Abstract
Thrombin is a multifunctional protease that plays a key role in hemostasis, thrombosis, and inflammation. Most thrombin inhibitors currently used as antithrombotic agents target thrombin's active site and inhibit all of its myriad of activities. Exosites 1 and 2 are distinct regions on the surface of thrombin that provide specificity to its proteolytic activity by mediating binding to substrates, receptors, and cofactors. Exosite 1 mediates binding and cleavage of fibrinogen, proteolytically activated receptors, and some coagulation factors, while exosite 2 mediates binding to heparin and to platelet receptor GPIb-IX-V. The crystal structures of two nucleic acid ligands bound to thrombin have been solved. Previously Padmanabhan and colleagues solved the structure of a DNA aptamer bound to exosite 1 and we reported the structure of an RNA aptamer bound to exosite 2 on thrombin. Based upon these structural studies we speculated that the two aptamers would not compete for binding to thrombin. We observe that simultaneously blocking both exosites with the aptamers leads to synergistic inhibition of thrombin-dependent platelet activation and procoagulant activity. This combination of exosite 1 and exosite 2 inhibitors may provide a particularly effective antithrombotic approach.
Keywords: thrombin, aptamer, exosite 1, exosite 2, coagulation, platelet activation
INTRODUCTION
Thrombin is a serine protease involved in platelet activation and aggregation, endothelial cell activation, and coagulation (Coughlin 2000; Celikel et al. 2003). Prothrombin (FII) is converted to thrombin (FIIa) upon activation of coagulation via activation of tissue factor and Factor VIIa complex (Coughlin 2000). Thrombin, in turn, converts fibrinogen into fibrin, the matrix of a blood clot. Moreover, it triggers platelet activation, releasing activators such as ADP, serotonin, thromboxane A2, and expressing P-selectin and CD40 ligand on the platelet surfaces (Coughlin 2000). Thrombin's procoagulant activity on the platelet surface also acts through several positive feedback loops to generate additional thrombin. The pathological manifestation of excess or inappropriately localized thrombin generation is thrombosis. Clinical manifestations include myocardial infarction, deep vein thrombosis, stroke, and peripheral vascular disease, which are leading causes of morbidity and mortality in the United States today (Cavendish et al. 2004).
Thrombin has three main binding sites: the catalytic or active site, where its primary plasma inhibitor, antithrombin (AT), binds and inhibits its coagulation activity; and two anion-binding sites, exosite 1 and exosite 2. Exosite 1 binds to fibrinogen, thrombomodulin, hirudin, and the amino terminus of heparin cofactor II (HCII) (Becker et al. 1999). Exosite 2 mediates binding of thrombin to heparin and to platelet surface glycoprotein Ib-alpha (GPIba). Ligand binding to either exosite 1 or exosite 2 may also influence reactivity of the active site of thrombin (Fredenburgh et al. 1997). Recently, two groups independently solved the crystal structure of thrombin bound to its platelet receptor GPIbα (Celikel et al. 2003; Dumas et al. 2003). Based on their structure work, Celikel et al. (2003) postulated that exosite 2 binding of thrombin was essential in order to potentate exosite 1-dependent modulation of fibrinogen clotting.
Aptamers are single-stranded nucleic acid molecules that can directly inhibit protein function by binding to their targets with high affinity and specificity (Nimjee et al. 2005). A number of aptamers have been isolated that bind to thrombin and a G-quartet containing DNA aptamer, termed ARC 183, that binds to thrombin on exosite 1 (Holland et al. 2000) has shown its potential as an anticoagulant in CABG surgery (Bock et al. 1992; Griffin et al. 1993; DeAnda et al. 1994). TOG25 is 2′-fluoropyrimidine-modified RNA aptamer that binds to exosite 2 (Jeter et al. 2004) and has a nominal effect on clotting times but appears to have a considerable effect on thrombin-mediated platelet activation (White et al. 2001).
Given the different roles of exosites 1 and 2 binding sites on thrombin during the coagulation process and platelet activation, we hypothesize that these aptamers might have different clinical benefits. Moreover, we theorize that utilizing both aptamers as a “drug cocktail” may provide advantages over using each one alone and elicit the desired effects of AT therapy, namely, anticoagulation and anti-platelet activity.
RESULTS
DNA and RNA aptamers bind to thrombin on different exosites
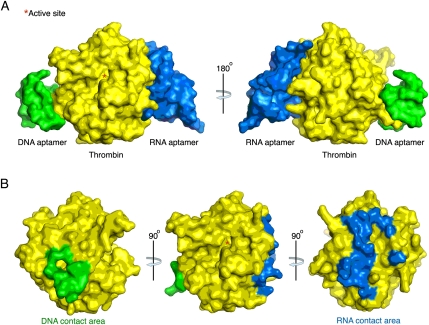
The crystal structure of a DNA aptamer bound to thrombin was published in 1993 and 1996, and the structure provided the first example of how a DNA aptamer could interact with a protein that does not naturally recognize a nucleic acid (Bock et al. 1992; Padmanabhan et al. 1993; Padmanabhan and Tulinsky 1996) Moreover, mutagenesis studies on thrombin confirmed that the DNA aptamer binds exosite 1 (Padmanabhan et al. 1993). Similar thrombin mutagenesis studies indicated that the RNA aptamer TOG25 binds to thrombin near exosite 2 (Jeter et al. 2004). Moreover, we recently solved the crystal structure of this aptamer bound to thrombin, and it demonstrated that the RNA aptamer does directly contact the exosite 2 portion of thrombin (Long et al. 2008). Using the coordinates from this study and from the literature, we developed a proposed structure of both aptamers bound to thrombin. It supports the assertions made by others about the landscape of exosites 1 and 2 and the potential for ligands that bind to either epitope to do so without interfering with one another (Fig. 1).
FIGURE 1.
Space-filling structure of the DNA and RNA aptamers bound to thrombin. The structure illustrates the mutually exclusive binding of the DNA aptamer (ARC183) for exosite 1 and RNA aptamer (TOG25) for exosite 2. The active site represented by (*) sits between the exosites.
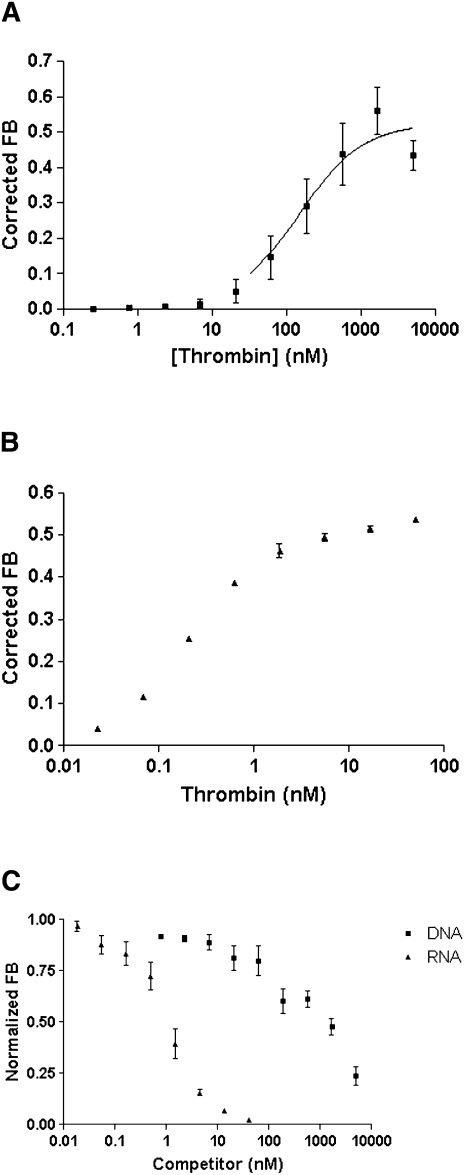
Binding experiments carried out using the double-filter nitrocellulose method showed that the DNA aptamer ARC183, which has a Kd of 113 ± 20 nM (Fig. 2A), exhibited a much lower affinity for thrombin than the RNA aptamer TOG25 with dissociation constant (Kd) of 540 ρM (Fig. 2B; White et al. 2001). The competition assay between the DNA and RNA aptamer revealed there was no competition between the two aptamers for binding to thrombin (Fig. 2C).
FIGURE 2.
Binding affinity (Kd) and competition between aptamers for thrombin. (A) The DNA aptamer binds to thrombin with an affinity (Kd) of 112 ± 7 nM. (B) The RNA aptamer binds to thrombin with an affinity (Kd) of 562 ± 7 ρM. (C) Competition data reveals that no binding inhibition occurs between the DNA and RNA aptamer.
DNA aptamer and RNA aptamer function differently in APTT, PT, and TCT: when used together, these aptamers have a synergistic effect in these clotting assays
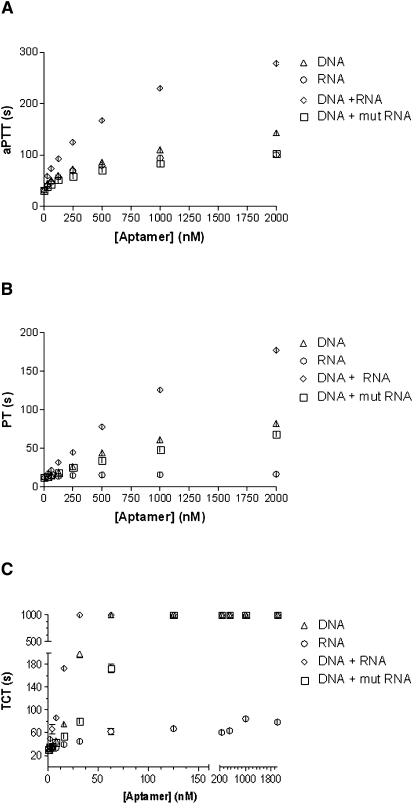
Having verified the binding of DNA and RNA aptamers to human α-thrombin, we next sought to evaluate the effect of each aptamer in three thrombin-sensitive clotting assays, activated partial thromboplastin time (aPTT), prothrombin time (PT), and thrombin clot time (TCT). For all three experiments, one or both drugs were added to platelet poor plasma at concentrations ranging from 31.25 to 2000 nM (concentrations indicate the sum of equal amounts of the two aptamers; for example, 2000 nM corresponds to 1000 nM of ARC183 + 1000 nM of TOG25). In experiments where only one aptamer was tested, a nonsense oligonucleotide was also added to serve as a negative control. A significant difference in clotting times as measured in aPTT assays was observed between DNA, RNA, and DNA + RNA aptamers together (P < 0.0001) (Fig. 3A). At a concentration of 2000 nM, the DNA aptamer increased the aPTT from a baseline of 32.3 ± 0.1 sec to 143.5 ± 4.5 sec, which decreased in a dose-dependent manner to 44.8 ± 0.3 sec at an aptamer concentration of 31.25 nM. At a concentration of 2000 nM, the RNA aptamer increased the aPTT to 100.7 ± 0.8 sec, and like the DNA aptamer, decreased in a dose-dependent manner to 41 sec.
FIGURE 3.
Clotting activity with the DNA + RNA aptamers better than DNA or RNA alone. (A) Activated partial thromboplastin time (aPTT). (B) Prothrombin time (PT) and (C) thrombin clot time (TCT). Triangles = DNA aptamer; circles = RNA aptamer; diamonds = DNA + RNA; squares = DNA + mutant RNA aptamer. Statistically significant difference between groups in all assays except DNA versus DNA + mutant RNA (P < 0.0001).
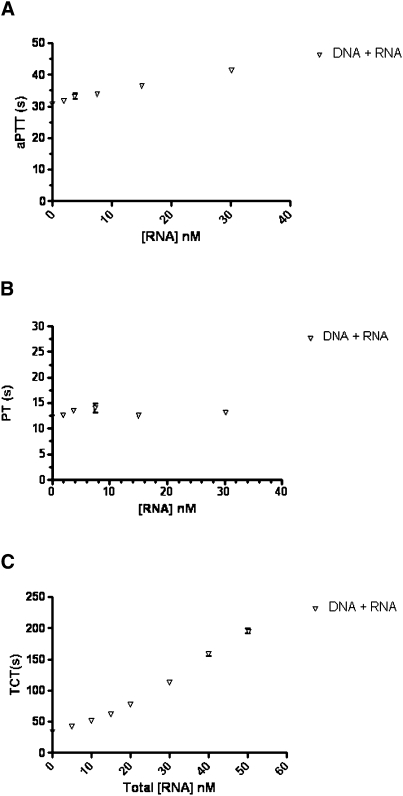
When both aptamers were used together, each at a concentration of 1000 nM for a total aptamer concentration of 2000 nM, the aPTT increased to 278 ± 0.8 sec, which was greater than the aPTT of each aptamer individually or the DNA aptamer in conjunction with a mutant version of the TOG25 RNA aptamer (White et al. 2001) that contains a single nucleotide substitution (Fig. 3A). This apparent synergistic effect of both aptamers was also seen at total concentration of 1000 nM. At 500 nM, however, there was no significant difference between the values of the sum of the aPTT of the DNA and RNA and both compounds tested together (166.4 ± 2.6 sec versus 167.5 ± 0.4 sec, P = 0.72). At doses below 500 nM, the effect of both the DNA and RNA ligands in the aPTT remains greater than each aptamer alone, but the synergistic effect is no longer observed (Fig. 3A). At concentrations below 32 nM the aptamers have minimal effect (Fig. 4A).
FIGURE 4.
Clotting activity with the DNA + RNA aptamers at concentrations <40 nM. (A) Activated partial thromboplastin time (aPTT). (B) Prothrombin time (PT) and (C) thrombin clot time (TCT).
The results from the PT share similar trends with the aPTT; however, the results are even more pronounced. There was a statistically significant difference in the PT between the DNA (or DNA + mutRNA), RNA, and DNA + RNA aptamer in the assay (P < 0.0001) (Fig. 3B). At a concentration of 2000 nM, the DNA aptamer increased the PT from a baseline of 13.2 ± 0.1 sec to 82.0 ± 0.8 sec. This decreased in a dose-dependent manner to 14.1 ± 0.2 sec, which was statistically insignificant from baseline (P = 0.06). At a concentration of 2000 nM, the RNA aptamer nominally increased the PT to 16.4 sec and at a concentration of 62.5 nM, had essentially returned to baseline (13.4 sec, P = 0.18 compared with baseline). However, when both aptamers were tested together at 2000 nM total concentration, the PT was 177.6 ± 0.4 sec. At concentrations below 31.25 nM, the PT for the DNA + RNA did not significantly change (Fig. 4B).
The TCT is a specific assay that measures the conversion of fibrinogen to fibrin in the presence of thrombin and is therefore sensitive to inhibitors that interfere with the catalytic activity of thrombin. A statistically significant difference was observed between the DNA (or DNA + mutRNA), RNA, and DNA + RNA aptamer on the TCT (P < 0.0001); however, the picture was quite different from that observed in the other two thrombin-sensitive clotting assays. The effect of the DNA aptamer on TCT was quite pronounced, and at concentrations of aptamer above 62.5 nM, the clotting time exceeded the upper limit of the assay (>999 sec) (Fig. 3C). The RNA aptamer, on the other hand, did not have a potent effect on TCT, with a clot time of 79.1 ± 0.7 sec at a concentration of 2000 nM and decreased in a dose-dependent manner to baseline at a concentration of 7.8 nM (Fig. 3C). The effect of DNA + RNA aptamers on the TCT was similar to that seen in the DNA group; however, it was interesting to observe that the effect of the DNA was enhanced by the addition of the RNA aptamer, whereas the upper limit of the assay was exceeded only when >62.5 nM of the DNA aptamer was added, 31.25 nM of the DNA + RNA aptamer was able to achieve this level of anticoagulation. Moreover, the addition of both nucleic acids, even at the low concentration of 1.95 nM, resulted in TCT times that are significantly above baseline (49.7 ± 0.4 sec, P < 0.005 versus baseline) even though this concentration is well below the Kd of the DNA aptamer for thrombin (Fig. 4C).
DNA aptamer and RNA aptamer function differently in the CD62P platelet activation assay: when used together, these aptamers have a pronounced synergistic effect in this assay
As previously mentioned, thrombin is a potent activator of platelets and upon binding to the protease-activated receptor (PAR) on the surface of the platelet, induces the up-regulation of p-selectin (Coughlin 2000). In order to measure the effect of the DNA and RNA aptamers on platelet activity, we utilized a flow cytometry assay with a P-selectin antibody (CD62P) to assess platelet activation.
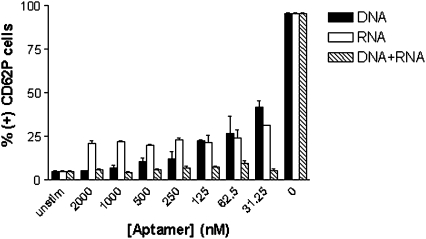
In experiments in which platelets were activated by the direct addition of exogenous thrombin, the DNA aptamer, RNA aptamer, and DNA + RNA aptamers together all had a significant effect on down-modulating platelet activation, although statistically significant differences existed between all three groups (P < 0.0001) (Fig. 5). At the highest concentration, the DNA aptamer inhibited platelet activation to that equal to unstimulated platelets (P = 0.65), and this effect responded in a dose-dependent manner to dilutions of DNA aptamer to 41.7 ± 3.7% (+) CD62P cells at a concentration of 31.25 nM DNA aptamer. The RNA aptamer inhibited platelet activation with much lower variation than the DNA aptamer, ranging from 21.2 ± 1.6% to 31.5 ± 0.5% at the highest and lowest concentration tested. As with the clotting assays, platelet activation was most potently inhibited by DNA + RNA, remaining close to baseline at all concentrations tested. This was particularly interesting given that in the clotting assays, this was not the case, and indicates the role of exosite 1 and 2 inhibition on effective control of platelet activation for potential clinical application.
FIGURE 5.
Platelet activation by direct addition of thrombin is suppressed most effectively with DNA + RNA. Solid bar = DNA aptamer; open bar = RNA aptamer; hatched bar = DNA + RNA. Statistically significant differences between all three groups (P < 0.0001).
DISCUSSION
We have described the inhibitory effects on several aspects of the coagulation process of blocking thrombin's exosites 1 and 2 using aptamers. The structural data illustrate that these nucleic acid ligands used in this experiment bind to mutually exclusive sites and do not overlap. Furthermore, using both DNA and RNA aptamers to inhibit thrombin's activity, we showed a synergistic effect in the aPTT, PT, and TCT plasma clotting assays. Finally, we were able to also show that this synergistic inhibition of thombin also inhibits platelet activity, as demonstrated in a platelet activation assay.
In the clotting assays, the endpoint is the formation of a fibrin clot after thrombin cleaves the fibrinogen. The explanation for the synergistic effects of the two aptamers may have to do with the role that exosite 2 plays in fibrinogen cleavage. While the majority of thrombin binding to fibrinogen is mediated by exosite 1, Pospisil et al. (2003) found that binding of thrombin to a variant of the gamma chain of fibrinogen, gamma′, is mediated by exosite 2. Fibrinogen bearing the gamma′ chain represents up to about 15% of circulating fibrinogen (Lovely et al. 2002). Thus, the aptamer that binds exosite 2 might reduce thrombin binding to a subpopulation of fibrinogen molecules to cause a modest increase in clotting times and exert a synergistic effect with the aptamer that binds exosite 1.
The two aptamers also exert a synergistic inhibitory effect on platelet activation. Our current understanding of thrombin-mediated platelet activation is that its initial high-affinity binding to platelet surface GPIb-alpha is via exosite 2, which facilitates subsequent binding of a second thrombin molecule via exosite 1 (Li et al. 2001; Celikel et al. 2003; Dumas et al. 2003). The interaction with GPIb-alpha also promotes thrombin binding to its proteolytically activated receptor-1 (PAR-1) via exosite 1, which leads to receptor cleavage (De Candia et al. 2001; De Cristofaro et al. 2001). Both GPIb-alpha binding and PAR-1 cleavage play roles in thrombin-mediated platelet activation (Coughlin 2000; Dicker et al. 2001). We found that blocking exosite 1 with the DNA aptamer was more effective in inhibiting platelet activation, but that the RNA aptamer could also inhibit platelet activation to a lesser extent. Thus, our findings are consistent with the literature in indicating that interactions mediated both by exosite 1 and exosite 2 are involved in platelet activation.
Recently, Kim and Tan (Kim et al. 2008) presented two bivalent activity of two aptamers to thrombin. They tested the DNA aptamer used in this study (Bock et al. 1992), and a DNA aptamer that binds to exosite 2 in the same fashion as Tog25 (Kim et al. 2008). In contrast to our data, their results showed that mixing the two aptamers had no additive effect on clotting inhibition. There was a synergistic effect in their study only when they constructed a linker between the aptamers to exosites 1 and 2 (Kim et al. 2008). The DNA aptamer for exosite 1 are the same in both cases. The exosite 2 aptamer used in their study was similar in size and binding affinity. The main difference was that they used a DNA ligand while ours is an RNA aptamer. While they provided no structural data to explain their results, it is possible that their exosite 2 aptamer bound in such a way that it caused steric or charge repulsion to the exosite 1 aptamer. Similarly, Muller et al. (2007) has shown that when two DNA aptamers to exosites 1 and 2 on thrombin are linked with an appropriate sized spacer (15 dA oligonucleotides), improved binding and additive anticoagulant effects could be achieved. They did not report clinically used clotting inhibition studies as is presented here. Moreover, although the effects of the linked aptamers were more modest compared with the current study, their results did indicate the importance of spacing when linking two functional aptamer molecules.
In testing the activity of the DNA aptamer in the aPTT, PT, and TCT clotting assays, in the presence of mutant RNA aptamer for thrombin, we observed no synergistic activity of the aptamers. On the contrary, the clotting times were mildly decreased. Possible explanation of this includes thrombin protein integrity or reagent activity. Alternatively, it is possible that the mutant RNA aptamer for thrombin interacted with the protein target or in some other way, obstructed the DNA aptamer from binding to exosite 1, limiting its inhibitory activity.
There are numerous thrombin inhibitors clinically available today. Heparin, the most common anticoagulant, functions by acting as a cofactor to AT and facilitating its binding to thrombin's active site. Other thrombin inhibitors either bind solely to the active site or span the active site and exosite 1 (Hirsh et al. 2005; Linkins and Weitz 2005). Current thrombin inhibitors do not address platelet activation; moreover, development has not exploited exosite 2 inhibition as a more effective means to control thrombosis. The high degree of specificity of aptamers for their targets, and the ability to modify their bioavailability, circulating half-life and clearance makes them attractive compounds to tailor as drug candidates for specific clinical indications (Rusconi et al. 2004). We believe that the approach of combining two specific aptamers with synergistic inhibitory effects may provide a highly effective approach to preventing thrombosis in a variety of clinical settings.
MATERIALS AND METHODS
Synthesis of DNA and RNA aptamers
The DNA aptamer ARC183 (5′-GGTTGGTGTGGTTGG-3′) was synthesized by Trilink Biotechnologies. The 2′-fluoromodified RNA aptamer TOG25 (5′-GGGAACAAAGCUGAAGUACUUACCC-3′) was synthesized by Dharmacon Research, Inc.
Binding studies
Dissociation constants (Kd) of both ARC183 and TOG25 were determined using a double-filter, nitrocellulose binding method (Wong and Lohman 1993). Briefly, DNA and RNA were dephosphorylated using bacterial alkaline phosphatase (Gibco BRL) and end-labeled at the 5′ with T4 polynucleotide kinase (New England Biolabs) and [γ32P]ATP (Amersham Pharmacia Biotech) (Fitzwater and Polisky 1996). Direct binding was performed by incubating 32P-ARC 183 with thrombin (Hematologic Technologies) concentrations ranging from 5 μM to 85 ρM in binding buffer (20 mM HEPES at pH 7.4, 150 mM NaCl, 2 mM CaCl2, 0.01% BSA) at 37°C. The fraction bound of the nucleic acid–protein complex was quantified with a PhosphorImager (Molecular Dynamics). The nonspecific binding of radiolabeled nucleic acid was subtracted (Wong and Lohman 1993) and reported as the mean and standard error of the mean from three experiments.
Clotting assays
Activated partial thromboplastin time assays were carried out using a model ST4 mechanical coagulometer (Diagnostica Stago). Aptamer in Dulbecco's phosphate-buffered saline (DPBS; Invitrogen) (5 μL) or DPBS alone was added to pooled normal human plasma (50 μL) (George King Biomedical), and incubated for 5 min at 37°C. Then MDA Platelin (50 μL) (bioMérieux) was added to activate plasma and incubated for 5 min at 37°C. Addition of 25 mM CaCl2 (50 μL) (bioMérieux) to initiate the clotting reaction followed, and clot formation time was recorded. All reactions were performed in duplicate, and only duplicates differing by <10% were used in the analysis.
Prothrombin time assays were carried out using a model ST4 mechanical coagulometer (Diagnostica Stago). Aptamer in DPBS (Invitrogen) (5 μL) or DPBS alone was added to pooled normal human plasma (50 μL) (George King Biomedical), and incubated for 5 min at 37°C. Then MDA Simplastin L (bioMeriux) was added to initiate the clotting reaction, and clot formation time was recorded. All reactions were performed in duplicate, and only duplicates differing <10% were used in the analysis.
Thrombin clot time assays were carried out using model ST4 mechanical coagulometer (Diagnostica Stago). Aptamer in DPBS (Invitrogen) (5 μL) or DPBS alone was added to pooled normal human plasma (50 μL) (George King Biomedical), and incubated for 5 min at 37°C. Clotting was initiated using 50 μL of TCT solution (50 U/mL thombin in PBS) and clot formation time was recorded. All reactions were performed in duplicate, and only duplicates differing <10% were used in the analysis.
Platelet activation assay
Platelets were purified from freshly drawn blood from healthy volunteers (Hoffman et al. 1992). Briefly, platelets were isolated by density gradient centrifugation, then separated from plasma proteins by gel-filtration over a 50 mL column of Sepharose Cl-2B in Tyrodes buffer (15 mM HEPES at pH 7.4, 3.3 mM Na2PO4, 138 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 5.5 mM dextrose) with 1 mg/mL bovine serum albumin. In each experiment, 25 μL of platelets were incubated with 25 μL of aptamer in PBS or PBS alone and 50 μL of thrombin to a final concentration of 1 nM. Platelet activation was measured by platelets with α-granule protein phycoerythrin antibody CD62P (Becton Dickenson) and analyzed by flow cytometry (FACScan, Becton Dickenson) (Stenberg et al. 1985). Platelet activation was expressed as the mean and standard error of the mean of percent positive cells from two experiments.
Statistical analysis
We expressed most data as the mean ± standard error. We used two-way ANOVA to compare aPTT, PT, TCT, and platelet activation between ARC 183 and TOG25 and each compound compared with administering them together in each assay at various doses. We compared individual points using a two-tailed Student t test. We considered differences statistically significant when the P value was 0.05 or less.
ACKNOWLEDGMENTS
We would like to thank Dr. Dougald (Mac) Monroe and Dr. Frank Church for helpful discussions. This work was supported in part by the National Institute of Health to B.A.S. and by the US Department of Veterans Affairs (to Z.V. and M.H.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1240109.
REFERENCES
- Becker DL, Fredenburgh JC, Stafford AR, Weitz JI. Exosites 1 and 2 are essential for protection of fibrin-bound thrombin from heparin-catalyzed inhibition by antithrombin and heparin cofactor II. J Biol Chem. 1999;274:6226–6233. doi: 10.1074/jbc.274.10.6226. [DOI] [PubMed] [Google Scholar]
- Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- Cavendish JJ, Fugit RV, Safani M. Role of antiplatelet therapy in cardiovascular disease I: Acute coronary syndromes. Curr Med Res Opin. 2004;20:1839–1843. doi: 10.1185/030079904X10665. [DOI] [PubMed] [Google Scholar]
- Celikel R, McClintock RA, Roberts JR, Mendolicchio GL, Ware J, Varughese KI, Ruggeri ZM. Modulation of α-thrombin function by distinct interactions with platelet glycoprotein Ibalpha. Science. 2003;301:218–221. doi: 10.1126/science.1084183. [DOI] [PubMed] [Google Scholar]
- Coughlin SR. Thrombin signaling and protease-activated receptors. Nature. 2000;407:258–264. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- De Candia E, Hall SW, Rutella S, Landolfi R, Andrews RK, De Cristofaro R. Binding of thrombin to glycoprotein Ib accelerates the hydrolysis of Par-1 on intact platelets. J Biol Chem. 2001;276:4692–4698. doi: 10.1074/jbc.M008160200. [DOI] [PubMed] [Google Scholar]
- De Cristofaro R, De Candia E, Landolfi R, Rutella S, Hall SW. Structural and functional mapping of the thrombin domain involved in the binding to the platelet glycoprotein Ib. Biochemistry. 2001;40:13268–13273. doi: 10.1021/bi010491f. [DOI] [PubMed] [Google Scholar]
- DeAnda A, Jr, Coutre SE, Moon MR, Vial CM, Griffin LC, Law VS, Komeda M, Leung LL, Miller DC. Pilot study of the efficacy of a thrombin inhibitor for use during cardiopulmonary bypass. Ann Thorac Surg. 1994;58:344–350. doi: 10.1016/0003-4975(94)92206-3. [DOI] [PubMed] [Google Scholar]
- Dicker IB, Pedicord DL, Seiffert DA, Jamieson GA, Greco NJ. Both the high affinity thrombin receptor (GPIb-IX-V) and GPIIb/IIIa are implicated in expression of thrombin-induced platelet procoagulant activity. Thromb Haemost. 2001;86:1065–1069. [PubMed] [Google Scholar]
- Dumas JJ, Kumar R, Seehra J, Somers WS, Mosyak L. Crystal structure of the GpIbalpha-thrombin complex essential for platelet aggregation. Science. 2003;301:222–226. doi: 10.1126/science.1083917. [DOI] [PubMed] [Google Scholar]
- Fitzwater T, Polisky B. A SELEX primer. Methods Enzymol. 1996;267:275–301. doi: 10.1016/s0076-6879(96)67019-0. [DOI] [PubMed] [Google Scholar]
- Fredenburgh JC, Stafford AR, Weitz JI. Evidence for allosteric linkage between exosites 1 and 2 of thrombin. J Biol Chem. 1997;272:25493–25499. doi: 10.1074/jbc.272.41.25493. [DOI] [PubMed] [Google Scholar]
- Griffin LC, Tidmarsh GF, Bock LC, Toole JJ, Leung LL. In vivo anticoagulant properties of a novel nucleotide-based thrombin inhibitor and demonstration of regional anticoagulation in extracorporeal circuits. Blood. 1993;81:3271–3276. [PubMed] [Google Scholar]
- Hirsh J, O'Donnell M, Weitz JI. New anticoagulants. Blood. 2005;105:453–463. doi: 10.1182/blood-2003-12-4195. [DOI] [PubMed] [Google Scholar]
- Hoffman M, Monroe DM, Roberts HR. A rapid method to isolate platelets from human blood by density gradient centrifugation. Am J Clin Pathol. 1992;98:531–533. doi: 10.1093/ajcp/98.5.531. [DOI] [PubMed] [Google Scholar]
- Holland CA, Henry AT, Whinna HC, Church FC. Effect of oligodeoxynucleotide thrombin aptamer on thrombin inhibition by heparin cofactor II and antithrombin. FEBS Lett. 2000;484:87–91. doi: 10.1016/s0014-5793(00)02131-1. [DOI] [PubMed] [Google Scholar]
- Jeter ML, Ly LV, Fortenberry YM, Whinna HC, White RR, Rusconi CP, Sullenger BA, Church FC. RNA aptamer to thrombin binds anion-binding exosite-2 and alters protease inhibition by heparin-binding serpins. FEBS Lett. 2004;568:10–14. doi: 10.1016/j.febslet.2004.04.087. [DOI] [PubMed] [Google Scholar]
- Kim Y, Cao Z, Tan W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc Natl Acad Sci. 2008;105:5664–5669. doi: 10.1073/pnas.0711803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CQ, Vindigni A, Sadler JE, Wardell MR. Platelet glycoprotein Ibα binds to thrombin anion-binding exosite II inducing allosteric changes in the activity of thrombin. J Biol Chem. 2001;276:6161–6168. doi: 10.1074/jbc.M004164200. [DOI] [PubMed] [Google Scholar]
- Linkins LA, Weitz JI. New anticoagulant therapy. Annu Rev Med. 2005;56:63–77. doi: 10.1146/annurev.med.56.082103.104708. [DOI] [PubMed] [Google Scholar]
- Long SB, Long MB, White RR, Sullenger BA. Crystal structure of an RNA aptamer bound to thrombin. RNA. 2008;14:2504–2512. doi: 10.1261/rna.1239308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovely RS, Falls LA, Al-Mondhiry HA, Chambers CE, Sexton GJ, Ni H, Farrell DH. Association of γA/γ′ fibrinogen levels and coronary artery disease. Thromb Haemost. 2002;88:26–31. [PubMed] [Google Scholar]
- Muller J, Wulffen B, Potzsch B, Mayer G. Multidomain targeting generates a high-affinity thrombin-inhibiting bivalent aptamer. ChemBioChem. 2007;8:2223–2226. doi: 10.1002/cbic.200700535. [DOI] [PubMed] [Google Scholar]
- Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: An emerging class of therapeutics. Annu Rev Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- Padmanabhan K, Tulinsky A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr D Biol Crystallogr. 1996;52:272–282. doi: 10.1107/S0907444995013977. [DOI] [PubMed] [Google Scholar]
- Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. The structure of α-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J Biol Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- Pospisil CH, Stafford AR, Fredenburgh JC, Weitz JI. Evidence that both exosites on thrombin participate in its high affinity interaction with fibrin. J Biol Chem. 2003;278:21584–21591. doi: 10.1074/jbc.M300545200. [DOI] [PubMed] [Google Scholar]
- Rusconi CP, Roberts JD, Pitoc GA, Nimjee SM, White RR, Quick G, Jr, Scardino E, Fay WP, Sullenger BA. Antidote-mediated control of an anticoagulant aptamer in vivo. Nat Biotechnol. 2004;22:1423–1428. doi: 10.1038/nbt1023. [DOI] [PubMed] [Google Scholar]
- Stenberg PE, McEver RP, Shuman MA, Jacques YV, Bainton DF. A platelet α-granule membrane protein (GMP-140) is expressed on the plasma membrane after activation. J Cell Biol. 1985;101:880–886. doi: 10.1083/jcb.101.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R, Rusconi C, Scardino E, Wolberg A, Lawson J, Hoffman M, Sullenger B. Generation of species cross-reactive aptamers using ‘toggle’ SELEX. Mol Ther. 2001;4:567–573. doi: 10.1006/mthe.2001.0495. [DOI] [PubMed] [Google Scholar]
- Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: Application to protein-nucleic acid interactions. Proc Natl Acad Sci. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]