Summary
Extracellular DNA (eDNA), a by-product of cell lysis was recently established as a critical structural component of the Enterococcus faecalis biofilm matrix. Here, we describe fratricide as the governing principle behind gelatinase (GelE) mediated cell death and eDNA release. GFP reporter assays confirmed that GBAP quorum non-responders (GelE−SprE−) were a minority subpopulation of prey cells susceptible to the targeted fratricidal action of the quorum responsive predatorial majority (GelE+SprE+). The killing action is dependent on GelE, and the GelE producer population is protected from self-destruction by the co-production of SprE as an immunity protein. Targeted gene inactivation and protein interaction studies demonstrate that extracellular proteases execute their characteristic effects following downstream interactions with the primary autolysin, AtlA. Finally, we address a mechanism by which GelE and SprE may modify the cell wall affinity of proteolytically processed AtlA resulting in either a pro- or anti-lytic outcome.
Keywords: Enterococcus faecalis, fratricide, autolysis, AtlA, gelatinase, serine protease
Introduction
Opportunistic enterococcal infections have traditionally been perceived as serious clinical threats due to rising incidence of antibiotic resistance and lateral transfer of resistance traits (Chang et al., 2003; Noble et al., 1992). More recently, these same threats have been associated with the existence of bacteria as surface adherent communities called biofilms (Fux et al., 2005). The close residential proximity of organisms within biofilms and altered genetic responses primarily driven by density dependent mechanisms may aid the observed heightened tolerance to antibiotics and dissemination of resistance traits (Jayaraman and Wood, 2008). Observations of clinical enterococcal biofilms on endodontic surfaces, bilary stents, urinary catheters, heart valves and tissue surfaces suggest a correlation of this pathogen’s community lifestyle and virulence (Carniol and Gilmore, 2004).
Analogous to developmental processes (biofilm development, competence regulation and sporulation) displayed by some bacteria, biofilm formation by enterococci involves quorum signaling (Hancock and Perego, 2004; Mohamed et al., 2004). The quorum signal is derived from the fsr gene cluster of E. faecalis (fsrABDC) and results from the processing and secretion of FsrD by FsrB to yield an 11-amino acid cyclized peptide lactone termed gelatinase biosynthesis-activating pheromone (GBAP) capable of density dependent signal transduction via the FsrAC two-component histidine kinase-response regulator pathway (Nakayama et al., 2006). Unlike the peptide pheromones involved in conjugal mating and plasmid transfer in Enterococcus, the formation of a cyclized lactone is thought to protect GBAP from proteolytic turnover by enterococcal proteases (Nakayama et al., 2006). Transcriptional profiling has indicated strong expression of two co-transcribed secreted extracellular proteases, gelatinase (GelE) and serine protease (SprE) following Fsr signal activation (Bourgogne et al., 2006; Qin et al., 2000). A number of independent studies have observed a critical role for gelatinase in enterococcal biofilm development (Hancock and Perego, 2004; Kristich et al., 2004; Mohamed et al., 2004). Gelatinase is proposed to activate lysis of a sub-population of bacteria and thereby catalyze the release of genomic DNA (eDNA), a critical component of the biofilm matrix (Thomas et al., 2008). Intriguingly the phenotypic effects of SprE opposed those of GelE with higher rates of lysis, eDNA release and increased biofilm development observed upon its inactivation (Thomas et al., 2008). Earlier observations by Shockman and others suggested the cellular target of GelE to be an autolysin (Shockman and Cheney, 1969; Waters et al., 2003). Although a direct interaction of SprE with a cell surface autolysin is yet to be established, evidence suggests it can modify rates of autolysis (Thomas et al., 2008). At least three autolysins (AtlA, AtlB and AtlC) have been identified to be secreted by E. faecalis of which AtlA is thought to be crucial for biofilm development (Kristich et al., 2008; Mesnage et al., 2008). Based on sequence similarity, AtlA was proposed to be made up of three domains. The central catalytic domain is responsible for the glucosaminidase activity (Eckert et al., 2006). The C-terminal domain is composed of six LysM modules that afford peptidoglycan affinity and possibly target autolysins to the division septum and poles (Steen et al., 2008). No known function yet exists for the T/E rich N-terminal domain (Eckert et al., 2006).
The existence of a bimodal bacterial population was hypothesized to explain the nature of enterococcal cell death leading to biofilm development (Thomas et al., 2008). The first sub-population would be a predatory population that produces GelE as an effector of cell death and is immune to self-inflicted GelE toxicity by virtue of the co-transcribed SprE protease while the second, a lysis susceptible prey population that does not produce either extracellular protease. Such bimodality in an otherwise isogenic bacterial population during growth is plausible given that cell density dictates those cells that respond to the GBAP quorum peptide and activate protease expression. Bacteria that do not respond to quorum signaling would then be susceptible to GelE mediated cell death due to their inability to produce the immunity factor, SprE. Although the above hypothesis is consistent with bacterial fratricide (Claverys and Havarstein, 2007), wherein a specific cell is responsible for predation of its clonal neighbor, an alternate hypothesis of altruistic suicide or programmed cell death of the quorum responder remains a possibility (Bayles, 2007; Rice et al., 2007).
In the current study, we show that extracellular proteases regulate enterococcal fratricide leading to eDNA release and biofilm development. Further AtlA is demonstrated to be the target of both GelE and SprE and this interaction is critical to the regulation of enterococcal fratricide and biofilm development.
Results
SprE prevents altruistic suicide in enterococcal populations
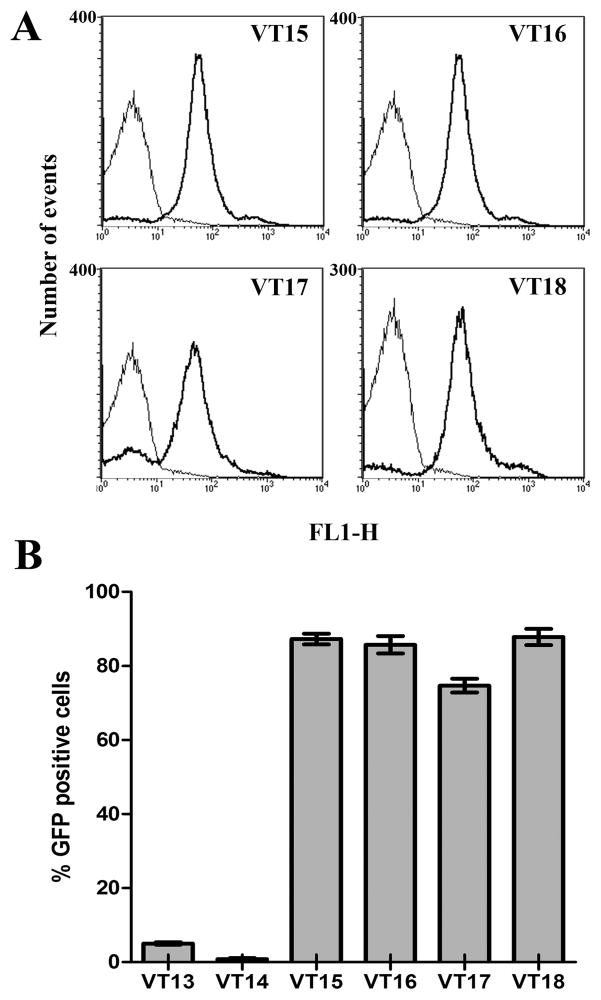
Programmed cell death was recently hypothesized to be critical for S. aureus biofilm development (Bayles, 2007; Rice et al., 2007). To investigate whether extracellular proteases (GelE and SprE) controlled an altruistic nature of cell death and lysis in E. faecalis populations, a GBAP responsive GFP-reporter construct (pVT31) was introduced into V583, VT01(ΔgelE), VT02 (ΔsprE) and VT03 (ΔgelE sprE) isogenic protease deletion mutant backgrounds resulting in strains VT15, VT16 (ΔgelE), VT17 (ΔsprE) and VT18 (ΔgelEsprE) respectively. The plasmid pVT31 contains a 1055-bp fragment composed of a gelE promoter-gfp gene fusion, in a high copy vector (pAT28) background. As the expression of GelE in native circumstances is dependent on GBAP levels, the coupling of the gelE promoter to the gfp gene in pVT31 would enable the expression of GFP to be dependent on the extracellular concentration of GBAP. Hence enterococci (bearing plasmid pVT31) that respond to the quorum peptide (GBAP) would switch on the expression of GFP. However cells that do not respond to GBAP would retain a GFP− phenotype. This phenotypic segregation in GFP expression effectively facilitated tracking of cells that responded to the quorum peptide within the wild type and each of the mutant populations by flow cytometry. The stringency of pVT31 as a reporter construct was determined using E. faecalis VT14, a derivative of V583 harboring pVT30 (reporter construct lacking functional promoter fusion) and VT13, an fsrA insertion mutant that contains pVT31. The absence of a functional FsrA would prevent VT13 from responding to the accumulation of quorum peptide (GBAP). Accordingly figure 1A depicts a clear shift in fluorescence intensities of VT15, VT16, VT17 and VT18 relative to VT14. Both the above controls (VT13 and VT14) exhibited a signal noise less than ~6% relative to VT15 (V583 pVT31) (Fig 1B). Fluorescent signal generated by VT13 likely arises from a basal level of expression from the gelE promoter, which is known to occur independent of FsrA (Singh et al., 2005). The relatively weak signal observed in strain VT14 is likely attributed to use of a weak promoter in the vector sequence, as plasmid pVT30 lacks the sequence for the gelE promoter.
Figure 1.
Estimation of altruistic suicide in enterococcal populations by flow cytometry. A. Representative flow histograms of GBAP responders from wild type VT15, and isogenic protease mutants- VT16 (ΔgelE), VT17 (ΔsprE) and VT18 (ΔgelEsprE) cultured to stationary phase. The pVT31 reporter plasmid in these isogenic strains activates GFP expression in response to GBAP quorum signaling. GFP fluorescence (FL-1H) corresponding to VT15, VT16, VT17 and VT18 are plotted (thick solid lines) and are contrasted in each instance with autofluorescence (thin solid lines) from VT14 (V583 pVT30, reporter construct lacking functional gelE promoter fusion). B. Percent GFP+ cells (GBAP responders) following overnight growth of VT13 (V583::fsrA), VT14, VT15, VT16, VT17 and VT18. The percent estimates were derived from histograms of seven independent trials ± SE using the CELLQuest program (version 3.1, Beckton and Dickinson).
Quantitative estimates of GFP+ cells (GBAP responders) in wild type and extracellular protease mutant backgrounds were achieved by flow cytometry. The settings were adjusted such that GFP− cells were within the first log decade while cells with higher intensities were considered GFP+ (Fig 1A). Estimation of GFP+ cells (GBAP responders) by flow cytometry indicate (Fig 1B) that although the majority of the wild-type (VT15) E. faecalis population under the present assay conditions responded to quorum signaling at the stationary phase of growth, approximately 15% correspond to GBAP non-responders. The independent effects of GelE and SprE on GBAP responders were characterized with respect to relative numbers of GFP+ responders within isogenic extracellular protease mutant populations and their geometric mean fluorescence intensities (GMFI). Relative numbers of GFP+ cells (Fig 1B) in VT16 (85.68 ± 7.02%) and VT18 (87.09 ± 6.5%) were similar to those found in E. faecalis VT15 (87.26 ± 4.37%). Interestingly, the absence of SprE expression in VT17 resulted in a significant decrease in GFP+ responder cells to 74.66 ± 5.63% as compared to parental strain (P< 0.05, Bonferronis test). This suggested that GBAP responders within a population underwent significant lysis only in the absence of SprE. Consistent with this observation the geometric mean fluorescence intensity (GMFI) of GFP+ responder cells of the VT17 (ΔsprE) population was significantly lower (59.23 ± 2.64) compared to that of V583 and mutants deficient in GelE expression (V583, 74.64 ± 13.11, VT16, 81.61 ± 6.97 and VT18, 83.22 ± 3.55). These observations are also consistent with earlier reports suggesting a pro-lytic activity for GelE and an anti-lytic activity for SprE (Thomas et al., 2008).
Interplay of GelE and SprE in enterococcal fratricide
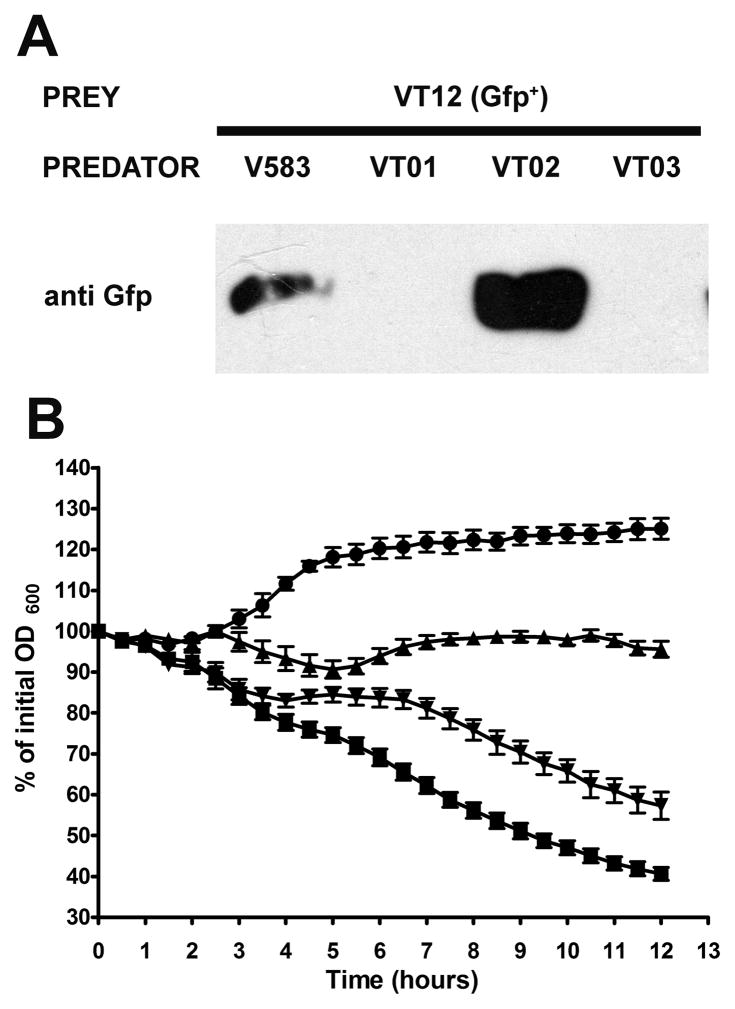
In order to assess the role of extracellular proteases of Enterococcus faecalis in fratricide, predator-prey co-culture lysis assays were performed. Parental and isogenic protease deletion mutants (predators) – V583, VT01 (ΔgelE), VT02 (ΔsprE) and VT03 (ΔgelEsprE) were co-cultivated in the presence of VT12, a double protease mutant strain that constitutively expressed GFP from plasmid pMV158GFP (Nieto and Espinosa, 2003). This previously characterized prey strain is deficient in lysis due to its inability to produce GelE (Thomas et al., 2008). However as the downstream cellular target of GelE that affects autolysis is still intact within these cells; their lysis may be activated when they are exposed to diffusible GelE. Consequently we reasoned that fratricidal control of the GFP+ prey population by diffusible extracellular proteases produced by the co-cultivated predator strains may be assayed by tracking the levels of GFP released into the culture supernatant. Neither of the purified extracellular proteases (GelE or SprE) exhibited an effect on GFP turnover at the concentrations assayed (data not shown) and hence any signal detected in culture supernatants should be a true reflection of bacterial lysis. Western blot analysis of culture supernatants, using anti-GFP antibody clearly showed that the lysis of the prey (VT12) population is dependent on the diffusible gelatinase (GelE) produced by the predator (Fig 2A; compare lanes 1 and 3 to lanes 2 and 4). Whereas co-cultivation of the parental V583 (GelE+SprE+) predators resulted in wild type levels of target lysis, GelE mutant predators (VT01 and VT03) demonstrated no detectable lysis of VT12 (prey). Interestingly, the use of VT02 (SprE−) as the predator resulted in an increased lysis of prey (Fig 2A, compare lanes 1 and 3), suggesting that diffusible SprE was able to modulate GelE dependent fratricide. As expected, spectrofluorometric measures of GFP fluorescence emanating from the prey population that survived the fratricidal action mediated by the predators showed a significant reduction in cell fluorescence when co-cultivated with VT02 (ΔsprE) compared to V583, VT01 and VT03 (data not shown).
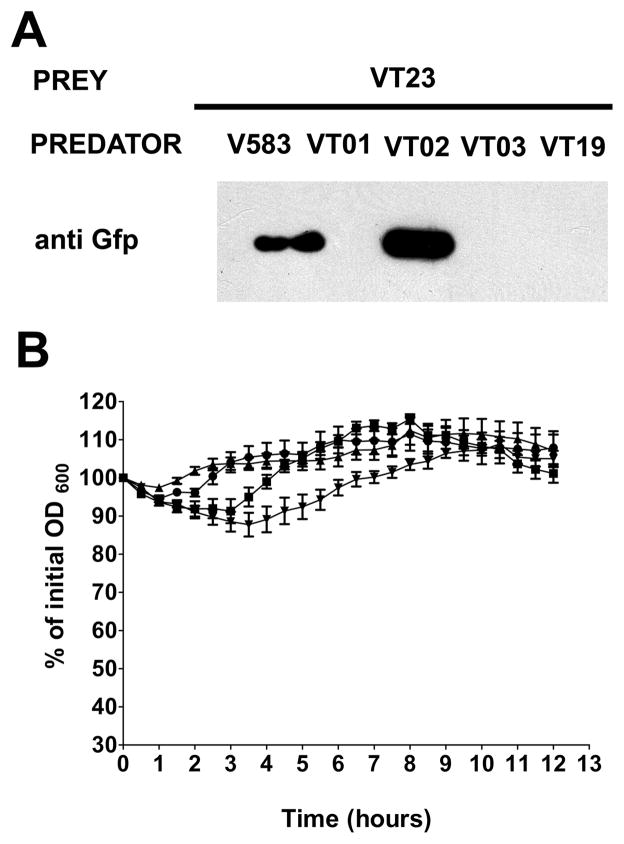
Figure 2.
Extracellular proteases regulate enterococcal fratricide. A. Detection of GFP immunoreactivity in stationary phase predator-prey co-culture supernatants. Predator (V583, GelE+SprE+; VT01, GelE−SprE+; VT02, GelE+SprE−; and VT03, GelE−SprE−) and prey (VT12, GelE−SprE− GFP+) were co-inoculated at a 20:1 ratio of prey: predator and grown overnight at 37°C prior to immunodetection of GFP. B. Effect of extracellular proteases on lysis of VT03 (GelE−SprE−). Differences in lysis rates of VT03 in the absence or presence of extracellular proteases (GelE and SprE) are exhibited as percent values of initial optical density at 600 nm. VT03 was incubated alone (●) or in the presence of 100 ng of GelE (■), SprE (▲) or GelE and SprE (▼;) together over a period of 12 hours at 37°C. Data are mean ± SE of three independent trials with each performed in triplicate.
Consistent with these results, we also observed that the addition of purified extracellular proteases (GelE and SprE) in physiological concentrations (~30nM) (Hancock and Perego, 2004; Makinen et al., 1989) resulted in the differential lysis of VT03 (ΔgelEsprE) (Fig 2B). Incubation with purified GelE alone caused a rapid increase in the lysis-rate of VT03 (ΔgelEsprE). While SprE by itself did not affect VT03 (ΔgelEsprE) rates of lysis, it prevented the initial increase in optical density observed in the VT03 strain without exogenous proteases. Furthermore, co-incubation of SprE with an equal concentration of purified GelE reduced the rate of GelE mediated lysis. These findings not only point to the ability of GelE to initiate fratricide but also of SprE to modulate GelE activity.
Comparison of autolysin profiles among enterococcal strains
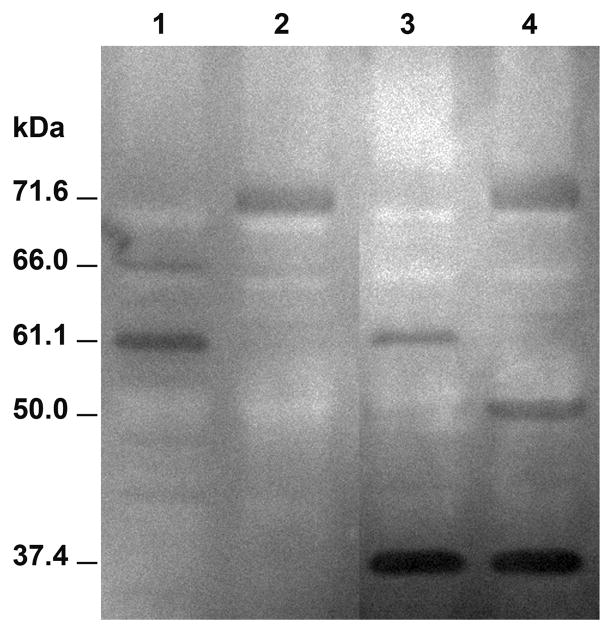
Based on previously reported autolytic rates of protease mutants (Thomas et al., 2008) and on observations by Shockman reflecting the role of GelE in activating an autolysin (Shockman and Cheney, 1969), we hypothesized enterococcal cell death to be a direct consequence of extracellular proteases manipulating cell surface associated autolysin(s). To determine potential autolysins regulated by extracellular proteases, we subjected cell wall protein extracts of GelE mutants from E. faecalis V583 and OG1RF strain backgrounds to zymography (Fig 3). Surface autolysin profiles of V583 and OG1RF exhibited significant differences. While a minor high molecular weight autolysin (~66kDa) was apparent in protein extracts derived from OG1RF, this activity was not visualized in VT20 (OG1RFΔgelE) or V583 cell wall extracts (Fig 3; compare lane 1 to lanes 2, 3 and 4). However a novel 38kDa autolytic band that seemed to be immune to the proteolytic activity of GelE was uniquely found in the V583 strain background and not in OG1RF (Figure 3; compare lane 1 and 2 to lanes 3 and 4). Similarly, a 50kDa autolytic band could only be detected in VT01 cell wall extracts, suggesting sensitivity toward GelE activity (Figure 3; compare lanes 3 and 4). In light of a recent study by Bourgogne et al. (Bourgogne et al., 2008) the absence of the 38kDa and 50kDa autolytic bands from OG1RF is not surprising as this strain lacks the prophage-associated endolysins present in V583 (Paulsen et al., 2003).
Figure 3.
Enterococcus faecalis cell surface autolysin profiles. Surface proteins were extracted from overnight grown cultures by boiling bacterial pellets in SDS-PAGE sample loading buffer. Samples were resolved on an 8% SDS polyacrylamide gel containing 0.1% purified OG1RF cell wall as substrate. Lanes 1 through 4 represents cell wall protein extracts from OG1RF (GelE+SprE+), VT20 (OG1RFΔgelE; GelE−SprE+), V583 (GelE+SprE+) and VT01 (V583ΔgelE; GelE−SprE+), respectively. Calculated molecular weights are indicated on the left.
Interestingly, only two autolytic bands (71.6 kDa, found in VT01 (V583ΔgelE) and VT20 (OG1RFΔgelE); 61.1 kDa in V583 and OG1RF) remained common among the two strains. Consistent with previous reports (Eckert et al., 2006; Mesnage et al., 2008), we identified and mapped the 71.6 kDa activity to AtlA by MALDI-TOF mass spectrometry. Targeted mutagenesis of atlA by insertion inactivation (see Materials and Methods) in V583 and OG1RF strain backgrounds (VT19 and VT21, respectively) demonstrated the 61.1 kDa activity to correspond to the proteolytically processed form of AtlA (see supplementary Fig S1). Interestingly, autolysin profiles of VT02 (ΔsprE) when compared with V583 appeared to display a considerable increase in the intensity of the 61.1 kDa band (see supplementary Fig S2). These results collectively suggest that in parental strains, GelE and SprE are capable of differentially regulating the turnover of AtlA.
AtlA is critical for eDNA release and biofilm development
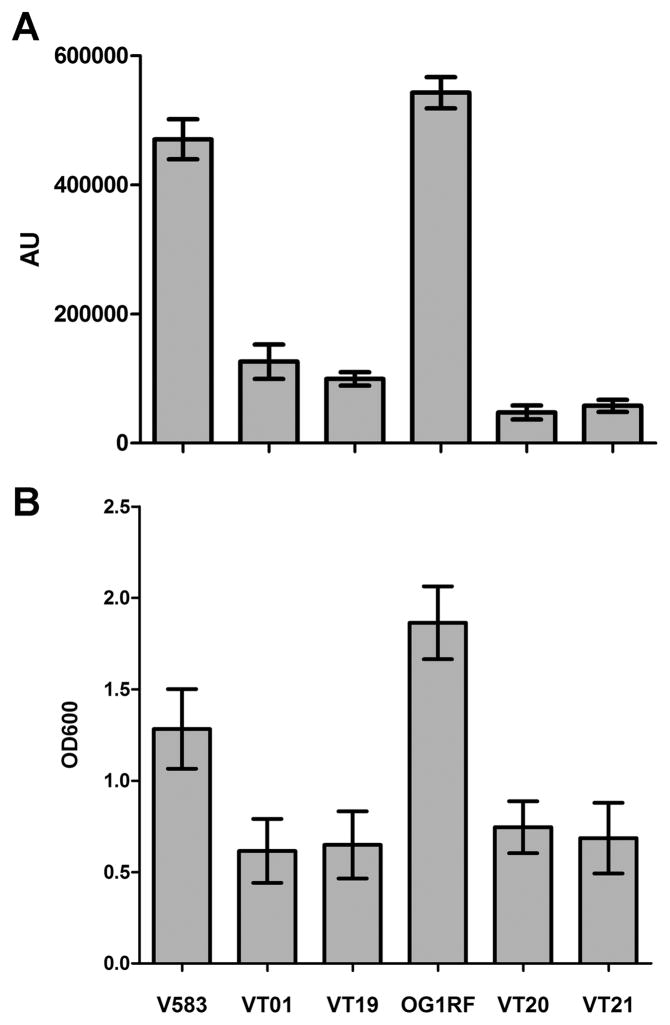
Recent studies have observed that inactivation of AtlA was detrimental to biofilm development of E. faecalis OG1RF (Kristich et al., 2008). Given that, AtlA was the sole autolysin whose activity was demonstrated to be regulated by both proteases, we tested for differences in eDNA release and biofilm forming abilities of VT19 (V583::atlA) and VT21 (OG1RF::atlA) against parental strains. Culture supernatants of overnight grown E. faecalis V583 and OG1RF parental strains and their corresponding AtlA mutants were assayed for eDNA using a DNA specific dye, SYTOX green (Molecular Probes). As expected and confirming our previous observations (Thomas et al., 2008) deletion of gelE in V583 (VT01) and OG1RF (VT20) backgrounds resulted in a significant 4- and 12-fold decrease in eDNA release as compared to their respective parental strains (Fig. 4A, P< 0.0001, t-test). More importantly, the decrease in eDNA release due to the inactivation of atlA paralleled GelE mutants from both strain backgrounds. This strongly suggested that AtlA may be the major target of extracellular proteases on the cell surface, that mediates fratricide and eDNA release.
Figure 4.
AtlA is critical for eDNA release and biofilm formation. A. Detection of extracellular DNA in culture supernatants. Cell free culture supernatants of V583 (GelE+), VT01 (V583ΔgelE), VT19 (V583::atlA), OG1RF (GelE+), VT20 (OG1RF ΔgelE) and VT03 (OG1RF::atlA) were supplemented with the DNA specific dye, SYTOX green to a final concentration of 1 μM before being assayed spectrofluorometrically as described. Fluorescence intensity data are represented as arbitrary units (AU). B. Biofilm formation of E. faecalis strains on polystyrene. Biofilm formation was assayed as a function of crystal violet stain (measured at 550 nm) retained by the biofilm biomass grown for 24 hours. Data are mean ± SE of three independent trials. The strain designations shown in panel B correlate with panel A.
Because AtlA mutants are deficient in eDNA release, we further hypothesized these strains to be defective in biofilm formation. Quantitative analysis of biofilms grown on polystyrene surfaces revealed that AtlA mutants, similar to GelE mutants formed significantly less biofilm biomass as compared to parental strains (Fig. 4B, P< 0.0001, t-test). The observed deficiencies in biofilm formation were not a result of growth defects as all mutants displayed similar growth kinetics compared to the parental strains (data not shown). These observations are consistent with a critical role for AtlA in eDNA release and biofilm formation.
Role of AtlA in fratricide
Given that extracellular proteases and AtlA contribute to similar biological pathways in E. faecalis, we hypothesized that fratricidal effector- modulator functions of GelE and SprE, are channeled through their downstream interactions with AtlA. However, prey-predator co-culture lysis assays using VT23 (an atlA mutant derivative of VT12 (ΔgelE-sprE, GFP+) as prey showed no abrogation of V583 and VT02 (ΔsprE) predator-mediated fratricide (Fig 5A; lanes 1 and 3), although maintaining GelE dependency (Fig 5A; lanes 2 and 4).
Figure 5.
Enterococcus faecalis AtlA is a critical determinant of protease mediated fratricide. A. GFP immunoreactivity from predator-prey co-culture supernatants. Predators represented by V583 (GelE+SprE+), VT01 (GelE−SprE+), VT02 (GelE+SprE−), VT03 (GelE−SprE−) and VT19 (GelE+SprE+AtlA−) were each individually co-cultured with prey (VT23, GelE+SprE+AtlA−GFP+). B. Effect of extracellular proteases on lysis of VT22 (VT03::atlA, GelE−SprE−AtlA−). Lysis rates are exhibited as percent values of initial optical density at 600 nm. VT22 was incubated alone (●) or in the presence of 100 ng of GelE (■), SprE (▲) or GelE and SprE together (▼;) over a period of 9 hours at 37°C. Data are mean ± SE of three independent trials with each performed in triplicate.
In an attempt to address these seemingly confounding observations, we tested the autolytic rates of the atlA mutant derivative of VT03, designated VT22, in the presence of purified enterococcal extracellular proteases. Addition of physiological concentrations of GelE or SprE (~30 nM) individually or together did not lead to any significant differences in lysis of VT22 as compared to protease negative controls (Fig 5B). This suggested that GelE dependent fratricide of VT23 observed earlier was possibly due to predator specific factors (in addition to GelE) that interacted with the prey population. As AtlA has previously been shown to be a diffusible enzyme capable of hydrolyzing the cell walls of Micrococcus lysodeikticus (Qin et al., 1998) and E. faecalis (Mesnage et al., 2008), we hypothesized that AtlA may fulfill the role of a predator factor responsible for the lysis of prey cells in the population. Such diffusible AtlA may possibly be released from a subpopulation of predator cells (~15%, refer Fig 1B) that have not responded to the quorum signal and hence are susceptible to the effector functions of GelE. To test this hypothesis we co-cultured VT19 (V583::atlA) as a predator population against VT23 (ΔgelEsprE,::atlA, GFP+) prey population. Figure 5A (lane 5) clearly indicates the absence of any detectable fratricide in the prey population upon inactivation of the predator AtlA, suggesting a crucial role for soluble (diffusible) forms of AtlA in fratricide.
Characterizing the effects of GelE and SprE on recombinant AtlA
To further investigate the interaction between extracellular proteases and AtlA, we subjected recombinant histidine tagged AtlA (rAtlA, Fig 6A) expressed and purified from E. coli cultures to GelE and SprE treatment. GelE treated rAtlA was multiply processed into minor forms within 30 minutes (Fig 6B; lane 2) and was completely turned over by 5 hours (data not shown). Zymogram analysis showed that most processed forms of rAtlA were active on V583 cell wall (Fig 6C; lane 2) suggesting that GelE preferentially cleaves initially at non-catalytic sites within AtlA. The full-length 72.1 kDa autolytic band seemed to show activity only after extended periods of incubation (data not shown), suggesting an enhancement in activity upon proteolytic cleavage. Interestingly SprE action on rAtlA resulted in a single major 62kDa form and a minor higher molecular weight form (Fig 6B and 6C, lane 3). From the current experimental conditions, although it would seem that rAtlA has a faint auto-catalytic activity and processes itself into the 62kDa form, the action of SprE clearly augments the rate of this processing while that of gelatinase seemed to affect the turnover of rAtlA.
Figure 6.
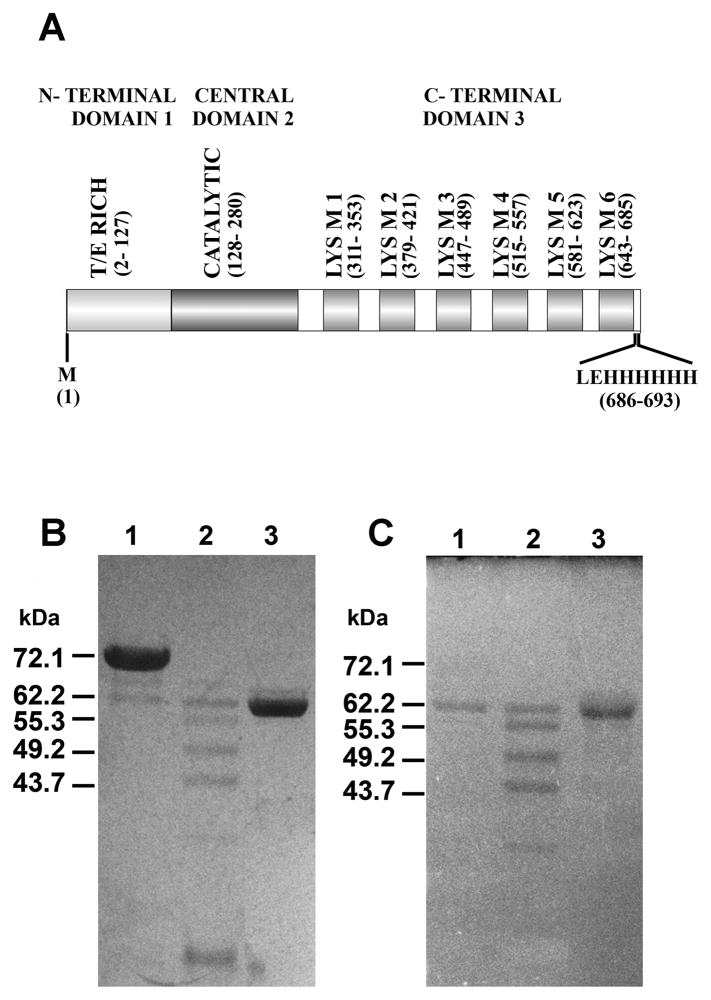
GelE and SprE differentially process AtlA. A. Schematic representation of domains present within recombinant AtlA. Domain 1, Threonine and glutamic acid rich N-terminal domain; domain 2, N-acetyl glucosaminidase central catalytic domain; domain 3, C-terminal domain containing six LysM modules predicted to be crucial for peptidoglycan adherence. The recombinant protein was engineered to start with the residue, methionine and end with a 6 × histidine tag. Schematic was graphed using protein domain illustrator program(Ren et al., 2009), DOG 1.0.3 B. SDS PAGE analysis of proteolytically processed forms of AtlA. 10 μg of purified AtlA was incubated alone (lane 1) or in the presence of 30 nM GelE (lane 2) or SprE (lane 3) for 30 minutes at 37°C. C. Zymogram analysis of proteolytically processed AtlA. Lanes represent AtlA alone (lane 1), AtlA incubated with GelE (lane 2) or SprE (lane 3). Calculated molecular weights are indicated on the left.
Comparison of the molecular weights of GelE and SprE processed recombinant AtlA fragments (Fig 6B) to their corresponding spectrograms (generated from MALDI TOF MS/MS peptide mass mapping) empirically identified regions within rAtlA that were susceptible to protease attack. As compared to the full length recombinant AtlA (72.3 kDa, includes 6XHis tag, but devoid of signal peptide), the 62kDa (Fig 6B, lanes 2 and 3) form maintained an intact C-terminal domain inclusive of the histidine tag, suggesting that both GelE and SprE specifically cleaved within the N-terminal T/E rich domain of rAtlA. Additionally, GelE also displayed cleavage specificities within the C- terminal domain. This was evident as the smaller rAtlA forms (55.3, 49.2, and 43.7kDa; Fig 6B, lane 2) shared a common N-terminus but exhibited clear differences in their C- terminal peptide fingerprints (data not shown).
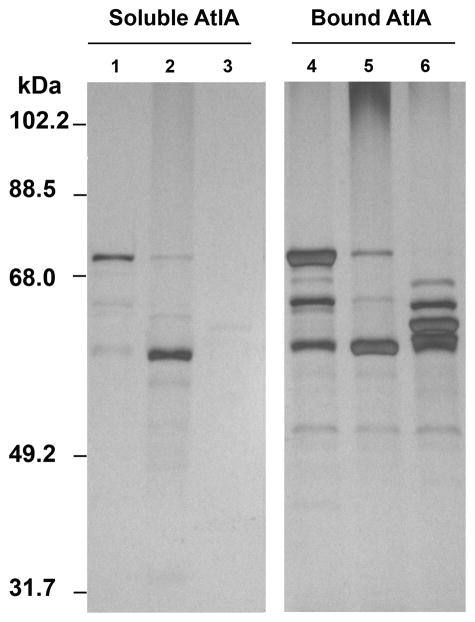
Given that the C-terminal domain of AtlA is composed of multiple units of LysM modules (Fig 6A) it seemed plausible that the action of GelE on the C-terminal domain of AtlA may catalyze its release from the cell surface. To test this hypothesis and to investigate any effect SprE may have toward rAtlA’s affinity to the cell surface, we assayed the cell wall affinities of full-length and proteolytically processed forms of rAtlA. Recombinant AtlA was initially allowed to bind E. faecalis V583 cell walls, followed by individual treatments with GelE and SprE proteases. Relative affinities of rAtlA forms processed by GelE and SprE to the cell wall was determined as a function of their ability to remain in the supernatant fraction on centrifugation as opposed to the pelleted cell wall bound fraction. Although it was clearly evident from the analysis that cell wall bound rAtlA displayed altered processing by GelE compared to the unbound form (compare proteolytic pattern from Fig 7, lane 5 and Fig 6B, lane 2), the supernatant fractions revealed that GelE treatment resulted in processed soluble forms of rAtlA (Fig 7, lane 2). Notably, SprE treatment resulted in the absence of any detectable soluble forms of AtlA in the supernatant fraction. Complementing these observations, comparison of the GelE and SprE processed cell wall bound forms of rAtlA displayed significant differences (Fig 7; compare lanes 5 and 6). Cleavage of rAtlA by SprE resulted in processed forms with molecular masses approximately equal to and greater than 62kDa (Fig 7, lane 6) that showed significantly higher affinity to peptidoglycan compared to the ~ 62kDa band resulting from GelE treatment (Fig 7, lane 5). Given that cleavage of rAtlA by SprE did not result in the loss of LysM modules (a known peptidoglycan binding and localization domain) it stands to reason that AtlA may be shuttled to the septum optimally after SprE processing.
Figure 7.
Differential affinities of GelE and SprE processed recombinant AtlA forms to cell wall peptidoglycan. Following incubation with purified cell wall peptidoglycan, rAtlA was treated with GelE (lanes 2 and 5) and SprE (lanes 3 and 6). Lanes 1 and 4 contained rAtlA that was not treated with either GelE or SprE. Lanes 1–3 represent the soluble fraction of rAtlA after cell wall recovery, and lanes 4–6 represents the cell wall bound forms of rAtlA.
We reasoned that optimal proteolytic activity of SprE toward rAtlA may prevent GelE from subsequently mediating its (rAtlA’s) release from the cell surface, thus arguing for the observed resistance of attacker subpopulations (SprE+) to GelE mediated lysis. To test this hypothesis, we pretreated rAtlA bound to peptidoglycan with physiological concentrations of SprE and then subjected it to the proteolytic action of an equimolar concentration of GelE. Supplemental figure S3 clearly demonstrates the inability of GelE to release SprE processed rAtlA from the peptidoglycan fraction into the supernatant. Further, SprE was unable to prevent the release of peptidoglycan bound rAtlA into the supernatant if GelE acted on the autolysin first. This suggests that SprE prevents lysis of attacker populations by efficiently competing with GelE for the primary autolysin, AtlA. However, the exact mechanism of competition by the extracellular proteases toward AtlA remains to be elucidated. Collectively these results suggest that in wild-type populations the role of SprE may be to prevent altruistic suicide of quorum responders by limiting the release of AtlA from the cell surface.
Discussion
The results presented in this study demonstrate extracellular protease mediated fratricide to be responsible for governing eDNA release and biofilm development of E. faecalis. Death of sibling cells (fratricide) mediated by isogenic cells within the same population has previously been implicated in developmental processes (competence and sporulation) of Streptococcus pnuemoniae and Bacillus subtilis (Ellermeier et al., 2006; Gilmore and Haas, 2005; Gonzalez-Pastor et al., 2003; Guiral et al., 2005; Havarstein et al., 2006). Cell-cell signaling during these developmental processes results in the coordinated production of specific killing factors and immunity proteins within a subpopulation of cells (predators). Killing factors which include bacteriocins and murein hydrolases target the death of a small susceptible isogenic population (prey) (Claverys and Havarstein, 2007). Susceptibility of prey to the killing factors is largely due to the absence of immunity proteins in this population, a cost these cells possibly pay for not participating in the signaling process. Death of the prey ultimately benefits the surviving population either in the form of nutrients in nutritionally stressed B. subtilis or as released genomic DNA for naturally competent S. pneumonia (Claverys and Havarstein, 2007). In analogy with these model systems, we have identified two prominent driving forces of cell death (killing factors) responsible for biofilm development in enterococci; secreted gelatinase (GelE) and the soluble autolysin, AtlA (Fig 8). Although cell wall peptidoglycan is refractory toward GelE’s ability to affect turnover of cell surface localized AtlA as compared to a relatively quick turnover in solution, it nevertheless allows limited cleavage potentially resulting in its release from the cell surface. We propose that released AtlA may result in bystander cell death especially in high cell density biofilms, although arguably the extent of such bystander effects will be limited by GelE to small sub-populations within the biofilm. Consistent with this hypothesis, we earlier reported the development of pockets filled with dead/lysed cells within the primary biofilm matt (Thomas et al., 2008).
Figure 8.
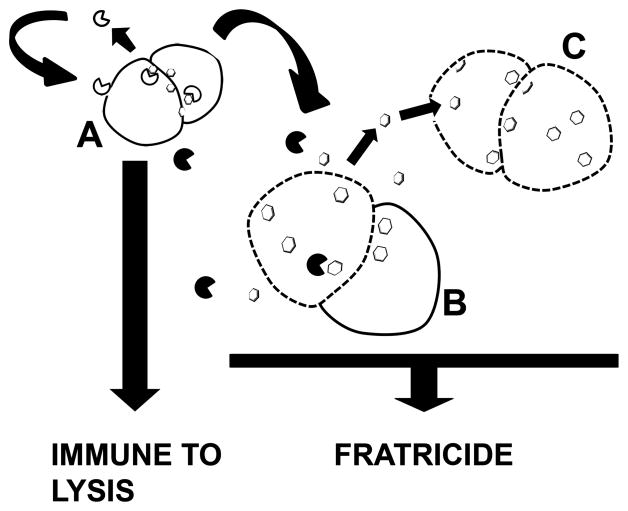
Model of GelE mediated fratricide in Enterococcus faecalis. In response to GBAP quorum sensing, predators (A) co-secrete GelE ( ) and SprE (
) and SprE ( ) extracellular proteases into the immediate environment. GelE and SprE diffuse from the producer cell and if GelE hits the target prior to SprE this event may result in lysis of sibling cells that have not responded to the quorum peptide, prey (B) resulting in the release of AtlA (
) extracellular proteases into the immediate environment. GelE and SprE diffuse from the producer cell and if GelE hits the target prior to SprE this event may result in lysis of sibling cells that have not responded to the quorum peptide, prey (B) resulting in the release of AtlA ( ) from their surface. Alternatively, unbound, but enzymatically active AtlA could bind and lyse neighboring cells (C). The extent of such bystander cell lysis may be directly dependent on the rate of AtlA turnover by GelE. SprE primarily acts as an immunity proteins in predator (protected cells depicted as solid lines) cells by increasing affinity of AtlA to localized subcellular locations on the cell surface (septum) to prevent GelE from activating autolysis. Cells that are damaged by AtlA are depicted with discontinuous lines. Cells that are protected against the lytic action of AtlA are depicted with solid lines.
) from their surface. Alternatively, unbound, but enzymatically active AtlA could bind and lyse neighboring cells (C). The extent of such bystander cell lysis may be directly dependent on the rate of AtlA turnover by GelE. SprE primarily acts as an immunity proteins in predator (protected cells depicted as solid lines) cells by increasing affinity of AtlA to localized subcellular locations on the cell surface (septum) to prevent GelE from activating autolysis. Cells that are damaged by AtlA are depicted with discontinuous lines. Cells that are protected against the lytic action of AtlA are depicted with solid lines.
Biofilm development in Staphylococcus aureus was recently demonstrated to be dependent on cell death and eDNA release (Rice et al., 2007). However, the nature of cell death observed in this case was proposed to be due to altruistic suicide or programmed cell death (PCD) (Bayles, 2007). Altruistic suicide of GelE+ enterococci would be conceptually improbable as over 85% of the population in stationary phase show evidence of their participation in Fsr signaling (Fig 1A) and the Fsr quorum response is known to differentially regulate over 300 genes involved in secondary regulatory cascades, virulence and metabolism (Bourgogne et al., 2006).
How do cells producing GelE ensure their own safety against self-inflicted lysis? Several lines of evidence suggest the co-transcribed serine protease, SprE, to be an immunity protein. First, GFP reporter assays confirm a significant increase in death of predator populations in an isogenic SprE mutated strain compared to the parental strain. Consistent with this observation, co-culture lysis assays which detect solubilized GFP from lysed cells also indicate that secreted SprE has significant trans-protective activities toward prey cells. Second, purified SprE was able to significantly reduce the rate of GelE induced cell lysis. Finally, pretreatment of peptidoglycan bound AtlA with purified SprE significantly reduced its GelE mediated release (Figure S3), a necessity for cell death. Consequently, it may be reasoned that SprE protects predator cells from lysis by modifying surface localized AtlA against further proteolysis by GelE.
Autolysins have consistently been described as regulators of cell death and biofilm development in different gram-positive species, although mechanistic details at a molecular level remain vague. Inactivation of the primary autolysins of Streptococcus mutans, Staphylococcus aureus and Staphylococcus epidermidis have been shown to decrease their abilities to form biofilms, presumably due to a defect in eDNA release (Ahn and Burne, 2006; Biswas et al., 2006; Heilmann et al., 1997; Qin et al., 2007; Shibata et al., 2005). GelE has previously been implicated in the proteolytic processing of a latent high molecular weight E. hirae muramidase 1 (137 kDa) to the active 87 kDa form (Shockman and Cheney, 1969). Although E. faecalis muramidase activities have been characterized for AtlB and AtlC (Mesnage et al., 2008), their prophage origins and existence in the active state as low molecular weight lysins with predicted molecular masses (50 kDa and 47 kDa respectively) suggest that GelE is not required for their activation from latency. On the contrary, zymogram analysis suggests that GelE targets a ~ 50 kDa autolysin of V583 (Fig 3, compare lanes 3 and 4). Whether AtlB identified by Mesnage et al. (Mesnage et al., 2008) corresponds to the ~ 50 kDa band that is only present in the absence of GelE in strain V583 awaits further characterization. However it is tempting to speculate based on protein size, as well as enzymatic activity that GelE may be turning over AtlB. Furthermore, the affect of protease processing on AtlB and AtlC by GelE or SprE would not have been observed in the study by Mesnage et al. (Mesnage et al., 2008) as this study used strain JH2-2, known to be deficient in protease production because of the absence of the fsr locus (Zeng et al., 2005). In spite of a recent study that suggested AtlA, the primary N-actyl glucosaminidase of E. faecalis as not being prone to proteolytic activation (Eckert et al., 2006), we clearly demonstrate that purified GelE activates cellular autolysis in an AtlA dependent manner (compare Fig 2B and Fig 5B). We propose that the observed activation of cellular autolysis may possibly be due to the altered affinity of AtlA to the cell wall of E. faecalis in the presence of GelE and SprE, rather than the activation of latent AtlA. In agreement with this hypothesis, our observations suggest that soluble forms of AtlA are critical to the process of enterococcal fratricide. Of the three domains within AtlA, the C-terminal LysM domain is critical for cell wall localization. Consistent with this observation, we noted that all C-terminal truncated forms of AtlA (that lost one or more LysM modules) resulting from GelE proteolysis also displayed reduced affinity to cell wall peptidoglycan (data not shown). It is noteworthy that the C-terminal truncated forms of AtlA still appear to retain at least four functional LysM modules, based on detectable tryptic fragments from MS analysis. Intriguingly, we also observed a 62 kDa N-terminal form of AtlA with an intact C-terminus that resulted from GelE proteolysis, but displayed significantly decreased peptidoglycan affinity. Although at physiological concentrations, SprE was capable of generating the 62kDa form of AtlA, the presence of cell wall seemed to significantly alter this processing into higher molecular weight truncations (> 62kDa). This is consistent with the predicted cleavage specificity of SprE, given the high glutamic acid content of the N-terminal TE-rich domain.
In light of the present findings, it is likely that in E. faecalis the N-terminal domain of AtlA is also required for efficient adherence to peptidoglycan and that GelE mediates the release of active AtlA from prey cell surfaces by proteolytically processing the N- and C-terminus of AtlA. Furthermore, processing of AtlA by GelE to generate soluble forms of the autolysin may allow localization to cell regions other than the septum leading to the lysis of prey cells. The ability of SprE to process AtlA on predator cell walls may alter its structural conformation potentially targeting AtlA to the septum where its activity would govern cell division rather than lysis. The affinity of SprE-processed AtlA for the cell wall would also prevent GelE from further processing AtlA. (Fig 8). Future studies will address whether proteolytic processing affects AtlA localization on the cell wall and this may provide additional clues as to the role of SprE and GelE in triggering a pro or anti-lytic response. It may also be noted that the relative concentration of GelE in a biofilm would be highest in the vicinity of the predators, and solubilized forms of AtlA have been shown to be much more susceptible to GelE turnover, which may provide an additional control point to ensure that the GelE producer population is not killed by soluble forms of AtlA.
The fsr-dependent quorum sensing may be considered a coordinate cooperative behavior that enterococci employ to produce costly public goods (e.g. proteases) that benefit the whole population. However as with any population that employs cooperative strategies, a threat in the emergence of cheaters that does not contribute to the cost of public goods, but benefit from them is very high (Diggle et al., 2007; West et al., 2006). The occurrences of quorum sensing cheaters have previously been reported among different bacterial communities (Diggle et al., 2007; Sandoz et al., 2007). Within the limits of our experimental conditions (see Materials and Methods) we have estimated approximately 15% of the population in the stationary phase of growth to be comprised of cheaters (Fig 1A) that do not respond to GBAP (as these quorum non-responders potentially benefit from nutrients generated from proteolytic activity of the remaining majority). This relatively high percentage of GBAP non responders is surprising considering the fact that by the time enterococci enter into the log phase, they have already secreted nano-molar concentrations of GBAP enough to activate fsr signaling within every cell of the population (Nakayama et al., 2001). Although, previous studies have implied the absence of a 23.9 kB region within the fsr locus (Eaton and Gasson, 2001; Nakayama et al., 2002)as a possible contributor to the rise of cheaters, this is unlikely to be the case in our experimental set up and hence more investigations are necessary to further characterize the mechanisms of cheater development among enterococcal populations. Although arguably cheaters may enjoy a fitness advantage over their cooperative siblings (as they do not share the same metabolic burden), our observations suggest the contrary wherein co-culturing the wild-type V583 with its isogenic FsrA quorum sensing mutant (cheaters) or a double protease mutant, led to ~12% decrease in the cheater population after overnight growth (Table S1). Based on these observations, it is tempting to speculate that fratricide may have evolved among cooperative enterococci as a way to police cheaters and prevent them from taking over the entire population.
Experimental procedures
Bacterial strains, plasmids and growth conditions
Relevant bacterial strains and plasmids used in the present study are listed in Table 1. Strains were cultured in Todd-Hewitt Broth (THB) or M17 media and grown as standing cultures at 37°C unless otherwise indicated. Escherichia coli Electro-Ten Blue was used for maintenance and propagation of plasmid constructions. Clones were cultured aerobically in Luria-Bertani broth at 37°C. The antibiotics used for selection included chloramphenicol at 10 μg/ml and spectinomycin at 750 μg/ml. Electrotransformations of E. faecalis were performed as previously described (Cruz-Rodz and Gilmore, 1990).
Table 1.
Bacterial strains and plasmids
| Strains | Genotype/relevant phenotype | Origin |
|---|---|---|
| E. faecalis | ||
| OG1RF | Clinical isolate | (Murray et al., 1993) |
| V583 | Clinical isolate, TIGR sequence strain; VnR | (Thomas et al., 2008) |
| VT01 | V583ΔgelE; SprE+ | (Thomas et al., 2008) |
| VT02 | V583ΔsprE; GelE+ | (Thomas et al., 2008) |
| VT03 | V583ΔgelE-sprE | (Thomas et al., 2008) |
| VT12 | VT03 (pMV158GFP); GFP+ TetR | (Thomas et al., 2008) |
| VT13 | V583::fsrA (pVT31); CmR, SpcR | This study |
| VT14 | V583 (pVT30); SpcR | This study |
| VT15 | V583 (pVT31); GFP+ SpcR | This study |
| VT16 | VT01 (pVT31); GFP+ SpcR | This study |
| VT17 | VT02 (pVT31); GFP+ SpcR | This study |
| VT18 | VT03 (pVT31); GFP+ SpcR | This study |
| VT19 | V583:: atlA; CmR | This study |
| VT20 | OG1RFΔgelE; SprE+ | This study |
| VT21 | OG1RF:: atlA; CmR | This study |
| VT22 | VT03::atlA; CmR | This study |
| VT23 | VT12::atlA (pMV158GFP); GFP+, CmR, TetR | This study |
| VT24 | VT03 (pVT08); SprE+ | This study |
| KS10 | V583ΔfsrA | Gorman, M. and Hancock, L.E., (unpublished) |
| E. coli | ||
| Electro Ten Blue | General plasmid maintenance strain | Stratagene |
| BL21 (DE3) | Protein expression strain | Stratagene |
| VT25 | BL21(DE3) (pVT21; AtlA+ * AmpR | This study |
| Plasmids | ||
| pAT28 | Broad host range shuttle vector; SpcR | (Trieu-Cuot et al., 1990) |
| p3CAT | p3TET derivative (Hancock and Perego, 2004), suicide vector; CmR, used for targeted gene inactivation | Hancock, LE (unpublished) |
| pMV158GFP | Gram-positive replicative vector expressing a GFP reporter | (Nieto and Espinosa, 2003) |
| pML115 | p3CAT derivative carrying an internal fragment of fsrA; generates fsrA::cm mutation in E. faecalis | Hancock, L.E (unpublished) |
| pVI01 | Low copy gelE promoter-gfp fusion reporter construct | Iyer, V and Hancock, L.E (unpublished) |
| pVT08 | pAT28 derivative carrying a 2123-bpEcoRI/BamHI fragment (native gelE promoter and full-length sprE); leads to the over expression of SprE | (Thomas et al., 2008) |
| pVT10 | p3CAT derivative carrying a 1006-bp EcoR1/BamH1 internal fragment of atlA; generates atlA::cm mutation in E. faecalis | This study |
| pVT21 | pET21b derivative carrying a 2056-bp Nde1/Xho1 atlA fragment devoid of signal peptide; used for the expression of AtlA in E. coli | This study |
| pVT30 | pAT28 derivative carrying a 759-bp SalI/SalI gfp fragment; promoterless gfp | This study |
| pVT31 | pAT28 derivative carrying a gelE promoter-gfp fusion; reporter construct that allows discrimination of predators within E. faecalis population | This study |
Resistant
Vn, vancomycin; Cm, chloramphenicol; Spc, spectinomycin; Tet, tetracycline; Amp, ampicllin
AtlA devoid of signal peptide (53 amino acid residues from the N-terminus)
Targeted gene mutagenesis
Construction of VT01, VT02 and VT03 are as described previously (Thomas et al., 2008). AtlA mutants were constructed by targeted insertional mutagenesis. An internal fragment of atlA was PCR amplified using primers Aut2f and Aut2r (Table 2) and cloned EcoRI/BamHI into the suicide vector (p3CAT). The resulting construct was verified by restriction analysis and was electroporated into E. faecalis V583 and OG1RF. Growth of transformants on antibiotic selective media (chloramphenicol, 10 μg/ml) followed by colony PCR with primers Auto2Up and M13R, confirmed the targeted mutation.
Table 2.
Primers used in this study
| Primer name | Sequence (5′ →3′) |
|---|---|
| M13R | CAGCTATGACCATGATTACG |
| Aut2-f | GAGAGAATTCTGGGGAGCAAGTACGCTATC |
| Aut2-r | CTCTGGATCCCCACCGGTGTTACCTGAAGT |
| Auto2Up | GAGACACCAACAACAGAA |
| Ef0799-SP-NdeI | GAGACATATGACAGAAGAGCAGCCAACAAATGC |
| Ef0799XhoI | GAGACTCGAGACCAACTTTTAAAGTTTGAC |
| VLAC1 | GTTGAATAACACTTATTCCTATC |
| Gfp3′SalI | CTCTGTCGACTACGAATGCTATTTGTATAG |
Flow Cytometry analysis
A 1119-bp fragment composed of a gelE promoter gfp fusion, was PCR amplified from pVI01 (Table 1) using primers VLAC1 and Gfp3′SalI (Table 2). The fragment was either restricted with EcoRI resulting in a 1055-bp fragment or with SalI giving rise to a 759-bp fragment. The 759-bp fragment containing the gfp was cloned into SalI restricted pAT28 resulting in pVT30, and the 1055-bp fragment was cloned into EcoR1/SmaI restricted pAT28 giving rise to pVT31. E. faecalis V583, VT01, VT02 and VT03 were transformed with pVT31 to track cells within the population that responded to GBAP and expressed GFP. The resulting strains were designated VT15, VT16, VT17 and VT18 respectively. pVT30 which contains promoterless gfp was electroporated into V583 as a negative control. Flow cytometric analysis was performed using one-day old stationary phase cultures of E. faecalis on a FACSCalibur flow cytometer (Beckton and Dickinson, San Jose, California). Cell samples were washed twice and diluted to a final concentration of 106 cells per ml in PBS. Cells were stained for 10 minutes with propidium iodide (2 μM) and analyses was carried out at a flow rate of ~2000 cells per s. In total 50,000 events were collected for each sample and FSC, SSC, FL-1 and FL-2 signals were measured using logarithmic amplifications. Bacteria were discriminated from background using a combination of FSC and SSC. Data were analyzed with the CELLQuest program (version 3.1, Beckton and Dickinson).
Zymography
An 8% SDS PAGE gel containing 0.1% (w/v) of prepared E. faecalis V583 cell wall as substrate was used to analyze autolysin activity. Gels were run under constant voltage (200V). Following electrophoresis, gels were washed extensively with deionized water to remove SDS and then in renaturation buffer (25 mM sodium phosphate pH 7.0, 1 mM MgCl2 and 1% TritonX-100) four times at intervals of one hour each and then incubated in fresh buffer overnight at 37°C. Activity was detected as clear bands after counter-staining the gel with 0.1% methylene blue in 0.01% KOH solution followed by destaining in deionized water.
Protein expression and purification
Gelatinase (GelE) was purified as previously described (Hancock and Perego, 2004). For purification of serine protease (SprE), two liters of THB was inoculated with 20 ml of an overnight culture of E. faecalis strain VT24 (ΔgelEsprE, pVT08, SprE+) and allowed to grow as a standing culture at 37°C for 24 h. Following separation of bacteria by centrifugation (30 min at 27,500 × g at 4°C), SprE was precipitated from the cell free culture supernatant by gradual addition of ammonium sulfate to a final saturation of 70% and incubated overnight at 4°C with constant agitation. The precipitants were removed by centrifugation (30 min at 27,500 × g at 4°C) and resuspended in 200 ml of 200 mM Tris-HCl and 5 mM CaCl2, pH 7.6. The 200 ml sample was filter-sterilized (0.2μm) and extensively dialyzed (2 weeks) against dialysis buffer (50 mM Tris-HCl and 5 mM CaCl2, pH 7.6). The sample was concentrated (Pierce ICON™ concentrator) down to 2 ml and was resolved at room temperature on a TSK G3000SW gel filtration column (7.8 mm × 300 mm; Tosoh Bioscience, Montgomeryville, PA) at a flow rate of 1 ml/min, using TSK buffer (0.06 M sodium phosphate, 0.1 M sodium sulfate, pH 7.0) and stored at 4°C.
Histidine tagged AtlA (devoid of signal sequence) was purified from Escherichia coli BL21 (DE3) harboring the expression construct pVT21 and grown in 1 L LB medium containing ampicillin (100μg/ml). Expression of recombinant protein was induced by the addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) to the E. coli cultures at an optical density (at 600 nm) of 0.6. Following IPTG treatment cultures were incubated for an additional 3 hours at 37°C after which bacterial cells were harvested by centrifugation and resuspended in 40 ml lysis buffer (20 mM Tris-HCl, 0.5 M NaCl, 10 mM imidazole, 20 mM-mercaptoethanol (pH 7.9). Cells were disrupted by ultra sonicaton (5 cycles of intermittent 20 sec pulse and 20 sec cooling) and the supernatant retrieved after ultracentrifugation at 45,000 rpm for 1 h at 4°C to remove cell debris. The supernatant was passed through a 0.2μm filter prior to being loaded onto a nickel nitrilotriacetic acid-agarose column (Qiagen). The His-tagged AtlA was eluted with a step-wise gradient of imidazole (20, 40, 100, and 250 mM) in lysis buffer and optimal purity obtained at 100 mM concentration of imidazole in lysis buffer. Purified AtlA was concentrated and buffer exchange (25 mM sodium phosphate buffer, pH 7) carried out using an ICON™ concentrator (Pierce). Purified protein was stored at 4°C for short term or −80°C for long term storage. Protein purity was analyzed by sodium dodecyl sulfate (SDS)-acrylamide gel electrophoresis (>95% for all purified proteins) and tryptic digests of the proteins followed by MALDI-TOF mass spectrometry confirmed their identities.
MS and MS/MS analysis
After staining gels with Coomassie R-250, selected gel bands were excised manually as 1–2 mm slices and transferred to 1.5 ml Eppendorf tubes. An equivalent slice from the protein-free region of the gel was excised as a background control. The control and test gel sections were then destained using three 30 min washes of 60 μl 1:1 acetonitrile: water at 30°C. Gel pieces were then dried for 10 min under vacuum. The gels were subjected to reduction and alkylation using 50 mM Tris (2-carboxyethyl) phosphine (TCEP) at 55°C for 10 min followed by 100 mM iodoacetamide in the dark at 30°C for 60 min. The carboxymethylated samples were thoroughly washed and redried in vacuo, then incubated with sequencing grade trypsin (Trypsin Gold, Promega, Madison, WI), 20 ng/μl in 40 mM ammonium bicarbonate, in 20 μl. Upon rehydration of the gels, an additional 15 μl of 40 mM ammonium bicarbonate and 10% acetonitrile was added, and gel sections incubated at 30°C for 17 h in sealed Eppendorf tubes. The aqueous digestion solutions were transferred to 1.5 ml clean Eppendorf tubes, and those tryptic fragments remaining within the gel sections were recovered by a single extraction with 50 μl of 50% acetonitrile in water containing 2% trifluoracetic acid (TFA) at 30°C for 1 h. The acetonitrile-water fractions were combined with the previous aqueous fractions and the liquid removed by speed vacuum concentration. The dried samples were resuspended in 10 μl of 30 mg/ml 2,5-dihydroxylbenzonic acid (DHB) (Sigma, St. Louis, MO) dissolved in 33% acetonitrile/0.1% TFA and 2 μl of peptide/matrix solution was applied to a Bruker aluminum target plate for MALDI-TOF and TOF/TOF analysis.
Mass spectra and tandem mass spectra were obtained on a Bluker Ultraflex II TOF/TOF mass spectrometer. Positively charged ions were analyzed in the reflector mode. MS and MS/MS spectra were analyzed with Flex analysis 3.0 and Bio Tools 3.0 software (Bruker Daltonics). Measurements were externally calibrated with 8 different peptides ranging from 757.39 to 3147.47 (Peptide Calibration Standard I, Bruker Daltonics) and internally re-calibrated with peptides from the autoproteolysis of trypsin.
Peptide ion search was performed using MASCOT software (Matrix Science) against the expected recombinant AtlA protein sequence. The following parameters were used for the database search: MS and MS/MS accuracies were set to < 0.6 Da. Trypsin as an enzyme, missed cleavages 1, carbamidomethylation of cysteine as fixed modification and oxidation of methionine as a variable modification.
GelE and SprE mediated proteolysis of AtlA
GelE or SprE (30nM) were incubated with 10μg of AtlA separately in 100 μl of reaction buffer (50 mM Tris-HCl and 5 mM CaCl2, pH 7.6). After 10, 30, 60 and 300 mins of incubation at 37°C, aliquots (25μl) were withdrawn and the reaction was stopped by the addition of SDS-PAGE sample loading buffer and boiling for 10 minutes. Samples were analyzed by sodium dodecyl sulfate (SDS)-acrylamide gel electrophoresis and silver staining. MALDI-TOF MS and MS/MS analysis was carried out as previously described, to empirically map regions of AtlA that were susceptible to proteolytic attack.
Biofilm assay
Biofilm assays were carried out as described previously (Thomas et al., 2008).
eDNA release assay
Measurements of eDNA release were carried out on stationary phase cultures using the nucleic acid stain SYTOX green (Invitrogen). Overnight grown cultures of E. faecalis (37°C) were centrifuged (13,000 × g; 3 minutes) to pellet cells and 200 μl of the culture supernatants were transferred to microtiter plate wells in triplicate. SYTOX green was supplemented to these wells to a final concentration of 1 μM and incubated for 10 minutes before being spectrofluorometrically measured with excitation at 485 nm and emission at 535 nm on a Perkin Elmer Victor 3 fluorescent plate reader.
Fratricide assay
Co-culture lysis assays were carried out using V583, VT01 (ΔgelE), VT02 (ΔsprE), VT03 (ΔgelEsprE) and VT19 (V583::atlA) as attacker strains and either VT12 (ΔgelEsprE, GFP+) or VT23 (ΔgelEsprE,::atlA, GFP+) as targets. Attacker strains grown overnight were diluted 1:1000 in fresh Todd Hewitt broth (THB) media and were co-inoculated with target strains diluted 1:20 from an overnight culture grown for an equivalent period at 37°C in THBG (THB + 2% glycine). Following 24 hours of co-culture at 37°C, GFP (released as a result of lysis of target) from 250 μL of supernatant was precipitated using 4 volumes of cold acetone (−20°C) and resolved by SDS-PAGE. Detection of GFP in the supernatant was carried out by western blot analysis using anti-GFP antibodies.
Lysis assay
Lysis assays were carried out as described previously with the following modifications (Thomas et al., 2008). Briefly, pre-cultures of VT03 (ΔgelEsprE)or VT22 (ΔgelEsprE,::atlA) were grown overnight at 37°C in 2.5 ml THB and diluted 100-fold in SM17 media supplemented with 3% glycine. Following overnight growth at 37°C, 1.5 mL cultures were centrifuged at 13,000 rpm for 3 minutes and washed thoroughly, thrice in ice-cold sterile distilled water. After the third wash, the cells were resuspended in 10 mM sodium phosphate buffer (pH 6.8) and supplemented with 1 mM CaCl2. Two hundred microliters of the suspended cells were dispensed into a 96 well plate and the optical density at 600 nm was monitored for 9 h at 30-min intervals either in the presence or absence of protease treatments (30 nM of either GelE or SprE or both).
Peptidoglycan affinity assay
Purified recombinant AtlA (10 μg) was allowed to bind with 20 mg wet-weight of isolated E. faecalis V583 cell wall for 15 minutes at 37°C in 100 μL of buffer (10 mM Tris-HCl and 5 mM CaCl2, pH 7.6). Approximately, 30 nM of either GelE or SprE was supplemented separately into each sample mix. For co-protease treatments recombinant AtlA bound to V583 peptidoglycan (20mg wet-weight) were either pretreated with 30 nM GelE or SprE for 10 minutes. This was followed by the immediate addition of the second complementary protease (30 nM; SprE or GelE) for 30 minutes. Samples (25 μL) were withdrawn at intervals of 10, 30, 60 and 300 minutes. Supernatants containing unbound proteolytically processed AtlA derivatives were retrieved after centrifugation (13,000 × g; 10 minutes). The pellet was washed twice in fresh buffer to remove loosely adherent AtlA from pellet and the cell wall bound fraction eluted in SDS PAGE sample loading buffer. Unbound and bound AtlA derivatives were resolved by SDS PAGE and visualized by silver staining.
Acknowledgments
We are very grateful to Tammy Koopman, Melinda J Wilkerson and Sherry Fleming for support with flow cytometry. We also thank John M Pfeffer and Anthony J Clarke for help with zymography. Finally we are indebted to Yu Xue and Jian Ren for help with using the protein domain visualization software DOG 1.0. This study was supported by a Heartland Affiliate Beginning Grant-in-Aid 0660072Z and 0860084Z from the American Heart Association (to L.E.H), a grant-in-aid from the Terry C. Johnson Cancer Center at Kansas State University (to V.C.T), NSF-MRI grant-0521587 (to J.T) and the Targeted Excellence Program at K-State, Functional Genomics Consortium (to J.T), and a training award (to N.H.) from NIH Grant # P20 RR16475 from the INBRE program of the National Center for Research Resources.
References
- Ahn SJ, Burne RA. The atlA operon of Streptococcus mutans: role in autolysin maturation and cell surface biogenesis. J Bacteriol. 2006;188:6877–6888. doi: 10.1128/JB.00536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayles KW. The biological role of death and lysis in biofilm development. Nat Rev Microbiol. 2007;5:721–726. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- Biswas R, Voggu L, Simon UK, Hentschel P, Thumm G, Gotz F. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol Lett. 2006;259:260–268. doi: 10.1111/j.1574-6968.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- Bourgogne A, Hilsenbeck SG, Dunny GM, Murray BE. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J Bacteriol. 2006;188:2875–2884. doi: 10.1128/JB.188.8.2875-2884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgogne A, Garsin DA, Qin X, Singh KV, Sillanpaa J, Yerrapragada S, Ding Y, Dugan-Rocha S, Buhay C, Shen H, Chen G, Williams G, Muzny D, Maadani A, Fox KA, Gioia J, Chen L, Shang Y, Arias CA, Nallapareddy SR, Zhao M, Prakash VP, Chowdhury S, Jiang H, Gibbs RA, Murray BE, Highlander SK, Weinstock GM. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 2008;9:R110. doi: 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carniol K, Gilmore MS. Signal transduction, quorum-sensing, and extracellular protease activity in Enterococcus faecalis biofilm formation. J Bacteriol. 2004;186:8161–8163. doi: 10.1128/JB.186.24.8161-8163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC, Downes FP, Shah S, Rudrik JT, Pupp GR, Brown WJ, Cardo D, Fridkin SK. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med. 2003;348:1342–1347. doi: 10.1056/NEJMoa025025. [DOI] [PubMed] [Google Scholar]
- Claverys JP, Havarstein LS. Cannibalism and fratricide: mechanisms and raisons d’etre. Nat Rev Microbiol. 2007;5:219–229. doi: 10.1038/nrmicro1613. [DOI] [PubMed] [Google Scholar]
- Cruz-Rodz AL, Gilmore MS. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol Gen Genet. 1990;224:152–154. doi: 10.1007/BF00259462. [DOI] [PubMed] [Google Scholar]
- Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol. 2001;67:1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert C, Lecerf M, Dubost L, Arthur M, Mesnage S. Functional analysis of AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis. J Bacteriol. 2006;188:8513–8519. doi: 10.1128/JB.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Gilmore MS, Haas W. The selective advantage of microbial fratricide. Proc Natl Acad Sci U S A. 2005;102:8401–8402. doi: 10.1073/pnas.0503828102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- Guiral S, Mitchell TJ, Martin B, Claverys JP. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci U S A. 2005;102:8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock LE, Perego M. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J Bacteriol. 2004;186:5629–5639. doi: 10.1128/JB.186.17.5629-5639.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock LE, Perego M. Systematic inactivation and phenotypic characterization of two-component signal transduction systems in Enterococcus faecalis V583. J Bacteriol. 2004;186:7951–7958. doi: 10.1128/JB.186.23.7951-7958.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havarstein LS, Martin B, Johnsborg O, Granadel C, Claverys JP. New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol Microbiol. 2006;59:1297–1307. doi: 10.1111/j.1365-2958.2005.05021.x. [DOI] [PubMed] [Google Scholar]
- Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- Jayaraman A, Wood TK. Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Annu Rev Biomed Eng. 2008;10:145–167. doi: 10.1146/annurev.bioeng.10.061807.160536. [DOI] [PubMed] [Google Scholar]
- Kristich CJ, Li YH, Cvitkovitch DG, Dunny GM. Esp-independent biofilm formation by Enterococcus faecalis. J Bacteriol. 2004;186:154–163. doi: 10.1128/JB.186.1.154-163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristich CJ, Nguyen VT, Le T, Barnes AM, Grindle S, Dunny GM. Development and use of an efficient system for random mariner transposon mutagenesis to identify novel genetic determinants of biofilm formation in the core Enterococcus faecalis genome. Appl Environ Microbiol. 2008;74:3377–3386. doi: 10.1128/AEM.02665-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen PL, Clewell DB, An F, Makinen KK. Purification and substrate specificity of a strongly hydrophobic extracellular metalloendopeptidase (“gelatinase”) from Streptococcus faecalis (strain 0G1-10) J Biol Chem. 1989;264:3325–3334. [PubMed] [Google Scholar]
- Mesnage S, Chau F, Dubost L, Arthur M. Role of N-acetylglucosaminidase and N-acetylmuramidase activities in Enterococcus faecalis peptidoglycan metabolism. J Biol Chem. 2008;283:19845–19853. doi: 10.1074/jbc.M802323200. [DOI] [PubMed] [Google Scholar]
- Mohamed JA, Huang W, Nallapareddy SR, Teng F, Murray BE. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect Immun. 2004;72:3658–3663. doi: 10.1128/IAI.72.6.3658-3663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray BE, Singh KV, Ross RP, Heath JD, Dunny GM, Weinstock GM. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J Bacteriol. 1993;175:5216–5223. doi: 10.1128/jb.175.16.5216-5223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Cao Y, Horii T, Sakuda S, Akkermans AD, de Vos WM, Nagasawa H. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol Microbiol. 2001;41:145–154. doi: 10.1046/j.1365-2958.2001.02486.x. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Kariyama R, Kumon H. Description of a 23.9-kilobase chromosomal deletion containing a region encoding fsr genes which mainly determines the gelatinase-negative phenotype of clinical isolates of Enterococcus faecalis in urine. Appl Environ Microbiol. 2002;68:3152–3155. doi: 10.1128/AEM.68.6.3152-3155.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Chen S, Oyama N, Nishiguchi K, Azab EA, Tanaka E, Kariyama R, Sonomoto K. Revised model for Enterococcus faecalis fsr quorum-sensing system: the small open reading frame fsrD encodes the gelatinase biosynthesis-activating pheromone propeptide corresponding to staphylococcal agrD. J Bacteriol. 2006;188:8321–8326. doi: 10.1128/JB.00865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C, Espinosa M. Construction of the mobilizable plasmid pMV158GFP, a derivative of pMV158 that carries the gene encoding the green fluorescent protein. Plasmid. 2003;49:281–285. doi: 10.1016/s0147-619x(03)00020-9. [DOI] [PubMed] [Google Scholar]
- Noble WC, Virani Z, Cree RG. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett. 1992;72:195–198. doi: 10.1016/0378-1097(92)90528-v. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- Qin X, Singh KV, Xu Y, Weinstock GM, Murray BE. Effect of disruption of a gene encoding an autolysin of Enterococcus faecalis OG1RF. Antimicrob Agents Chemother. 1998;42:2883–2888. doi: 10.1128/aac.42.11.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Singh KV, Weinstock GM, Murray BE. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect Immun. 2000;68:2579–2586. doi: 10.1128/iai.68.5.2579-2586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. DOG 1.0: illustrator of protein domain structures. Cell Res. 2009;19:271–273. doi: 10.1038/cr.2009.6. [DOI] [PubMed] [Google Scholar]
- Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoz KM, Mitzimberg SM, Schuster M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci U S A. 2007;104:15876–15881. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Kawada M, Nakano Y, Toyoshima K, Yamashita Y. Identification and characterization of an autolysin-encoding gene of Streptococcus mutans. Infect Immun. 2005;73:3512–3520. doi: 10.1128/IAI.73.6.3512-3520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockman GD, Cheney MC. Autolytic enzyme system of Streptococcus faecalis. V. Nature of the autolysin-cell wall complex and its relationship to properties of the autolytic enzyme of Streptococcus faecalis. J Bacteriol. 1969;98:1199–1207. doi: 10.1128/jb.98.3.1199-1207.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KV, Nallapareddy SR, Nannini EC, Murray BE. Fsr-independent production of protease(s) may explain the lack of attenuation of an Enterococcus faecalis fsr mutant versus a gelE-sprE mutant in induction of endocarditis. Infect Immun. 2005;73:4888–4894. doi: 10.1128/IAI.73.8.4888-4894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen A, Buist G, Kramer NE, Jalving R, Benus GF, Venema G, Kuipers OP, Kok J. Reduced lysis upon growth of Lactococcus lactis on galactose is a consequence of decreased binding of the autolysin AcmA. Appl Environ Microbiol. 2008;74:4671–4679. doi: 10.1128/AEM.00103-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas VC, Thurlow LR, Boyle D, Hancock LE. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J Bacteriol. 2008;190:5690–5698. doi: 10.1128/JB.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 1990;18:4296. doi: 10.1093/nar/18.14.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Antiporta MH, Murray BE, Dunny GM. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J Bacteriol. 2003;185:3613–3623. doi: 10.1128/JB.185.12.3613-3623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- Zeng J, Teng F, Murray BE. Gelatinase is important for translocation of Enterococcus faecalis across polarized human enterocyte-like T84 cells. Infect Immun. 2005;73:1606–1612. doi: 10.1128/IAI.73.3.1606-1612.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]