Abstract
Cap-independent initiation of translation is thought to promote protein synthesis on some mRNAs during times when cap-dependent initiation is down-regulated. However, the mechanism of cap-independent initiation is poorly understood. We have previously reported the secondary structure within the yeast minimal URE2 IRES element. In this study, we sought to investigate the mechanism of internal initiation in yeast by assessing the functional role of nucleotides within the minimal URE2 IRES element, and delineating the cis-sequences that modulate levels of internal initiation using a monocistronic reporter vector. Furthermore, we compared the eIF2A sensitivity of the URE2 IRES element with some of the invasive growth IRES elements using ΔeIF2A yeast. We found that the stability of the stem–loop structure within the minimal URE2 IRES element is not a critical determinant of optimal IRES activity, and the downstream sequences that modulate URE2 IRES-mediated translation can be defined to discrete regions within the URE2 coding region. Repression of internal initiation on the URE2 minimal IRES element by eIF2A is not dependent on the stability of the secondary structure within the URE2 IRES element. Our data also indicate that eIF2A-mediated repression is not specific to the URE2 IRES element, as both the GIC1 and PAB1 IRES elements are repressed by eIF2A. These data provide valuable insights into the mRNA requirements for internal initiation in yeast, and insights into the mechanism of eIF2A-mediated suppression.
Keywords: IRES, eIF2A, cap-independent, URE2, cis sequences
INTRODUCTION
Eukaryotic cells are capable of initiating protein synthesis by both cap-dependent and cap-independent (internal initiation or IRES-mediated) mechanisms. Cap-dependent initiation relies on the presence of an m7G cap structure for recruitment of the 40S ribosomal subunit to the 5′ end of the mRNA (Gebauer and Hentze 2004). In contrast, cap-independent initiation typically depends on an RNA secondary structure within the mRNA molecule that functions in lieu of the cap structure to recruit the 40S ribosomal subunit (Komar and Hatzoglou 2005). Protein synthesis initiated by the cap-independent mechanism is thought to be less efficient than cap-dependent translation, but is thought to be a competitive form of initiation during times when cap-dependent translation initiation is down-regulated, such as during cellular stresses (Merrick 2004; Spriggs et al. 2008). Cap-independent initiation is prominent during cell stresses including: hypoxia, viral infection, and glucose starvation (Gilbert et al. 2007; Spriggs et al. 2008). As such, it is important to understand the mechanisms responsible for cap-independent initiation of protein synthesis because it will provide insight into the mechanism for cellular responses to various stresses.
Ure2p is involved in nitrogen assimilation and formation of prion-like aggregates responsible for the [URE3] phenotype (Bousset et al. 2001; Cooper 2002). These two functions are connected in the cell because of the production of both long and short versions of the protein. The balance of these two forms affects formation of prion aggregates, since the expression of the short form of the protein can cure cells of the [URE3] phenotype (Bousset et al. 2001). The ability to respond to nitrogen limiting conditions is also affected by levels of the two forms of Ure2p because proteins involved in prion aggregates associated with [URE3] are not active in signaling during nitrogen assimilation (Wickner et al. 2000). Our laboratory has demonstrated that the short form of the protein results from internal initiation of translation in yeast (Komar et al. 2003). Several other yeast IRES elements have been described based on activity during glucose starvation (Gilbert et al. 2007). Many of these IRES elements rely on poly(A) stretches and PABP for activity. However, little work has been done to understand the sequence and structure requirements for these IRES elements.
We have previously shown that the URE2 IRES element is repressed by eukaryotic initiation factor 2A (eIF2A) (Komar et al. 2005). In an effort to understand the mode of action of eIF2A, we previously defined the minimal URE2 IRES element (Reineke et al. 2008). The minimal URE2 IRES element is located between nucleotides 205 and 309 in the URE2 coding region (where the adenosine within the AUG codon for the long form of Ure2p is nucleotide 1). We have shown that the minimal URE2 IRES element is also capable of folding into a small stem–loop structure that encompasses the internal AUG codon (Reineke et al. 2008). This feature may shield the AUG codon from base-pair interactions with incoming initiator methionyl tRNA (met-tRNAi) necessary for translation initiation. However, the role of eIF2A in regulating the URE2 IRES element remains unclear.
In this study, we have extended our understanding of the functionally relevant nucleotides and structure within the URE2 minimal IRES element. To this end, we have investigated the in vivo relevance of various regions of the URE2 minimal IRES element using site-directed mutagenesis. Furthermore, we have determined that the absolute stability of the URE2 minimal IRES stem is not linearly related to the in vivo activity. Since all of the mutants and deletions of the URE2 minimal IRES element are capable of being repressed by eIF2A, we also examined the ability of eIF2A to repress activity of some of the invasive growth IRES elements described previously (Gilbert et al. 2007).
RESULTS
Little nucleotide sequence variation exists within the minimal URE2 IRES element
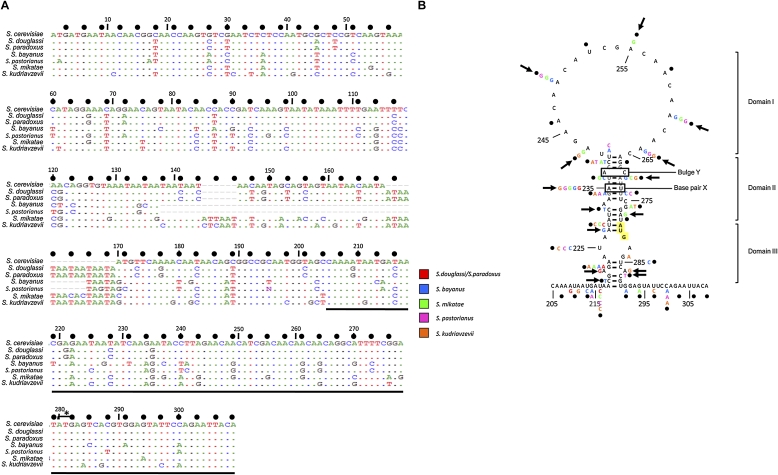
We have previously described the minimal IRES element within the yeast URE2 mRNA, and conducted an initial phylogenetic comparison of the URE2 IRES element (Reineke et al. 2008). To extend this analysis, we investigated the variation of the URE2 minimal IRES element more intensively and found that the URE2 minimal IRES sequence is highly conserved within the Saccharomyces genus, with the highest variation appearing in Saccharomyces bayanus and the lowest variation in Saccharomyces douglassi and paradoxus (Fig. 1A). Saccharomyces douglassi and paradoxus are 100% identical to each other in this region of the URE2 gene, so these sequences are not useful as independent sequences for comparison to the Saccharomyces cerevisiae sequence (Fig. 1A). While the idea that there is little codon variation in this region of URE2 suggests that the amino acid sequence is important in biology, it has been demonstrated that the prion characteristics conferred by the amino acid sequence are not highly conserved (Talarek et al. 2005). Of course, the gene is not highly conserved outside of the Saccharomyces genus, so this analysis is limited as a result of the small number of sequences available.
FIGURE 1.
Phylogenetic comparison of the URE2 minimal IRES element in seven Saccharomyces genera. (A) ClustalW comparison of the nucleotide sequences of the prion-forming domain and the minimal URE2 IRES element in the seven genera listed. The minimal URE2 IRES element is underlined. Identical nucleotides are illustrated as color dots, while variable nucleotides are indicated by colored letters. Wobble nucleotides are highlighted with bold dots (●) above the Saccharomyces cerevisiae sequence with the nucleotide numbers. The internal AUG codon is highlighted with a bracket and star. (B) The sequence comparison in A is mapped onto the previously determined secondary structure of the Saccharomyces cerevisiae minimal URE2 IRES element (Reineke et al. 2008). The variation in each genus at each nucleotide is indicated by colored letters that correspond to the genus (red, Saccharomyces douglassi and paradoxus; blue, Saccharomyces bayanus; green, Saccharomyces mikatae; fuchsia, Saccharomyces pastorianus; orange, Saccharomyces kudriavzevii). Bold arrows indicate nucleotide positions that vary without perturbing the base-pairing status. The internal AUG codon is highlighted in yellow, and numbers correspond to nucleotide numbers listed in A. Domains 1–3 of the minimal URE2 IRES element are indicated in brackets, and base-pair X and bulge Y are indicated with boxes.
Wobble nucleotides are the most valuable for phylogenetic comparison because the flexibility in which nucleotides are present has much less of an effect on aminoacyl-tRNA selection. However, the frequency of variation in wobble positions of the URE2 minimal IRES element is similar to what is observed in the rest of the prion forming domain of URE2 (Fig. 1A, “●” indicates wobble nucleotides). Furthermore, the frequency of variation at wobble positions in one of the GAPDH genes (TDH1) is similar to what is observed in both the minimal IRES element and the prion forming domain of URE2 (Supplemental Fig. 1). Nonetheless, the variation between the seven genera that do occur exists primarily in the wobble position of the codons in the URE2 coding region (Fig. 1A).
Despite the possibility that the high conservation within the Saccharomyces genus may be an artifact related to the few sequences useful for comparison, the variation that does exist may provide some insight into the nucleotides that may be important for activity. When the variable nucleotides are mapped onto the secondary structure within the Saccharomyces cerevisiae minimal URE2 IRES element, some covariation is observed. For example, in domain 2, the nonwobble nucleotide A227 and wobble nucleotide C231 are mutated from canonical Watson–Crick base pairs to noncanonical G:U base pairs in Saccharomyces bayanus, which represents partial covariation at these nucleotides (Fig. 1B). Additionally, the nonwobble nucleotide A235 is partially covariant in all the sequences compared (Fig. 1B). The base pair between nucleotides U237 and A270, which are both very flexible by virtue of being located in wobble positions, is either fully or partially conserved in all the genera examined (Fig. 1B). In domain 1, nucleotides 243, 249, 255, 261, and 264 all vary between adenosine and guanosine residues, which introduce no obvious secondary structure that may affect internal initiation (Fig. 1B). Assuming that all these sequences function as IRES elements, the variation in these nucleotides supports previous evidence that the poly(A)-rich nature of the loop (Fig. 1B, domain 1) is not an important feature for internal initiation for this IRES element (Reineke et al. 2008).
The poly(A)-rich nature of the domain 1 loop is not important for efficient internal initiation
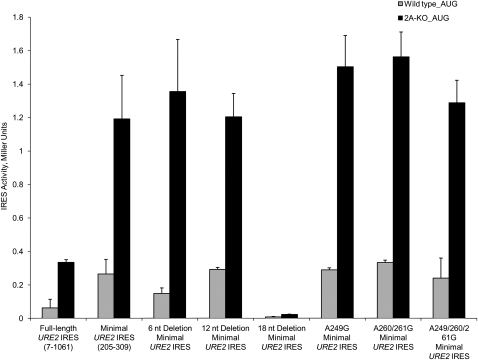
As mentioned above, we have shown that either the size of the domain 1 loop or the sequence is important for efficient internal initiation (Reineke et al. 2008). Since previous work has suggested that poly(A) stretches in some invasive growth, IRES elements are important for activity (Gilbert et al. 2007), we sought to delineate whether the size or poly(A)-rich nature of the domain 1 loop in the URE2 IRES element is important for activity. This was done using the same monocistronic reporter construct containing a stable stem–loop upstream of a fusion between the URE2 coding region and LacZ, which our laboratory has used in the past (Komar et al. 2005; Reineke et al. 2008). In combination, we employed adenosine to guanosine mutants at nucleotide positions 249, 261, and 262, which severely breaks up the poly(A)-rich nature of the loop. Interestingly, domain 1 single A249G, double A260/261G, and triple A249/260/261G mutants of the URE2 IRES element all have activity comparable to what is observed with the wild-type minimal URE2 IRES element (Fig. 2). Furthermore, these mutants occur naturally in Saccharomyces bayanus, mikatae, and pastorianus (Fig. 1B), which, assuming these sequences function as IRES elements, further supports the idea that the poly(A)-rich nature is not an important factor in internal initiation of the URE2 IRES element.
FIGURE 2.
The poly(A)-rich nature of the domain 1 apical loop is not critical for optimal activity of the URE2 minimal IRES element. Using the monocistronic p281-4 reporter plasmid harboring a stable stem–loop upstream of a fusion between the URE2 and LacZ open reading frames (Altmann et al. 1993), deletions and mutations within the domain 1 apical loop were tested in wild-type (gray bars) and ΔeIF2A (2A-KO; black bars) yeast strains. Reporter activity is presented in Miller Units [(OD420 × 1000)/(OD600 × reaction time)] on the y-axis, while the identity of deletions and mutations present in the minimal URE2 IRES element are indicated on the x-axis. Minimal URE2 IRES deletions of 6, 12, and 18 nt in the domain 1 loop were reported previously (Reineke et al. 2008).
Our laboratory has shown that deletions within the URE2 coding region do not affect the repression of the URE2 IRES element by eIF2A (Reineke et al. 2008), which suggests eIF2A affects internal initiation in a general manner because we have been unable to identify a sequence responsible for eIF2A-mediated suppression. However, the possibility that eIF2A affects translation initiation through the minimal URE2 IRES element has not been explored extensively. As such, each of the mutations in domain 1 were tested in both wild-type and ΔeIF2A yeast cells. In each case, levels of eIF2A-mediated repression are not affected by adenosine to guanosine mutations (Fig. 2). These data suggest that the A-rich nature of the apical loop located in domain 1 of the URE2 minimal IRES element is not important for URE2 IRES-mediated translation or for eIF2A-mediated repression. Furthermore, these data imply that PABP may not be involved in the pathway for eIF2A-mediated repression of translation initiation. However, this has not been directly tested.
Nucleotides within domain 2 are important for efficient cap-independent initiation of the Saccharomyces cerevisiae URE2 IRES element
Previous data suggest that the size of the domain 1 loop is important for internal initiation, since deletion of 18 nucleotides (nt), but not 12 nt, results in a reduction in IRES activity to levels comparable to background (Fig. 2; Reineke et al. 2008). It has been documented that stem stability is optimal with loops between 4 and 7 nt in size (Groebe and Uhlenbeck 1988), which implies that reducing the size of the domain 1 loop from 12 to 6 nt (Fig. 2, 12 nt and 18 nt loop deletions) may stabilize the stem in such a way that prevents unfolding necessary for efficient internal initiation. To test the importance of the stability of the URE2 IRES stem, we utilized two point mutations in the domain 2 stem (A235C and U272G) that abrogate formation of an A:U base pair (Fig. 1B, base-pair X) in the middle of the stem. The A235C mutation does not abrogate levels of internal initiation as compared to the wild-type minimal URE2 IRES element (Fig. 3A). In contrast, the U272G mutant resulted in levels of activity ∼20-fold lower (Fig. 3A). Restoration of this base pair by making an A235C/U272G double mutant did not result in restoration of IRES activity to wild-type levels (Fig. 3A). To exclude the possibility that changes in steady state mRNA levels are responsible for the changes in β-galactosidase activity, we employed quantitative real-time PCR to analyze levels of reporter mRNA (Supplemental Fig. 2). The results of this analysis indicate that changes in mRNA levels are not responsible for the reduction in activity observed with the U272G mutant. Therefore, uracil at domain 2 position 272 is important for efficient internal initiation on the URE2 IRES element, which is further supported by phylogenetic data indicating this nucleotide is invariant in all seven sequences (Fig. 1B).
FIGURE 3.
Nucleotides within domain 2 are important for efficient internal initiation of translation on the URE2 minimal IRES element. (A) Base-pair X in domain 2 (nucleotides A235 and U272) was mutated, and IRES activity was measured. (B) Bulge Y in domain 2 (nucleotides A238 and C269) was mutated, and IRES activity was measured. In A and B, activity measurements were conducted using the p281-4 reporter plasmid in both wild-type (gray bars) and ΔeIF2A yeast (2A-KO; black bars) cells, and represented as in Figure 2 (Altmann et al. 1993). Mutations in the minimal URE2 IRES element are indicated.
Since destabilizing the URE2 IRES stem had unexpected effects on IRES activity, we sought to use stabilizing mutations to test the role of stem stability on IRES activity in a reciprocal fashion to the experiments described in Figure 3A. We have previously shown that introduction of an A238G mutation in domain 2 results in an eightfold reduction in IRES activity (Reineke et al. 2008). Quantitative real-time PCR indicates that the reduction in β-galactosidase activity observed for the A238G mutant is not a result of reductions in reporter mRNA levels (Supplemental Fig. 2). Furthermore, we have shown that this mutation results in stabilization of the URE2 IRES stem in vitro using RNase protection assays (Reineke et al. 2008). The A238G mutant stabilizes the stem by converting an internal bulge (Fig. 1B, internal bulge Y) to a G:C Watson–Crick base pair. The reduction in activity that results from the A238G mutation suggests that either the nucleotide is important for recognition or introduction of a stabilizing G:C base pair generates a stem–loop that is too stable for efficient internal initiation of translation. To address this question, we introduced a single C269A mutation into the IRES, and then combined this mutation with the A238G mutant. The C269A mutant is not expected to alter internal bulge Y in terms of base pairing, while the A238G/C269A double mutant maintains internal bulge Y within the URE2 minimal IRES element. Interestingly, preserving internal bulge Y in the minimal URE2 IRES stem using the A238G/C269A double mutation results in low levels of IRES activity comparable to what is observed with the A238G single mutation (Fig. 3B). Again, the importance of A238 in IRES-mediated translation initiation of the URE2 minimal IRES element is supported in phylogenetic data indicating that this nucleotide is invariant within the Saccharomyces genera. However, the nucleotides in both base-pair X and internal bulge Y are not in wobble positions, so the organism does not have as much freedom to alter these nucleotides. In terms of IRES function, these data suggest that the nucleotide identity, rather than the base-pairing status at this position, is important for efficient internal initiation of translation, thereby supporting the conclusion from the experiment in Figure 3A.
The role of eIF2A in repression of URE2 IRES-mediated translation was investigated using mutations in both base-pair X and internal bulge Y of domain 2. When either nucleotide contributing to base-pair X was mutated, there was no effect on the ability of eIF2A to repress internal initiation of translation (Fig. 3A). Furthermore, mutation of either nucleotide in internal bulge Y did not affect repression by eIF2A (Fig. 3B). These data suggest that the stability of domain 2 of the URE2 minimal IRES element is not important, per se, for repression of IRES-mediated translation initiation by eIF2A. In addition, these nucleotides are not important for the repressive effect since increased initiation is observed in ΔeIF2A yeast cells.
Mutations that alter the base-pairing status with the internal AUG codon do not abrogate levels of internal initiation of translation
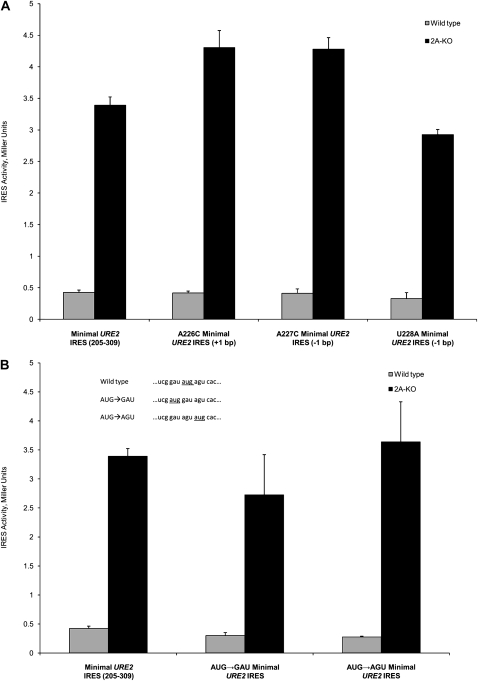
The above data indicate that the stability of the domain 2 stem is not important for internal initiation of translation on the URE2 minimal IRES element. However, it is still possible that the stability of domain 3, around the internal AUG codon, is important for efficient internal initiation. The stability in domain 3 was anticipated to be important during a step prior to recognition of met-tRNAi because the AUG codon must be available to interact with the anticodon. As such, mutations that abolish base-pair interactions with the internal AUG codon, thus rendering the AUG more available to interact with met-tRNAi, would be expected to result in increased levels of internal initiation. To test this hypothesis, we introduced three point mutations in domain 3 that had varying effects on base-pair interactions with the internal AUG codon. One mutation (A226C) is predicted to result in formation of an additional base pair, while two mutations (A227C and U228A) are predicted to result in one less base-pair interaction with the internal AUG codon. Interestingly, none of these mutations had significant effects on internal initiation of translation (Fig. 4A, cf. each mutant and the wild-type minimal URE2 IRES).
FIGURE 4.
Base-pairing status and location of the internal AUG codon are not imperative for IRES activity. (A) Altering the base-pairing status by making mutations in domain 3 of the minimal URE2 IRES element does not affect levels of cap-independent translation initiation. Identity and location of the mutations are indicated on the x-axis. (B) The location of the internal AUG codon within the minimal URE2 IRES element is not a strict requirement for internal initiation of translation. The AUG codon was translocated by switching its position either one codon upstream (AUG→GAU) or one codon downstream (AUG→AGU). The sequences surrounding the AUG codon (for the wild-type, AUG→GAU, and AUG→AGU constructs) are shown in the inset, with the internal AUG codon underlined. The values in A and B are measured using the p281-4 reporter plasmid (Altmann et al. 1993) in both the wild-type (gray bars) and ΔeIF2A (2A-KO; black bars) yeast cells, and represented as in Figure 2.
Since no mutation that abolished interactions with the internal AUG codon resulted in reduced cap-independent translation, we hypothesized that the location of the internal AUG codon may not be a strict requirement for efficient internal initiation. To address this question, we switched the AUG codon with the codon directly before (AUG→GAU) and after (AUG→AGU) using site-directed mutagenesis (Fig. 4B, inset). In addition to altering the order of the codons, these mutations are predicted to result in a loss of three base pairs or a gain of one base pair, respectively, in the overall stem–loop structure. Neither mutation resulted in a significant change in IRES-mediated translation from the URE2 IRES element (Fig. 4B, cf. either mutation and the wild-type minimal URE2 IRES).
While the above data show that parts of domains 1 and 2 are not important for the repressive effect of eIF2A, we hypothesized that the base-pairing status or location of the internal AUG codon may be determinants for eIF2A-mediated repression. This hypothesis was tested by analyzing the effect of the A226C, A227C, U228A, AUG→GAU, and AUG→AGU mutants in the ΔeIF2A yeast cells. As in wild-type cells, there was no change in levels of internal initiation for any of these mutants of the minimal URE2 IRES element (Fig. 4A,B). These data indicate that the base-pairing status of the internal AUG codon and the location are not requirements for repression by eIF2A. The lack of change in IRES-mediated translation in both the wild-type and ΔeIF2A cells suggests that the steps that lead up to met-tRNAi delivery are the same for delivery by both eIF2 and eIF2A.
Stem stability is not an absolute requirement for URE2 IRES-mediated translation initiation
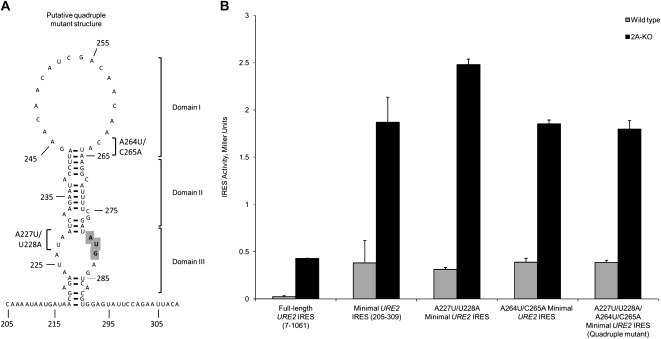
Removal of a base pair using the U272G mutation (Fig. 1B) resulted in a decrease in internal initiation (Fig. 3A). Furthermore, altering the base-pairing status of the internal AUG codon did not abrogate levels of URE2 IRES-mediated translation (Fig. 4A,B). As such, we sought to confirm the hypothesis that stem stability is not a requirement for efficient internal initiation on the URE2 IRES element. This was accomplished by introducing two mutations in domain 1 of the URE2 minimal IRES element that would be expected to result in formation of two additional base pairs, and disrupting two base pairs with the internal AUG codon with site-directed mutagenesis (a total of four mutations) (Fig. 5A). This strategy is valuable since it preserved the overall number of base pairs throughout domains 1–3. When these sets of mutations were tested by themselves and in combination, there was no obvious effect on internal initiation from the URE2 IRES element (Fig. 5B, cf. the two single mutants, as well as the quadruple mutant, and the wild-type URE2 IRES element). Furthermore, in the absence of eIF2A, the levels of internal initiation are similar when compared to the wild-type URE2 minimal IRES element (Fig. 5B). These data support previous data indicating that the stability of the stem in domains 2 and 3 is not important for efficient internal initiation of translation from the URE2 minimal IRES element, since adjusting the stability of each region, while maintaining the overall stability of the IRES element, did not result in a change in IRES-mediated translation initiation.
FIGURE 5.
Alterations in both the size of the domain 2 stem and base pairs with the internal AUG codon do not affect levels of cap-independent initiation. (A) The putative structure of the minimal URE2 IRES element resulting from the quadruple mutant consisting of the addition of two base pairs in domain 1 (A264U and C265A) and removal of two base pairs in domain 3 (A227U and U228A) is shown. This structure was predicted using the Mfold structure prediction software (Mathews et al. 1999; Zuker 2003), and supported by biochemical data indicating the formation of the stem–loop structure for the wild-type minimal URE2 IRES element (Reineke et al. 2008). The mutations in domains 1 and 3 are indicated with brackets. Double mutants (A264U/C265A and A227U/U228A) are expected to result in alterations of only the domains in which the mutations are made (domains 1 and 3, respectively). (B) Activity of the double (A264U/C265A and A227U/U228A) and quadruple mutants (A264U/C265A/A227U/U228A) in the minimal URE2 IRES element were tested using the p281-4 reporter plasmid (Altmann et al. 1993) in both wild-type (gray bars) and ΔeIF2A (2A-KO; black bars) yeast cells, and are represented as in Figure 2.
To further confirm the result that the stability of the URE2 IRES is not an important factor in cap-independent initiation, we plotted the activity of each mutant in both wild-type and ΔeIF2A yeast strains as a percent of the wild-type minimal URE2 IRES in wild-type cells (Fig. 6A). This was done for mutants examined previously and in this study (Reineke et al. 2008). Interestingly, there was no linear correlation between IRES activity and URE2 IRES stability in either cell type (Fig. 6A). This indicates that, at least over the range of stabilities examined, the stability of the URE2 IRES element is not a critical factor for efficient internal initiation of translation in vivo. Furthermore, there are few mutants wherein the fold change as compared to the wild-type URE2 minimal IRES element did not differ between wild-type and ΔeIF2A cells (Fig. 6A, left panel; Fig. 6A, right panel, each letter identifies the mutant). Activity for each mutant is shown in both wild-type (Fig. 6A, left panel, gray boxes) and ΔeIF2A (Fig. 6A, left panel, black diamonds) yeast cells. These data lead to the formulation of a URE2 minimal IRES map in which nucleotides critical for activity are indicated (Fig. 6B, N represents a nucleotide that can be altered without affecting internal initiation, and X represents nucleotides that have not been tested for functional roles in cap-independent translation initiation). This map suggests that only a few nucleotides are important for activity. Many of these nucleotides are conserved between the different yeast species (Fig. 1B), which indicates an evolutionarily conserved function that may be required for IRES activity. Base pairs and single-stranded nucleotides that are partially or fully co-variable, as indicated by phylogenetic comparisons, are also denoted on this map (Fig. 1B, bold dots and box, respectively).
FIGURE 6.
Overall stability of the URE2 minimal IRES stem is not critical for efficient internal initiation. (A) The activity of each mutant used previously (Reineke et al. 2008) and in this study was plotted against the predicted stability of the mutant URE2 minimal IRES element (left panel). Activity is represented as a percent of the wild-type minimal URE2 IRES element in wild-type cells (0.20671 Miller Units). Activity of each mutant is indicated by the same letter in both wild-type (gray boxes) and ΔeIF2A (2A-KO; black diamonds) yeast cells (right panel). (B) Nucleotides shown to be important for optimal IRES-mediated translation in functional assays are plotted on the structure of the minimal URE2 IRES element, which was determined previously. Nucleotides, not critical for efficient cap-independent initiation, are depicted with “N,” while those that are important are depicted with the single letter nucleotide code. Nucleotides lacking data indicating a functional role in internal initiation are denoted with “X.” Black dots highlight nucleotides participating in base-pair interactions that are partially covariable, and the black box highlights the single base pair that is covariable in phylogenetic comparisons. Block arrows denote single stranded nucleotides that vary without perturbing secondary structure as predicted by mFold.
Analysis of the 3′ cis-regulatory elements suggest they are small and distance dependent
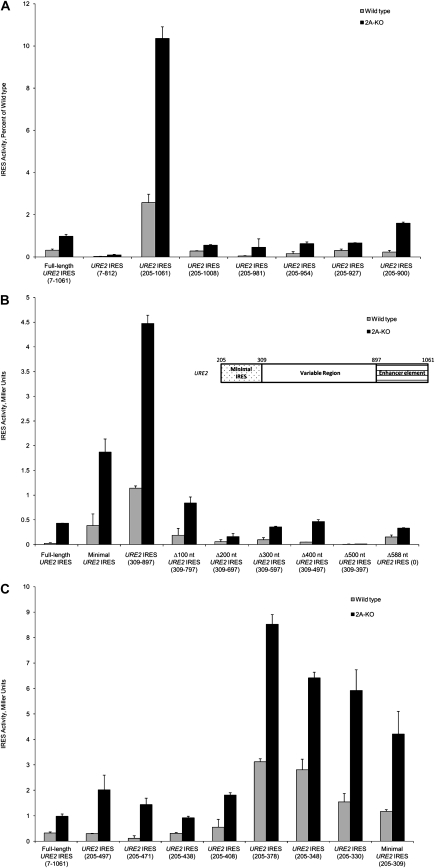
We have previously described the presence of two cis-regulatory sequences downstream from the internal AUG codon. An enhancer sequence located between nucleotides 812 and 1061 of the URE2 coding region was described (Reineke et al. 2008). The effect of this enhancer sequence is evident upon deletion of the region between nucleotides 812 and 1061 (Fig. 7A, cf. 7-1061 and 7-812). In an effort to define the 3′ boundary of this enhancer element, we made truncations 3′ of the enhancer element to delineate the sequence important for modulating activity of the minimal URE2 IRES element. Characterization of the enhancer element was done in the absence of the previously described inhibitor element 5′ of the minimal URE2 IRES element (Reineke et al. 2008). Deletion of 53 nt from the 3′ end of the enhancer completely abolished the effect on the minimal IRES element (Fig. 7A, cf. 205-1061 and 205-1008). Further deletions up to nucleotide 900 maintained the low level of internal initiation observed for the 205-1008 deletion. These data indicate that the enhancer sequence is not functional in the absence of the region between nucleotides 1008 and 1061.
FIGURE 7.
The downstream enhancer and inhibitory elements can be isolated to smaller regions within the URE2 coding region. (A) Small deletions within the URE2 coding region were made from the 3′ end to determine the 3′ boundary of the enhancer element downstream from the minimal URE2 IRES element. Nucleotide numbers correspond to the URE2 nucleotides present in the reporter constructs being examined. Nucleotide 1 is the adenosine nucleotide in the AUG initiation codon in the prion forming domain of URE2. (B) Consecutive 100 nt internal deletions within the URE2 coding region were used to examine the distance dependence of the enhancer element between nucleotides 1008 and 1061. These deletions were then tested using the previously described p281-4 construct (Altmann et al. 1993). The deletions were made in the “variable region” illustrated in the inset. For each deletion, the minimal IRES (dotted region, nucleotides 205–309; inset) and the enhancer element (lined region, nucleotides 897–1061; inset) remained constant. The nucleotide numbers on the x-axis indicate which nucleotides within the variable region are present in the respective construct. (C) Small deletions were made from the 3′ end of the inhibitory element directly downstream from the minimal URE2 IRES element. In A–C, activity of each deletion was measured using the p281-4 reporter plasmid (Altmann et al. 1993) in both wild-type (gray bars) and ΔeIF2A (2A-KO; black bars) yeast cells, and is represented as in Figure 2. Furthermore, the nucleotides within the URE2 coding region present in the constructs represented in A–C are depicted on the x-axis.
Since it was possible that the activity of the enhancer is distance-dependent, we sought to investigate this possibility using internal deletions. As such, we made consecutive deletions of 100 nt between nucleotides 309 and 897—i.e., deletions were made in the variable region illustrated in the inset of Figure 7B in 100 nt increments. The largest deletion of 588 nt resulted in the enhancer sequence being directly downstream from the minimal URE2 IRES element. Interestingly, deletion of only 100 nt was enough to abrogate the enhancer effect of this sequence [Fig. 7B, cf. URE2 IRES (309-897) and Δ100 nt URE2 IRES (309-797)]. Further deletions did not result in recovery of the enhancer effect (Fig. 7B). These data indicate that the enhancer sequence either requires some sequence between it and the minimal URE2 IRES element, or there are spatial restraints that enable the enhancer to be functional. These data suggest that delineation of the 5′ boundary of the enhancer sequence would be difficult because any deletion at the 5′ end of the enhancer would result in altering the distance between the enhancer and the minimal URE2 IRES element.
We then attempted to further characterize the inhibitory sequence directly downstream from the minimal URE2 IRES element (Reineke et al. 2008). The effect of this inhibitory sequence can be observed upon deletion of the region between 309 and 497 nt (Fig. 7C, cf. 205-497 and 205-309). Characterization of the inhibitory element was done by making consecutive deletions from the 3′ end of the inhibitory sequence starting at nucleotide 497 and ending at the 3′ boundary of the minimal URE2 IRES element. In an effort to isolate the functional effects of the inhibitory sequence, the deletions were done in the absence of regulatory elements upstream of the minimal URE2 IRES element. This deletion analysis indicated that the sequence necessary for the inhibitory effect is located between nucleotides 378 and 408 because the inhibitory effect disappeared upon deletion of that region of the URE2 gene (Fig. 7C, cf. 205-378 and 205-408). Prediction of secondary structure within nucleotides 378-408 of the URE2 gene using Zuker's Mfold server suggest that the inhibitory sequence is capable of forming a small stem–loop structure, and 6 out of 9 nt within the loop are capable of forming base pairs with the minimal IRES element (data not shown). Subsequent deletions result in levels of internal initiation comparable to what is observed with the minimal URE2 IRES element.
The repressive effect of eIF2A on each of these constructs was examined based on the possibility that an alternative conformation of the IRES element may be adopted in the presence of functional enhancer or inhibitory sequences, thus rendering the IRES insensitive to eIF2A. Each enhancer sequence and internal deletion demonstrated that there is no change in the ability of eIF2A to repress internal initiation from the URE2 IRES element (Fig. 7A,B). Furthermore, IRES-mediated translation is still repressed by eIF2A in the presence of various deletions of the inhibitory sequence (Fig. 7C). These data suggest that eIF2A may act on IRES elements in a general manner.
eIF2A preferentially represses other IRES elements
Based on the idea that many variations of the URE2 IRES element have been tested, and all are repressed by eIF2A, we considered the possibility that eIF2A may act in a general manner on cap-independent initiation in yeast. Since few IRES elements have been identified in yeast, we chose to examine the effect of eIF2A on some of the invasive growth IRES elements previously described (Gilbert et al. 2007). These elements were investigated using the monocistronic IRES reporter construct used in the experiments presented throughout this manuscript. However, we were only able to examine IRES elements in GIC1 and PAB1 because several other invasive growth IRES elements (FLO8, TIF4632, NCE102, and YMR181c) displayed cryptic promoter activity in our system (data not shown). These data indicate that repression by eIF2A is not specific to the URE2 IRES element, and raises the possibility that the presence or absence of poly(A) stretches may be a determinant for eIF2A-mediated repression, since neither the URE2, GIC1, nor PAB1 contain functional poly(A)-stretches. However, more IRES elements need to be evaluated to more definitively characterize the RNA requirements for eIF2A-mediated repression.
DISCUSSION
Little is known about what RNA structural elements or motifs are required for efficient cap-independent initiation for cellular mRNAs. Our data show that the yeast translational machinery relies on the presence of a couple of nucleotides in domain 2 for efficient internal initiation of the URE2 IRES element (Fig. 1B, A238, U272). Interestingly, these nucleotides are completely conserved amongst the seven Saccharomyces genera examined (Fig. 1B). Despite the high level of conservation of the URE2 minimal IRES element, the overall stability of the URE2 minimal IRES stem–loop structure does not appear to be a critical factor for IRES-mediated translation (Fig. 6A). This is consistent with previous reports that have shown that many strong eukaryotic IRES elements contain little secondary structure (Xia and Holcik 2009). In fact, the yeast YMR181c IRES element has been shown to be extremely active, but few double-stranded regions exist in vitro (Gilbert et al. 2007). Of course, the active forms of both the URE2 and YMR181c IRES elements are not known, but it is possible that there are proteins important for the stabilization of certain secondary structures that are required for efficient internal initiation.
We have shown that several cis-elements are capable of regulating internal initiation from the URE2 IRES element. Both an enhancer and inhibitory element are present downstream from the internal AUG codon within the URE2 minimal IRES element (Fig. 7A,C). Some information is available about the role of upstream sequences in modulating levels of internal initiation. For example, the Cat-1 IRES element is regulated by the presence of an upstream open reading frame. Translation of this upstream open reading frame is thought to result in formation of an active IRES element that directs synthesis of the Cat-1 protein (Yaman et al. 2003). Furthermore, some IRES elements rely on downstream sequences for efficient internal initiation. For example, mutations in the 264-nt capsid coding region, which is located downstream from the internal AUG codon of Giardiavirus, are capable of abrogating internal initiation of translation (Garlapati and Wang 2004). Additionally, recruitment of the 48S complex to the HIV-2 mRNA relies on sequences downstream from the internal AUG codon (Herbreteau et al. 2005). However, the Giardiavirus and HIV-2 IRES elements are viral IRES elements, which unlike the URE2 minimal IRES element, are known to rely on complex secondary structure. Often the regions downstream from the internal AUG codon in most viral IRES elements and all cellular IRES elements are not examined. As such, the role of downstream sequences in the regulation of IRES-mediated translation is not well understood.
Baird et al. (2007) have suggested that the XIAP, EGL1, and the AQP4 IRES elements share binding sites for trans-acting factors that regulate IRES activity. Hence, they propose that cellular IRES elements are not defined by overall structure, but rely on short motifs and trans-acting factors for activity (Baird et al. 2007). Since the stability of the URE2 IRES element is not important for activity, binding of trans-acting factors during certain cell stresses may regulate translation from the URE2 minimal IRES element. It will be of interest to investigate which trans-acting factors bind the URE2 IRES element, and what conditions promote production of the short form of Ure2p. Since we have determined that domain 2 A238 and U272 are important for activity, mutants of these nucleotides can be used as negative controls in RNA affinity chromatography designed to identify proteins with binding sites encompassing these nucleotides.
Our data also suggest that the size of the domain 1 apical loop is important for optimal activity (Fig. 2; Reineke et al. 2008). However, the importance of the domain 1 loop is not expected to be due to the A-rich nature based on mutations that severely break up this characteristic without abrogating IRES activity (Fig. 2). This is in stark contrast to several of the invasive growth IRES elements that have been described to depend on A-rich stretches for efficient cap-independent initiation of translation (Gilbert et al. 2007). While the conventional use of poly(A)-binding protein for efficient internal initiation on the URE2 IRES element cannot be ruled out, it is not expected to be involved in eIF2A-mediated repression of internal initiation. This conclusion is supported by data indicating that adenine to guanine domain 1 mutants of the URE2 minimal IRES, which have significantly fewer continuous adenine residues, are suppressed by eIF2A. As such, we believe the domain 1 apical loop may be important for recognition of trans-acting factors, excluding PABP, which are important for efficient internal initiation on the URE2 IRES element. Since eIF2A and PABP seem to regulate IRES-mediated translation initiation in yeast, it is intriguing to consider the possibility of two pathways for internal initiation, wherein one involves eIF2A and the other includes PABP.
We have demonstrated that the repressive effect of eIF2A is not specific for the URE2 minimal IRES element (Fig. 8). Our data indicate that eIF2A acts in a general manner on a subset of IRES elements, and eIF2A-mediated repression does not rely on a particular sequence or structure within the IRES element. These data raise the question of how eIF2A may be repressing cap-independent initiation. There is evidence that the eIF2A mRNA is rapidly turned over during cell stress (Komar et al. 2005). Furthermore, our laboratory has data that indicate that eIF2A protein is turned over during some cell stresses, consistent with the short half-life determined previously (Komar et al. 2005; data not shown). These pieces of information suggest that expression from eIF2A-repressed IRES elements increases during cell stresses that promote eIF2A turnover. We anticipate that regulation of IRES-mediated protein synthesis by eIF2A does not result from direct binding of the protein to mRNAs. Rather, it is expected that eIF2A regulates translation of IRES-containing mRNAs by binding to the ribosome, which is supported by previous data indicating that mammalian eIF2A directs binding of met-tRNAi to the 40S ribosomal subunit in an AUG codon-dependent manner (Adams et al. 1975; Merrick and Anderson 1975).
FIGURE 8.
Eukaryotic initiation factor 2A-mediated repression is not specific to the URE2 IRES element. Using the p281-4 reporter plasmid (Altmann et al. 1993), the activity of the invasive growth IRES elements (Gilbert et al. 2007) was tested in both wild-type (gray bars) and ΔeIF2A (black bars) yeast cells. IRES activity is represented in Miller units as in Figure 2.
Overall, our data further define the nucleic acid requirements for internal initiation on the URE2 minimal IRES element, and provide some evidence for multiple internal initiation pathways in yeast. It is increasingly apparent that trans-acting factors likely play an integral role in the regulation of translation of URE2 and other IRES elements. Included in these factors is eIF2A, which we show is an important mediator of internal initiation of multiple yeast IRES elements. It will be important in the future to identify other factors that mediate internal initiation, and further define the role of eIF2A to fully understand cap-independent initiation of translation.
MATERIALS AND METHODS
Yeast strains and culture conditions
Yeast used for these studies were of the BY4741 background (Research Genetics). For all reporter assay experiments, wild-type BY4741 (MATa, his3-1, leu2-0, met15-0, ura3-0) and eIF2A knockout strains (MATa, his3-1, leu2-0, met15-0, ura3-0, ygr054∷KanMX) were inoculated from fresh synthetic dropout (SD) plates and grown overnight at 30°C in 5 mL SD medium containing histidine, methionine, leucine, and 2% glucose. After 20–22 h of growth, yeast were spun down at 3000 rpm for 5 min and washed with 10 mL sterile deionized water. Yeast were then resuspended in 5 mL SD medium containing the aforementioned amino acids and 2% galactose for induction of mRNA expression. Yeast were incubated with galactose at 30°C for 18–20 h before harvesting.
Plasmids and cloning
The yeast plasmid p281-4 harboring a GAL1/10 promoter, a stable stem–loop, and a β-galactosidase reporter gene (Altmann et al. 1993) was utilized for URE2 mutagenesis analyses and experiments comparing the invasive growth IRES elements described previously (Gilbert et al. 2007). Each URE2 mutant was produced by site-directed techniques in the previously described pBIISK+(URE2) plasmid (Komar et al. 1997). Invasive growth IRES elements were subcloned from the pT7-stem_Fluc(pA)62 series of plasmids described previously (Gilbert et al. 2007). Both the URE2 mutants and deletions and the invasive growth IRES elements were subcloned as described previously (Reineke et al. 2008). All constructs were verified by sequencing prior to use in reporter assays. Primer sequences are available upon request. Approximately 200 ng of sequenced p281-4 plasmids were then transformed into both BY4741 and BY4741 ΔeIF2A yeast strains for β-galactosidase measurements using standard lithium acetate methods (Ito et al. 1983).
Reporter assays
Spectrophotometric β-galactosidase assays were performed as described previously (Reineke et al. 2008). These experiments were repeated at least three times. Representative experiments with standard error are presented.
Reporter mRNA analysis
Cultures were grown and mRNA expression was induced as described previously (Reineke et al. 2008). Cultures were then split so that half could be used for RNA isolation, while the other could be used for reporter protein expression analysis. RNA was extracted as described previously (Reineke et al. 2008), and 100 ng total RNA were used in quantitative real-time PCR as described previously (Zhang and Stavnezer 2009).
Bioinformatic studies
All URE2 sequences were obtained by basic local alignment sequence analysis (BLAST) of the Saccharomyces cerevisiae minimal URE2 IRES element nucleotide sequence. Since some of the minimal URE2 IRES elements are a CAA repeat, some sequences appeared during the BLAST search by virtue of the presence of CAA repeats despite dissimilarity throughout the remaining sequence. Therefore, those sequences were discarded. Relevant sequences were subsequently subjected to ClustalW analysis. Variation at wobble positions was calculated as a percent of total positions that were variable in any of the other seven genera examined.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dr. Wendy Gilbert (Massachusetts Institute of Technology) for providing plasmids containing invasive growth IRES elements for these studies. This work was supported by the NIH (GM-68079 [to W.C.M.] and T32 GM-08056 [to L.C.R.]).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1722809.
REFERENCES
- Adams SL, Safer B, Anderson WF, Merrick WC. Eukaryotic initiation complex formation. Evidence for two distinct pathways. J Biol Chem. 1975;250:9083–9089. [PubMed] [Google Scholar]
- Altmann M, Muller PP, Wittmer B, Ruchti F, Lanker S, Trachsel H. A Saccharomyces cerevisiae homolog of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J. 1993;12:3997–4003. doi: 10.1002/j.1460-2075.1993.tb06077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird SD, Lewis SM, Turcotte M, Holcik M. A search for structurally similar cellular internal ribosome entry sites. Nucleic Acids Res. 2007;35:4664–4677. doi: 10.1093/nar/gkm483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousset L, Belrhali H, Janin J, Melki R, Morera S. Structure of the globular region of the prion protein Ure2 from the yeast Saccharomyces cerevisiae. Structure. 2001;9:39–46. doi: 10.1016/s0969-2126(00)00553-0. [DOI] [PubMed] [Google Scholar]
- Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: Connecting the dots. FEMS Microbiol Rev. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlapati S, Wang CC. Identification of a novel internal ribosome entry site in giardiavirus that extends to both sides of the initiation codon. J Biol Chem. 2004;279:3389–3397. doi: 10.1074/jbc.M307565200. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert WV, Zhou K, Butler TK, Doudna JA. Cap-independent translation is required for starvation-induced differentiation in yeast. Science. 2007;317:1224–1227. doi: 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- Groebe DR, Uhlenbeck OC. Characterization of RNA hairpin–loop stability. Nucleic Acids Res. 1988;16:11725–11735. doi: 10.1093/nar/16.24.11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbreteau CH, Weill L, Decimo D, Prevot D, Darlix JL, Sargueil B, Ohlmann T. HIV-2 genomic RNA contains a novel type of IRES located downstream from its initiation codon. Nat Struct Mol Biol. 2005;12:1001–1007. doi: 10.1038/nsmb1011. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: Mystery of their existence. J Biol Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- Komar AA, Lesnik T, Cullin C, Guillemet E, Ehrlich R, Reiss C. Differential resistance to proteinase K digestion of the yeast prion-like (Ure2p) protein synthesized in vitro in wheat germ extract and rabbit reticulocyte lysate cell-free translation systems. FEBS Lett. 1997;415:6–10. doi: 10.1016/s0014-5793(97)01082-x. [DOI] [PubMed] [Google Scholar]
- Komar AA, Lesnik T, Cullin C, Merrick WC, Trachsel H, Altmann M. Internal initiation drives the synthesis of Ure2 protein lacking the prion domain and affects [URE3] propagation in yeast cells. EMBO J. 2003;22:1199–1209. doi: 10.1093/emboj/cdg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Gross SR, Barth-Baus D, Strachan R, Hensold JO, Goss Kinzy T, Merrick WC. Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. J Biol Chem. 2005;280:15601–15611. doi: 10.1074/jbc.M413728200. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Merrick WC. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- Merrick WC, Anderson WF. Purification and characterization of homogeneous protein synthesis initiation factor M1 from rabbit reticulocytes. J Biol Chem. 1975;250:1197–1206. [PubMed] [Google Scholar]
- Reineke LC, Komar AA, Caprara MG, Merrick WC. A small stem–loop element directs internal initiation of the URE2 internal ribosome entry site in Saccharomyces cerevisiae. J Biol Chem. 2008;283:19011–19025. doi: 10.1074/jbc.M803109200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- Talarek N, Maillet L, Cullin C, Aigle M. The [URE3] prion is not conserved among Saccharomyces species. Genetics. 2005;171:23–34. doi: 10.1534/genetics.105.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Taylor KL, Edskes HK, Maddelein ML, Moriyama H, Roberts BT. Prions of yeast as heritable amyloidoses. J Struct Biol. 2000;130:310–322. doi: 10.1006/jsbi.2000.4250. [DOI] [PubMed] [Google Scholar]
- Xia X, Holcik M. Strong eukaryotic IRESs have weak secondary structure. PLoS One. 2009;4:e4136. doi: 10.1371/journal.pone.0004136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaman I, Fernandez J, Liu H, Caprara M, Komar AA, Koromilas AE, Zhou L, Snider MD, Scheuner D, Kaufman RJ, et al. The zipper model of translational control: A small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 2003;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- Zhang H, Stavnezer E. Ski regulates muscle terminal differentiation by transcriptional activation of Myog in a complex with Six1 and Eya3. J Biol Chem. 2009;284:2867–2879. doi: 10.1074/jbc.M807526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]