Abstract
Somatic hypermutation (SHM) normally occurs as a consequence of the expression of activation-induced cytidine deaminase (AID) by antigen-activated, mature B cells during T cell-dependent (Td) germinal center (GC) responses. Nonetheless, despite their inability to express CD154 and initiate GC responses, patients with type-I hyper IgM syndrome (HIGM1), support populations of IgM+IgD+CD27+ B cells that express mutated Ig genes. The origin of these mutated B cells is unknown; the IgM+IgD+CD27+ cells do not express AID and appear to acquire mutations independent of stringent selection by Ag. Here we demonstrate that immature/transitional 1 (im/T1) B cells from the bone marrow of CD154 deficient mice express AID and acquire Ig mutations that lack the hallmarks of antigenic selection via BCR signaling. Comparable levels of AID expression was found in developmentally immature B cells recovered from murine fetal liver and from human im/T1 B cells recovered from umbilical cord blood. AID expression in human fetal liver was also robust, approaching that of human tonsil tissue and the human GC B cell line, Ramos. These observations lead us to conclude that AID expression in developing human B cells is the origin of the mutated IgM+IgD+CD27+ B cells present in HIGM1 patients and we propose that both mice and human share a latent, AID-dependent pathway for the pre-immune diversification of B lymphocytes that is more prominent in chicken, sheep, and rabbits.
Keywords: immunodeficiency diseases, B cells, AID, antibodies, repertoire development
Introduction
Somatic hypermutation (SHM)3 mediated by activation-induced cytidine deaminase (AID) introduces high frequencies of point mutations into the rearranged Ig genes of activated B cells (1–4). Remarkably, AID also mediates Ig class-switch recombination (CSR) and gene conversion (3, 4).
AID expression, SHM and CSR are thought to be confined largely to GC B cells present in the follicles of secondary lymphoid tissues (4) where they are crucial for adaptive immunity. Ig mutations introduced by AID are selected in GC for increased affinity to Ag and promote entry into the B-cell memory compartments by clonal competition (5, 6). Importantly, AID activity incurs significant costs of genomic damage that can lead to cell death and even oncogenic transformation (7, 8).
There is evidence, however, for SHM outside of the GC microenvironment. For example, Shlomchik and colleagues have reported active SHM in plasmablast-like cells located in the splenic bridging channels of autoimmune mice (9). Other laboratories (10, 11), including ours (12), have detected low levels of AID and evidence for SHM (and CSR) in immature and transitional 1 (im/T1) mouse B cells; evidence for Ig SHM and CSR has been recovered from human fetal liver, as well (13, 14). The most persuasive evidence for SHM outside of GC, however, comes from patients with type 1 hyper IgM syndrome (HIGM1) (15, 16). HIGM1 patients do not express functional CD154 and can not support cognate T:B interactions. Consequently, these patients have normal/elevated levels of serum IgM but little IgG, are unable to generate T-cell dependent (Td) Ab responses, lack GC, and show little or no capacity for antigen dependent SHM, CSR, and humoral immune memory (15, 16).
The GC reaction is characteristic of Td humoral responses and relies on continuing interactions between Ag-specific B- and T cells mediated by CD40 and CD154 (17–20). Interruption of CD40:CD154 interactions prevents or abrogates the GC reaction (20–22) as does genetic blocks of CD154 expression (18). Humans with HIGM1 carry genetic defects that impair CD154 expression (15, 16) and are susceptible to opportunistic infections (23). Surprisingly, HIGM1 patients often exhibit systemic autoimmunity (23), suggesting that B-cell tolerance may be improperly regulated (24).
Despite the virtual absence of GC in secondary lymphoid tissues (16), many HIGM1 patients possess a subset of circulating IgM+IgD+CD27+ B cells carrying mutated V(D)J rearrangements (25–28), a cellular phenotype associated with IgM memory B cells (29, 30). Nonetheless, the origin(s) of these B cells and their acquisition of V(D)J mutations are controversial.
Some investigators have concluded that IgM+IgD+CD27+ B cells in HIGM1 patients represent un-switched memory B cells derived from cryptic GC (31, 32), a conclusion consistent with the general restriction of SHM to GC B cells and identification of CD27 as a marker of post-GC memory B lymphocytes (29, 30). In contrast, Weller et al. (28) have suggested that these mutated IgM+IgD+CD27+ B cells are not memory cells but circulating precursors of marginal zone (MZ) B cells, noting that the mutated IgM+IgD+CD27+ B cells from HIGM1 patients exhibit a highly diverse Ig repertoire and mutations that do not appear to be Ag-selected (33). Recent evidence indicates that SHM in HIGM1 patients arises independently of both Td (14) and T-cell independent (Ti) humoral responses (33) and that mutated IgM+IgD+CD27+ B cells may be recovered from human fetal spleen (14), observations that suggest the possibility of physiologic SHM during the development of human B-lineage cells.
Programmed AID expression during B-cell development is found in a diverse collection of unrelated vertebrate species and the primary repertoires of B cells in birds (34–36), rabbits (37, 38), and sheep (39) are significantly modified by AID-dependent SHM or gene conversion. Although the primary B-cell repertoires of mice and humans are generated virtually wholly by combinatorial association, the low levels of AID observed in im/T1 B cells (10–12, 40) may represent a conserved developmental program that is active in some species (e.g., birds and sheep) and latent in others (mouse and human).
Here we show that im/T1 B cells from the bone marrow (BM) of CD154−/− mice constitutively express AID and exhibit significant SHM in rearranged VH gene segments. These mutant B cells do not display evidence for antigen-driven clonal selection and are unlikely to represent GC B lymphocytes given the absence of serum Ab responses to Td Ag and lack of splenic and Peyer’s patch GC in CD154−/− mice. Parallel studies of human im/T1 B cells from umbilical cord blood and fetal liver also demonstrated significant levels of AID expression at levels comparable to or above that of murine im/T1 B cells.
We propose that developmentally-regulated AID expression in im/T1 B cells represents a phylogenetically conserved program for Ig diversification that is normally latent in mice and humans but may be revealed in the absence of CD154-dependent regulation.
Materials and Methods
Human cord blood samples
Fresh cord blood samples were harvested from healthy term placentas and were provided without identifiers for the quantification of human AID mRNA by quantitative RT-PCR. Informed consent for donation to a public cord blood bank was obtained from mothers as part of a protocol approved by the Duke University Institutional Review Board.
Mice
Female C57BL/6 (BL/6), congenic AID−/− (4) and CD154−/− (19) mice were bred and maintained under specific pathogen-free conditions at the Duke University Animal Care Facility. Mice used in experiments were 6- to 12-wk of age. All experiments involving animals were approved by the Duke University Institutional Animal Care and Use Committee.
Cell lines
The Abelson murine leukemia virus (AMuLV)-transformed cell lines SJLxB6 (gift of Y. Zhuang), C12, PD31 (41), KG7, 63-12 (42) and 220-8 (43) (gifts of M. Schlissel), or Ramos cell lines (44) (gifts of M. Scharff) were blindly subcloned; paired clones were subsequently used to quantify AID expression by quantitative RT-PCR.
Ag and immunizations
Mice were immunized i.p. with 100 μg (3-hydroxy-4-nitrophenyl)acetyl (NP)- chicken γ-globulin (CGG) in alum. NP-CGG contained 10–15 mol NP/mol CGG. Mice were killed 16 days after immunization, and BM and spleen were harvested for flow cytometry and/or histology.
Flow cytometry
Analysis of cell phenotype and cell sorting were performed by flow cytometry. Human B cells from umbilical cord blood were enriched by negative selection using the RosetteSep B-cell enrichment reagent (StemCell Technologies, Vancouver, BC, Canada) and labeled with fluorochrome-conjugated mAb specific for human cell surface molecules. FITC-, PE-, PECy5-, APC-, APC-Cy7-, Pacific Blue-conjugated mAb specific for human IgM, IgD, CD3, CD16, CD235a, CD34, CD19, CD27 (BD Bioscience), and CD14 (CalTag) were used to identify human T1, transitional 2 (T2), and mature follicular (MF) B cell compartments (45). Murine fetal liver samples were prepared from embryo 16 days and 19 days of gestation. Murine pro-/pre-, im/T1, MF, MZ, and GC B cells from fetal liver, BM, Peyer’s patch (PP) and spleen were identified and sorted as described (12). Labeled cells were analyzed/sorted in a FACS Vantage with DIVA option (BD Bioscience) or FACS Aria (BD Bioscience). Flow cytometric data were analyzed with FlowJo software (Treestar Inc., Palo Alto, CA).
Immunohistochemistry
Tissues from naïve and immunized mice were embedded in Tissue-Tek O.C.T. compound (Sakura Finetek, Torrance, CA) and snap-frozen in 2-methylbutane cooled with liquid nitrogen. Frozen tissues were stored at −80°C. Serial 5-μm- thick cryosections were cut on a LEICA CM3050 S cryostat (Leica Microsystems, Bannockburn, IL) and thaw mounted onto Superfrost Plus slides (Thermo scientific, Portsmouth, NH). After air drying, sections were fixed with ice-cold acetone/methanol (1:1) at −20°C for 10 min and stored at −80°C until use. Frozen sections were rehydrated and then blocked with PBS containing 5% normal goat serum, 5% rat serum and 100 μg/ml of rat IgG (Pierce) before immunolabeling. Sections were stained with a combination of fluorochrome-conjugated mAb specific for mouse IgD, IgM, GL-7, TCRβ, and/or CD93 in blocking buffer at 4°C overnight. After washing, stained sections were mounted in Fluoromount-G (Southern Biotech) and examined with an Axiovert 200M microscope. Representative images were photographed and analyzed with Axiocam MRm and AxioVision AC 4.5 software (Carl Zeiss, Thornwood, NY).
ELISA
Serum Ab to [(4-hydroxy-5-iodo-3-nitrophenyl)acetyl (NIP) from naïve and immunized mice were determined by ELISA (6, 46). Briefly, 96-well plates were coated with 3 μg/ml of NIP5-BSA, NIP25-BSA or BSA in 0.1M carbonate buffer (pH 9.0) at 4°C overnight and blocked with 1%BSA in PBS. After washing with PBS containing 0.1%Tween 20, serially diluted samples or mAb standards were added to each well and incubated at room temperature for 2 h. For the dissociation of pentameric IgM into monomers, we treated serum samples and mAb standards with 2-ME as described (6, 46). After washing, HRP-conjugated goat anti-mouse IgM or HRP-conjugated goat anti-mouse IgG (both, Southern Biotech) was added and incubated at room temperature for 1 h. HRP activity was visualized using a TMB peroxidase kit (Bio-Rad Laboratories) and optical densities were determined at 450 nm. The NP/NIP mAb B1-8μ (47) and H33Lγ1 (22) Ab were used as ELISA standards.
Amplification of VHDJH2 rearrangements
Genomic DNA was isolated by phenol-chloroform extraction from sorted B-cell subsets from BL/6, CD154−/−, and AID−/− mice. Specific VDJ rearrangements were amplified by a nested PCR by Pfu polymerase (Stratagene, La Jolla, CA) (48) and forward primers specific for the V3 subfamily of VH1 gene segments and a reverse primer specific for JH2 [(1) and supplemental Table S1].4 After an initial 20 cycles of amplification, a second round of 20 cycles was performed using internal primers (1). Amplified VDJ products were ligated into pBS II SK(+) plasmid (Stratagene) and cloned by bacterial transformation (1). Cloned VDJ inserts were sequenced in an Applied Biosystems automated DNA sequencer and analyzed by IMGT/V-QUEST (http://imgt.cines.fr) and NCBI blast search software. All sequence data are available at GenBank (http://www.ncbi.nlm.nih.gov/Genbank, accession number: GQ162485-GQ162778).
RT-PCR
Expression of murine and human AID mRNA was quantified by quantitative RT-PCR (12). Briefly, total RNA was extracted from various B-cell populations, subcloned Ramos cells, and AMuLV-transformants. Total RNA libraries of human adult tonsil (Biochain) and fetal liver (Biochain and Stratagene), were purchased; cDNA from mouse and human tissues was prepared by standard methods (12, 49). PCR primers used in this study are listed in Table S1. AID cDNA was amplified using a nested PCR method (12). The relative expression levels of AID message to Igβ message were calculated by the comparative threshold cycle method (12).
Statistics
Statistical significance of data was determined by Mann-Whitney’s U test (for mutation frequencies), or Student’s t test (for serum Ab titers and AID mRNA levels). A p value of <0.05 was defined as significant.
Results
No Ab or GC responses in immunized CD154−/− mice
To determine the capacity, if any, of CD154−/− mice to respond to Td Ag, we immunized CD154−/− mice and congenic BL/6 controls with NP-CGG in alum (1, 48) and quantified serum IgM and IgG Ab (6, 46); total and high-affinity Ab were differentiated by binding to NIP25-BSA and NIP5-BSA (6, 46). In addition, treatment of serum samples with 2-ME to dissociate IgM Ab (6, 46), allowed us to minimize low affinity, avidity dependent binding by IgM (6, 46).
The serum of naïve BL/6 and CD154−/− mice contained undetectable levels of IgM reactive with NIP5 but ≈300 μg/ml of NIP25-binding IgM (Fig. 1A). This binding, however, was abolished completely by exposure to 2-ME (Fig. 1A), indicating avidity-dependent binding and demonstrating that neither naïve knockout nor control mice express detectable levels of NIP-specific serum IgM Ab. Low levels, 20–30 μg/ml, of IgG reactive to NIP25 were present in both naïve CD154−/− and BL/6 mice but IgG reactive with NIP5 could not be detected in either group (Fig. 1B).
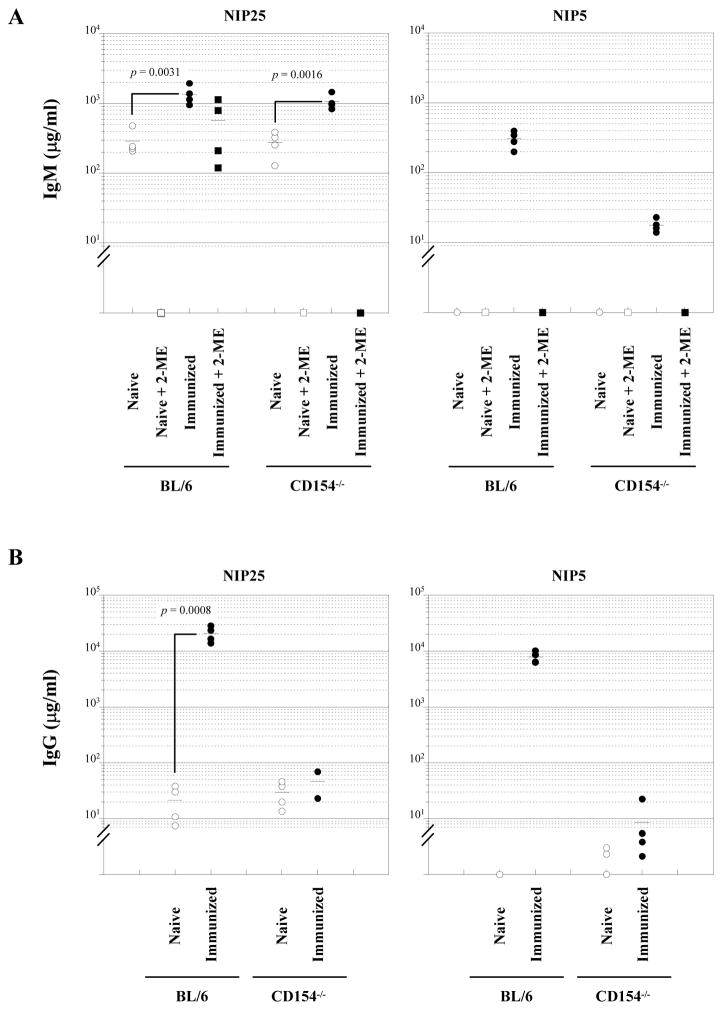
Figure 1.
Absence of specific Td-Ab production in CD154−/− mice. NIP-binding IgM (A) and IgG (B) titers in sera from naïve (n = 4, open symbols) and immunized (day 16, n = 4, closed symbols) BL/6 and CD154−/− mice were measured by ELISA. Mice were immunized with NP-CGG/alum via i.p. and sera were collected day 16 after immunization. Sera were treated with 2-ME (squares) or without 2-ME (circles). NIP5-BSA and NIP25-BSA were used as coating Ag to detect high affinity and total NIP-binding Ab, respectively. Statistical significance was determined using Student’s t test.
Immunization of BL/6 mice with NP-CGG/alum elicited significant IgM responses. At 16 days after immunization, serum IgM [1348 (±426) μg/ml] reactive with NIP25 was increased ≈4-fold over naïve controls (p = 0.0031) and in contrast to naïve mice, >50% of this IgM Ab retained NIP25-binding after reduction with 2-ME (Fig. 1A).
NP-CGG, however, did not induce Ag-specific IgM in CD154−/− mice. Although immunization resulted in a >3-fold increase [1063 (±269) μg/ml] of NIP25-binding IgM (p = 0.0016), this activity was completely abolished by 2-ME (Fig. 1A). We conclude that immunization did not elicit specific IgM production in CD154−/− mice but increased serum IgM levels non-specifically. Indeed, CD154−/−mice injected with adjuvant alone also exhibited similar increases in serum IgM levels (data not shown).
Immunization elicited robust IgG responses in BL/6 mice; by 16 days post- immunization (18); NIP25-binding serum IgG levels increased 1000-fold above naïve controls (p = 0.0008) (Fig. 1B). In contrast, NIP-specific IgG did not increase in the sera of immunized CD154−/− mice (Fig. 1B).
The lack of serum Ab in immunized CD154−/− mice was correlated with the lack of splenic GC responses (Fig. 2). Naïve BL/6 mice contained few (0.23 ± 0.1%) B220highGL-7high GC B cells (22) (Fig. 2A) but by day 16 after immunization, their frequency increased ≥4-fold (0.93 ± 0.18%; Fig. 2B). In contrast, only rare (<0.1%) splenic B cells with the GC phenotype could be detected in naïve CD154−/− mice and their frequency was not increased by immunization (Figs. 2C, D).
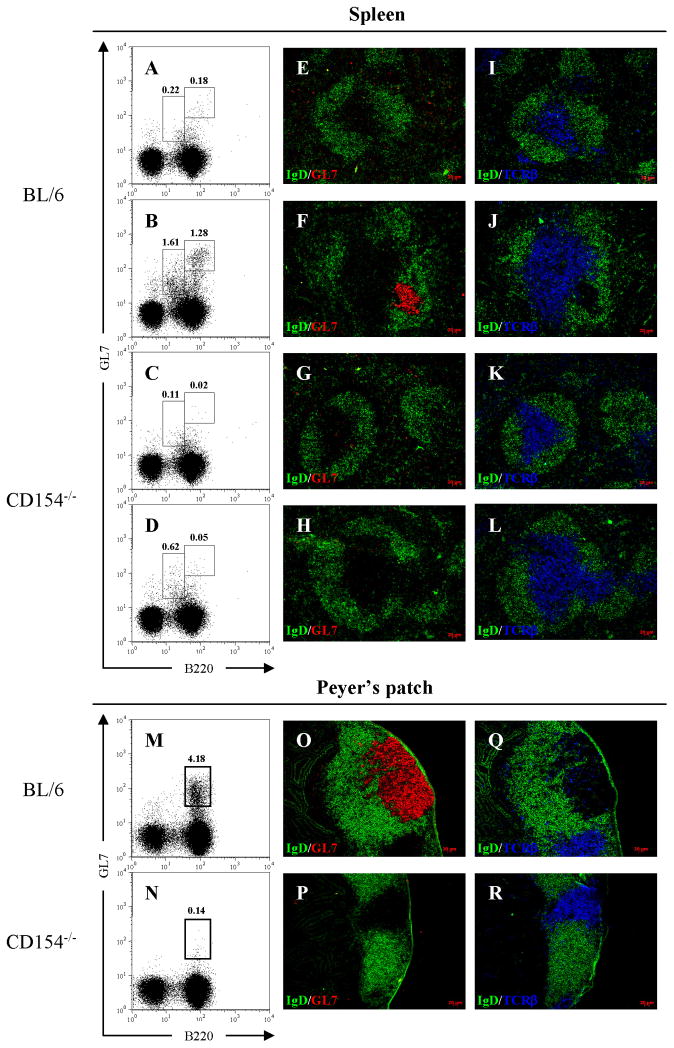
Figure 2.
Absence of GC in CD154−/− mice. GC formations in the spleen and PP of naïve and immunized BL/6 and CD154−/− mice were assessed by flow cytometry and immunofluorescence microscopy. Mice were immunized with NP-CGG/alum via i.p. and cells and tissues were harvested 16 days after immunization. A–D, splenocytes from naïve (A and C) and immunized (B and D) animals were labeled with mAb specific for B220 and GL-7. M and N, PP cells from naïve animals were labeled with mAb specific for B220 and GL-7. Numbers represent percentages of B220highGL-7high GC B cells and B220lowGL-7int developing B cells in PI− live cells. Serial frozen sections of naïve (E, I, G and K) and immunized (F, J, H and L) spleen or PP (O–R) were labeled with a combination of mAb specific for IgD (green; FITC) and GL-7 (red; PE), or TCRβ (blue, PE) as indicated. Original magnification: ×100. Data shown is representative of at least three independent experiments.
Immunohistochemical labeling with mAb for the GL-7 antigen and IgD revealed typical GL-7+IgD− splenic GC (50, 51) in immunized BL/6 mice (Fig. 2F), whereas no GL-7+IgD− B cell clusters were present in the spleens of naïve and immunized CD154−/− mice (Figs. 2G, H). Thus, immunization of CD154−/− mice with NP-CGG/alum neither elicits Ag-specific serum Ab nor splenic GC responses.
No PP GC in CD154−/− mice
Immunization with Ti type II Ag can elicit “abortive GC” in CD154−/− mice (52) that have been linked to low frequencies of Ig SHM in some (53), but not all (54), studies. To determine whether Ti GC might provide an alternative source for SHM in CD154−/− mice, we determined whether PP GC were present in CD154−/− mice. PP sample the Td and Ti Ag of the intestinal flora to trigger humoral responses (55, 56) and the PP GC are constitutively present in normal mice may be suppressed by antibiotics that reduce the gut flora (55, 56). PP GC, therefore, provide a significant test of the capacity for Ti GC in CD154−/− mice.
To enumerate PP GC B cells, we determined the frequencies of GL- 7highCD93−B220high cells by flow cytometry. In naïve BL/6 mice, a substantial fraction (5.25± 0.78%; Fig. 2M) of PP B cells expressed the phenotype of GC B lymphocytes, whereas no or few (0.30± 0.23%; Fig. 2N) GC B cells were present in the PP of CD154−/− mice. Although the PP of BL/6 mice contained prominent IgD−GL-7+ GC (Fig. 2O), GC-like structures could not be demonstrated in the PP of CD154−/− mice (Fig. 2P).
The absence of GC in CD154−/− mice was not due to any generalized disorganization of lymphoid architecture in the spleen or PP. Characteristic T-cell zones and B-cell follicles were present in both BL/6 (Figs. 2I, J, Q) and CD154−/− (Figs. 2K, L, R) mice. We conclude that neither Td nor Ti GC are formed in CD154−/− mice under physiologic conditions.
Immunization increases the numbers of developing B cells in the spleens of CD154−/− mice
Whereas immunization with NP-CGG/alum did not induce GC responses in CD154−/− mice (Figs. 2D, H, L), immunization comparably mobilized developing (GL-7intCD93+B220low) B cells from the BM to the spleen in both BL/6 and CD154−/− animals [(57); Figs. 2B, D]. In naïve BL/6 mice, GL-7intCD93+B220low cells comprised 0.2% of splenocytes (Fig. 2A), but immunization increased their frequency 7-fold by day 16 (Fig. 2B). Similarly, GL-7intCD93+B220low cells represented 0.1% of splenocytes in naïve CD154−/− mice (Fig. 2C); immunization increased their frequency 6-fold (Fig. 2D).
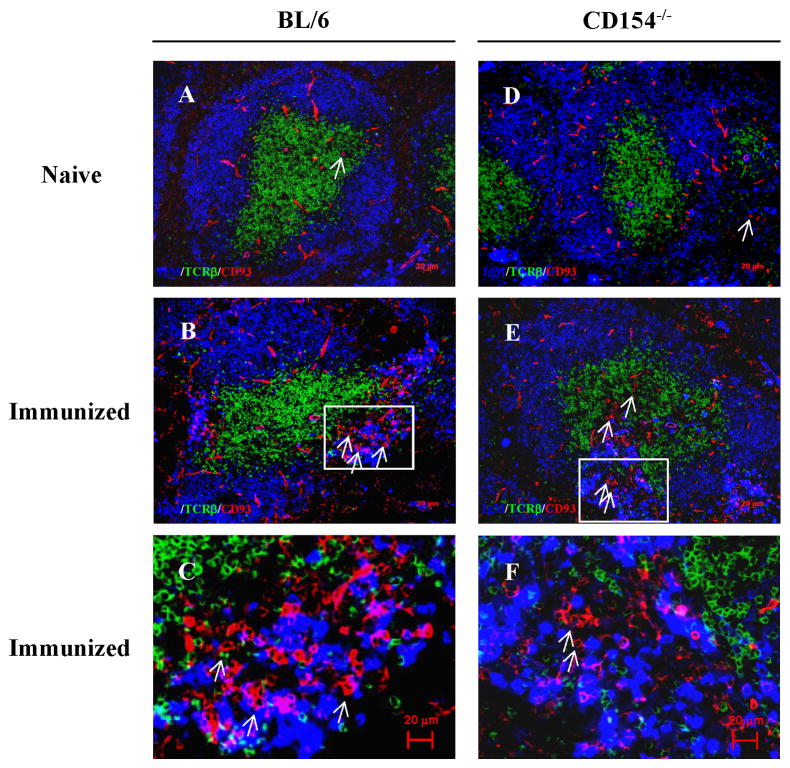
To locate CD93+ B cells in naïve and immunized BL/6 and CD154−/− mice, we labeled histologic sections of spleen with fluorochrome-tagged mAb specific for CD93, IgM, and TCRβ (Fig. 3). Whereas all sections (naïve, Figs. 3A, D) contained populations of spindle-shaped CD93+ endothelial cells (58, 59), immunization obviously increased the numbers of CD93+ B lymphocytes (compare Figs. 3A, B), consistent with our flow cytometric analyses (Figs. 2A, B). This increase in CD93+ splenic lymphocytes was most obvious in the splenic bridging channels [(60); Figs. 3B, C] where plasmacytes expressing intracellular IgM were also present [(61, 62) and Fig. 3C]. Immunization comparably increased the numbers of CD93+ splenic lymphocytes in the bridging channels of CD154−/− mice (Figs. 3D, E) where they accumulated along with IgM+ plasmacytes (Fig. 3F). We note that SHM in autoreactive plasmablasts present in splenic bridging channels has been reported (9).
Figure 3.
Immunization with alum adjuvant mobilizes CD93+ cells from the BM and localizes them in splenic bridging channels. Mice were immunized with NP-CGG/alum via i.p. and 16 days after immunization, the location of mobilized CD93+ BM cells was determined by immunofluorescence microscopy. Frozen spleen sections from naïve (A and D) and immunized (B, C, E, F) BL/6 (A–C) or CD154−/− (D–F) mice were labeled with mAb specific for IgM (blue; AlexaFluor350), TCRβ (green; FITC) and CD93 (red; PE). Arrows indicate CD93+ lymphocyte-like cells. Bridging channel areas indicated by rectangles in panels B and E are shown at higher magnification in panels C and F, respectively. Original magnification: ×100. Data shown are typical of at least two independent experiments.
In the BM of BL/6 and CD154−/− mice, the patterns of GL-7, IgM, CD21, and CD23 expression on developing (CD93+) and recirculating mature (CD93−) B cells are identical (Fig. S1A). After immunization, CD93+B220+ splenic emigrants in BL/6 mice comprised similar numbers of IgM− and IgM+ cells [(12, 63); Fig. S1B] whereas >80% of CD93+B220+ splenocytes in immunized CD154−/− mice expressed IgM (Fig. S1B). The spleens of immunized CD154−/− mice, therefore, contain fewer (IgM−CD93+) pro-/pre-B cells compared to BL/6 controls; inflammatory mobilization of BM B-lineage cells in CD154−/− may preferentially target im/T1 B cells.
AID expression by CD154−/− im/T1 B cells
We (12) and Imanishi-Kari (10, 11) have demonstrated levels of AID expression in im/T1 B cells from BL/6 mice sufficient to support limited CSR and Igκ SHM. Although AID expression by im/T1 B cells occurs in the absence of T lymphocytes or CD154 (10–12), it is unclear whether SHM is equally independent of CD40 signals triggered by T cells or other CD154+ cell compartments (64–68). To compare AID expression in B-lineage cells from CD154−/− and normal mice, we quantified AID message in pro-/pre-B, im/T1, MF, and MZ B cells from naïve and immunized BL/6 and CD154−/− mice (12). AID message in GC B cells from BL/6 mice and im/T1 and MF B cells from congenic AID−/− mice served as positive and negative comparators, respectively (3, 4) (Fig. 4). In addition, we compared AID expression by im/T1 B cells to that of AMuLV-transformed B-cell lines (41–43).
Figure 4.
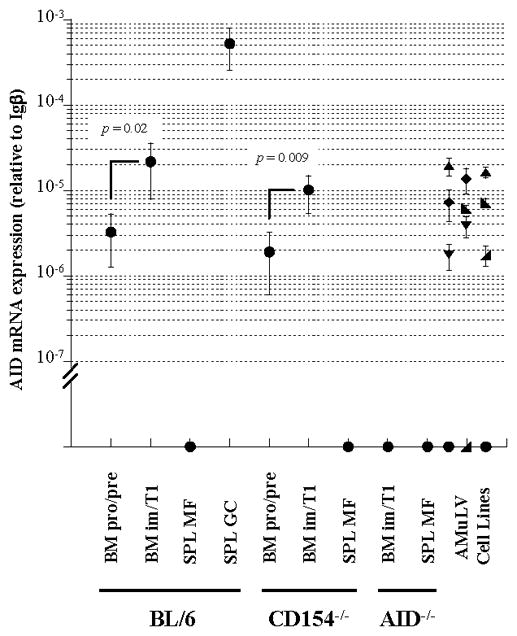
Developmentally regulated levels of AID expression in murine B-cell compartments. Levels of AID mRNA in pro-/pre-, im/T1, MF and GC B cell compartments from BM or spleen of BL/6, CD154−/− and AID−/− mice, and subcloned AMuLV-transformed cell lines, were determined by a quantitative RT-PCR. AID mRNA levels are shown as relative values to Igβ mRNA levels (mean ± SD) in each sample. B cell subsets (●, n = 3–6); AMuLV-transformed cell lines: S/B6 (●), C12 ( ), PD31 (▲), KG7 (▼;), 63-12 (
), PD31 (▲), KG7 (▼;), 63-12 ( ), and 220-8 (◆), n = 2. Statistical significance was determined using Student’s t test
), and 220-8 (◆), n = 2. Statistical significance was determined using Student’s t test
GC B cells from immunized BL/6 mice expressed the highest levels (≈500 × 10−6 relative to Igβ) of AID transcript; reciprocally, AID message in MF B cells from BL/6 and CD154−/− mice or im/T1 and MF B cells from AID−/− mice (4) was undetectable (Fig. 4). Consistent with our earlier study (12), pro-/pre-B cells from BL/6 and CD154−/− mice contained low, but significant quantities (≈0.6% of GC B cells; Fig. 4) of AID message. In both strains, AID message levels rose 5- to 7-fold in im/T1 B cells (p≤ 0.02; Fig. 4).
Expression of AID by im/T1 B cells, 2% to 4% of GC B cells, was not quantitatively different (p = 0.71), from that of six AMuLV-transformed pre-B cell lines (Fig. 4). Indeed, AID expression by BL/6 and CD154−/− im/T1 B cells (range: 6.0–37.8 × 10−6, relative to Igβ) and AID+ AMuLV-transformants (range: 1.8–19.3 × 10−6, relative to Igβ) overlapped (Fig. 4).
SHM in CD154−/− im/T1 B cells
To determine whether AID expression in developing B cells is sufficient to support IgH SHM, we amplified genomic VDJ rearrangements from highly purified (>95%) populations of pro-/pre-, im/T1 B cells from the BM and MF B cells from the spleens of naïve BL/6, CD154−/−, and AID−/− mice using Pfu polymerase (48, 69). In addition, GC B cells were recovered from immunized BL/6 mice 16 days after immunization with NP-CGG/alum. Amplicands comprised 11 related VH1 gene segments (V3 subgroup) rearranged to JH2 (70); VDJ products were cloned, sequenced (48, 69), and VH mutations were identified by comparison to germline (1, 69). Mutation frequencies were estimated from unique VDJ rearrangements (n = 22–59) as defined by CDR3 sequence and matched B-cell subsets from congenic AID−/− mice (4) established a baseline for mutation frequencies (Table I). Rare mutations (≤1/VH gene segment) recovered from AID−/− B cells indicated methodological errors of ≈ 2.2× 10−4 mutations/bp sequenced (range: 1.3–4.5 × 10−4/bp; Table I).
Table I.
SHM in im/T1 B cells from CD154−/− mice
| B Cell Type | Mutations in unique VH1(V3)DJH2 Rearrangements | ||||||
|---|---|---|---|---|---|---|---|
| Nos. VDJ rearrangements (in frame) | Basepairs sequenced | Nos. mutations | Nos. mutated rearrangements (% total) | Mutation freqa. (× 10−4) | RGYW/WRCYbΔhotspot/Δtotal (%) | ΔR:ΔSc | |
| BL/6 | |||||||
| SPL GCd | 22 (19) | 5.9 × 103 | 95 | 20 (91%) | 161*** | 31/95 (33%) | 70:25 |
| SPL MF | 31 (25) | 8.3 × 103 | 3 | 3 (10%) | 3.6 | 1/3 (33%) | 2:1 |
| BM pro/pre | 24 (19) | 6.4 × 103 | 2 | 2 (8%) | 3.1 | 0/2 (0%) | 2:0 |
| BM im/T1 | 22 (20) | 5.9 × 103 | 3 | 3 (14%) | 5.1 | 1/3 (33%) | 3:0 |
| CD154−/− | |||||||
| SPL MF | 29 (25) | 7.8 × 103 | 3 | 3 (10%) | 3.9 | 2/3 (67%) | 1:2 |
| BM pro/pre | 34 (27) | 9.1 × 103 | 3 | 3 (9%) | 3.3 | 0/3 (0%) | 1:2 |
| BM im/T1 | 48 (41) | 12.9 × 103 | 21 | 13 (27%) | 16.3*** | 5/21 (24%) | 11:10 |
| AID−/− | |||||||
| BM pro/pre | 25 (18) | 6.7 × 103 | 3 | 3 (12%) | 4.5 | 1/3 (33%) | 1:2 |
| BM im/T1 | 59 (49) | 15.8 × 103 | 2 | 2 (3%) | 1.3 | 0/2 (0%) | 1:1 |
| Total AID−/− | 84 (67) | 22.5 × 103 | 5 | 5 (6%) | 2.2 | 1/5 (20%) | 2:3 |
VH1(V3)DJH2 rearrangements from B-cell subsets were cloned into bacteria and sequenced.
The mutation frequency is calculated by numbers of mutation/basepairs sequenced.
Numbers of mutation in RGYW/WRCY motifs (Δhotspot), total numbers of mutation (Δtotal), and percentage of Δhotspot in Δtotal are shown.
Numbers of replacement mutations (ΔR) and silent mutations (ΔS) are shown.
p<0.001; mutation frequency of B-cell subset is significantly different from the mutation frequency of total AID−/− B cells calculated by Mann-Whitney’s U test.
No excess VH mutations were observed in VDJ fragments recovered from any pro-/pre-B or MF B-cell population (3.1–3.9× 10−4 mutations/bp; p = 0.43 to 0.68), whether from BL/6 or CD154−/− mice (Table I). In contrast, numerous VH mutations were present in GC B cells (161 × 10−4 mutations/bp; p < 0.001) from immunized BL/6 animals (Table I).
Significantly elevated numbers of VH mutations were, however, recovered from im/T1 B cells from CD154−/− mice (16.3 × 10−4, p < 0.001). Indeed, the VH mutation frequency in this im/T1 B-cell cohort is 10% of that observed in GC B cells from immunized BL/6 mice and demonstrates a capacity for developmental SHM in the absence of CD40:CD154 interaction.
In contrast to CD154−/− mice, we found that the number of VH mutations in im/T1 B cells from naïve BL/6 mice (5.1 × 10−4/bp) was modestly, but not significantly (p = 0.23), elevated above AID−/− controls (2.2 × 10−4/bp; Table I).
SHM in GC B cells exhibits a characteristic bias for transition mutations; transitions typically constitute some 50% – 60% of AID driven V(D)J mutations (71). In our samples, transitions accounted for 48% of VH mutations in CD154−/− im/T1 B cells and 61% of the mutations recovered from GC B cells (Fig. S2). Mutation at RGYW/WRCY motifs is also a hallmark of AID-mediated SHM (72) and constitutes≈30% of VH mutations in human B cells (73, 74). The distribution of VH mutations in im/T1 B cells from CD154−/− mice revealed a preference (24%) for these “hotspots” similar to that of GC B cells (33%) and consistent with AID dependent SHM (Table I) (72).
VH mutations in im/T1 B cells are unselected by antigen
Replacement (R) mutations are strongly linked to affinity-driven selection (69, 75) and ≈ 75% (70/95) of the VH mutations recovered from GC B cells were R substitutions (Table I). In contrast, R mutations constituted 52% (11:10) of the VH mutations recovered from the im/T1 B cells of CD154−/− mice (Table I). In addition, the distribution of VH mutation sites differed substantially between GC B cells and im/T1 B cells. Mutations in GC B cells were frequent (35% of total) in CDR1 and -2 whereas CDR mutations in im/T1 B cells constituted ≤5% of total mutations (data not shown). Reduced frequencies of R- and CDR1/2 mutations in im/T1 B cells indicate a lack of stringent Ag-driven selection.
VH gene usage in mutated im/T1 B cells differed significantly from that of GC B cells elicited by NP-CGG. GC B-cell populations were dominated by rearrangements of the V186.2 (50%) and C1H4 (29%) gene segments (Fig. S3); these frequencies underestimate Ag selection in GC as we excluded clonally related sequences from our analysis (1, 69). Instead, VH gene usage by BM im/T1 B cells from CD154−/− mice was diverse (Fig. S3). The VH mutations recovered in CD154−/− im/T1 B cells were distributed uniformly among the various VH amplicands (data not shown).
AID expression in murine fetal liver
In both mice and humans, the fetal liver supports lymphopoiesis and, as gestation proceeds, contains increasingly mature B-lineage cells (76, 77). The fetus is normally isolated from infection and exposure to exogenous Ag (78) and any AID expression in the murine fetal liver is, therefore, unlikely to be induced by extrinsic factors. To determine whether murine fetal B-lineage cells express AID, we amplified AID message from fetal liver B cells recovered at 16 and 19 days of gestation (E16 and E19; Fig. 5).
Figure 5.
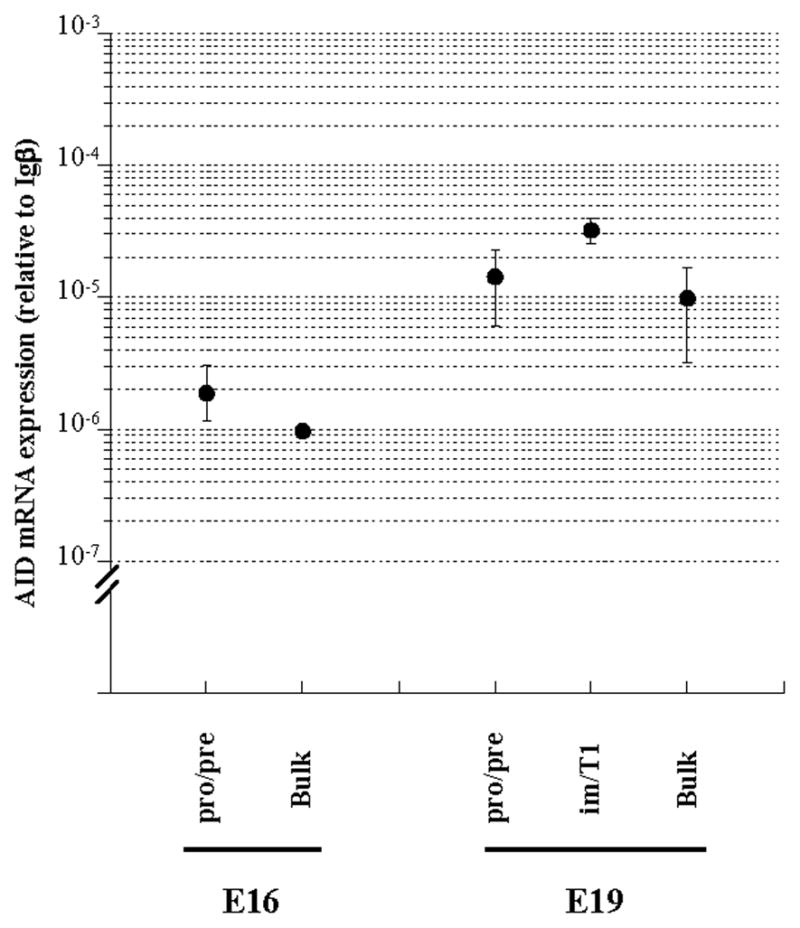
AID expression in developing B-lineage cells from murine fetal liver. AID mRNA expression in pro-/pre- B (B220lowCD93+IgM−IgD−) and im/T1 B (B220lowCD93+IgM+IgD−) cell compartments from murine fetal liver samples (16 days and/or 19 days of gestation, Fig. S4) were measured by quantitative RT-PCR. AID mRNA levels are shown as relative values to Igβ mRNA levels (mean ± SD) in each sample (n = 2–4).
As reported (79, 80), only pro-/pre-B cells were present in E16 fetal liver but pro-/pre-B and im/T1 B-cells could be recovered from E19 samples. Some 2% of E16 fetal liver cells were B220lowCD93+ (2.4± 0.6%) and of these, all (99.8± 0.1%) expressed the IgM−IgD− phenotype (Fig. S4). In E19 samples, B220lowCD93+ cells comprised ≈ 20% of fetal liver cells (18.9 ± 1.4%) and while most (99.1± 0.2%) of these were IgM−IgD− pro-/pre-B cells, a small minority (0.8± 0.1%) expressed IgM (Fig. S4).
Pro-/pre-B cells from E16 fetal liver expressed quantities of AID message comparable to that of pro-/pre-B cells from adult BM (Figs. 4, 5). In E19 fetal liver, however, AID expression in pro-/pre-B cells was 7-fold higher (Fig. 5), reaching levels that approached AID expression in im/T1 B cells from adult BM (Fig. 4). AID message levels in E19 im/T1 B cells were also elevated; with expression levels nearly double that of im/T1 B cells from BM (Figs. 4, 5).
AID expression in human im/T1 B cells and fetal liver
To determine whether developmentally regulated AID expression might exist in humans, we sorted im/T1 (CD19+CD34−IgMintIgD−/lowCD27−), T2 (CD19+CD34−IgMhighIgDhighCD27−) and MF (CD19+CD34−IgMintIgDintCD27−) B cells from human umbilical cord blood [Fig. S5; (45, 81)] and quantified AID message levels (Fig. 6). In addition, we quantified AID message in commercial RNA libraries made from human fetal liver and tonsil, and from the Ramos human GC B-cell line (44). Human tonsil is rich in AID+ GC B cells (82, 83) and Ramos cells express AID and exhibit Ig SHM (44).
Figure 6.
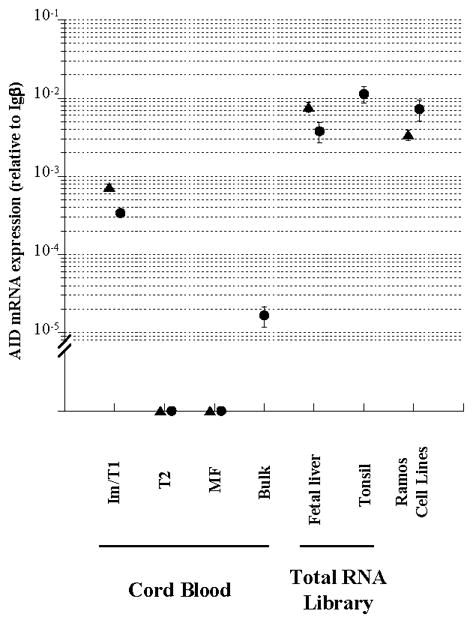
AID expression in human cord blood im/T1 B cells and fetal liver. AID mRNA expression in im/T1, T2, and MF B cell compartments from human cord blood (Fig. S5), and fetal liver samples were measured by quantitative RT-PCR. Unsorted cord blood cells (bulk) were compared to ensure that these B-cell compartments contained the majority of recovered AID message. Tonsil and Ramos cell lines served as positive controls. AID mRNA levels are shown as relative values to Igβ mRNA levels (mean ± SD, 4 independent measurements) in each sample. Each symbol represents an individual donor, subclone, or commercial source.
Human AID expression levels were highest in tonsil (1.1× 10−2 relative to Igβ) with Ramos cells expressing about half as much AID message (0.5× 10−2 relative to Igβ; Fig. 6). Consistent with AID expression by murine B cells [Fig. 4; (12)], human im/T1 B cells expressed low but significant levels of AID message (3%–6% of tonsil; Fig. 6) while AID was undetectable in T2 and MF B cells (Fig. 6). AID expression in im/T1 B cells accounted for virtually all AID message present in unsorted cord blood cells (Fig. 6) and no AID message was detected in the CD19− compartments of cord blood (data not shown). By this relative measure, AID message levels in human cord blood im/T1 B cells are comparable to that of murine im/T1 B cells capable of CSR (11, 12) and SHM [Table I; (10, 11)].
AID mRNA in two, independent fetal liver libraries (20 wks gestation, both) was substantially more abundant than in cord blood samples (Fig. 6). Remarkably, AID expression levels (3.0× 10−3 and 6.0× 10−3 relative to Igβ) in human fetal liver equaled that of Ramos cells (Fig. 6). The shared patterns of AID expression in mouse and human B-lineage cells suggest a conserved developmental program.
Discussion
HIGM1 patients support populations of IgM+IgD+CD27+ B cells carrying mutated V(D)J rearrangements (27, 28, 33). This cellular compartment is generally categorized as un-switched (IgM) memory B cells (29, 84) or, alternatively, “pre-diversified” progenitors of marginal zone (MZ) B cells (28, 85). In HIGM1 patients, the mutated IgM+IgD+CD27+ compartment develops relatively soon after birth (33), and perhaps even in the fetus (14), in the absence of detectable GC responses. Taken together, these observations are sufficiently surprising to imply a “second pathway” for VDJ SHM in the absence of cognate T:B interaction (27).
Despite reports of active SHM outside of GC (9–11, 14), examples of SHM and AID activity in non-GC B cells remain controversial as genetically modified mice that co-express fluorescent marker proteins and AID contain no marked immature B cells and few labeled cells not located in GC (86, 87). The absence of labeled developmentally immature B cells could reflect the low levels of AID message expressed by non-GC B cells [Figs. 4, 5 and (12, 86)], the relative brevity of AID expression by im/T1 B cells, or both (11). Conversely, reports of AID expression and activity by developmentally immature B cells in humans and mice may reflect artifactual inclusion of (Td or Ti) GC B cells in isolated populations of immature and transitional B cells and/or atypical distributions of GC B cells in tissue.
In an attempt to understand the conditions that lead to SHM in the absence of GC, we previously demonstrated that both pro-/pre-B and im/T1 B cells of CD154−/− mice express elevated levels of AID and that in im/T1 B cells at levels sufficient to support limited CSR (12). These observations raised the possibility that CD154−/− im/T1 cells might undergo SHM as well. If so, CD154 deficient mice could offer insight into the origin(s) of the hypermutated IgM+IgD+CD27+ B cells in HIGM1 patients and more generally, SHM by in non-GC B cells.
We found that in CD154−/− mice a potent Td immunogen does not elicit serum Ab (Fig. 1). In contrast to the robust Ab responses of normal congenic controls, CD154−/− mice did not mount specific IgM or IgG Ab responses after immunization with NP-CGG/alum. Whereas immunization did increase serum IgM in CD154−/− mice, these increases were non-specific and likely represent a general immune activation driven by the inflammatory properties of alum (88).
In addition to absent serum Ab responses, immunization of CD154−/− mice neither elicited GC nor increased the frequency of splenic B cells with the phenotype (B220hiGL-7hi) of GC B cells (Fig. 2). Thus, CD154 was required not only for the generation of GC architecture but also for the characteristic activation phenotype of GC B cells. Although we did not immunize with Ti Ag, CD154−/− mice respond to both type-I and -II Ti Ag (18, 19) and it remained possible that any AID expression in these mice might be derived from cells recruited to Ti “GC” (16, 54, 89). This concern was mitigated by the absence of PP GC in CD154−/−, demonstrating that even prolonged exposure to the (Td and) Ti Ag of the gut flora is insufficient to generate GC or B cells with a GC phenotype in the absence of CD154 expression (Fig. 2). In this, the immunodeficiency of CD154−/− mice appears to be more severe than some human HIGM1 patients (16). We conclude that any AID expression and activity in the B cell compartments of CD154−/− mice (Fig. 4) are highly unlikely to result from contamination by incidental Td- or Ti GC B cells.
Despite these differences in Ab production, immunization of BL/6 and CD154−/− mice with alum mobilizes BM CD93+ im/T1 B cells to the spleen [(63, 90); Fig. 2]. The mobilized CD93+ cells localize to the border of the red pulp and splenic T-cell zone in bridging channels [Fig. 3; (60)], extra follicular sites where short lived plasmacytes accumulate after immunization with Td Ag (61) and autoreactive B cells have been reported to undergo SHM (9). Similar mobilizations appear to occur in humans with systemic autoimmune disease, as these patients often exhibit inflammatory responses and the appearance of developmentally immature B cells in the periphery (91). The appearance of developing B cells in the periphery suggests a potential role for im/T1 B cells in autoreactive Ab production (92) and we speculate that autoreactive im/T1 B cells may persist or mature in autoimmune patients. Indeed, im/T1 B cells in autoimmune-prone mice are resistant to apoptosis after IgM cross-linking in vitro (93), whereas normal im/T1 B cells are exquisitely sensitive to BCR-induced apoptosis (94, 95).
As expected (12), pro-/pre-B and im/T1 B cells from adult CD154−/− and BL/6 mice expressed low but significant levels of AID whereas message was undetectable in the MF B-cell compartment of both control and knockout animals (Fig. 4). Quantitative RT-PCR results indicate that AID expression by im/T1 B cells ranged from 2% to 4% of that observed in primary, splenic GC B cells and was greater than or equal to a collection of AMuLV transformed B-cell lines (Fig. 4).
Significant AID expression by pro-/pre-B and im/T1 B cells from murine fetal liver was also observed (Fig. 5). Pro-/pre-B cells from E16 fetal liver expressed AID message at levels equivalent to pro-/pre-B cells from adult BM while AID expression in E19 fetal liver pro-/pre-B cells was 7-fold and E19 im/T1 B cells 2-fold higher than in pro-/pre-B and im/T1 cells from adult BM (Figs. 4, 5).
AID expression by human im/T1, T2 and MF B cells from umbilical cord blood mirrored the patterns observed in mice with significant AID expression present in im/T1 cells but not the T2 and MF compartments (Fig. 6). Human im/T1 B cells from cord blood express quantities of AID (3%–6% of tonsil tissue; Fig. 6) that are similar to those of murine im/T1 B cells (2%–4% of GC; Fig. 4). In mice, this level of AID is sufficient to support both CSR (11, 12) and SHM [Table I and (10, 11)]. AID message was also abundant in two human fetal liver RNA libraries (Fig. 6). Indeed, AID expression (relative to Igβ) in 20 wk human fetal liver was equivalent to that of the Ramos human GC B-cell line and about half that of human tonsil, a tissue rich in GC B cells (82, 83) (Fig. 6).
In contrast to our findings [Figs. 4–6; (12)] and those of Imanishi-Kari (10, 11), Papavasiliou and colleagues (96, 97) reported negligible levels of AID expression in developing B cells but significantly increased AID expression in developing B lymphocytes after viral transduction/infection. We also observed significant levels of AID message in cloned AMuLV-transformed B-cell lines (Fig. 4) but the levels of AID message in AMuLV transformants were similar to im/T1 B cells freshly recovered from BM (Fig. 4). Interestingly, mutation frequencies for the JH4 and λ5 genes [6.8- and 11.9 × 10−4/bp, respectively; (96)] in AMuLV-transformed pre-B cells said to express high levels of AID are below that of VH mutations in CD154−/− im/T1 B cells (16.3× 10−4; Table I) and comparable to those observed in BL/6 im/T1 B cells (5.1× 10−4; Table I). Thus, by either the measure of quantitative AID expression (Fig. 4) or Ig mutation frequencies (Table I), we were unable to confirm virally induced elevations of AID expression and SHM in AMuLV transformed B cells (96).
Although AID expression was equivalent in BL/6 and CD154−/− mice, significantly elevated VDJ mutation frequencies were observed only in CD154 deficient mice (Table I). Whereas this restriction of VH mutation may mirror the relationship of HIGM1 patients and normal individuals, the mechanism(s) responsible for this bias is unclear. In im/T1 B cells, the measured VH mutation frequency (16.3 × 10−4) that was 10% of GC B cells (161 × 10−4) (Table I) and, unlike GC B cells, mutations were present in only about a third (27%) of CD154−/−im/T1 B cells and showed little or no evidence for Ag driven selection (Table I). VH mutations in CD154−/− im/T1 B cells were characterized by low R:S ratios (Table I), broad dispersal within the VH gene segments, and appearance in diverse VH gene segments (Fig. S3). Together, these characteristics lead us to conclude that Ag does not expand specific (autoreactive?) clones of im/T1 B cells, although Ig signaling may promote SHM (11).
The numbers of VH mutations in mutated im/T1 sequences (average 1.6; range,1–4) was about one-third that observed in GC B cells (average 4.8; range, 1–12) but, nonetheless, easily distinguished from matched AID−/− controls, pro-/pre-B, and MF B cells (Table I). The absence of a CD27-like marker in mice precludes a directed search for any descendants of mutated im/T1 B cells but it is clear that the great majority of IgM+IgD+ MF B cells, even in CD154−/− mice, do not carry mutated Ig genes (Table I). It is possible that mutated im/T1 B cells enter a compartment that we did not directly sample (e.g., the peritoneum or the splenic MZ). If BCR signaling promotes SHM in im/T1 B cells (11), physiologic selection for im/T1 B cells with low affinity for self-antigens (98) could bias im/T1 B-cell differentiation to the MZ (95) or peritoneal B-cell compartments (99).
It is possible that AID+ im/T1 B cells have few or no descendants. Activated im/T1 B cells are highly sensitive to apoptotic signals and it is plausible that some CD154-dependent process normally eliminates AID+ im/T1, perhaps as a consequence of genotoxic stress (100). Of note, the survival and/or proliferation of im/T1 B cells is augmented by BAFF (12), a cytokine that is often elevated in autoimmune patients (101). The concatenation of BCR signaling and BAFF-enhanced survival of autoreactive im/T1 B cells may represent a pre-pathologic condition that promotes autoimmunity in peripheral tissues.
It remains unclear when and where human IgM+IgD+CD27+ B cells acquire Ig mutations. The mutation frequency of IgM+IgD+CD27+ B cells increases during first year of life (28), indicating that these B cells undergo SHM during that period. Recently, Scheeren et al. have provided evidence that human IgM+IgD+CD27+ B cells become mutated in utero when they recovered mutated IgM+IgD+CD27+ B cells from fetal spleen (14). Among fetal tissues, AID message is detected in mesenteric lymph node and liver [Fig. 6; (14, 83)] but not in spleen or BM (14); thereby, it is likely that B cells undergo SHM in fetal liver or fetal mesenteric lymph node during ontogeny.
AID expression and SHM in mouse CD154−/− im/T1 B cells (Fig. 4, Table I) suggest a potential relationship between murine im/T1 B cells and the mutated IgM+IgD+CD27+ B cells in HIGM1 patients (27, 28). These B-cell subsets share characteristically diverse Ig repertoires and, to lesser and greater degrees, Ig SHM that is independent of Td and Ti immune responses [(10, 33); Figs. 1, 2; Table I]. AID message is present in mouse im/T1 B cells from fetal liver, BM, and spleen [Figs. 4, 5; (10–12)] and in human im/T1 B cells from cord blood (Fig. 6) and in fetal hematopoietic tissue [Fig. 6; (14)]. We conclude that it is entirely plausible that human im/T1 B cells - or even earlier B-cell types in fetal tissues -could undergo SHM to produce the mutated IgM+IgD+CD27+ B cells present in HIGM1 patients.
The similar patterns of AID expression during B-cell development in mice and humans (10–12, 14, 83) suggest that developmentally regulated AID expression is not confined to birds (34–36), rabbits (37, 38), and sheep (39) but is a general phenomenon. Indeed, this pattern of AID expression may represent a developmental program shared by most, and perhaps all, vertebrate species (102). It is interesting to consider the possibility that the Ag-driven GC response evolved from this primitive developmental program. If so, understanding the physiologic onset, site, triggers, and significance for developmentally regulated AID expression constitute significant and novel goals in understanding B-cell biology.
Supplementary Material
Acknowledgments
We are grateful to T. Matt Holl and Kathleen O’Hara for comments on our manuscript. The Carolinas Cord Blood Bank at Duke University Medical Center provided anonymous cord blood samples.
Footnotes
This work was supported in part by the Bill and Melinda Gates Foundation and NIH grants AI024335, AI056363 (to G.K.) and AI066106 (to M.L.).
Abbreviations used in this paper: 2-ME, 2-mercaptoethanol; Ab, antibody; Ag, antigen; AID, activation-induced cytidine deaminase; AMuLV, Abelson murine leukemia virus; BCR, B-cell antigen receptor; CDR, complementarity-determining region; BL/6, C57BL/6; CGG, chicken γ globulin; CSR, class-switch recombination; FWR, framework region; GC, germinal center; HIGM1, type-1 hyper IgM syndrome; im/T1, immature/transitional 1; MF, mature follicular; MZ, marginal zone; NIP, (4-hydroxy-5-iodo-3-nitrophenyl)acetyl; NP, (3-hydroxy-4-nitrophenyl)acetyl; PI, propidium iodide; PP, Peyer’s patch; R/S, ratio of replacement and silent mutation; SHM, somatic hypermutation; Td, T cell- dependent; Ti, T cell-independent; T2, transitional 2.
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Jacob J, Kelsoe G, Rajewsky K, Weiss U. Intraclonal generation of antibody mutants in germinal centres. Nature. 1991;354:389–392. doi: 10.1038/354389a0. [DOI] [PubMed] [Google Scholar]
- 2.Berek C, Berger A, Apel M. Maturation of the immune response in germinal centers. Cell. 1991;67:1121–1129. doi: 10.1016/0092-8674(91)90289-b. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu M, V, Sankaranand S, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 5.Weiss U, Rajewsky K. The repertoire of somatic antibody mutants accumulating in the memory compartment after primary immunization is restricted through affinity maturation and mirrors that expressed in the secondary response. J Exp Med. 1990;172:1681–1689. doi: 10.1084/jem.172.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dal Porto JM, Haberman AM, Kelsoe G, Shlomchik MJ. Very low affinity B cells form germinal centers, become memory B cells, and participate in secondary immune responses when higher affinity competition is reduced. J Exp Med. 2002;195:1215–1221. doi: 10.1084/jem.20011550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Takizawa M, Tolarova H, Li Z, Dubois W, Lim S, Callen E, Franco S, Mosaico M, Feigenbaum L, Alt FW, Nussenzweig A, Potter M, Casellas R. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J Exp Med. 2008;205:1949–1957. doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.William J, Euler C, Christensen S, Shlomchik MJ. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 2002;297:2066–2070. doi: 10.1126/science.1073924. [DOI] [PubMed] [Google Scholar]
- 10.Mao C, Jiang L, Melo-Jorge M, Puthenveetil M, Zhang X, Carroll MC, Imanishi-Kari T. T cell-independent somatic hypermutation in murine B cells with an immature phenotype. Immunity. 2004;20:133–144. doi: 10.1016/s1074-7613(04)00019-6. [DOI] [PubMed] [Google Scholar]
- 11.Han JH, Akira S, Calame K, Beutler B, Selsing E, Imanishi-Kari T. Class switch recombination and somatic hypermutation in early mouse B cells are mediated by B cell and Toll-like receptors. Immunity. 2007;27:64–75. doi: 10.1016/j.immuni.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueda Y, Liao D, Yang K, Patel A, Kelsoe G. T-independent activation-induced cytidine deaminase expression, class-switch recombination, and antibody production by immature/transitional 1 B cells. J Immunol. 2007;178:3593–3601. doi: 10.4049/jimmunol.178.6.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milili M, Fougereau M, Guglielmi P, Schiff C. Early occurrence of immunoglobulin isotype switching in human fetal liver. Mol Immunol. 1991;28:753–761. doi: 10.1016/0161-5890(91)90118-4. [DOI] [PubMed] [Google Scholar]
- 14.Scheeren FA, Nagasawa M, Weijer K, Cupedo T, Kirberg J, Legrand N, Spits H. T cell-independent development and induction of somatic hypermutation in human IgM+IgD+CD27+ B cells. J Exp Med. 2008;205:2033–2042. doi: 10.1084/jem.20070447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korthauer U, Graf D, Mages HW, Briere F, Padayachee M, Malcolm S, Ugazio AG, Notarangelo LD, Levinsky RJ, Kroczek RA. Defective expression of T-cell CD40 ligand causes X-linked immunodeficiency with hyper-IgM. Nature. 1993;361:539–541. doi: 10.1038/361539a0. [DOI] [PubMed] [Google Scholar]
- 16.Facchetti F, Appiani C, Salvi L, Levy J, Notarangelo LD. Immunohistologic analysis of ineffective CD40-CD40 ligand interaction in lymphoid tissues from patients with X-linked immunodeficiency with hyper-IgM. Abortive germinal center cell reaction and severe depletion of follicular dendritic cells. J Immunol. 1995;154:6624–6633. [PubMed] [Google Scholar]
- 17.Foy TM, Shepherd DM, Durie FH, Aruffo A, Ledbetter JA, Noelle RJ. In vivo CD40-gp39 interactions are essential for thymus-dependent humoral immunity. II. Prolonged suppression of the humoral immune response by an antibody to the ligand for CD40, gp39. J Exp Med. 1993;178:1567–1575. doi: 10.1084/jem.178.5.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J, Foy TM, Laman JD, Elliott EA, Dunn JJ, Waldschmidt TJ, Elsemore J, Noelle RJ, Flavell RA. Mice deficient for the CD40 ligand. Immunity. 1994;1:423–431. doi: 10.1016/1074-7613(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 19.Renshaw BR, Fanslow WC, 3rd, Armitage RJ, Campbell KA, Liggitt D, Wright B, Davison BL, Maliszewski CR. Humoral immune responses in CD40 ligand-deficient mice. J Exp Med. 1994;180:1889–1900. doi: 10.1084/jem.180.5.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han S, Hathcock K, Zheng B, Kepler TB, Hodes R, Kelsoe G. Cellular interaction in germinal centers. Roles of CD40 ligand and B7–2 in established germinal centers. J Immunol. 1995;155:556–567. [PubMed] [Google Scholar]
- 21.Foy TM, Laman JD, Ledbetter JA, Aruffo A, Claassen E, Noelle RJ. gp39-CD40 interactions are essential for germinal center formation and the development of B cell memory. J Exp Med. 1994;180:157–163. doi: 10.1084/jem.180.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi Y, Dutta PR, Cerasoli DM, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. V. Affinity maturation develops in two stages of clonal selection. Journal of Experimental Medicine. 1998;187:885–895. doi: 10.1084/jem.187.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy J, Espanol-Boren T, Thomas C, Fischer A, Tovo P, Bordigoni P, Resnick I, Fasth A, Baer M, Gomez L, Sanders EA, Tabone MD, Plantaz D, Etzioni A, Monafo V, Abinun M, Hammarstrom L, Abrahamsen T, Jones A, Finn A, Klemola T, DeVries E, Sanal O, Peitsch MC, Notarangelo LD. Clinical spectrum of X-linked hyper-IgM syndrome. J Pediatr. 1997;131:47–54. doi: 10.1016/s0022-3476(97)70123-9. [DOI] [PubMed] [Google Scholar]
- 24.Herve M, Isnardi I, Ng YS, Bussel JB, Ochs HD, Cunningham-Rundles C, Meffre E. CD40 ligand and MHC class II expression are essential for human peripheral B cell tolerance. J Exp Med. 2007;204:1583–1593. doi: 10.1084/jem.20062287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brezinschek HP, Dorner T, Monson NL, Brezinschek RI, Lipsky PE. The influence of CD40-CD154 interactions on the expressed human V(H) repertoire: analysis of V(H) genes expressed by individual B cells of a patient with X-linked hyper-IgM syndrome. Int Immunol. 2000;12:767–775. doi: 10.1093/intimm/12.6.767. [DOI] [PubMed] [Google Scholar]
- 26.Monson NL, Foster SJ, Brezinschek HP, Brezinschek RI, Dorner T, Lipsky PE. The role of CD40-CD40 ligand (CD154) interactions in immunoglobulin light chain repertoire generation and somatic mutation. Clin Immunol. 2001;100:71–81. doi: 10.1006/clim.2001.5049. [DOI] [PubMed] [Google Scholar]
- 27.Weller S, Faili A, Garcia C, Braun MC, Le Deist FF, de Saint Basile GG, Hermine O, Fischer A, Reynaud CA, Weill JC. CD40-CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc Natl Acad Sci U S A. 2001;98:1166–1170. doi: 10.1073/pnas.98.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weller S, Braun MC, Tan BK, Rosenwald A, Cordier C, Conley ME, Plebani A, Kumararatne DS, Bonnet D, Tournilhac O, Tchernia G, Steiniger B, Staudt LM, Casanova JL, Reynaud CA, Weill JC. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood. 2004;104:3647–3654. doi: 10.1182/blood-2004-01-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med. 1998;188:1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agematsu K, Hokibara S, Nagumo H, Komiyama A. CD27: a memory B-cell marker. Immunol Today. 2000;21:204–206. doi: 10.1016/s0167-5699(00)01605-4. [DOI] [PubMed] [Google Scholar]
- 31.Willenbrock K, Jungnickel B, Hansmann ML, Kuppers R. Human splenic marginal zone B cells lack expression of activation-induced cytidine deaminase. Eur J Immunol. 2005;35:3002–3007. doi: 10.1002/eji.200535134. [DOI] [PubMed] [Google Scholar]
- 32.Tangye SG, Good KL. Human IgM+CD27+ B cells: memory B cells or “memory” B cells? J Immunol. 2007;179:13–19. doi: 10.4049/jimmunol.179.1.13. [DOI] [PubMed] [Google Scholar]
- 33.Weller S, Mamani-Matsuda M, Picard C, Cordier C, Lecoeuche D, Gauthier F, Weill JC, Reynaud CA. Somatic diversification in the absence of antigen-driven responses is the hallmark of the IgM+IgD+CD27+ B cell repertoire in infants. J Exp Med. 2008;205:1331–1342. doi: 10.1084/jem.20071555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynaud CA, Anquez V, Dahan A, Weill JC. A single rearrangement event generates most of the chicken immunoglobulin light chain diversity. Cell. 1985;40:283–291. doi: 10.1016/0092-8674(85)90142-4. [DOI] [PubMed] [Google Scholar]
- 35.Reynaud CA, Anquez V, Grimal H, Weill JC. A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell. 1987;48:379–388. doi: 10.1016/0092-8674(87)90189-9. [DOI] [PubMed] [Google Scholar]
- 36.Reynaud CA, Dahan A, Anquez V, Weill JC. Somatic hyperconversion diversifies the single Vh gene of the chicken with a high incidence in the D region. Cell. 1989;59:171–183. doi: 10.1016/0092-8674(89)90879-9. [DOI] [PubMed] [Google Scholar]
- 37.Becker RS, Knight KL. Somatic diversification of immunoglobulin heavy chain VDJ genes: evidence for somatic gene conversion in rabbits. Cell. 1990;63:987–997. doi: 10.1016/0092-8674(90)90502-6. [DOI] [PubMed] [Google Scholar]
- 38.Knight KL. Restricted VH gene usage and generation of antibody diversity in rabbit. Annu Rev Immunol. 1992;10:593–616. doi: 10.1146/annurev.iy.10.040192.003113. [DOI] [PubMed] [Google Scholar]
- 39.Reynaud CA, Garcia C, Hein WR, Weill JC. Hypermutation generating the sheep immunoglobulin repertoire is an antigen-independent process. Cell. 1995;80:115–125. doi: 10.1016/0092-8674(95)90456-5. [DOI] [PubMed] [Google Scholar]
- 40.Hsu HC, Wu Y, Yang P, Wu Q, Job G, Chen J, Wang J, Accavitti-Loper MA, Grizzle WE, Carter RH, Mountz JD. Overexpression of activation-induced cytidine deaminase in B cells is associated with production of highly pathogenic autoantibodies. J Immunol. 2007;178:5357–5365. doi: 10.4049/jimmunol.178.8.5357. [DOI] [PubMed] [Google Scholar]
- 41.Alt F, Rosenberg N, Lewis S, Thomas E, Baltimore D. Organization and reorganization of immunoglobulin genes in A-MULV-transformed cells: rearrangement of heavy but not light chain genes. Cell. 1981;27:381–390. doi: 10.1016/0092-8674(81)90421-9. [DOI] [PubMed] [Google Scholar]
- 42.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 43.Lewis S, Rosenberg N, Alt F, Baltimore D. Continuing kappa-gene rearrangement in a cell line transformed by Abelson murine leukemia virus. Cell. 1982;30:807–816. doi: 10.1016/0092-8674(82)90285-9. [DOI] [PubMed] [Google Scholar]
- 44.Zhang W, Bardwell PD, Woo CJ, Poltoratsky V, Scharff MD, Martin A. Clonal instability of V region hypermutation in the Ramos Burkitt’s lymphoma cell line. Int Immunol. 2001;13:1175–1184. doi: 10.1093/intimm/13.9.1175. [DOI] [PubMed] [Google Scholar]
- 45.Sims GP, Ettinger R, Shirota Y, Yarboro CH, Illei GG, Lipsky PE. Identification and characterization of circulating human transitional B cells. Blood. 2005;105:4390–4398. doi: 10.1182/blood-2004-11-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dal Porto JM, Haberman AM, Shlomchik MJ, Kelsoe G. Antigen drives very low affinity B cells to become plasmacytes and enter germinal centers. J Immunol. 1998;161:5373–5381. [PubMed] [Google Scholar]
- 47.Kelsoe G, Reth M, Rajewsky K. Control of idiotope expression by monoclonal anti-idiotope and idiotope- bearing antibody. Eur J Immunol. 1981;11:418–423. doi: 10.1002/eji.1830110513. [DOI] [PubMed] [Google Scholar]
- 48.Han S, Dillon SR, Zheng B, Shimoda M, Schlissel MS, Kelsoe G. V(D)J recombinase activity in a subset of germinal center B lymphocytes. Science. 1997;278:301–305. doi: 10.1126/science.278.5336.301. [DOI] [PubMed] [Google Scholar]
- 49.Ueda Y, Kondo M, Kelsoe G. Inflammation and the reciprocal production of granulocytes and lymphocytes in bone marrow. J Exp Med. 2005;201:1771–1780. doi: 10.1084/jem.20041419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cervenak L, Magyar A, Boja R, Laszlo G. Differential expression of GL7 activation antigen on bone marrow B cell subpopulations and peripheral B cells. Immunol Lett. 2001;78:89–96. doi: 10.1016/s0165-2478(01)00239-5. [DOI] [PubMed] [Google Scholar]
- 51.Christoph T, Rickert R, Rajewsky K. M17: a novel gene expressed in germinal centers. Int Immunol. 1994;6:1203–1211. doi: 10.1093/intimm/6.8.1203. [DOI] [PubMed] [Google Scholar]
- 52.Gaspal FM, McConnell FM, Kim MY, Gray D, Kosco-Vilbois MH, Raykundalia CR, Botto M, Lane PJ. The generation of thymus-independent germinal centers depends on CD40 but not on CD154, the T cell-derived CD40-ligand. Eur J Immunol. 2006;36:1665–1673. doi: 10.1002/eji.200535339. [DOI] [PubMed] [Google Scholar]
- 53.Toellner KM, Jenkinson WE, Taylor DR, Khan M, Sze DM, Sansom DM, Vinuesa CG, MacLennan IC. Low-level hypermutation in T cell-independent germinal centers compared with high mutation rates associated with T cell-dependent germinal centers. J Exp Med. 2002;195:383–389. doi: 10.1084/jem.20011112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lentz VM, Manser T. Cutting edge: germinal centers can be induced in the absence of T cells. J Immunol. 2001;167:15–20. doi: 10.4049/jimmunol.167.1.15. [DOI] [PubMed] [Google Scholar]
- 55.Casola S, Otipoby KL, Alimzhanov M, Humme S, Uyttersprot N, Kutok JL, Carroll MC, Rajewsky K. B cell receptor signal strength determines B cell fate. Nat Immunol. 2004;5:317–327. doi: 10.1038/ni1036. [DOI] [PubMed] [Google Scholar]
- 56.Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–1427. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- 57.Gartner F, Alt FW, Monroe RJ, Seidl KJ. Antigen-independent appearance of recombination activating gene (RAG)-positive bone marrow B cells in the spleens of immunized mice. J Exp Med. 2000;192:1745–1754. doi: 10.1084/jem.192.12.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petrenko O, Beavis A, Klaine M, Kittappa R, Godin I, Lemischka IR. The molecular characterization of the fetal stem cell marker AA4. Immunity. 1999;10:691–700. doi: 10.1016/s1074-7613(00)80068-0. [DOI] [PubMed] [Google Scholar]
- 59.Dean YD, McGreal EP, Gasque P. Endothelial cells, megakaryoblasts, platelets and alveolar epithelial cells express abundant levels of the mouse AA4 antigen, a C-type lectin-like receptor involved in homing activities and innate immune host defense. Eur J Immunol. 2001;31:1370–1381. doi: 10.1002/1521-4141(200105)31:5<1370::AID-IMMU1370>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 60.Mitchell J. Lymphocyte circulation in the spleen. Marginal zone bridging channels and their possible role in cell traffic. Immunology. 1973;24:93–107. [PMC free article] [PubMed] [Google Scholar]
- 61.Jacob J, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3- nitrophenyl)acetyl. II. A common clonal origin for periarteriolar lymphoid sheath-associated foci and germinal centers. J Exp Med. 1992;176:679–687. doi: 10.1084/jem.176.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garcia De Vinuesa C, Gulbranson-Judge A, Khan M, O’Leary P, Cascalho M, Wabl M, Klaus GG, Owen MJ, MacLennan IC. Dendritic cells associated with plasmablast survival. Eur J Immunol. 1999;29:3712–3721. doi: 10.1002/(SICI)1521-4141(199911)29:11<3712::AID-IMMU3712>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 63.Ueda Y, Yang K, Foster SJ, Kondo M, Kelsoe G. Inflammation Controls B Lymphopoiesis by Regulating Chemokine CXCL12 Expression. J Exp Med. 2004;199:47–58. doi: 10.1084/jem.20031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gauchat JF, Henchoz S, Mazzei G, Aubry JP, Brunner T, Blasey H, Life P, Talabot D, Flores-Romo L, Thompson J, et al. Induction of human IgE synthesis in B cells by mast cells and basophils. Nature. 1993;365:340–343. doi: 10.1038/365340a0. [DOI] [PubMed] [Google Scholar]
- 65.Gauchat JF, Henchoz S, Fattah D, Mazzei G, Aubry JP, Jomotte T, Dash L, Page K, Solari R, Aldebert D, et al. CD40 ligand is functionally expressed on human eosinophils. Eur J Immunol. 1995;25:863–865. doi: 10.1002/eji.1830250335. [DOI] [PubMed] [Google Scholar]
- 66.Grewal IS, Flavell RA. CD40 and CD154 in cell-mediated immunity. Annu Rev Immunol. 1998;16:111–135. doi: 10.1146/annurev.immunol.16.1.111. [DOI] [PubMed] [Google Scholar]
- 67.Henn V, Slupsky JR, Grafe M, Anagnostopoulos I, Forster R, Muller-Berghaus G, Kroczek RA. CD40 ligand on activated platelets triggers an inflammatory reaction of endothelial cells. Nature. 1998;391:591–594. doi: 10.1038/35393. [DOI] [PubMed] [Google Scholar]
- 68.Wykes M, Poudrier J, Lindstedt R, Gray D. Regulation of cytoplasmic, surface and soluble forms of CD40 ligand in mouse B cells. Eur J Immunol. 1998;28:548–559. doi: 10.1002/(SICI)1521-4141(199802)28:02<548::AID-IMMU548>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 69.Jacob J, Przylepa J, Miller C, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3- nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J Exp Med. 1993;178:1293–1307. doi: 10.1084/jem.178.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gu H, Tarlinton D, Muller W, Rajewsky K, Forster I. Most peripheral B cells in mice are ligand selected. J Exp Med. 1991;173:1357–1371. doi: 10.1084/jem.173.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 72.Rogozin IB, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim Biophys Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 73.Dorner T, Foster SJ, Farner NL, Lipsky PE. Somatic hypermutation of human immunoglobulin heavy chain genes: targeting of RGYW motifs on both DNA strands. Eur J Immunol. 1998;28:3384–3396. doi: 10.1002/(SICI)1521-4141(199810)28:10<3384::AID-IMMU3384>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 74.Martin A, Bardwell PD, Woo CJ, Fan M, Shulman MJ, Scharff MD. Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature. 2002;415:802–806. doi: 10.1038/nature714. [DOI] [PubMed] [Google Scholar]
- 75.Allen D, Cumano A, Dildrop R, Kocks C, Rajewsky K, Rajewsky N, Roes J, Sablitzky F, Siekevitz M. Timing, genetic requirements and functional consequences of somatic hypermutation during B-cell development. Immunol Rev. 1987;96:5–22. doi: 10.1111/j.1600-065x.1987.tb00506.x. [DOI] [PubMed] [Google Scholar]
- 76.Dorshkind K, Montecino-Rodriguez E. Fetal B-cell lymphopoiesis and the emergence of B-1-cell potential. Nat Rev Immunol. 2007;7:213–219. doi: 10.1038/nri2019. [DOI] [PubMed] [Google Scholar]
- 77.Gathings WE, Lawton AR, Cooper MD. Immunofluorescent studies of the development of pre-B cells, B lymphocytes and immunoglobulin isotype diversity in humans. Eur J Immunol. 1977;7:804–810. doi: 10.1002/eji.1830071112. [DOI] [PubMed] [Google Scholar]
- 78.Ducluzeau R. Implantation and development of the gut flora in the newborn animal. Ann Rech Vet. 1983;14:354–359. [PubMed] [Google Scholar]
- 79.Owen JJ, Cooper MD, Raff MC. In vitro generation of B lymphocytes in mouse foetal liver, a mammalian ‘bursa equivalent’. Nature. 1974;249:361–363. doi: 10.1038/249361a0. [DOI] [PubMed] [Google Scholar]
- 80.Raff MC, Megson M, Owen JJ, Cooper MD. Early production of intracellular IgM by B-lymphocyte precursors in mouse. Nature. 1976;259:224–226. doi: 10.1038/259224a0. [DOI] [PubMed] [Google Scholar]
- 81.Marie-Cardine A, Divay F, Dutot I, Green A, Perdrix A, Boyer O, Contentin N, Tilly H, Tron F, Vannier JP, Jacquot S. Transitional B cells in humans: characterization and insight from B lymphocyte reconstitution after hematopoietic stem cell transplantation. Clin Immunol. 2008;127:14–25. doi: 10.1016/j.clim.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 82.Pascual V, Liu YJ, Magalski A, de Bouteiller O, Banchereau J, Capra JD. Analysis of somatic mutation in five B cell subsets of human tonsil. J Exp Med. 1994;180:329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Muto T, Muramatsu M, Taniwaki M, Kinoshita K, Honjo T. Isolation, tissue distribution, and chromosomal localization of the human activation-induced cytidine deaminase (AID) gene. Genomics. 2000;68:85–88. doi: 10.1006/geno.2000.6268. [DOI] [PubMed] [Google Scholar]
- 84.Klein U, Kuppers R, Rajewsky K. Variable region gene analysis of B cell subsets derived from a 4-year- old child: somatically mutated memory B cells accumulate in the peripheral blood already at young age. J Exp Med. 1994;180:1383–1393. doi: 10.1084/jem.180.4.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weller S, Reynaud CA, Weill JC. Splenic marginal zone B cells in humans: where do they mutate their Ig receptor? Eur J Immunol. 2005;35:2789–2792. doi: 10.1002/eji.200535446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crouch EE, Li Z, Takizawa M, Fichtner-Feigl S, Gourzi P, Montano C, Feigenbaum L, Wilson P, Janz S, Papavasiliou FN, Casellas R. Regulation of AID expression in the immune response. J Exp Med. 2007;204:1145–1156. doi: 10.1084/jem.20061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eisenbarth SC, Colegio OR, O’Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008 doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Vinuesa CG, Cook MC, Ball J, Drew M, Sunners Y, Cascalho M, Wabl M, Klaus GG, MacLennan IC. Germinal centers without T cells. J Exp Med. 2000;191:485–494. doi: 10.1084/jem.191.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Monroe RJ, Seidl KJ, Gaertner F, Han S, Chen F, Sekiguchi J, Wang J, Ferrini R, Davidson L, Kelsoe G, Alt FW. RAG2:GFP knockin mice reveal novel aspects of RAG2 expression in primary and peripheral lymphoid tissues. Immunity. 1999;11:201–212. doi: 10.1016/s1074-7613(00)80095-3. [DOI] [PubMed] [Google Scholar]
- 91.Girschick HJ, Grammer AC, Nanki T, Vazquez E, Lipsky PE. Expression of recombination activating genes 1 and 2 in peripheral B cells of patients with systemic lupus erythematosus. Arthritis Rheum. 2002;46:1255–1263. doi: 10.1002/art.10264. [DOI] [PubMed] [Google Scholar]
- 92.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 93.Roy V, Chang NH, Cai Y, Bonventi G, Wither J. Aberrant IgM signaling promotes survival of transitional T1 B cells and prevents tolerance induction in lupus-prone New Zealand black mice. J Immunol. 2005;175:7363–7371. doi: 10.4049/jimmunol.175.11.7363. [DOI] [PubMed] [Google Scholar]
- 94.Carsetti R, Kohler G, Lamers MC. Transitional B cells are the target of negative selection in the B cell compartment. J Exp Med. 1995;181:2129–2140. doi: 10.1084/jem.181.6.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wen L, Brill-Dashoff J, Shinton SA, Asano M, Hardy RR, Hayakawa K. Evidence of marginal-zone B cell-positive selection in spleen. Immunity. 2005;23:297–308. doi: 10.1016/j.immuni.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 96.Gourzi P, Leonova T, Papavasiliou FN. A role for activation-induced cytidine deaminase in the host response against a transforming retrovirus. Immunity. 2006;24:779–786. doi: 10.1016/j.immuni.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 97.Gourzi P, Leonova T, Papavasiliou FN. Viral induction of AID is independent of the interferon and the Toll-like receptor signaling pathways but requires NF-{kappa}B. J Exp Med. 2007;204:259–265. doi: 10.1084/jem.20061801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hayakawa K, Asano M, Shinton SA, Gui M, Allman D, Stewart CL, Silver J, Hardy RR. Positive selection of natural autoreactive B cells. Science. 1999;285:113–116. doi: 10.1126/science.285.5424.113. [DOI] [PubMed] [Google Scholar]
- 99.Lam KP, Rajewsky K. B cell antigen receptor specificity and surface density together determine B-1 versus B-2 cell development. J Exp Med. 1999;190:471–477. doi: 10.1084/jem.190.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang J, Roschke V, Baker KP, Wang Z, Alarcon GS, Fessler BJ, Bastian H, Kimberly RP, Zhou T. Cutting edge: a role for B lymphocyte stimulator in systemic lupus erythematosus. J Immunol. 2001;166:6–10. doi: 10.4049/jimmunol.166.1.6. [DOI] [PubMed] [Google Scholar]
- 102.Marr S, Morales H, Bottaro A, Cooper M, Flajnik M, Robert J. Localization and differential expression of activation-induced cytidine deaminase in the amphibian Xenopus upon antigen stimulation and during early development. J Immunol. 2007;179:6783–6789. doi: 10.4049/jimmunol.179.10.6783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.