∼109 copies of mature miR-427, making it possible to detect the mature miRNA by direct labeling with 32P-GTP (data not shown). Moreover, quantitative Northern blot analysis of RNAs from dissected embryo pieces showed that miR-427 was uniformly distributed throughout early embryos both before (data not shown) and after MBT (Fig. 1C) and turned over with a half-life of ∼15 h following its peak levels during gastrulation (data not shown).
The robust accumulation of miR-427 within a few hours of fertilization resulted, in large part, from the presence of hundreds of copies of a ∼1.2-kb DNA repeat sequence in the X. laevis genome (Fig. 1D) encoding four isotypes of miR-427 (Fig. 1E, a–d). From Southern blot analyses, we estimated that each X. laevis cell contains about 1000 miR-427 coding regions (see Materials and Methods) and inferred that many of these repeats are in tandem arrays (data not shown). The presence of pri-miR-427 transcripts that are much longer than 28S rRNA (∼4–8 kb) (Fig. 1A) indicated that multimeric pri-miRNA transcripts may be generated by read-through between adjacent 1.2-kb repeat units, consistent with the existence of X. laevis cDNA clones that encompass tandemly arrayed repeats (Fig. 1E).
miR-427 is necessary for deadenylation of cyclin B2 mRNA
The maternal cyclin B2 and A1 mRNAs, which become polyadenylated during egg maturation and shortly after fertilization (Sheets et al. 1994), undergo rapid deadenylation and destabilization at MBT in a process that depends on specific 3′ UTR sequences and requires transcription by RNA Pol II (Audic et al. 2001). The α-amanitin-sensitive accumulation of miR-427 (Fig. 1B) raised the possibility that this miRNA promotes the inactivation and destruction of these maternal cyclin mRNAs. Previous deletion mapping of the cis-acting regions needed for this deadenylation revealed no extended sequences shared between the two mRNAs (Audic et al. 2001, 2002). However, a re-examination showed that the required region in each mRNA contained a single short miRNA recognition element (referred to as MRE427) comprised of sequences complementary to the seed sequence of miR-427 plus short sequences that could pair with additional regions of the miRNA (Figs. 2A, 5A, see below; Weigel and Izaurralde 2006).
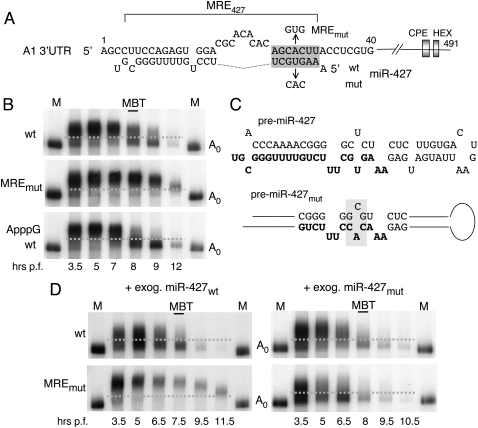
FIGURE 5.
MRE•miRNA base pairing in the deadenylation of cyclin A1 3′ UTR RNA. (A) Nucleotide sequences in the cyclin A1 3′ UTRs (top strand) showing the predicted seed-match (shaded box) and upstream 3′ UTR sequences that allow for functional pairing with miR-427 (bottom strand; cf. Fig. 6). The inactivating mutation of the MRE (MREmut) and the compensatory change in the seed sequence miR-427 (mut) are indicated. (B) Deadenylation of 32P-labeled A1 3′ UTR reporter RNAs lacking a coding region and containing either a wild-type (wt) or mutant MRE (MREmut), or an ApppG- cap, to ensure lack of translation. (C) Sequences of wild-type and mutant forms of pre-miR-427. Nucleotide changes in pre-miRmut (shaded) were designed to encode miR-427mut with an altered seed sequence; (bold) mature miRNA sequences. (D) Rescue of MREmut by the compensatory mutation of the miR-427 seed sequence. Kinetics of polyadenylation and deadenylation of cyclin A1 3′ UTR reporter RNAs with wild-type (top panels) or mutant (bottom panels) seed-matches; the reporters were co-injected with exogenous pre-miRNAs generating either wild-type (left panels) or mutant (right panels) miR-427.
To test directly if miR-427 contributes to the destabilization or deadenylation of cyclin B2 mRNA, we first inactivated the miRNA with a complementary 2′-O-methylated oligonucleotide (Hutvánger et al. 2004; Meister et al. 2004). Injection of this oligonucleotide into one-cell embryos led to stabilization of the mRNA after MBT (Fig. 2B, cf. top and middle panels), whereas a control 2′-O-methylated oligonucleotide (complementary to let-7a RNA) had no effect (Fig. 2B, bottom panel). The complementary 2′-O-methyl oligonucleotide (but not the control oligonucleotide) also slowed the rate of deadenylation of the cyclin B2 mRNA, as monitored by differences in the electrophoretic mobilities of a 3′ end fragment of the mRNA generated by treatment with RNase H (Fig. 2C). Thus, miR-427 function is required for normal deadenylation and turnover of cyclin B2 mRNA after MBT.
Next, we asked if exogenous miR-427 could promote deadenylation of cyclin B2 mRNA prior to MBT. For this, we generated the miRNA prematurely (but not in excessive amounts) by injecting titrated amounts of in vitro made pre-miR-427 (data not shown; Fig. 5C) into one-cell embryos (Fig. 2D; Lund and Dahlberg 2006). The early production of mature miR-427 led to accelerated deadenylation and destabilization of the cyclin B2 mRNA (Fig. 2E); in contrast, injection of a control pre-miRNA (see below, Fig. 5C, pre-miR-427mut) that generated a variant miRNA with an altered seed sequence had no detectable effect on the mRNA turnover (data not shown; Fig. 2E). This result shows both that exogenous miR-427 can function in deadenylation prior to MBT, and that the onset of deadenylation is likely determined by the availability of the miRNA.
The MRE427 is necessary for deadenylation of mRNA at MBT
To test if the predicted MRE427 can function in the deadenylation of cyclin B2 mRNA, we used in vitro synthesized 32P-labeled reporter RNAs (Gb•B2), in which a β-globin coding region was fused to the 180-nt 3′ UTR of cyclin B2 mRNA (Fig. 3A; Audic et al. 2001). The cyclin B2 3′ UTR contains two cytoplasmic polyadenylation elements (CPEs) and an AAUAAA hexanucleotide polyadenylation signal (HEX) that are required for cytoplasmic poly(A) addition (Sheets et al. 1994). Either CPE can function in polyadenylation in the absence of the other (Guo et al. 2008). As expected, upon injection into one- or two-cell embryos, Gb•B2 RNA became polyadenylated, as evidenced by a decrease in gel mobility (Fig. 3B). Like endogenous cyclin B2 mRNA, the Gb•B2 RNA is subject to deadenylation and destabilization after MBT (Audic et al. 2001). In contrast, a variant reporter RNA with a 3-nt substitution in MRE427 (MREmut) that disrupts base pairing with the seed sequences of miR-427 remained polyadenylated and was stable (Fig. 3B, bottom panel). Moreover, a 180-nt transcript consisting solely of the full-length cyclin B2 mRNA 3′ UTR also underwent deadenylation in an MRE427-dependent manner (Fig. 4B). Thus, the wild-type MRE427 is necessary for deadenylation and rapid decay, but translation of the target RNA is unlikely to be required (see below).
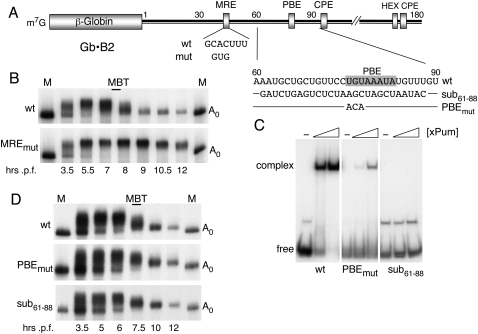
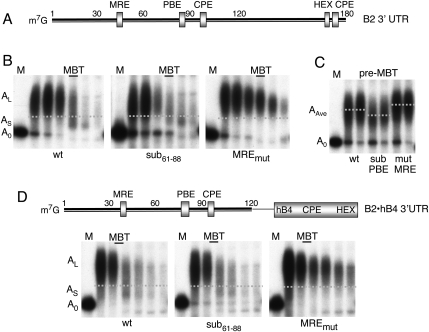
FIGURE 3.
Deadenylation of exogenous cyclin B2 reporter RNAs. (A) Schematic representation of the chimeric β-globin•cyclin B2 3′ UTR reporter mRNA (Gb•B2), indicating the seed-match for miR-427 (MRE), cytoplasmic polyadenylation elements (CPE), hexanucleotide polyadenylation signal (HEX), and pumilio binding element (PBE); the sequence of the PBE and substitution mutants of it are shown on the right. (B) Kinetics of polyadenylation and deadenylation of Gb•B2 reporter RNAs harboring wild-type (top) or mutant seed-matches (MREmut, bottom). Polyacrylamide gel analyses of 32P-labeled reporter RNAs that were injected into two-cell embryos (∼2 h p.f.) and reisolated at the indicated times. Marker lanes (M) show the gel mobilities of the injected, nonpolyadenylated (A0) transcripts. (C) PBE-dependent binding of Xenopus Pumilio protein (xPum). The electrophoretic mobility shift assays monitor in vitro complex formation between recombinant xPum (at 120 and 500 nM) and the indicated wild-type (wt) or substituted (PBEmut, sub61-88) 31-nt 32P-labeled RNAs. (D) PBE-independent deadenylation. Kinetics of polyadenylation and deadenylation of Gb•B2 reporter RNAs containing wild-type or substituted PBE regions were analyzed as in B.
FIGURE 4.
Functions of MRE and PBE in poly(A) metabolism of cyclin B2 3′ UTR reporter RNAs. (A) Schematic representation of the cyclin B2 3′ UTR. (B) Kinetics of polyadenylation and deadenylation of wild-type RNA or substitution variants altering the MRE (MREmut) or PBE (sub61–88). 32P-labeled reporter RNAs were injected and assayed as in Figure 3B; samples were taken at 5, 6.5, 8, 10.5, 12.5, and ∼15 h p.f. AS and AL denote short and long poly(A) tail length, respectively (dotted lines). (C) Less extensive polyadenylation of the reporter RNA lacking the PBE. The pre-MBT samples (5 and 6.5 h p.f.) from the experiment shown in B were analyzed in the same gel, for direct comparison of the average poly(A) tail lengths (AAve, dotted lines). (D) Normal deadenylation kinetics of chimeric B2 3′ UTR reporter RNAs deleted of nucleotides 120–180 and the fused to hB4 3′ UTR sequences, which furnish CPE plus HEX functions. The wild-type (1–120 nt) and PBE or MRE substituted reporter RNAs were assayed as in B; samples were taken at 4.5, 6, 8, 9, 10.5, and 12 h p.f.
A Pumilio binding element does not enhance deadenylation
The region in the 3′ UTR of cyclin B2 mRNA encompassing the deadenylation element (Audic et al. 2001) also contains the sequence UGUAAAUA (Fig. 4A), which conforms to a Pumilio protein binding element (PBE) (Zamore et al. 1997; Wickens et al. 2002). The pumilio protein of X. laevis (xPum) is orthologous to Puf proteins that recruit the mRNA deadenylase complex CCR4-NOT in yeast (Goldstrohm et al. 2006) and contribute to miRNA function in C. elegans (Nolde et al. 2007). Because the CCR4-NOT complex is required for miRNA-mediated deadenylation of mRNAs in Drosophila cells (Behm-Ansmant et al. 2006), we asked if the xPum binding site contributed to deadenylation of B2 3′ UTR reporter RNAs in X. laevis embryos.
The presumptive PBE sequence, but not variants with substitutions of 3 nt or the entire site plus flanking sequences, was able to promote binding of recombinant xPum to a 30-nt RNA in an in vitro gel shift assay (Fig. 3C), indicating that this PBE is functional. However, introduction of these substitutions into the Gb•B2 (Fig. 3D) or B2 3′ UTR (Fig. 4B) reporter RNAs had little, if any, effect on the timing or efficiency of deadenylation, making it unlikely that xPum participates in deadenylation of these RNAs. In contrast, disruption of the PBE reduced the efficiency of the poly(A) addition to the reporter RNAs, as evidenced by an increase in the amount of Gb•B2 RNA that remained nonpolyadenylated at 5 or 6 h p.f. (Fig. 3D), and by the faster gel mobility [shorter poly(A) tail length] of the B2 3′ UTR reporter RNA lacking a PBE (Fig. 4C, AAve). Thus, the PBE appears to facilitate poly(A) addition onto these reporter RNAs, perhaps through interaction with Gld-2 protein (Barnard et al. 2004; Kwak et al. 2004). However, we note that the stimulating activity of this PBE was context-dependent (Piqué et al. 2008), as substitution of the PBE was inconsequential for polyadenylation of a chimeric 3′ UTR reporter RNA that uses the CPE/HEX of histone B4 3′ UTR (Fig. 4D).
The miR-427 target in the cyclin B2 3′ UTR is short and compact
To identify sequences other than MRE427 that are required for deadenylation of the 180-nt-long B2 3′ UTR, we generated additional variants by deletion or substitution. Polyadenylation of these reporter RNAs was assured even in the absence of native CPE and HEX elements by appending an RNA region containing CPE plus HEX sequences derived from histone B4 mRNA (Fig. 4D; Paris and Philippe 1990; Audic et al. 2002). Like the full-length cyclin B2 3′ UTR, a transcript lacking the 3′ terminal 60 nt underwent MRE427-dependent deadenylation, as did a transcript with a substitution of nucleotides 61–88 (Fig. 4D). Likewise, a shortened reporter RNA, containing only nucleotides 1–60 of the cyclin B2 3′ UTR, was subject to MRE427-mediated deadenylation (data not shown), but this transcript was less stable overall. Thus, nucleotides within the first 60 residues of the 3′ UTR (which include the MRE427, but not the PBE) suffice for the deadenylation.
Translation is not required for MRE427-promoted deadenylation
Like the 3′ UTR of cyclin B2 mRNA, the 3′ UTR of cyclin A1 mRNA supports deadenylation of a chimeric Gb•A1 mRNA at MBT (Audic et al. 2001). A 491-nt-long A1 3′ UTR reporter RNA containing the putative MRE427 (near the 5′ terminus) plus CPE and HEX sequences, but no PBE (Fig. 5A), also underwent rapid polyadenylation and was subject to deadenylation and destabilization after MBT (Fig. 5B, top and middle panels). The deadenylation of the two reporter RNAs containing cyclin A1 and B2 3′ UTR sequences, but lacking coding regions, indicated that translation of the substrate is not required. This was tested by the use of a cyclin A1 3′ UTR reporter bearing an ApppG cap rather than m7GpppG cap. The cap substitution did not interfere with deadenylation, confirming that translation of the substrate is not required. A similar conclusion was reached previously in studies of other systems (Giraldez et al. 2006; Wu et al. 2006; Eulalio et al. 2007).
The MRE427 of cyclin A1 mRNA interacts directly with miR-427
Mutation of the sequence in the cyclin A1 mRNA that is complementary to the seed sequence of miR-427 led to stabilization of the reporter RNA, showing that this region functions as an MRE for deadenylation (Fig. 5B). To demonstrate a direct interaction of this region with miR-427, we generated a variant of pre-miR-427 that would produce miR-427mut (Fig. 5C), with a seed sequence complementary to the nucleotides substituted in the mutant, MREmut (Fig. 5A). Wild-type or mutant pre-miRNAs were then co-injected with A1 3′ UTR reporter RNAs bearing either the wild-type or mutant MRE, and deadenylation of the reporter RNAs was monitored. As expected from the results shown in Figure 2E, exogenous wild-type miR-427 accelerated deadenylation of the wild-type reporter RNA (Fig. 5B,D, cf. top panels) but not that of the reporter with MREmut (Fig. 5D, bottom-left panel). In contrast, exogenous miR-427mut accelerated deadenylation of the reporter RNA containing MREmut much more than that of the reporter with the wild-type MRE427 (Fig. 5D, right panels). We conclude that MRE427 functions through base-pairing with the seed sequence of miR-427.
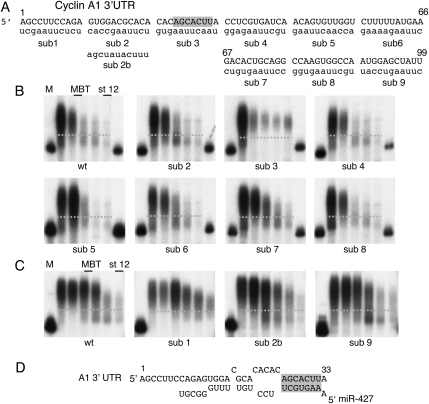
Few sequences of the A1 3′ UTR other than the MRE427 are important for deadenylation
Hartley and co-workers (Audic et al. 2002) previously showed that the first 99 nt of the cyclin A1 3′ UTR are sufficient for deadenylation of Gb•A1 3′ UTR reporter mRNAs. That region contains the miR-427-seed match defined above. To identify the minimal region of the A1 3′ UTR required for MRE427 function, we generated a series of substitution mutations throughout this 99-nt-long sequence in a 3′ UTR reporter RNA that contained the hB4 CPE/HEX cassette (Fig. 6). As expected from the results of Figure 5B, the region containing the sequences complementary to the seed sequence was essential for deadenylation (Fig. 6B, sub 3). In addition, the region containing nucleotides 1–11, which could potentially pair with the 3′ end of miR-427 (Fig. 5A), was needed for full efficiency of deadenylation (Fig. 6C, cf. wt and sub 1). In contrast, substitution of the other 11-nt sections throughout the reporter RNA had no appreciable effect on deadenylation (Fig. 6B,C), including the region containing nt 12–22 (Fig. 6B,C, sub 2, 2b), which also had the potential of pairing with parts of miR-427 (Fig. 6D). Thus MRE427 is a compact structure that consists of seed-match sequences plus noncontiguous sequences, which likely interact with other regions of miR-427. Consistent with these results, a truncated reporter RNA containing only the 5′ proximal 33 nt of the cyclin A1 3′ UTR (plus appended hB4 CPE and HEX sites) (Fig. 7A) was efficiently deadenylated in an MRE427- and miR-427-dependent manner (Fig. 7B).
FIGURE 6.
Identification of regions in the cyclin A1 3′ UTR needed for deadenylation. (A) Nucleotide sequences of region 1–99 of the cyclin A1 3′ UTR and 11-nt substitutions (lowercase) that were tested for their effects on deadenylation. (B,C) Kinetics of deadenylation of A1 3′ UTR (1–99)•hB4 chimeric reporter RNAs with wild-type sequences (wt) or 11-nt substitutions (sub 1–9). Injected 32P-labeled RNAs were analyzed as in Figure 3B, and samples were taken at 4, 5.5, 7.5, 8.5, 10.5, and 12 h p.f. (B) or 4, 5.5, 7.5, 8.5, 10.5, and 12 h p.f. (C). (D) A possible base-paired structure between miR-427 and nucleotides in region 12–22 (plus the seed-match) of wild-type A1 3′ UTR. Figure 5A shows a more likely alternative structure between miR-miR-427 and A1 3′ UTR that involves region 1–11.
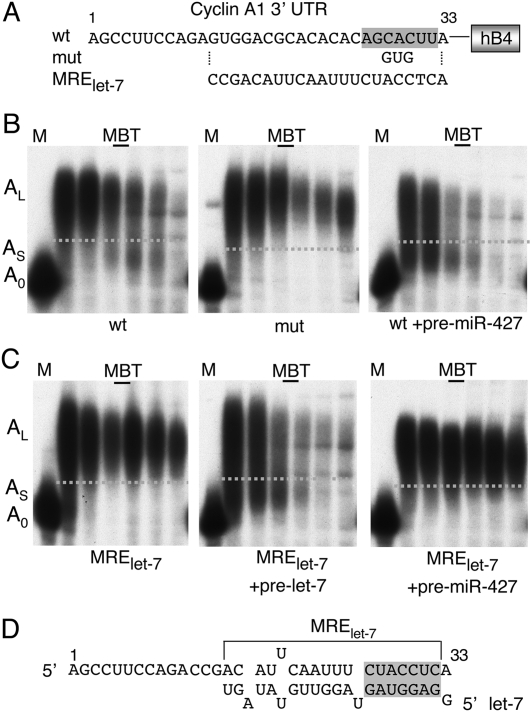
FIGURE 7.
Compact structure and sequence-specific nature of MREs. (A) Sequences of the “minimal” cyclin A1 3′ UTR (1–33) capable of supporting deadenylation, and substitutions that inactivate the seed-match of MRE427 (mut) or introduce an MRElet-7. (B) Deadenylation of the wild-type A1 3′ UTR (1–33)•hB4 chimeric reporter RNA (left) is blocked upon substitution of MRE427 (mut) and accelerated upon premature expression of miR-427 from co-injected pre-miR-427 (right). The samples were taken at 4, 6, 7.5, 8,5, 10.5, and 12.5 h p.f. (C) Deadenylation of the chimeric MRElet-7 reporter RNA depends on expression of exogenous let-7 miRNA; pre-miRNAs that encode let-7 RNA or miR-427 were co-injected with the MRElet-7 reporter, as indicated. The samples were taken at 4.5, 6, 7.5, 8.5, 9.5, and 12 h p.f. (D) The sequence of MRElet-7 and its predicted base pairing with let-7 RNA (Mayr et al. 2007).
Pairing between MRE and miRNA, not miR-427 identity, directs deadenylation
Finally, we asked if complexes containing miR-427-RISC and the cyclin A1 MRE427 had unique properties that promote deadenylation, or if other miR-RISC/MRE complexes would suffice. For this, we replaced nucleotides 12–32 of the “minimal” A1 3′ UTR reporter RNA (Fig. 7A) with a verified target site for let-7 RNA (here referred to as MRElet-7), derived from the 3′ UTR of human HMGA2 mRNA (Mayr et al. 2007). Early embryos are reported not to contain mature let-7 RNA (Watanabe et al. 2005) and, as expected, this substitution abolished deadenylation (Fig. 7C, left panel). However, co-injection of pre-let-7 RNA, but not pre-miR-427, led to efficient deadenylation and destabilization of the MRElet-7 reporter RNA prior to MBT (Fig. 7C, middle and right panels). Thus, while pairing between MRE and miRNA is essential, the identity of the miRNA does not appear to be crucial for mediating deadenylation in early X. laevis embryos.
DISCUSSION
We have shown that miR-427, a miRNA that rapidly accumulates to high levels very early in X. laevis development, mediates the efficient deadenylation of maternal cyclin A1 and B2 mRNAs at the midblastula transition (MBT). The 3′ UTRs of both mRNAs contain a single compact miR-427 target site that is required for this process. The ability of a single MRE427 to direct rapid deadenylation may be a consequence of the high concentration of miR-427-containing RISCs present in the embryo at MBT.
Although neither miR-427 nor its precursors are present at fertilization, ∼109 copies of the mature miRNA are produced within a few hours of X. laevis early embryogenesis, evenly distributed throughout the embryo. This rapid accumulation of high amounts of miR-427 by the time of MBT can be attributed to a high number of gene copies (500–1000 potential miR-427 coding regions per cell) and very efficient synthesis and processing of precursors (Fig. 1). Long pri-miR-427 precursors, which encode multiple copies of miR-427 sequences, are among the earliest transcripts synthesized by RNA polymerase II after fertilization, as revealed by their abundant accumulation well before the time of transcriptional activation of most zygotic mRNAs (Davidson 1986). The factor(s) responsible for this very early onset of expression, or for its cessation during gastrulation, remains unknown. However, β-catenin, which promotes the unusually early zygotic transcription of the nodal-related Xnr5 and Xnr6 genes in Xenopus cleavage embryos (Yang et al. 2002), is unlikely to be required, as an antisense morpholino oligonucleotide directed against β-catenin mRNA (Heasman et al. 2000) did not affect the accumulation of pre-miR-427 (data not shown).
High amounts of mature miR-427 can be generated earlier than normal by injecting exogenous pre-miR-427 into one- or two-cell embryos (Fig. 2D; Lund and Dahlberg, 2006). The resulting premature accumulation of miR-427 accelerates the onset of deadenylation of both endogenous and exogenous reporter mRNAs, showing that all other components needed for the deadenylation are present in the cleavage embryo (Figs. 2E, 5D). Thus, the key step that controls deadenylation of the maternal cyclin mRNAs is the generation of miR-427.
The 3′ UTR of cyclin B2 mRNA also contains two UGCUGCU sequences (nucleotides 51–57 and 64–70) (Fig. 2A), which are complementary to the seed sequence of miR-16, a maternally derived miRNA that is present in early embryos (Watanabe et al. 2005; E Lund, unpubl.). However, it is unlikely that miR-16 contributes to the turnover of cyclin B2 mRNA, as B2 3′ UTR reporter RNAs are stable and remain polyadenylated in the absence of functional miR-427 or MRE427. We note that miR-16 is at least two orders of magnitude less abundant than miR-427 (Watanabe et al. 2005; E Lund, unpubl.), suggesting that the amounts of miR-16•RISC in the embryo might be insufficient to mediate detectable deadenylation. Alternatively, the predicted MRE16 sequences may be inaccessible within the structure of the 3′ UTR, or maternal miR-16 may be sequestered in inactive complexes. A specific requirement for miR-427 or MRE427 sequences can be ruled out, as an artificial pair of let-7 miRNA and MRElet-7 was able to elicit deadenylation of a reporter RNA (Fig. 7C).
Generally, interactions of miRNAs with cognate MREs in targeted mRNAs can have a variety of consequences that include translational silencing and/or deadenylation plus destabilization of the mRNA. Which one of these processes predominates is likely to be determined both by the protein components of RISCs and by accessory factors that may be recruited to particular miR-RISC complexes (for review, see Filipowicz et al. 2008). The identities of the Ago proteins available for assembly into RISCs in Xenopus early embryos are unknown, but we have found that Ago2 activity is not detectable at this stage (E Lund, M Sheets, and JE Dahlberg, unpubl.). This raises the possibility that RISCs assembled de novo in early embryos have functions distinct from maternally assembled RISCs. Thus, the consequences of miRNA•MRE interaction may depend on the pool of Ago proteins available during RISC assembly (Wu et al. 2008), rather than the particular miRNA/mRNA pair.
For several reasons, it was appealing to consider PUF proteins as factors that would contribute to miR-427-promoted deadenylation of mRNAs. PUF proteins participate in poly(A) metabolism in both vertebrates and yeast, interacting with CCR4-NOT1 (Goldstrohm et al. 2006), and they facilitate certain miRNA functions in Caenorhabditis elegans (Nolde et al. 2007). Also, PUF binding elements (PBEs) and miRNA target sites tend to be clustered within 3′ UTR sequences of mRNAs bound by human PUF proteins (Galgano et al. 2008). A canonical PBE that is recognized in vitro by Xenopus Pumilio (xPum) protein is close to the MRE427 of cyclin B2 mRNA, but inactivation of this PBE was without observable effects on deadenylation or destabilization of B2 3′ UTR reporter RNAs in vivo (Figs. 3, 4). Moreover, no PBE is present in the cyclin A1 3′ UTR, making it is unlikely that xPum interacts with this miRNA regulatory system. The PBE in the B2 3′ UTR reporter RNA is located close to a cytoplasmic polyadenylation element (CPE) (Fig. 4; see discussion by Piqué et al. 2008) and appears to increase the efficiency of poly(A) addition, but a role of xPum in controlling cyclin B2 gene expression in early embryos is unclear.
The ability of miR-427 to promote deadenylation of ApppG-capped RNAs indicates that the nuclease responsible for deadenylation is unlikely to be PARN, which has optimal activity on 5′-m7Gppp-capped substrates (Dehlin et al. 2000; Wu et al. 2009). A good candidate is the Caf1/CCR4-NOT1 complex, which is required for miR-mediated deadenylation in both Drosophila cells (Behm-Ansmant et al. 2006) and NIH3T3 mouse cells (Chen et al. 2009) or a related complex, which might be recruited in conjunction with other essential factors, such as the P-body component GW182 (Eulalio et al. 2009a,b). The sizes of the degradation products decrease progressively with time (e.g., Figs. 2E, 3B), indicating that the Xenopus deadenylase acts exonucleolytically. However, this nuclease does not appear to remove the entire poly(A) tail, as shown by poly(A) tail lengths of endogenous cyclin B2 mRNAs (Fig. 2) and by the difference in gel mobilities of reporter RNAs prior to poly(A) addition and after extensive miR-mediated deadenylation in the early embryos (e.g., Fig. 4, cf. A0 and AS). We are currently testing the involvement of various X. laevis deadenylases in this process.
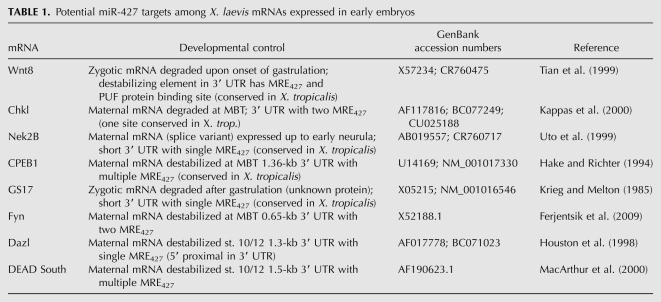
The numbers and identities of other X. laevis mRNAs whose expression might be controlled by the abundant miR-427 are not known. A limited bioinformatics search of X. tropicalis EST sequences previously identified mRNAs with potential miR-427 target sites in their predicted (but not verified) 3′ UTRs (Chen et al. 2005). However, it is unclear whether all of these mRNAs are expressed at the same time as miR-427, or if their MRE427 sequences are accessible to miR-427. Experimentally, we have identified Arrdc1, a maternally inherited mRNA that contains six potential MRE427 sites in a 1.8-kb 3′ UTR, as another candidate target in X. laevis (M Liu, E Lund, and JE Dahlberg, in prep.). This mRNA is subject to deadenylation after gastrulation but in a miR-427-independent manner, suggesting that these MRE427 sites are not accessible to the miRNA at MBT. However, in stage 12.5 embryos, several of the predicted sites function in translation repression of a chimeric luciferase reporter mRNA containing the Arrdc1 3′ UTR. Thus, MRE427 sequences may influence the fate of mRNAs in ways other than through deadenylation. Other Xenopus mRNAs that are deadenylated or destabilized shortly after MBT, and which contain one or more potential (but not validated) MRE427 sites in their 3′ UTRs, include GS17, CPEB, xWnt8, Chk1, Nek2b, Fyn, Dazl, and DEADSouth (summarized in Table 1).
TABLE 1.
Potential miR-427 targets among X. laevis mRNAs expressed in early embryos
In several ways, X. laevis miR-427 resembles the orthologous zebrafish miR-430 (Chen et al. 2005). Zebrafish depleted of miRNA-430 have developmental defects (Giraldez et al. 2005), and we have observed developmental anomalies in tadpoles developed from X. laevis embryos deficient in miR-427 (E Lund, M Sheets, and JE Dahlberg, unpubl.). Both miRNAs are encoded by large multigene families expressed very early in embryogenesis, and both target several maternal mRNAs for deadenylation and destabilization (Figs. 1, 2, 5; Giraldez et al. 2005, 2006). We note that zebrafish cyclin A1 mRNA, which is destabilized in a miR-430-dependent manner (Giraldez et al. 2006), contains a potential, noncanonical miR-430 target site in its 3′ UTR that was not recognized by the search algorithm used because the seed-match contains a single G–U base pair. However, other miR-target sites that lack a perfect seed-match have been identified in functional studies (Vella et al. 2004; Didiano and Hobert 2008). Thus, zebrafish cyclin A1 mRNA may well be a direct target of miR-430, suggesting an evolutionarily conserved mechanism of miR-regulation. The human embryonic cyclin A1 3′ UTR also has a potential (but not yet validated) target site for the orthologous human miRNAs.
Clusters of genes encoding miR-427 (Fig. 1) or miRNAs with the same seed sequence are present in the genomes of several other vertebrates, including X. tropicalis (miR-427; Tang and Maxwell 2008), zebrafish (miRNA-430; Giraldez et al. 2005), humans (miR-301, -302, -371-373; Suh et al. 2004; Cao et al. 2008), mouse (miR-290-295; Houbaviy et al. 2003), and primates (miR-519 and -520 families; Bentwich et al. 2005; Bortolin-Cavaillé et al. 2009). Most of the orthologous mammalian miRNAs are expressed very early in development in embryonic stem (ES) cells (Houbaviy et al. 2003; Suh et al. 2004) or primarily in placenta (Bentwich et al. 2005).
Recently, zebrafish miR-430 (Choi et al. 2007) and both Xenopus miR-427 and human miR-302 (Rosa et al. 2009) were shown to modulate the expression of genes in the Nodal signaling pathways that function in early germ-layer development, indicating that this family of orthologous miRNAs have related, phylogenetically conserved functions very early in development. Thus, evidence is emerging that members of this class of orthologous miRNAs play crucial roles in the development of early embryos by regulating the expression of maternal and early zygotic mRNAs.
MATERIALS AND METHODS
DNA constructs
The chimeric reporter constructs pGbA1, pGbA1Δ384, and pGbB2 have been described previously (Audic et al. 2001, 2002). pT7A1, containing the cyclin A1 3′ UTR, was constructed from pGbA1 by deletion of the β-globin coding region, and pT7A1 and pGbB2 variants with mutations in the MRE427 or PBE regions were generated by QuikChange Mutagenesis (Stratagene) (all primers and synthetic or mutagenic oligonucletides are listed in Table 2). Linear templates with T7 promoters encoding wild-type (wt) and variant B2 3′ UTRs or shorter regions of the A1 and B2 3′ UTRs fused to the histone B4 CPE plus HEX region (Paris and Philippe 1990) were generated by PCR from plasmid templates and/or synthetic oligonucleotides. The multi-step PCR scheme for substitution analysis of the A1 3′ UTR (nucleotides 1–99) (Fig. 6), is described in detail elsewhere (M Liu, E Lund, and JE Dahlberg, in prep.). Linear T7 templates for in vitro synthesis of chimeric (5′)-hammerhead ribozyme-pre-miR-427 or -pre-miR-427mut were constructed from sets of eight overlapping oligonucleotides (Zhang et al. 2004).
TABLE 2.
Oligonucleotides (all shown 5′–3′)
Primary miR-427 cDNA clones (GenBank accession numbers CA791305, BU911438, BU910064, and BU9067618 corresponding to IMAGE clones 5161839, 6637415, 6635823, and 6631749, respectively) were identified by BLAST search of X. laevis EST sequences using the xla-miR-427 sequence (Watanabe et al. 2005) as query. Plasmid clones pXlA1, pXlB2 (Minshull et al. 1990), pGS17 (Krieg and Melton 1985), and pri-miR-427 IMAGE clones 6637415 and 6631749 were used for generation of hybridization probes.
In vitro RNA synthesis
32P-labeled, m7GpppG- or ApppG-capped reporter RNAs were synthesized in vitro using T7 RNA polymerase (Promega; Stratagene) and α[32P]-UTP (PerkinElmer) as described (Pasquinelli et al. 1995), except that transcripts were not gel-purified prior to injection. Unlabeled hammerhead-ribozyme-pre-miR-427 RNAs were transcribed in the presence of 1 mM NTPs and excess MgCl2 (12 mM) to ensure efficient ribozyme cleavage (Price et al. 1995), and released pre-miR-427 RNA was gel-purified prior to injection.
Embryo injection
X. laevis embryos were obtained according to standard protocols (Keller 1991) and maintained at 21°–25°C in 0.25× Marc's modified Ringer's (MMR) (100 mM NaCl, 2 mM KCl, 1 mM MgSO4, 2 mM CaCl2, 5 mM Hepes at pH 7.4), supplemented with 5% Ficoll for injected embryos and staged according to Nieuwkoop and Faber (1967). 32P-labeled reporter RNAs (∼1–2 fmol in 10–15 nL of H2O), 2′-OMe-containing oligonucleotides (Integrated DNA Technologies) (Table 2) (∼0.5 pmol/embryo), or unlabeled pre-miRNAs (∼25 fmol/embryo) were injected alone or together into one- or two-cell embryos.
RNA analyses
For each time-point, pools of three to five embryos injected with 32P-labeled reporter RNAs and/or unlabeled pre-miRNAs were homogenized in proteinase K solution, and after digestion at 37°C total RNAs were isolated by phenol extraction and ethanol precipitation (Pasquinelli et al. 1995; Audic et al. 2001). For analyses by Northern blot and/or autoradiography, 0.5–2 embryo equivalents of total RNA were resolved by electrophoresis in denaturing 0.5× TEB, 7 M urea polyacrylamide gels (4% or 6% [30:0.8] for reporter RNAs; 20% [19:1] for miRNAs). For analyses of endogenous mRNAs or pri-miR-427 transcripts, pools of 10 embryos were homogenized in 1 mL of Trizol (Invitrogen), and total RNAs were isolated according to the manufacturer's instructions. For analysis of poly(A) tail length (Sheets et al. 1994), 1–2 embryo equivalents of total RNA (in 20 μL) were incubated with 5 μM DNA oligonucleotides complementary to sequences ∼500 nt from the 3′ ends of cyclin A1 or B2 mRNAs (Table 2), ±100 μM oligo-dT15, and treated with RNase H (gift of Dr. JM Keck, University of Wisconsin–Madison). For Northern blots, 0.5–2 embryo equivalents of untreated or RNase H-treated total RNAs were denatured with glyoxal and resolved by electrophoresis in 1× BPTE and 1.2% or 2% agarose gels, respectively (Sambrook and Russell 2001).
Northern blots
Small or large RNAs were transferred to Zeta Probe (Bio-Rad) nylon membranes by electrophoresis or capillary action, respectively, and the membrane-bound, UV-fixed RNAs were hybridized in buffer H (0.25–0.3 M Na2HPO4, 7% SDS, 1 mM EDTA, and 1% RNase-free BSA) at 42°C (miRNA) or 65°C (mRNA/pri-miRNAs). For detection of miR-427, a uniformly α[32P]-UTP-labeled antisense RNA probe (Table 2) was generated according to the mirVana Probe Construction protocol (Ambion). For detection of cyclin A1 or B2 and GS17 mRNAs and pri-miR-427 transcripts, antisense single-stranded DNA probes were generated by linear amplification (40 cycles of 1–2 min extension at 72°C), using 0.1 μg of linearized plasmid DNA, 0.4 μM primer (Table 2), 50 μCi of [32P]-dCTP (3000 Ci/mmol; Amersham Biosciences) and 2.5 U of MasterAmp Tth DNA polymerase (Epicentre Biotechnologies) in a 25 μL reaction containing 20 μM dCTP and 200 μM dATP, dGTP, and dTTP.
Southern blots
Genomic DNA was isolated from X. laevis testis using MasterPure DNA Purification (Epicentre Technologies), and Southern blotting of restricted DNAs was done according to standard protocols (Sambrook and Russell 2001). miR-427 coding regions were detected by hybridization in buffer H at 65°C with uniformly labeled single-stranded RNA probes, generated by in vitro transcription of Stu1-cut IMAGE clone 6631749 DNA with SP6 (or T7) RNA polymerase, using a 10-μL reaction containing 0.1 mM UTP and 20 μCi [32P]-UTP (800Ci/mmol; PerkinElmer).
For determination of miR-427 gene copy numbers, the hybridization signals of the 1.2-kb miR-427 repeat DNA in 3.2 μg of EcoR1- (or Stu1-) digested X. laevis genomic DNA were compared with that of 30 pg of recombinant 1.2-kb fragment (Fig. 1C) by quantitative phosphorimager analyses. From these, the 1.2-kb repeat DNA in 3.2 × 106 pg of genomic DNA was estimated to comprise 80 pg, or ∼0.0025%. This fraction corresponds to ∼80 kb per X. laevis haploid genome (i.e., 0.25 × 10−4 × 3.1 × 109 bp), or about 65 copies of the 1.2-kb repeat DNA. Given that each 1.2-kb repeat encodes four copies of miR-427 sequences (Fig. 1D), we estimate that the tetraploid X. laevis genome contains ∼1000 miR-427 coding sequences.
Gel-shift analyses
GST-fused Xenopus Pumilio protein (xPum amino acids 824–1185) was expressed and purified as described (Rouhana and Wickens 2007). Uniformly α[32P]-UTP-labeled wt or mut PBE RNAs were made according to the miRVana protocol (Ambion) (Table 2) and gel-purified prior to use. For EMSA analyses, 0.1 pmol of labeled RNA (plus 10 μg of tRNA) and various amounts of xPum were incubated in 20 μL of binding buffer (10 mM Hepes at pH 8.0, 1 mM EDTA, 50 mM KCl, 2 mM DTT, 0.02% Tween-20, and 0.1 mg/mL BSA) for 30 min at room temperature prior to loading on a 0.5× TBE, 6% (29:1) native polyacrylamide gel.
ACKNOWLEDGMENTS
This work was supported by NIH grants GM-30220 (to J.E.D.) and HD-43996 (to M.D.S.), and American Cancer Society grant RSG-01-054-CCG (to R.S.H.). We thank Suzanne Blaser Imboden for excellent technical assistance.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1882009.
REFERENCES
- Audic Y, Anderson C, Bhatty R, Hartley RS. Zygotic regulation of maternal cyclin A1 and B2 mRNAs. Mol Cell Biol. 2001;21:1662–1671. doi: 10.1128/MCB.21.5.1662-1671.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic Y, Garbrecht M, Fritz B, Sheets MD, Hartley RS. Zygotic control of maternal cyclin A1 translation and mRNA stability. Dev Dyn. 2002;225:511–521. doi: 10.1002/dvdy.10191. [DOI] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Ce1l. 2004;19:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Bartel D. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Bortolin-Cavaillé ML, Dance M, Weber M, Cavaillé J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009;37:3464–3473. doi: 10.1093/nar/gkp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- Cao H, Yang CS, Rana TM. Evolutionary emergence of microRNAs in human embryonic stem cells. PLoS One. 2008;3:e2820. doi: 10.1371/journal.pone.0002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-YA, Zheng D, Xia Z, Shyu A-B. Ago–TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1709. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, Gaidatzis D, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes & Dev. 2005;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Davidson EH. Gene activity in early development. Academic Press; Orlando, FL: 1986. [Google Scholar]
- Dehlin E, Wormington M, Körner CG, Wahle E. Cap-dependent deadenylation of mRNA. EMBO J. 2000;19:1079–1086. doi: 10.1093/emboj/19.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3′ UTR. RNA. 2008;14:1297–1317. doi: 10.1261/rna.1082708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes & Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009a;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Tritschler F, Izaurralde E. The GW182 protein family in animal cells: New insights into domains required for miRNA-mediated gene silencing. RNA. 2009b;15:1433–1442. doi: 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjentsik Z, Sindelka R, Jonak J. Expression patterns of Src-family tyrosine kinases during Xenopus laevis development. Int J Dev Biol. 2009;53:163–168. doi: 10.1387/ijdb.072311zf. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Guo X, Gourronc F, Lyons-Levy G, Mitchell TA, Hartley RS. ElrA and AUF1 differentially bind the cyclin B2 mRNA. Biochem Biophys Res Commun. 2008;377:653–665. doi: 10.1016/j.bbrc.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- Hartley RS, Rempel RE, Maller JL. In vivo regulation of the early embryonic cell cycle in Xenopus. Dev Biol. 1996;173:408–419. doi: 10.1006/dbio.1996.0036. [DOI] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C. β-catenin signaling activity dissected in the early Xenopus embryo: A novel antisense approach. Dev Biol. 2000;222:124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific microRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Houston DW, Zhang J, Maines JZ, Wasserman SA, King ML. A Xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development. 1998;125:171–180. doi: 10.1242/dev.125.2.171. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappas NC, Savage P, Chen KC, Walls AT, Sible JC. Dissection of the XChk1 signaling pathway in Xenopus laevis embryos. Mol Biol Cell. 2000;11:3101–3108. doi: 10.1091/mbc.11.9.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R. Early embryonic development of Xenopus laevis. Methods Cell Biol. 1991;36:61–113. doi: 10.1016/s0091-679x(08)60273-3. [DOI] [PubMed] [Google Scholar]
- Krieg PA, Melton DA. Developmental regulation of a gastrula-specific gene injected into fertilized Xenopus eggs. EMBO J. 1985;4:3463–3471. doi: 10.1002/j.1460-2075.1985.tb04105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci. 2004;101:4407–4412. doi: 10.1073/pnas.0400779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb Symp Quant Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- MacArthur H, Houston DW, Bubunenko M, Mosquera L, King ML. DEADSouth is a germ plasm specific DEAD-box RNA helicase in Xenopus related to eIF4A. Mech Dev. 2000;95:291–295. doi: 10.1016/s0925-4773(00)00357-9. [DOI] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;31:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J, Blow JJ, Hunt T. Translation of cyclin mRNA is necessary for extracts of activated Xenopus eggs to enter mitosis. Cell. 1989;56:947–956. doi: 10.1016/0092-8674(89)90628-4. [DOI] [PubMed] [Google Scholar]
- Minshull J, Golsteyn R, Hill CS, Hunt T. The A- and B-type cyclin associated cdc2 kinases in Xenopus turn on and off at different times in the cell cycle. EMBO J. 1990;9:2865–2875. doi: 10.1002/j.1460-2075.1990.tb07476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. North-Holland Publishing; Amsterdam, The Netherlands: 1967. Normal table of Xenopus laevis (Daudin) [Google Scholar]
- Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Nolde MJ, Saka N, Reinert KL, Slack FJ. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′ UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev Biol. 2007;305:551–563. doi: 10.1016/j.ydbio.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris J, Philippe M. Poly(A) metabolism and polysomal recruitment of maternal mRNAs during early Xenopus development. Dev Biol. 1990;140:221–224. doi: 10.1016/0012-1606(90)90070-y. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Dahlberg JE, Lund E. Reverse 5′ caps in RNAs made in vitro by phage RNA polymerases. RNA. 1995;1:957–967. [PMC free article] [PubMed] [Google Scholar]
- Piqué M, López JM, Foissac S, Guigó R, Méndez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132:434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- Price SR, Ito N, Oubridge C, Avis JM, Nagai K. Crystallization of RNA-protein complexes. I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J Mol Biol. 1995;249:398–408. doi: 10.1006/jmbi.1995.0305. [DOI] [PubMed] [Google Scholar]
- Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Rouhana L, Wickens M. Autoregulation of GLD-2 cytoplasmic poly(A) polymerase. RNA. 2007;13:188–199. doi: 10.1261/rna.333507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Sheets MD, Fox CA, Hunt T, Vande Woude G, Wickens M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes & Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Tang GQ, Maxwell ES. Xenopus microRNA genes are predominantly located within introns and are differentially expressed in adult frog tissues via post-transcriptional regulation. Genome Res. 2008;18:104–112. doi: 10.1101/gr.6539108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Nakayama T, Dixon MP, Christian JL. Post-transcriptional regulation of Xwnt-8 expression is required for normal myogenesis during vertebrate embryonic development. Development. 1999;126:3371–3380. doi: 10.1242/dev.126.15.3371. [DOI] [PubMed] [Google Scholar]
- Uto K, Nakajo N, Sagata N. Two structural variants of Nek2 kinase, termed Nek2A and Nek2B, are differentially expressed in Xenopus tissues and development. Dev Biol. 1999;208:456–464. doi: 10.1006/dbio.1999.9231. [DOI] [PubMed] [Google Scholar]
- Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA:target interaction. Chem Biol. 2004;11:1619–1623. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Mise K, Okuno T, Suzuki T, Minami N, Imai H. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005;579:318–324. doi: 10.1016/j.febslet.2004.11.067. [DOI] [PubMed] [Google Scholar]
- Weigel D, Izaurralde E. A tiny helper lightens the maternal load. Cell. 2006;124:1117–1118. doi: 10.1016/j.cell.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′ UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. Importance of translation and nonnucleolytic ago proteins for on-target RNA interference. Curr Biol. 2008;18:1327–1332. doi: 10.1016/j.cub.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Nilsson P, Henriksson N, Niedzwiecka A, Lim MK, Cheng Z, Kokkoris K, Virtanen A, Song H. Structural basis of m7GpppG binding to poly(A)-specific ribonuclease. Structure. 2009;17:276–286. doi: 10.1016/j.str.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Yang J, Tan C, Darken RS, Wilson PA, Klein PS. β-catenin/Tcf-regulated transcription prior to the midblastula transition. Development. 2002;129:5743–5752. doi: 10.1242/dev.00150. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]