Abstract
Transcription of the mitochondrial genome in Dictyostelium discoideum gives rise to eight major polycistronic RNA species that can be detected by Northern hybridization. In order to determine whether these transcripts could possibly derive from processing of even larger transcripts, reverse transcriptase polymerase chain reactions (RT-PCRs) were performed in an attempt to amplify the intervening regions between the eight major transcripts. All but one intervening region were successfully reverse transcribed and amplified, indicating that even larger transcripts existed and that the eight major transcripts detected previously may be the products of transcript processing. Southern hybridization analyses of DNA fragments representing the sequences between the eight major transcripts with in vitro capped mitochondrial RNA identified the 5′ end of only one of the eight major transcripts as a genuine transcription start site. The ability to initiate transcription from DNA sequences upstream of the identified transcription initiation site was demonstrated in bacterial cells expressing the Dictyostelium mitochondrial RNA polymerase. We conclude that transcription of the Dictyostelium mitochondrial genome is initiated at a single site, generating a large polycistronic transcript that is very efficiently, probably cotranscriptionally processed into mature RNA species. This is the first report on a protist mitochondrial DNA that is, although much larger in size than its metazoan counterparts, transcribed from a single transcription initiation site.
Keywords: Dictyostelium discoideum, mitochondrial genome, mitochondrial transcription, transcription initiation, transcript processing
INTRODUCTION
Mitochondrial gene expression is as diverse as the size, structure, gene content, and organization of mitochondrial genomes in the different eukaryotic species. Although almost all organisms employ a surprisingly similar and well-conserved bacteriophage-like RNA polymerase (Cermakian et al. 1997), the transcription process itself may vary with regard to the number and sequence of mitochondrial promoters and the need for specificity factors. In mammalian mitochondria, for example, transcription of the rather small and compact genome (16.6 kb) (Anderson et al. 1981) is initiated at a single promoter on each strand of the mitochondrial DNA (mtDNA). Both promoters are divergently oriented and lie adjacent to each other in the same control region, the displacement loop region. From here, transcription gives rise to large polycistronic transcripts, which are cotranscriptionally cleaved to produce individual RNA molecules (Clayton 1991). Transcription from single uni- or bidirectional promoters seems to be common to the mitochondria of all vertebrata (Bogenhagen and Romanelli 1988; L'Abbe et al. 1991; Jaehning 1993). Less-derived organisms tend to have larger mitochondrial genomes, which, due to their size require multiple, if not many promoters for the expression of the more complex genomes. In fungi, the size of the mitochondrial genomes varies from 19.4 kb in Schizosaccharomyces pombe to more than 80 kb in Saccharomyces cerevisiae (Schäfer et al. 2005). Depending on the genome size, the number of promoters ranges from two to multiple and a simple consensus sequence consisting of a nine-nucleotide sequence ATATAAGTA has been shown to be sufficient for accurate transcription initiation (Schinkel et al. 1986; Costanzo and Fox 1990). Multiple promoters have also been identified in plant mitochondria, which are known to harbor the largest genomes studied so far (Newton et al. 1995; Tracy and Stern 1995; Brennicke et al. 1999). However, only limited similarities between the mitochondrial promoters both within and between species have been observed, and a truly conserved consensus sequence has not emerged (Hanic-Joyce and Gray1991; Binder and Brennicke 1993; Rapp et al. 1993; Binder et al. 1996). From the growing number of available mitochondrial genome sequences and the studies on mitochondrial transcription in the different organisms, it appears that larger mitochondrial genomes require the presence of multiple promoters. In contrast to this, we show here that in the simple protist Dictyostelium discoideum, the relatively large mitochondrial genome of 56 kb in size is transcribed from only a single, unidirectional promoter. We provide evidence for the existence of a large primary transcript in reverse transcriptase polymerase chain reactions (RT-PCRs) and in capping experiments, and we also demonstrate that the sequences upstream of the unique transcription initiation site can be bound by the Dictyostelium mitochondrial RNA polymerase to specifically initiate transcription.
RESULTS
In a previous study, we investigated transcription in the mitochondria of the cellular slime mould Dictyostelium discoideum by Northern hybridization. Using gene-specific probes directed against all genes present in the mitochondrial genome, we detected eight large polycistronic transcripts, some of which were further processed to form smaller mono-, di-, or tricistronic RNA molecules (Barth et al. 1999, 2001). We did not observe any larger transcripts in any of the hybridization studies, and from this we concluded the Dictyostelium mitochondrial genome to be transcribed into eight major transcripts. In an attempt to identify a consensus promoter sequence, the 5′ ends of these transcripts were mapped by primer extension analysis and the sequences upstream of the putative transcription initiation sites were aligned (Barth et al. 2001). This approach, however, may have been limited by the possibility that some of the 5′ ends determined by primer extension analysis represented sites of RNA processing rather than transcription initiation. To determine which of the major transcripts had 5′ ends that were genuine transcription initiation sites and which had been generated by RNA processing, we performed a series of RT-PCRs and 5′ capping experiments.
The previously detected eight major mitochondrial RNA transcripts are processing intermediates rather than primary transcripts
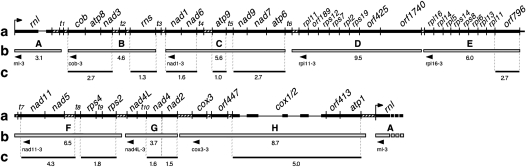
The eight major transcripts detected previously by Northern hybridization (Barth et al. 2001) may be derived from even larger primary transcripts. It is possible that Northern hybridization has not been sensitive enough to detect these primary transcripts if their levels had been too low due to rapid processing. We therefore employed RT-PCR to detect RNA fragments spanning the intervening sequences between the eight major transcripts. The sequences of the 5′ and 3′ primers and their exact position in the mitochondrial genome of Dictyostelium are listed in Table 1. The approximate binding sites for the 3′ primers used for reverse transcription are indicated in Figure 1. Negative controls in which the reverse transcription step has been omitted were conducted to monitor for any residual DNA contamination in the DNase I-treated RNA preparations used for the RT-PCR experiments. PCR amplifications using genomic DNA as template were performed as positive controls to demonstrate that the primers were able to amplify the regions of interest. As shown in Figure 2, RT-PCR products were obtained for all intervening regions, except for the region between transcript H and A upstream of the rnl sequence. The successful amplification of intervening regions in these experiments demonstrated the existence of RNA transcripts in Dictyostelium mitochondria that span the regions between the eight major RNAs observed in Northern blots. This suggests that the large polycistronic RNAs observed previously were themselves derived from processing of one or more even larger primary transcripts. The fact that all but one of the intervening regions were amplified in RT-PCR further suggests that the single nonamplifiable region between transcripts H and A contains the site of transcription initiation for a single, large primary transcript. This primary transcript may have been difficult to detect in the Northern hybridization studies due to rapid cotranscriptional processing of the primary transcript into smaller, mature RNA molecules. We did not find any evidence for a similar large primary transcript derived from the opposite strand, supporting the fact that all genes in the Dictyostelium mitochondrial genome are located on and transcribed from one strand.
TABLE 1.
Sequences of the 5′ and 3′ primers and their exact position in the mitochondrial genome of Dictyostelium
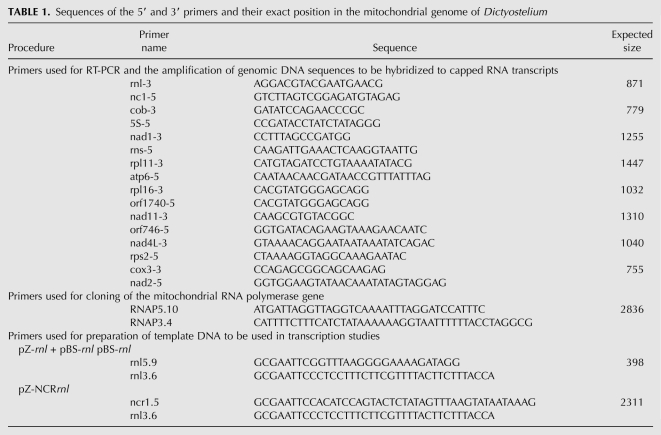
FIGURE 1.
Transcript map of the mitochondrial genome in Dictyostelium. Schematic representation of (a's) the primary transcript indicating the genes and their organization in the circular mitochondrial genome (55.5 kb) (Ogawa et al. 2000). All genes are oriented in the same direction. Intronic sequences are shown as thin lines, intergenic regions are depicted as hatched lines. t1–t10 represent clusters of tRNAs that may serve as processing signals. The large arrow marks the unique transcription initiation site upstream of the rnl gene. (b's) The eight major transcripts A–H detected by Northern hybridization (Barth et al. 2001). Arrowheads indicate the appropriate binding sites for the 3′ primers used for reverse transcription. Some of the transcripts shown in (b) are further processed to yield mono-, di- and tricistronic transcripts (c's). The sizes of the transcripts are not to scale, their actual sizes are indicated by the numbers below the transcripts.
FIGURE 2.
Amplification by RT-PCR of cDNA fragments spanning the intervening regions between transcripts H and A (panel 1), A and B (panel 2), B and C (panel 3), C and D (panel 4), D and E (panel 5), E and F, (panel 6), F and G (panel 7), and G and H (panel 8). Negative controls (reverse transcription step omitted) are shown in lane 1 of each panel, lane 2 of each panel represents the positive controls (PCR of genomic DNA). Lane 3 of each panel shows the RT-PCR results for the intervening regions. The size of DNA standards is indicated in kilobases (kb).
It was possible that one or more of the RT-PCR products obtained above were derived from inefficient termination of transcription of upstream transcripts. To rule this out and to determine how many genuine transcription initiation sites are present in the mitochondrial genome, we performed in vitro capping experiments of the mitochondrial transcripts and used the capped mitochondrial RNA as probes in subsequent Southern hybridization analyses. In vitro capping by guanylyl transferase with [α-32P]GTP allows the specific labeling of only those 5′ termini that originate from the initiation of transcription (Auchincloss and Brown 1989; Binder and Brennicke 1993). It can therefore be used to discriminate primary transcripts from those transcripts that are derived from post-transcriptional processing, since only primary 5′ RNA termini contain the di- or triphosphate required for the addition of the cap.
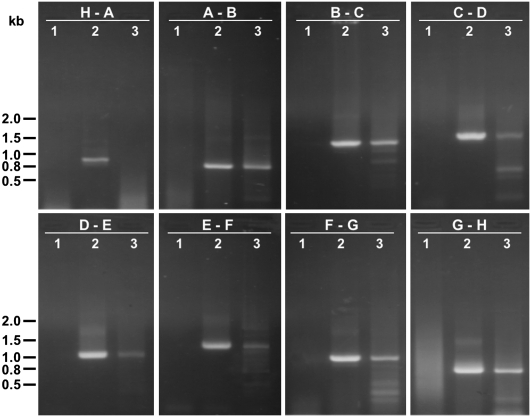
For the Southern hybridization experiments, DNA fragments corresponding to the intervening regions between the eight major transcripts (Fig. 1; Table 1) were amplified by PCR, separated by agarose gel electrophoresis, and transferred onto nylon membrane. The DNA fragments were then allowed to hybridize with the in vitro capped mitochondrial RNA. Only the DNA fragment spanning the region between transcript H and A was detected by the capped mitochondrial RNA (Fig. 3), indicating that only transcript A had been capped by guanylyl transferase. This result clearly identified the 5′ end of transcript A to be a genuine transcription start site. Since none of the other DNA fragments were found to hybridize to capped RNA, the 5′ end of transcript A must represent the only transcription start site; this finding complements the results obtained in the RT-PCR experiments, suggesting that the Dictyostelium mitochondrial genome is transcribed from a single initiation site.
FIGURE 3.
(a) Amplified DNA fragments representing the intervening regions between transcripts F and G (lane 1), G and H (lane 2), H and A (lane 3), A and B (lane 4), B and C (lane 5), C and D (lane 6), D and E (lane 7), and E and F (lane 8). The size of DNA standards (lane M) is indicated in kilobases (kb). (b) Hybridization of capped mitochondrial RNA to the DNA fragments shown in a after being transferred onto nylon membrane.
Transcription can be initiated from the transcription initiation site by a heterologously expressed Dictyostelium mitochondrial RNA polymerase
The exact 5′ end of transcript A, which was shown in the preceding section to represent the only genuine transcription start site, was mapped previously by primer extension analysis to position 18,514 in the Dictyostelium mitochondrial genome (Barth et al. 2001). This places the single transcription initiation site into a noncoding region upstream of the rnl gene encoding the large ribosomal subunit RNA (Fig. 1). The sequences upstream of the transcription initiation site in this noncoding region were therefore of particular interest as they may contain promoter and transcription factor binding sites. To demonstrate that these sequences serve as binding sites for proteins directly involved in the transcription process, we identified and cloned the nuclear gene for the Dictyostelium mitochondrial RNA polymerase (rpmA) based on the widespread sequence similarity among mitochondrial RNA polymerases (Cermakian et al. 1997). The rpmA gene contains a continuous open reading frame of 2850 base pairs (bp) in length and codes for a predicted protein of 950 amino acid residues. All catalytically essential residues, as determined on the basis of the T7 RNA polymerase crystal structure (Sousa et al. 1993), are conserved. As expected for a nuclear-encoded mitochondrial protein, the amino terminus of the Dictyostelium protein is predicted to form an amphipathic helix with the ability to target the protein to the mitochondria (data not shown). Mitochondrial targeting was also suggested by high probability scores obtained using the software programs Predotar (http://www.inra.fr/predotar/; probability score for mitochondrial targeting: 0.985) and MitoProt (probability score: 0.895) (Claros and Vincens 1996).
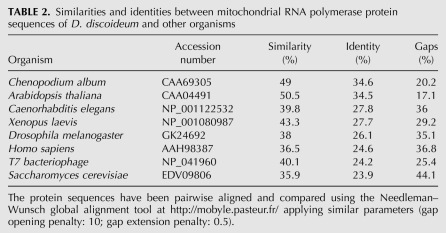
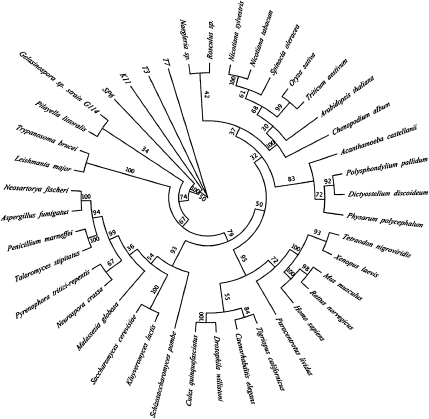
Sequence comparisons of the Dictyostelium protein with mitochondrial RNA polymerase sequences of other organisms demonstrated the high degree of similarity among the different mitochondrial RNA polymerases, with the overall degree of homology decreasing from the C-terminus to the N-terminus (Fig. 4). Pairwise alignments showed that the Dictyostelium protein was most similar to the mitochondrial RNA polymerases from the higher plants Chenopodium album and Arabidopsis thaliana (34.6% and 34.5% identity, 49.0% and 50.5% similarity, respectively), followed by those of Caenorhabditis elegans, Xenopus laevis, Drosophila melanogaster, Homo sapiens, T7 bacteriophage, and various yeasts (Table 2). A phylogenetic tree was constructed to investigate the evolutionary relationships between the different RNA polymerase sequences. Interestingly, the Dictyostelium RNA polymerase clearly associates with higher plant RNA polymerases, well separated from a branch containing the fungal, human, and T7 bacteriophage RNA polymerases (Fig. 5).
FIGURE 4.
Sequence alignment of the Dictyostelium mitochondrial RNA polymerase protein sequence (Dd, accession no. AAK73754) with RNA polymerase sequences of Xenopus laevis (Xl, accession no. NP_001080987), Homo sapiens (Hs, accession no. AAH98387), Drosophila melanogaster (Dm, accession no. GK24692), Chenopodium album (Ca, accession no. CAA69305), Saccharomyces cerevisiae (Sc, accession no. EDV09806), and Caenorhabditis elegans (Ce, accession no. NP_001122532). The unique asparagine- (N) and serine- (S) rich sequences in the Dictyostelium protein not present in other RNA polymerases are underlined. The predicted cleavage site for the N-terminal mitochondrial targeting sequence, as predicted by various online tools, is located just upstream of the N- and S-rich sequences between residues Ser18 (S) and Phe19 (F). Identical residues are indicated by (*), while (:) or (.) indicate either strong or weak similarities, respectively. Per line, 120 residues are shown.
TABLE 2.
Similarities and identities between mitochondrial RNA polymerase protein sequences of D. discoideum and other organisms
FIGURE 5.
Phlogenetic tree demonstrating the evolutionary relationship between mitochondrial RNA polymerases of 41 different organisms. The maximum likelihood phylogeny was generated using the aLRT-PHYML algorithm (Guindon and Gascuel 2003; Anisimova and Gascuel 2006) at LIRMM (Dereeper et al. 2008). The tree is based on multiple protein sequence alignments generated using MUSCLE (Edgar 2004). Statistical support for branches was assessed using the bootstrapping method (100 bootstrap replicates).
The amino terminus of the putative Dictyostelium protein contains a sequence of asparagine (N) and serine (S) residues, which is not present in other RNA polymerases (Fig. 4). To demonstrate that the repeat sequences are encoded in the mature rpmA mRNA and to exclude the presence of intronic sequences in the rpmA gene, we performed RT-PCR experiments, which demonstrated that this intron-like, A/T-rich sequence coding for the polyasparagine and polyserine residues is part of the coding sequence of the gene (data not shown). We therefore concluded that the Dictyostelium RNA polymerase gene is free of introns and that the sequence of asparagine and serine residues in the amino terminus of the protein is unique to the Dictyostelium protein. The cDNA sequence of the Dictyostelium rpmA gene has been deposited in GenBank under accession number AY040092.
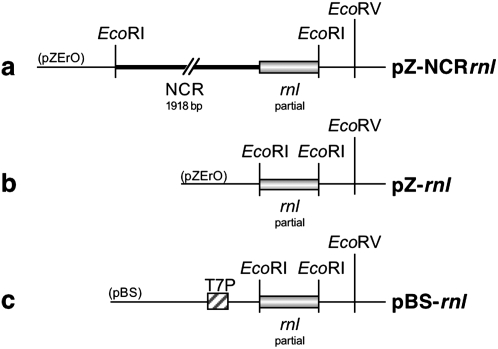
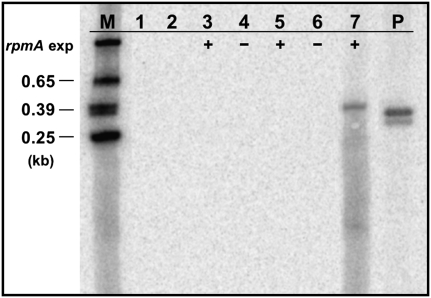
To verify that transcription occurs from the identified transcription initiation site, the rpmA cDNA was cloned into the Escherichia coli expression vector pQE30 (Invitrogen). The resulting construct, pQ-rpmA, was used to express the Dictyostelium protein in E. coli. Based on the assumption that the noncoding region containing the transcription start site also contains all regulatory elements required for transcription to occur, we prepared appropriate template DNA in the form of two different recombinant plasmids. The first plasmid, pZ-NCRrnl, contained the entire noncoding sequences and part of the downstream rnl gene cloned into the EcoRI site of the cloning vector pZErO-2 (Invitrogen) (Fig. 6A). The second plasmid, pZ-rnl, contained the same portion of the rnl gene cloned into pZErO-2, but without upstream sequences (Fig. 6B). Due to the absence of any upstream sequences, transcription from this construct was not expected to occur and the construct therefore served as a negative control in our studies. A third plasmid, pBS-rnl, served as a positive control. It contained the same 5′ portion of the rnl gene without any upstream sequences inserted into the EcoRI site of pBluescriptSK+ (Invitrogen). This vector contains a T7 promoter upstream of the multicloning site (Fig. 6C), which we used in a T7 RNA polymerase-mediated reaction to produce an rnl transcript as a positive control. For the transcription experiments, E. coli cells harboring the pQ-rpmA vector were cotransformed either with plasmid pZ-rnl or with plasmid pZ-NCRrnl. Expression of the Dictyostelium rpmA gene from pQ-rpmA was induced by growing exponential bacterial cultures (OD600: 0.4–0.6) for 4 h at 37°C in the presence of 1 mM IPTG, after which the cultures were shifted to 21°C for 1 h. The bacterial RNA was extracted, separated on agarose gels, and blotted onto nylon membrane to detect any rnl-transcripts using an rnl-specific probe. Bacterial cells harboring only a single type of plasmid, either plasmid pZ-rnl (Fig. 7, lane 1), pZ-NCRrnl (Fig. 7, lane 2), or pQ-rpmA (Fig. 7, lane 3) did not express any RNA that hybridized to the rnl-specific probe. An rnl-specific transcript was also not detected in bacterial cells harboring both plasmid pZ-rnl and pQ-rpmA, regardless of whether or not the expression of the rpmA gene was induced (Fig. 7, lanes 4,5). In fact, the rnl-specific probe detected an rnl-transcript only in bacteria containing both plasmids pZ-NCRrnl and pQ-rpmA and only after the expression of the rpmA gene had been induced with IPTG (Fig. 7, lane 7). The presence of a distinct and clear band with almost no smear above the detected transcript implies that transcription must have initiated consistently from the same initiation site, which was found in mapping experiments to be identical to the transcription initiation site identified in the D. discoideum mitochondrial genome (data not shown). The lack of smearing also suggests precise transcription termination by suitable vector sequences downstream from the cloned fragment; however, the exact 3′ end of the transcript was not determined. The detected rnl transcript was slightly larger than the transcript obtained in a T7 RNA polymerase mediated in vitro transcription reaction (Fig. 7, lane P), reflecting the presence of additional, noncoding sequences (66 bp) between the transcription start site and the rnl gene in plasmid pZ-NCRrnl. Thus, in the absence of a suitable E. coli promoter, heterologous expression of rnl in E. coli depended upon the provision (in trans) of the mitochondrial RNA polymerase encoded by rpmA and the presence (in cis) of the noncoding sequences upstream of the transcription initiation site. Expression from the same template DNA was not observed in in vitro experiments using a purified D. discoideum mitochondrial RNA polymerase, suggesting inappropriate reaction conditions or the lack of essential cofactors. Supplementing the in vitro reaction with mitochondrial protein extracts did not overcome the lack of expression, but led to the rapid degradation of the template DNA due to high nuclease activities present in the mitochondrial protein extracts (data not shown).
FIGURE 6.
DNA templates used in transcription experiments. (a) Plasmid pZ-NCRrnl, containing the entire noncoding region (NCR, 1913 bp) and a portion (398 bp) of the downstream rnl gene cloned into pZErO-2; (b) pZ-rnl, containing only the portion of the rnl gene but no upstream sequences; (c) pBS-rnl, containing the same portion of the rnl gene cloned into pBluescriptSK+ for in vitro expression by T7 RNA polymerase from the T7 promoter (T7P). Relevant restriction enzyme recognition sites are indicated.
FIGURE 7.
Specific transcription by the Dictyostelium mitochondrial RNA polymerase expressed in E. coli host cells. Northern blot analysis using a Dictyostelium rnl-specific probe against total RNA extracted from E. coli cultures harboring plasmid pZ-rnl (lane 1), plasmid pZ-NCRrnl (lane 2), plasmid pQE-rpmA (lane 3), plasmids pZ-rnl and pQE-rpmA (lanes 4,5), and plasmids pZ-NCRrnl and pQE-rpmA (lanes 6,7). Expression of the Dictyostelium mitochondrial RNA polymerase (rpmA exp) from pQE-rpmA is indicated by + (induced) or – (noninduced). As a positive control, a portion of the rnl gene was expressed from plasmid pBS-rnl under the control of the T7 promoter (lane P). The size of radioactively labeled RNA markers (M) is indicated in kilobases (kb).
The above experiments clearly demonstrated that transcription of the rnl gene can be initiated specifically by the mitochondrial RNA polymerase at the identified transcription initiation site. From the results obtained here and from those obtained in the RT-PCR and the capping experiments, we can confidently conclude that transcription of the Dictyostelium mitochondrial genome occurs from a single transcription initiation site.
DISCUSSION
This study was undertaken to establish whether previously observed transcripts in the mitochondria of the cellular slime mold Dictyostelium discoideum represented genuine primary transcripts or processing intermediates derived from larger transcripts. The results obtained clearly demonstrate that the D. discoideum mitochondrial genome is transcribed from a single transcription initiation site and that the previously observed transcripts were, in fact, the products of processing of a single, primary transcript. That transcripts larger than those detectable by Northern hybridization exist in D. discoideum mitochondria was shown in the RT-PCR experiments, where all but one of the intervening sequences between adjacent transcripts were amplified. Capping experiments identified the only 5′ end that was generated by transcription initiation and allowed us to distinguish it from those 5′ends that were created by transcript processing. The presence of only one initiation site implies that transcription from this site generates a single, polycistronic precursor RNA, which must be very efficiently, probably cotranscriptionally processed, since no transcripts larger than those observed in previous studies (Barth et al. 1999, 2001) have been detected. Considering the position of the tRNA genes, which are located in the vicinity of the maturation sites at the RNA level, it can be assumed that the processing of the precursor is, at least in part, achieved by the release of tRNA molecules. The excision of tRNAs has been described as an RNA maturation mechanism in a variety of organisms, including animals, fungi, and green algae (Ojala et al. 1980, 1981; Burger et al. 1985; Dyson et al. 1989; Schäfer et al. 2005; Wolff and Kück 1996).
A prerequisite for polycistronic transcription from a single initiation site is the uniform orientation of the genes to be transcribed. The Dictyostelium mitochondrial genome complies with this condition, as it is one of the few mitochondrial genomes sequenced so far, where all genes have been found to be located on and transcribed from the same DNA strand (Ogawa et al. 2000). Other organisms with a similar mitochondrial gene organization include Acanthamoeba castellanii (Burger et al. 1995), Chlamydomonas eugametos (Denovan-Wright et al. 1998), Monosiga brevicollis and Pedinomonas minor (Gray et al. 1998), and other social amoebae such as Dictyostelium citrinum, Dictyostelium fasciculatum, and Polysphondylium pallidium (Heidel and Glöckner 2008). It is therefore possible that the mitochondrial genomes of these organisms also contain single transcription units.
Transcription from the identified single transcription initiation site is further supported by the position of the transcription initiation site in the only longer noncoding region of the mitochondrial genome. Considering the importance of the length and structure of the sequences surrounding mitochondrial promoters, as demonstrated in yeast, for example, noncoding regions are expected to be large enough to contain promoter and regulatory elements essential for gene expression (Biswas 1999; Pfeuty et al. 2006). In the D. discoideum mitochondrial genome, most genes are tightly packed and some even overlap, and most of the few noncoding regions are likely to be too short to harbor individual promoter and regulatory elements. The noncoding region upstream of the rnl gene, however, is—with 1900 bp in length—the largest noncoding region, and therefore, well suited to carry genetic control elements. Direct proof for the presence of a functioning transcription initiation site located in the noncoding region upstream of the rnl gene was provided in expression studies. A truncated rnl transcript was detected in bacterial cells expressing the Dictyostelium mitochondrial RNA polymerase and harboring a construct (pZ-NCRrnl) containing the entire noncoding sequences and part of the downstream rnl gene. Expression of the rnl gene did not occur in the absence of the mitochondrial RNA polymerase nor in the absence of any noncoding sequences upstream of the rnl gene (from construct pZ-rnl). These results clearly demonstrated the importance of sequences within the noncoding region for mitochondrial transcription initiation, and that these sequences can be specifically bound by a mitochondrial RNA polymerase expressed in the same host cell.
The presence of a single transcription initiation site in the Dictyostelium mitochondrial genome puts the organism into an interesting position with regard to evolution. With a size of 55.6 kilobases (kb), the mitochondrial DNA shows the typical features of a protist mitochondrial genome. These include a high number of genes (33 protein-encoding genes, six open reading frames, two ribosomal RNA genes, and 18 transfer RNA genes), a high A + T content (72.6%), a small number of noncoding regions, and the presence of large and small subunit ribosomal protein genes, which are arranged in a bacterial-like gene cluster (Ogawa et al. 2000; Barth et al. 2007). Compared with this, mitochondrial genomes in all multicellular organisms, especially in animals (<20 kb) (Wolstenholme 1992) but also in some fungi (Schizosaccharomyces pombe, 19.4 kb) (Paquin et al. 1997; Schäfer et al. 2005) and some green (Chlamydomonas reinhardii, 15.8 kb) (Michaelis et al. 1990) and red algae (Chondrus crispus, 25.8 kb) (Leblanc et al. 1995) appear to be much smaller and more compact mitochondrial DNAs. These smaller mitochondrial genomes are commonly transcribed from either single, bidirectional promoters or from two or more unidirectional promoters into at least two polycistronic transcription units (Bogenhagen and Romanelli 1988; L'Abbe et al. 1991; Shadel and Clayton 1993). In contrast, transcription of mitochondrial genomes larger than the metazoan mitochondrial DNA requires several, if not many promoters (Kubelik et al. 1990; Tracy and Stern 1995; Unseld et al. 1997; Foury et al. 1998). The D. discoideum mitochondrial genome, however, poses an exception to this rule: although much larger in size (55.6 kb) than its metazoan counterparts, it has been shown in this study to be transcribed from a single transcription initiation site. The results presented here therefore give insight into the structural and functional diversity of mitochondrial genomes and their expression.
MATERIALS AND METHODS
Strains and culture conditions
Dictyostelium discoideum strain AX2 (Darmon et al. 1975), a derivative of wild-type strain NC4, was grown axenically in HL-5 medium (1% Difco proteose peptone, 0.5% Difco yeast extract, 0.043% Na2HPO4.2H2O, 0.035% KH2PO4, 1% Glucose) (Cocucci and Sussman 1970) at 21°C.
Isolation of genomic DNA
Isolation of D. discoideum genomic DNA (gDNA) was performed in large scale as described by Noegel et al. (1985). D. discoideum AX2 cells were harvested by centrifugation and lysed with nuclear lysis buffer (10 mM Mg acetate, 10 mM NaCl, 30 mM Hepes, 10% Sucrose, and 2% NP-40). Intact nuclei were treated with hot (65°C) EDTA/Sarkosyl buffer (0.2 M EDTA, 2% N-Lauroyl sarcosine sodium salt) to lyse the nuclei and release gDNA. The gDNA was then purified on a CsCl density gradient and spooled from solution after ethanol precipitation. It was used as template in PCR amplifications.
Isolation and preparation of total RNA
Total RNA was isolated from D. discoideum cells following instructions supplied by the manufacturer of TRIzol reagents (Invitrogen). The 5 × 106 cells were pelleted and lysed in 1 mL of TRIzol reagent. After the addition of 0.2 mL of chloroform and subsequent centrifugation, the aqueous phase was collected and the RNA was recovered by isopropanol precipitation. Contaminating DNA was removed by incubating the total RNA with DNase I (DNA-free kit, Ambion) following the supplier's recommendation. The DNase-treated RNA samples were used in RT-PCR reactions. Total RNA from bacterial cultures was extracted using the same protocol with the addition of a 30-min incubation step at 65°C after the cells have been resuspended in TRIzol in order to improve cell lysis.
Reverse transcriptase polymerase chain reaction
Prior to the RT reaction, the DNase-treated RNA and appropriate primers were denatured at 95°C for 5 min. The RNA template was reverse transcribed using Moloney murine leukemia virus reverse transcriptase (M-MLV RT; Promega) and the resulting cDNA was amplified by PCR. PCR cycles (30) were at 94°C for 1 min, 55°C for 1.5 min, and 72°C for 2.5 min. All gene-specific 3′ and 5′ primers used are listed in Table 1. PCR reactions using gDNA or DNase-treated RNA as template served as positive or negative controls. The amplified DNA was separated on 1% agarose gels in 1x Tris-acetate (TAE) buffer.
In vitro capping of total RNA
In vitro capping of total RNA followed the protocol supplied by Ambion. Approximately 15 μg of purified RNA were added to 30 μL of capping buffer containing 50 mM Tris-HCl at pH 7.9, 1.25 mM MgCl2, 6 mM KCl, 2.5 mM DTT, 0.1 mg/mL BSA, 0.1 mM S-adenosyl-methionine; 100 μCi [α-32P]GTP, 0.6 U of pyrophosphatase, and 8 U of guanylyl transferase. After incubation for 1 h at 37°C, removal of unincorporated [α-32P]GTP, as well as precipitation of capped RNA, was achieved by isopropanol precipitation. The pellet was dissolved in TE buffer (10 mM Tris, 1 mM EDTA at pH 8) and used as an RNA probe in Southern hybridization experiments.
Southern hybridization
All intervening regions were amplified via PCR using the primers listed in Table 1, the location of the 3′ primers is also indicated in Figure 1. PCR cycles (30) were at 94°C for 1 min, 55°C for 1.5 min, and 72°C for 3 min. Approximately 10 μL of each DNA fragment were separated on 1% TAE-agarose gels and transferred onto a positively charged nylon membrane (Roche Diagnostics). The blot was hybridized with capped RNA, exposed to a PhosphorImager screen, and radioactivity was detected using the Amersham Storm 860 Imaging System.
In vitro transcription by T7 RNA polymerase
The in vitro transcription reactions were performed using T7 RNA polymerase (Promega) under the following conditions: 5×Transcription Buffer: 10 μL, RNasin Ribonuclease Inhibitor (Promega): 0.5 μL, DTT (10 mM): 5 μL, BSA (5 mg mL−1): 1 μL, ATP, CTP, UTP (10 mM each): 5 μL, GTP (0.1 mM): 5 μL, [α-32P]GTP (10 mCi mL−1): 30 μCi–50 μCi, linearized DNA template (pBS-rnl): 1–2 μg, T7 RNA polymerase (Promega): 2 μL, DEPC-treated sdH2O: to final volume (50 μL). The reactions were incubated at 21°C or 30°C for 30 min to 1 h.
ACKNOWLEDGMENTS
P.L. was the recipient of an Australian postgraduate award and P.R.F. was the recipient of funds from the Thyne Reid Memorial Trusts.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1710309.
REFERENCES
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Auchincloss AH, Brown GG. Soybean mitochondrial transcripts capped in vitro with guanylyltransferase. Biochem Cell Biol. 1989;67:315–319. [Google Scholar]
- Barth C, Greferath U, Kotsifas M, Fisher PR. Polycistronic transcription and editing of the mitochondrial small subunit (SSU) ribosomal RNA in Dictyostelium discoideum. Curr Genet. 1999;36:55–61. doi: 10.1007/s002940050472. [DOI] [PubMed] [Google Scholar]
- Barth C, Greferath U, Kotsifas M, Tanaka Y, Alexander S, Alexander H, Fisher PR. Transcript mapping and processing of mitochondrial RNA in Dictyostelium discoideum. Curr Genet. 2001;39:355–364. doi: 10.1007/s002940100196. [DOI] [PubMed] [Google Scholar]
- Barth C, Le P, Fisher PR. Mitochondrial biology and disease in Dictyostelium. Int Rev Cytol. 2007;263:207–252. doi: 10.1016/S0074-7696(07)63005-8. [DOI] [PubMed] [Google Scholar]
- Binder S, Brennicke A. Transcription initiation sites in mitochondria of Oenothera berteriana. J Biol Chem. 1993;268:7849–7855. [PubMed] [Google Scholar]
- Binder S, Marchfelder A, Brennicke A. Regulation of gene expression in plant mitochondria. Plant Mol Biol. 1996;32:303–314. doi: 10.1007/BF00039387. [DOI] [PubMed] [Google Scholar]
- Biswas TK. Nucleotide sequences surrounding the nonanucleotide promoter motif influence the activity of the yeast mitochondrial promoter. Biochem. 1999;38:9693–9703. doi: 10.1021/bi982804l. [DOI] [PubMed] [Google Scholar]
- Bogenhagen DF, Romanelli MF. Template sequences required for transcription of Xenopus laevis mitochondrial DNA from two bidirectional promoters. Mol Cell Biol. 1988;8:2917–2924. doi: 10.1128/mcb.8.7.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennicke A, Zabaleta E, Dombrowski S, Hoffmann M, Binder S. Transcription signals of mitochondrial and nuclear genes for mitochondrial proteins in dicot plants. J Hered. 1999;90:345–350. doi: 10.1093/jhered/90.3.345. [DOI] [PubMed] [Google Scholar]
- Burger G, Helmer-Cittrich MH, Nelson MA, Werner S, Macino G. RNA processing in Neurospora crassa mitochondria: Transfer RNAs punctuate a large precursor transcript. EMBO J. 1985;4:197–204. doi: 10.1002/j.1460-2075.1985.tb02336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Plante I, Lonergan KM, Gray MW. The mitochondrial DNA of the amoeboid protozoon, Acanthamoeba castellanii: Complete sequence, gene content and genome organization. J Mol Biol. 1995;245:522–537. doi: 10.1006/jmbi.1994.0043. [DOI] [PubMed] [Google Scholar]
- Cermakian N, Ikeda TM, Miramontes P, Lang BF, Gray MW, Cedergren R. On the evolution of the single-subunit RNA polymerases. J Mol Evol. 1997;45:671–681. doi: 10.1007/pl00006271. [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Clayton DA. Replication an transcription of vertebrate mitochondrial DNA. Annu Rev Cell Biol. 1991;7:453–478. doi: 10.1146/annurev.cb.07.110191.002321. [DOI] [PubMed] [Google Scholar]
- Cocucci SM, Sussman M. RNA in cytoplasmic and nuclear fractions of cellular slime mold amebas. J Cell Biol. 1970;45:399–407. doi: 10.1083/jcb.45.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo MC, Fox TD. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- Darmon M, Brachet P, Pereira da Silva LH. Chemotactic signals induce cell differentiation in Dictyostelium discoideum. Proc Natl Acad Sci. 1975;72:3163–3166. doi: 10.1073/pnas.72.8.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denovan-Wright EM, Nedelcu AM, Lee RW. Complete sequence of the mitochondrial DNA of Chlamydomonas eugametos. Plant Mol Biol. 1998;36:285–295. doi: 10.1023/a:1005995718091. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson NJ, Brown TA, Ray JA, Waring RB, Scazzocchino C, Davies RW. Processing of mitochondrial RNA in Aspergillus nidulans. J Mol Biol. 1989;208:587–599. doi: 10.1016/0022-2836(89)90150-2. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F, Roganti T, Lecrenier N, Purnelle B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998;440:325–331. doi: 10.1016/s0014-5793(98)01467-7. [DOI] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage E, Tim G, et al. Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res. 1998;26:865–878. doi: 10.1093/nar/26.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hanic-Joyce PJ, Gray MW. Accurate transcription of a plant mitochondrial gene in vitro. Mol Cell Biol. 1991;11:2035–2039. doi: 10.1128/mcb.11.4.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel AJ, Glöckner G. Mitochondrial genome evolution in the social amoebae. Mol Biol Evol. 2008;25:1440–1450. doi: 10.1093/molbev/msn088. [DOI] [PubMed] [Google Scholar]
- Jaehning J. Mitochondrial transcription: Is a pattern emerging? Mol Microbiol. 1993;8:1–4. doi: 10.1111/j.1365-2958.1993.tb01197.x. [DOI] [PubMed] [Google Scholar]
- Kubelik AR, Kennell JC, Akins RA, Lambowitz AM. Identification of Neurospora mitochondrial promoters and analysis of synthesis of the mitochondrial small rRNA in wild-type and the promoter mutant [poky] J Biol Chem. 1990;265:4515–4526. [PubMed] [Google Scholar]
- L'Abbe D, Duhaime JF, Lang BF, Morais R. The transcription of DNA in chicken mitochondria initiates from one major bidirectional promoter. J Biol Chem. 1991;266:10844–10850. [PubMed] [Google Scholar]
- Leblanc C, Boyen C, Richard O, Bonard G, Grienenberger JM, Kloareg B. Complete sequence of the mitochondrial DNA of the Rhodophyte Chondrus crispus (Gigartinales): Gene content and gene organization. J Mol Biol. 1995;250:484–495. doi: 10.1006/jmbi.1995.0392. [DOI] [PubMed] [Google Scholar]
- Michaelis G, Vahrenholz C, Pratje E. Mitochondrial DNA of Chlamydomonas reinhardtii: The gene for apocytochrome b and the complete functional map of the 15.8 kb DNA. Mol Gen Genet. 1990;223:211–216. doi: 10.1007/BF00265056. [DOI] [PubMed] [Google Scholar]
- Newton KJ, Winberg B, Yamato K, Lupod S, Stern DB. Evidence for a novel mitochondrial promoter preceding the cox2 gene of Perennial teosintes. EMBO J. 1995;14:585–593. doi: 10.1002/j.1460-2075.1995.tb07034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noegel A, Welker DL, Metz BA, Williams KL. Presence of nuclear associated plasmids in the lower eukaryote Dictyostelium discoideum. J Mol Biol. 1985;185:447–450. doi: 10.1016/0022-2836(85)90416-4. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Yoshino R, Angata K, Iwamoto M, Pi M, Kuroe K, Matsuo K, Morio T, Urushihara H, Yanagisawa K, et al. The mitochondrial DNA of Dictyostelium discoideum: Complete sequence, gene content, and genome organization. Mol Gen Genet. 2000;263:514–519. doi: 10.1007/pl00008685. [DOI] [PubMed] [Google Scholar]
- Ojala D, Merkel C, Gelfand R, Attardi G. The tRNA punctuate the reading of genetic information in human mitochondrial DNA. Cell. 1980;22:393–403. doi: 10.1016/0092-8674(80)90350-5. [DOI] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- Paquin B, Laforest M-J, Forget L, Roewer I, Wang Z, Longcore J, Lang BF. The fungal mitochondrial genome project: Evolution of fungal mitochondrial genomes and their gene expression. Curr Genet. 1997;31:380–395. doi: 10.1007/s002940050220. [DOI] [PubMed] [Google Scholar]
- Pfeuty A, Dufresne C, Gueride M, Lecellier G. Mitochondrial upstream promoter sequences modulate in vivo the transcription of a gene in yeast mitochondria. Mitochondrion. 2006;6:289–298. doi: 10.1016/j.mito.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Rapp WD, Lupold DS, Mack S, Stern DB. Architecture of the maize mitochondrial atp1 promoter as determined by linker-scanning and point mutagenesis. Mol Cell Biol. 1993;13:7232–7238. doi: 10.1128/mcb.13.12.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS, Clayton DA. Mitochondrial transcription initiation, variation, and conservation. J Biol Chem. 1993;268:16083–16086. [PubMed] [Google Scholar]
- Schäfer B, Hansen M, Lang BF. Transcription and RNA-processing in fission yeast mitochondria. RNA. 2005;11:785–795. doi: 10.1261/rna.7252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinkel AH, Groot-Koerkamp MJA, Van der Horst GTJ, Touw EPW, Osinga KA, Van der Bliek AM, Veeneman GH, Van Boom JH, Tabak HF. Characterization of the promoter of the large ribosomal RNA gene in yeast mitochondria and separation of mitochondrial RNA polymerase into two different functional components. EMBO J. 1986;5:1041–1047. doi: 10.1002/j.1460-2075.1986.tb04320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa R, Chung YJ, Rose JP, Wang B-C. Crystal structure of bacteriophage T7 RNA polymerase at 3.3 Å resolution. Nature. 1993;364:593–599. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]
- Tracy RL, Stern DB. Mitochondrial transcription initiation: Promoter structures and RNA polymerases. Curr Genet. 1995;28:205–216. doi: 10.1007/BF00309779. [DOI] [PubMed] [Google Scholar]
- Unseld M, Marienfeld JR, Brandt P, Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet. 1997;15:57, 61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- Wolff G, Kück U. Transcript mapping and processing of mitochondrial RNA in the chlorophyte alga Prototheca wickerhamii. Plant Mol Biol. 1996;30:577–595. doi: 10.1007/BF00049333. [DOI] [PubMed] [Google Scholar]
- Wolstenholme DR. Animal mitochondrial DNA: Structure and evolution. Int Rev Cytol. 1992;141:173–216. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]