Abstract
Epigenetic silencing of genes in association with aberrant promoter DNA hypermethylation has emerged as a significant mechanism in the development of human cancers. Such genes are also often targets of the Polycomb group repressive complexes in embryonic cells. The Polycomb repressive complex (PRC) 2 has been best studied in this regard. We now examine a link between PRC1 and cancer specific gene silencing. Here we show a novel and direct association between a constituent of the PRC1 complex, CBX7, with gene repression and promoter DNA hypermethylation of genes frequently silenced in cancer. CBX7 is able to complex with DNA methyltransferase enzymes leading us to explore a role for CBX7 in maintenance and initiation of gene silencing. Knockdown of CBX7 was unable to relieve suppression of deeply silenced genes in cancer cells, however, in embryonal carcinoma (EC) cells, CBX7 can initiate stable repression of genes that are frequently silenced in adult cancers. Furthermore, we are able to observe assembly of DNA methyltransferases at CBX7 target gene promoters. Sustained expression of CBX7 in EC cells confers a growth advantage and resistance to retinoic acid induced differentiation. In this setting, especially, there is increased promoter DNA hypermethylation for many genes by analysis of specific genes as well as through epigenomic studies. Our results allow us to propose a potential mechanism, through assembly of novel repressive complexes, by which the Pc component of PRC1 can promote the initiation of epigenetic changes involving abnormal DNA hypermethylation of genes frequently silenced in adult cancers.
Keywords: Polycomb, CBX7, DNA Hypermethylation, Gene Silencing
INTRODUCTION
Epigenetic silencing in association with promoter DNA hypermethylation is a mechanism by which tumor suppressor genes (TSGs) are inactivated in adult cancer (1, 2). The mechanisms that underlie the targeting of such key genes for DNA hypermethylation remain poorly understood. Recent studies have shown that a large group of genes silenced in association with aberrant promoter DNA hypermethylation in cancer are frequent targets of the PcG repressive complexes in normal and neoplastic embryonic cells (3–5). These studies suggest a stem cell origin for cancer in that silencing of genes in adult cancer are linked to the epigenetic control of stem/precursor cell gene expression patterns regulated by PcG proteins. However, a direct link has not yet been established.
PcG proteins are typically found as constituents of one of several complexes including the Polycomb Repressive Complex (PRC) 2/3 or PRC1 (6, 7). The PRC2 complex contains the histone methyltransferase, EZH2, that can catalyze the trimethylation of H3K27 that can then serve as a docking site for the PRC1 complex (8). EZH2 was recently implicated in silencing of previously identified PRC2 target genes (9) and found to associate with DNA methylation machinery (10). Additional studies, however, demonstrated that knockdown of EZH2 does not usually re-express DNA hypermethylated genes in cancer (11) (12). The PRC1 complex contains several PcG proteins including a polycomb (Pc) component that contains a chromodomain that can interpret histone modifications associated with gene silencing (13). While Pc is encoded by a single gene in Drosophila, there are many Pc family members in mammalian cells, including CBX7 (6, 7).
Recent studies suggest that CBX7 may play an important role in tumorigenesis. CBX7 was identified in a functional screen for genes involved in senescence bypass (14) at least partially via repression of expression of the TSG, p16 (14, 15). CBX7 has also been shown to initiate lymphomagenesis and cooperate with c-myc in tumor progression in vivo (16). Analysis of the CBX7 chromodomain demonstrated that this protein can associate with repressive histone modifications including di- and tri-methylated H3K9 as well as tri-methylated H3K27 (17). Together, these studies suggest that a CBX7 containing complex may possess the ability to read histone modifications found in promoters of key genes including those susceptible to cancer specific DNA methylation (3, 4, 18).
In this study, we have explored the link between PRC1 and cancer specific gene silencing. We have found that CBX7 can exist in a complex with DNA methyltransferase enzymes (DNMTs) in cancer cells. We studied the impact of CBX7 on embryonal carcinoma (EC) cells and the potential role for CBX7 in targeting key genes frequently silenced in association with DNA hypermethylation in adult cancer. Our results allow us to propose a potential mechanism, whereby through assembly of repressive complexes, the Pc component of PRC1 can participate in and promote epigenetic changes involving abnormal DNA hypermethylation of genes frequently silenced in adult cancers.
MATERIALS AND METHODS
Plasmids
CBX7 (NM_175709.2) containing a C-terminal HA tag was cloned into pEF1alpha-IRES-puro…
Cell culture
Cell lines were maintained according to ATCC guidelines. Stable selection used 0.2µg/ml puromycin (Sigma) selection. Population doublings were calculated as described (14). All transfections were performed using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions.
Western analysis
Antibodies used were: CBX7 (Invitrogen, custom), HA (Santa Cruz), DNMT1 (Santa Cruz), and Flag (Sigma). Lamin-B (Santa Cruz) and β-Actin (Sigma) are used as loading controls where indicated.
Expression Analysis
Real-time PCR was performed using conditions and primers described previously including initial normalization to GAPDH (3). Microarray data was generated using Agilent whole human genome, 4×44K microarrays, as previously described (19).
Co-immunoprecipitation
Whole cell lysates were prepared using a modified RIPA buffer. Nuclear proteins (Pierce) were prepared using a standard RIPA buffer. Immunoprecipitation was performed with rotation overnight at 4°C. Protein A/G agarose beads (eBioscience) were added for 2 hours. Four washes were performed with TNE buffer. Complexes were eluted by boiling in 2x LDS loading buffer (Invitrogen).
siRNA
Tera-2 cells were transfected with a nontargeting control or DNMT1 targeting siRNA (sequences available upon request) as previously described (11).
Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described (11) using previously designed primers (3). Antibodies used were: DNMT1 (Imgenex), DNMT3a (Imgenex), DNMT3b (Imgenex), HA (Santa Cruz or Covance), IgG (Upstate).
Bisulfite Sequencing
Genomic DNA extraction and bisulfite modification, and sequencing from vector control, CBX7 expressing, or ATRA treated CBX7 expressing Tera-2 cells at passage 30 was performed as previously described (11, 20). Primers used were within promoters for CDH1, GATA4, sFRP4, and sFRP5, and sFRP1 (available upon request).
Infinium Analysis
DNA methylation was assessed using the Illumina Infinium platform (21). Each probe is assigned a β-value, indicating full methylation of a specific CpG site (β = 1), absence of methylation (β = 0), and every situation in between (0 <= β <= 1) using the signal of the methylated probe over the signal sum of the methylated plus unmethylated probe. Probes with insignificant or uncertain calls (p > 0.05) were removed from the subsequent analysis. In-vitro methylated DNA (IVD) and DNA from cells genetically deleted for DNA methyltransferases 1 and 3b (DKO) (22), were used as fully methylated and fully unmethylated controls, respectively. Probes that showed clear methylation (β > .75), or clear de-methylation (β < .25) in IVD and DKO were utilized.
Immunocytochemistry
Cells grown on cover slips were fixed in 3.7% formaldehyde and processed for staining as described previously (23).
RESULTS
CBX7 interacts with DNA methylation machinery
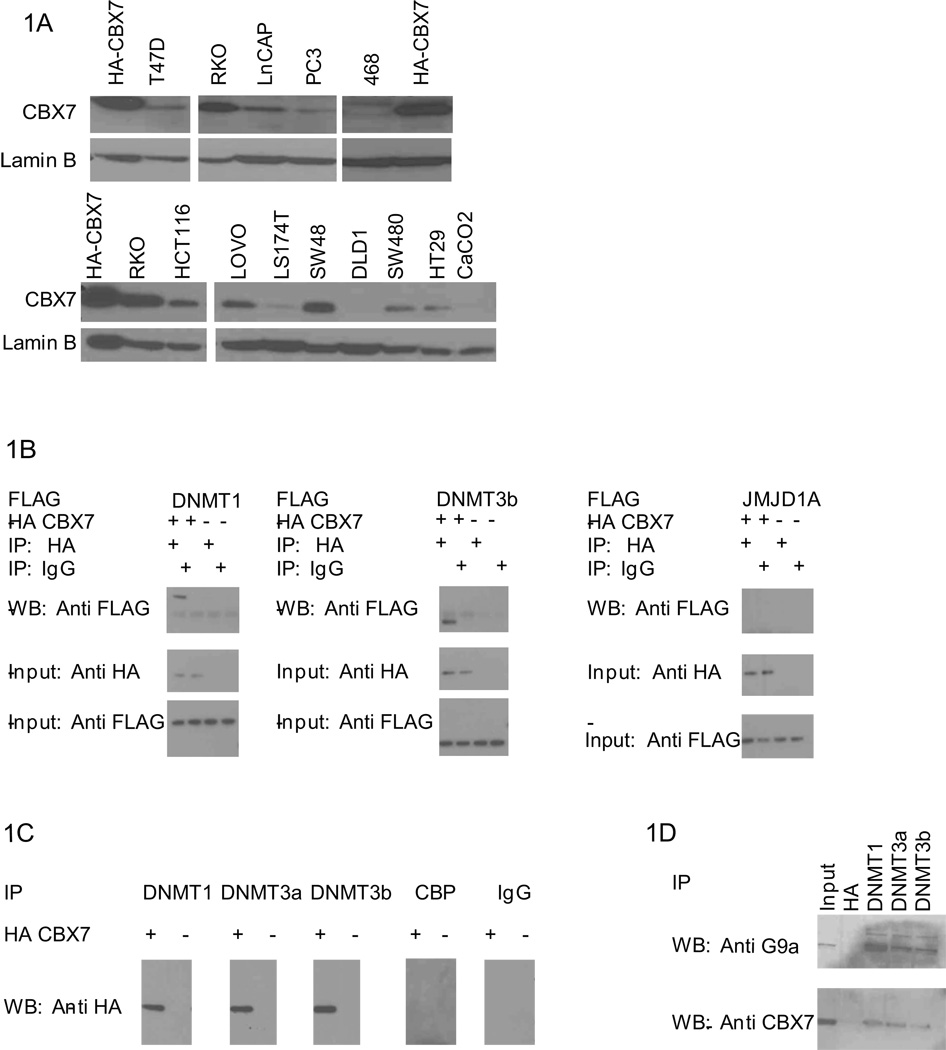
We first explored the links between CBX7 and stable silencing of key target genes in cancer by assessing expression of CBX7 in cancer cell lines. Colorectal cancer lines have the highest levels of CBX7 (Fig. 1a) and RKO cells, in particular, have high levels of CBX7 relative to other colorectal cancer lines surveyed (Fig. 1a). RKO has been previously characterized as having an unusually high number of genes silenced in association with DNA hypermethylation (19).
Figure 1. CBX7 interacts with DNMTs.
A, CBX7 is detected by anti-CBX7 antibody in breast, prostate, and colorectal cancer cell lines as indicated. HA-CBX7 cell lysate is used as a positive control. B, Co-IP and Western was performed in 293T cells expressing HA-CBX7 and FLAG constructs as indicated. C, Co-IP and Western was performed in NCCIT cells transfected with HA-CBX7 as indicated. Antibodies to CBP and IgG served as controls. D, Endogenous co- IP was performed in RKO cells with antibodies as indicated. HA was used as a negative control. Western analysis was performed using antibodies against endogenous CBX7 or G9a.
Previous studies have suggested that PRC1 complexes may be important in translating repressive chromatin modifications to gene repression (8, 13). CBX7 is known to interact with repressive chromatin modifications (17) and has been shown to repress expression of a known TSG (14), targeted for methylation in a variety of cancers (15). DNA methylation is mediated by DNMTs in normal (24–26) and cancer cells (22, 27, 28) leading us to search for interactions between CBX7 and these enzymes. We co-expressed FLAG-tagged DNMT1, and 3b with HA-tagged CBX7 in the 293T human embryonic kidney cell line and searched for interactions via co-immunoprecipitation (co-IP) analyses. We found co-IP between DNMT1, DNMT3b, and CBX7 (Fig. 1b) but not JMJD1A, a histone demethylase enzyme that would eradicate the silencing mark, dimethylated H3K9 (29), and thus is an unlikely component of a repressive complex. We found the above not only with the above tagged constructs, but also with all three endogenous DNMTs (Fig. 1c) in NCCIT cells, that have high levels of DNMTs (30). The transcriptional activator, CBP, not expected to reside in a repressive complex did not co-IP with CBX7. Finally, using as a positive control, G9a, a histone methyltransferase that establishes a key silencing mark, dimethylated H3K9 (29), and interacts with all three DNMTs (31), (32) we find that DNMTs are able to complex with CBX7 in RKO colorectal carcinoma (CRC) cells (Fig. 1d). Thus, CBX7/DNMT complexes exist in multiple cell types including cancer cells containing high levels of cancer specific DNA methylation, suggesting this PRC1 constituent may play a role in the establishment and/or maintenance of gene silencing.
CBX7 expression in cancer cells may not be the sole mechanism for maintaining gene silencing
To determine if CBX7 is necessary to maintain stable silencing of key target genes in cancer, we used siRNA to knockdown CBX7 in RKO cells (Fig. S1a). We did not find re-expression of the cancer related genes surveyed (Fig. S1b), suggesting that CBX7 does not solely maintain the deep silencing associated with cancer specific DNA hypermethylation. Similar results were observed with knockdown of the PRC2 protein, EZH2 (11, 12). Thus, while CBX7 may still be important in targeting genes for stable silencing involving DNA methylation, the resultant methylation may not be easily reversible through loss of a single PcG constituent.
CBX7 confers a growth advantage to EC cells
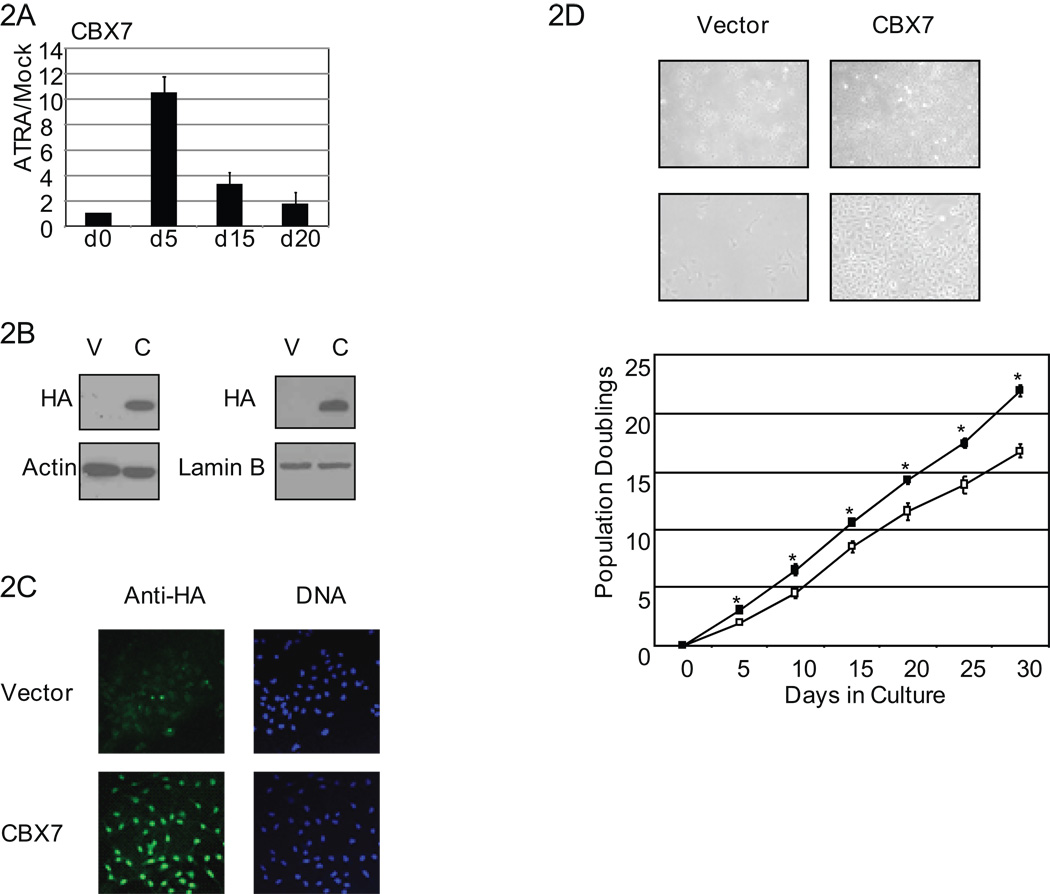
To explore the possibility that the CBX7 interactions observed may play a role in establishment of silencing of genes frequently associated with cancer specific DNA hypermethylation, we utilized embryonal carcinoma (EC) cells, Tera-2, that have less hypermethylation of cancer related genes than adult cancer cells (3). CBX7 expression is low in cycling Tera-2 cells, initially increases with ATRA treatment, which induces neural differentiation in these cells (33, 34), and decreases back to low expression as the induced cells fully differentiate and enter senescence (Fig. 2a). These data suggest that reduction in CBX7 expression may be important for EC cell terminal differentiation.
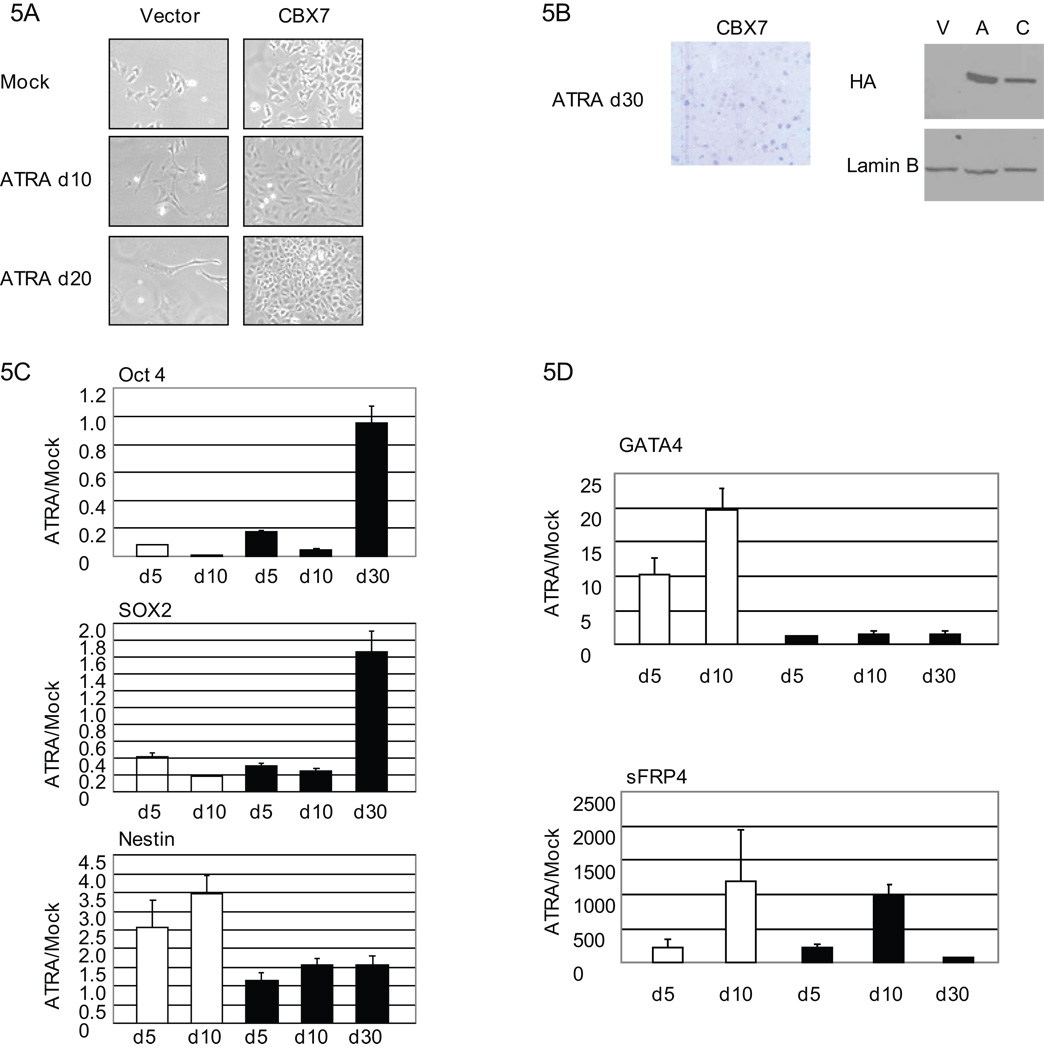
Figure 2. Stable overexpression of CBX7.
A, Tera-2 cells were treated with ATRA (2µM) for days as indicated. Real-time PCR expression for CBX7 is shown relative to mock treated cells. Values represent the mean fold change ± SEM for triplicates. B, Tera-2 cells were stably transfected with vector (V) or HA tagged CBX7 (C). CBX7 expression is detected by anti-HA antibody at passage 4 (left) and passage 21 (right). C, Anti-HA immunocytochemistry was used to detect CBX7 expression DNA by Hoescht stain. D, Vector control and CBX7 overexpressing cells are shown in culture at 4× (top) and 10× (bottom) magnification. Growth curve analysis of vector control (open boxes) and CBX7 cells (filled boxes). Values represent the mean ± SEM for triplicate wells. Asterisks indicate p<0.05 by Students t-test.
Our previous study shows that Tera-2 cells do possess, for many genes studied, repressive histone modifications even without dense promoter DNA methylation they exhibit in adult cancers (3). These histone modifications are those that might prime promoters for PRC1 docking, CBX7 interaction (17), and further movement toward stable silencing. To test this hypothesis we stably overexpressed hemagglutinin (HA) tagged CBX7 in Tera-2 cells. HA-tagged CBX7 was appropriately nuclear and stably expressed over twenty passages (Fig.2b, c). Similar to studies performed using MEFs and human fibroblasts (14), CBX7 overexpression was able to confer a growth advantage to Tera-2 cells when compared to vector control cells (Fig.2d) indicating that our CBX7 construct is functionally active in Tera-2 cells. However, unlike in the studies carried out in fibroblasts (14), p16 protein levels were unchanged in our system (data not shown) suggesting additional gene targets may be important in mediating the observed growth promotion.
CBX7 targets genes frequently silenced in association with hypermethylation in adult cancer
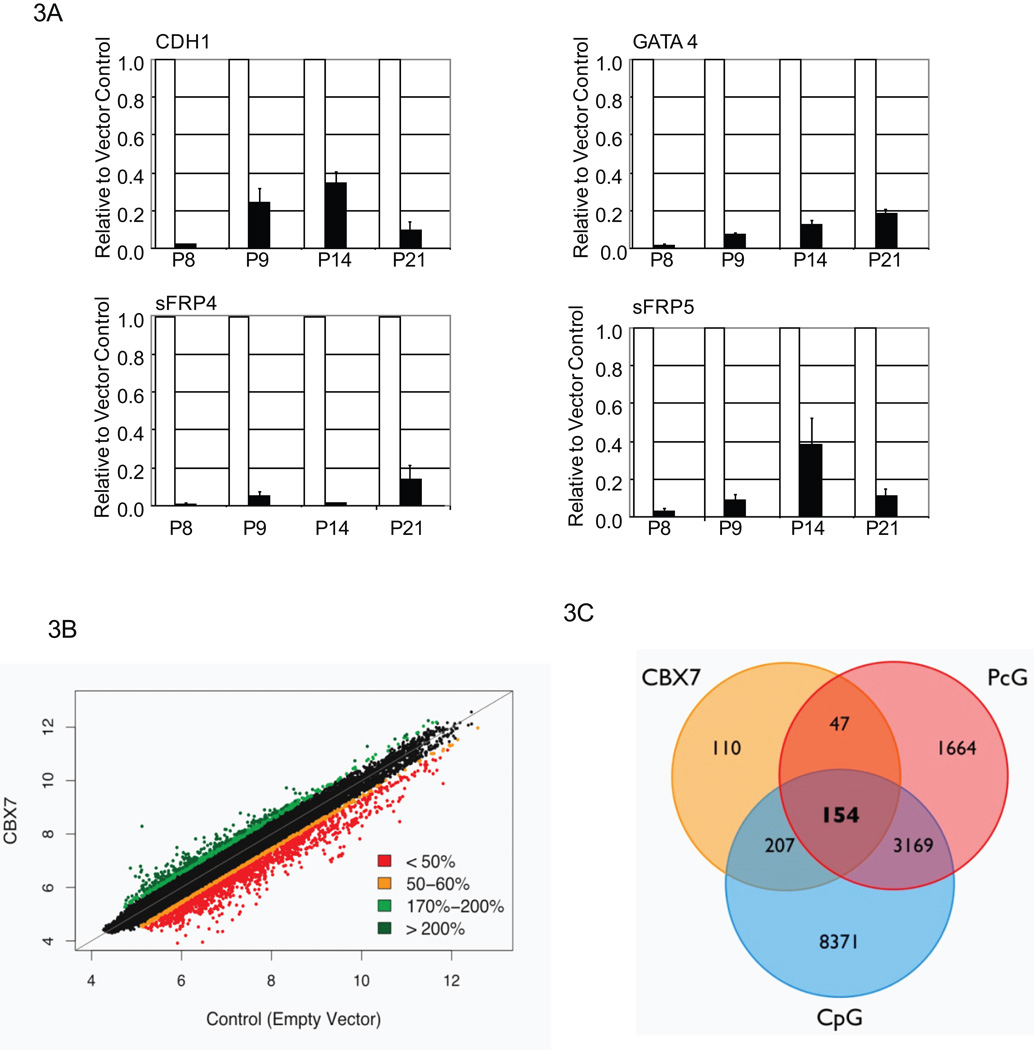
To identify targets of CBX7 mediated gene suppression that would be candidates for CBX7 in initiating stable gene silencing we carried out a real-time based gene survey. Specifically, we queried CpG-island containing genes marked by PcG in embryonic cells (35–37) and often silenced in association with DNA hypermethylation in cancer (1, 2) . We found that CDH1, TIMP3, TSP-1, sFRP4, sFRP5, GATA4, and GATA5 were repressed by CBX7 (data not shown). We selected CDH1, GATA4, sFRP4, and sFRP5, for further study. All four genes are are stably repressed over multiple passages (Fig. 3a).
Figure 3. CBX7 targets genes frequently silenced in adult cancer.
A, Real-time PCR expression in CBX7 cells (black bars) is shown relative to vector control cell expression (white bars), normalized to one. Values represent the mean fold change ± SEM for triplicates. Passage numbers are as indicated. B, Shown is a scatterplot of all probes on the Agilent microarray. Each spot represents the natural logarithm of the normalized single channel value in the empty vector (X-axis) versus the CBX7 overexpressing cells (Y-axis). The color code represents the relative expression level in CBX7 overexpressing cells versus control cells. C, Venn-diagram shows the distribution of CBX7 repressed genes with promoter region CpG-islands and genes regulated by PcG. A saturated generalized linear model fit to the nominal count data showed that associations between genes repressed by CBX7, CpG-island containing genes, and genes that are marked by PcG in progenitor cells are significant (p = 0.018406) as were the interactions between genes down regulated by CBX7 and genes known to be marked by PcG constituents in progenitor cells (p = 9.73e-05) and between genes down regulated by CBX7 containing CpG-islands (p = 0.002425).
We further addressed, on a global level, whether CBX7 may target an even larger pool of genes. Using an Agilent “Whole Human Genome” array (Fig. 3b) we found 518 genes are suppressed to less than fifty percent of control expression. Importantly, over 70 percent, of these genes also contain CpG-islands in their promoters. Finally, 154 genes, equaling almost 50 percent of CBX7 repressed genes that contain CpG-islands, have also previously been identified as PcG targets in stem or progenitor cells (35–38) (Fig. 3c).
Dependence of CBX7 mediated repression on DNMTs
We next utilized ChIP analysis to determine whether CBX7 was localized to its repressed gene targets. We found CBX7 at the promoter of all 4 target genes concomitant with enrichment of DNMT1, DNMT3a, and DNMT3b, (Fig. 4a). While there were no significant changes in repressive or activating histone modifications across all four genes (data not shown), our previously noted presence of “bivalent chromatin” (39) and additional repressive marks, dimethylated and trimethylated H3K9 (18, 40, 41) suggest that these resident histone modifications provide an appropriate setting for PRC1 docking via the CBX7 chromodomain (17) and recruitment of DNMTs
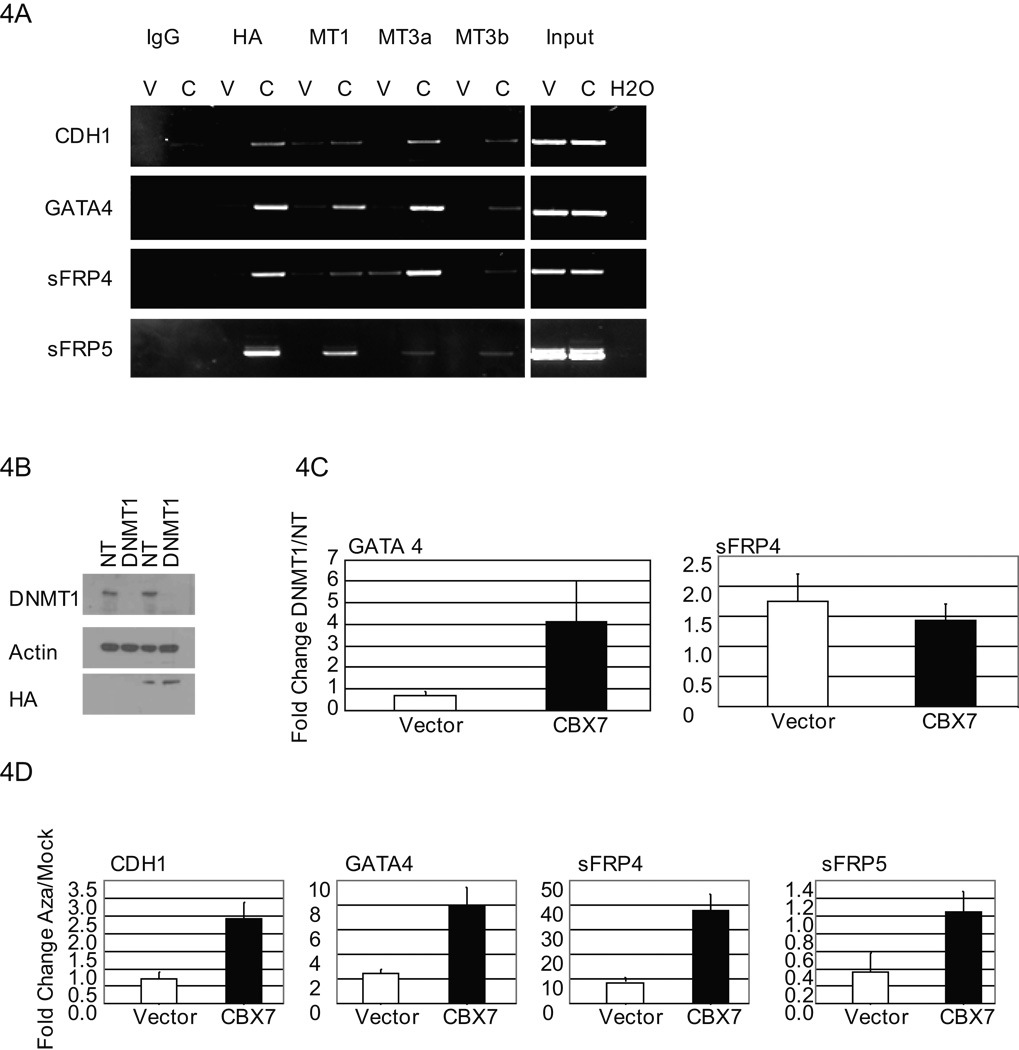
Figure 4. CBX7 repression of target genes is dependent on DNMTs.
A, ChIP was performed using antibodies as indicated on vector control (V) and CBX7 (C) overexpressing Tera-2 cells. PCR was performed for gene promoters as indicated. B, Shown are Western analyses of vector control (lanes 1, 2) and CBX7 overexpressing (lanes 3, 4) Tera-2 cells treated with a non-targeting (NT) siRNA control or DNMT1 targeted siRNA. CBX7 expression is detected by anti- HA antibody. C, Real-time PCR expression is shown as fold change for cells treated with DNMT1 targeted siRNA relative to NT siRNA control. White and black bars represent mean fold change ± SEM for vector control and CBX7 cells, respectively. D, Tera-2 cells stably expressing CBX7 (black bars) and empty vector control cells (white bars) were treated with 1 µM 5-aza-2'-deoxycytidine (5-aza) for 72 hours. Real-time PCR expression is shown as fold change ± SEM for 5-aza treated cells relative to mock treated controls.
The formation of a DNMT containing complex at CBX7 target gene promoters suggests that DNMTs may be required for CBX7 mediated gene suppression. We found that DNMT1 knockdown by siRNA resulted in a selective increase in GATA4 gene expression in CBX7 overexpressing cells only (Fig. 4b, c). sFRP4 gene expression increased somewhat in both vector control and CBX7 cells suggesting that DNMT1 may play a role in establishing the basal expression level for this gene in Tera-2 cells (Fig. 4c). CDH1 and sFRP5 expression were not affected by knockdown of DNMT1 (data not shown).
To further address the potential role for all three DNMTs in CBX7 mediated gene silencing, we treated CBX7 overexpressing and vector control cells with, 5-aza-2'-deoxycytidine, which inhibits all three DNMTs. We observed the most dramatic increase in gene expression of CDH1, GATA4, and sFRP4 selectively in CBX7 expressing cells (Fig. 4d), suggesting that all of the DNMTs are participating in CBX7 mediated repression at target gene promoters.
CBX7 overexpression attenuates response to differentiation by ATRA
PcG targets in embryonic or progenitor cells include many differentiation related genes that are frequently DNA hypermethylated in colon cancers (3–5) . Thus, we tested the effect of sustained CBX7 expression on differentiation in response to ATRA(33, 34). As expected, vector control cells treated with ATRA differentiated and ultimately senesced by day 21 (Fig. 5a). In sharp contrast, CBX7 cells adopted some morphological changes by day 10 but all cells did not senesce and a population actually began to undergo colony formation (Fig. 5b). CBX7 expression was stable throughout the ATRA treatment (Fig. 5b). CBX7 cells did respond to ATRA treatment during the first 10 days by reducing expression of stem cell genes, OCT4 and SOX2 (Fig. 5c). However, the persistent colony forming cells likely represent a selection of cells retaining embryonic properties since OCT4 and SOX2 remain at full or above basal levels by day 30 (Fig. 5c). Additionally, while vector control cells differentiate toward a neural lineage as evidenced by induction of the early neural precursor gene, NESTIN, CBX7 cells showed no change in NESTIN expression (Fig. 5c).
Figure 5. CBX7 overexpression attenuates ATRA responsiveness.
A, Vector control and CBX7 cells treated with ATRA (2µM) for 10 (d10) and 20 (d20) days versus mock treated cells at 10 days are shown in culture at 10× magnification. B, CBX7 cells were Giemsa stained at day 30 (d30). CBX7 expression is detected by anti-HA Western analysis of lysates collected at day 21. C, Stem cell (Oct4 and Sox2) and early neural progenitor marker (Nestin) gene expression was measured by real-time PCR during ATRA treatment of empty vector control (white bars) and CBX7 cells (black bars) for days shown. Expression is shown as fold change relative to Mock treated cells. Values represent the mean fold change ± SEM for triplicates. D, Real-time PCR expression is shown as fold change ± SEM relative to mock treated cells during 2 µM ATRA treatment of vector control cells (white bars) and CBX7 overexpressing cells (black bars) for days shown.
Two CBX7 target genes, GATA4 and sFRP4, have previously been shown to increase in expression upon ATRA induced differentiation in the Tera-2 cells (3). GATA4 has also been previously described as necessary for differentiation of gastric epithelium in mice (42). In vector control cells we saw a 20-fold increase in expression of this gene by 10 of ATRA treatment (Fig. 5d). However, throughout the entire course of ATRA induced differentiation and senescence, there was no change in GATA4 expression in CBX7 expressing cells. sFRP4 expression increased dramatically in the treated, vector control cells but, following a transient increase through day 10 in CBX7 cells, this level was not sustained by 30 days in ATRA (Fig. 5d). Together, these data suggest that CBX7 cells may be initially responsive to ATRA but do not terminally differentiate, can bypass senescence, and survive this process in a progenitor-like state.
CBX7 promotes initiation of DNA hypermethylation
As described, expression of sFRP4 and GATA4 either returned to or was held in a suppressed state in ATRA resistant CBX7 cells (Fig. 5d). This may reflect a selection process in which the CBX7 cells that were unable to induce expression of differentiation responsive genes were the population that escaped ATRA induced senescence. The studies with 5-azacytidine (Fig. 4c) suggest that promoter DNA hypermethylation provides a potential mechanism by which this may occur. Thus, we examined DNA methylation at the promoters of our four target genes that were stably repressed by CBX7.
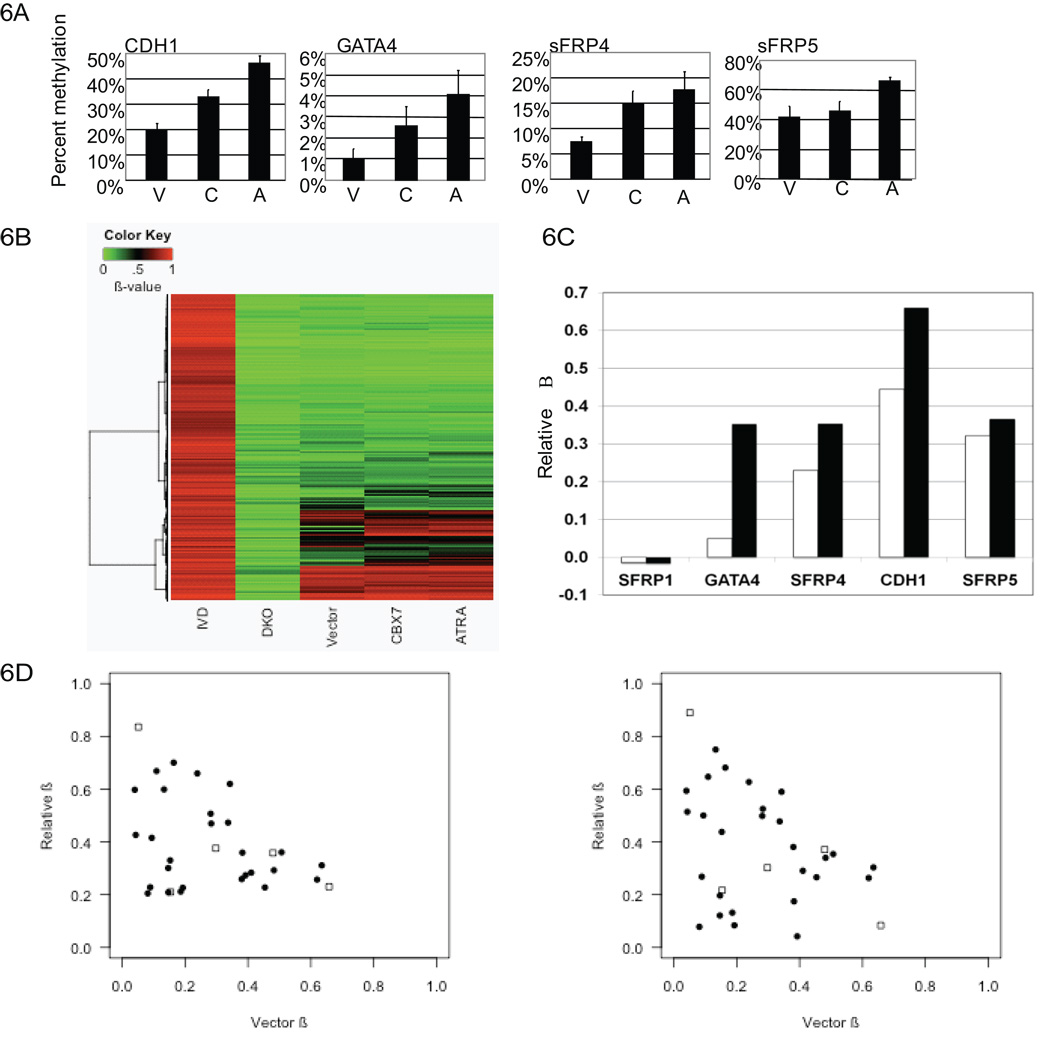
Using bisulfite sequence analysis, we observed a modest increase in methylation of the CDH1, GATA4, and sFRP4 promoters in CBX7 cells relative to parental cells and vector controls (Fig. 6a, S2, S4) DNA methylation became most prominent in all four genes in CBX7 cells that were exposed to ATRA for thirty days (Fig. 6a, S4). A control gene (sFRP1) that was not a target of CBX7 mediated gene repression also did not show any increase in promoter DNA methylation (Fig. S3). These data suggest that CBX7 can contribute to increasing promoter DNA methylation of key genes that may provide an additional layer of target gene suppression especially in cells with a block to differentiation induced cell senescence.
Figure 6. CBX7 promotes initiation of methylation.
A, Values represent numbers of methylated CpG’s relative to total CpG’s per allele for cell lines (passage 30). p-values for the differences between any of the three groups; vector (V) CBX7 (C), and ATRA resistant CBX7 (A) as determined by Kruskal-Wallis test are: CDH1: p = 1.259e-05; GATA4: p = 0.05337; SFRP4: p = 0.005834; SFRP5: p = 0.002336. B, Shown is a heat map depicting β-values for cell lines as indicated. DKO and IVD were used as unmethylated and fully methylated controls, respectively. The heat map is based on hierarchical clustering of β-values using Euclidean distance and Ward algorithm. Only probes within −1000 to +200 relative to the TSS were selected. C, Relative β-values for genes as indicated were calculated by subtracting the β-value for vector control cells from CBX7 overexpressing cells (white bars) or ATRA treated CBX7 cells black bars). D, Scatterplots showing relative β-values for genes that show increased methylation and decrease in expression in CBX7 cells (filled circles) or increased methylation and increased expression in CBX7 cells (open squares). Y-values indicate β-scores from CBX7 cells (left) and ATRA treated CBX7 cells (right) relative to vector control cells. X-values indicate β-scores as measured in vector control cells.
To further address a role for CBX7 in initiation of methylation, we utilized an epigenomic approach for analysis of DNA hypermethylation using the Illumina Infinium bead platform. We interrogated over 27,000 CpG loci that encompass over 14,000 genes (21, 43). While the majority of genes in the Tera-2 cell lines shown have relatively low levels of DNA methylation, we identified a progressive increase in promoter DNA methylation of 441 genes with CpG-islands in CBX7 cells and CBX7 cells that were exposed to ATRA for 30 days (Fig. 6b), independent of their expression status (Supplementary Table 1). 70% of these genes have been previously identified as PcG targets in embryonic cells (35–37), an even higher number than correlated with this parameter in all previous studies (3–5). Also, many genes showing the above increased methylation include genes known to be hypermethylated in human cancer including but not limited to MLH1, HIC1, SOX17 (reviewed in (1, 2, 44), as well as the four genes under study in this report (Fig. 6c).
We then focused our analysis to the subset of genes containing promoter CpG-islands that were both repressed by CBX7 as well as showed increased methylation as measured by Infinium. We identified 25 genes (Supplementary Table 2) that met these criteria whereas only 5 genes showed an increase in methylation that had increased expression upon CBX7 overexpression (Fig. 6d). Interestingly, most genes that showed an increase in methylation had relatively low baseline methylation in vector control cells (B≤0.4) (Fig. 6d). These data suggest that CBX7 mediated repression can promote methylation in a subset of target genes and that this methylation may often reflect a true initiation mechanism rather than expansion of already existing methylation.
DISCUSSION
It has been previously suggested that a stem-cell like chromatin state at the promoter regions of genes frequently targeted for DNA hypermethylation in cancer may predispose these genes for cancer specific silencing (3–5). While such hypotheses have included a potential role for PcG proteins in this process, a clear link has remained elusive. Our results now show a potential direct link between PRC1, stable suppression of gene expression, and the machinery involved in establishing cancer specific silencing in association with induction or expansion of promoter DNA hypermethylation.
We have shown that the RKO colorectal cancer cell line, previously characterized as having a substantial number of genes silenced in association with DNA hypermethylation (19), also contains the highest relative level of CBX7 expression. We have demonstrated that CBX7 can complex with DNMT1, DNMT3a, and DNMT3b. However, despite the presence of a complex containing CBX7 and DNMTs in RKO cells, we are unable to relieve gene suppression upon knockdown of CBX7. There are several potential explanations including one that has been observed for another PcG constituent. Our previous study, as well as those of others, have shown that while the trimethylated H3K27 mark, placed by the methyltransferase, EZH2, may be important in stem cells for targeting genes susceptible to cancer specific DNA methylation (3–5), knockdown of this protein in adult cancers is not sufficient to rescue gene silencing once such genes are DNA hypermethylated (11, 12). These previous findings as well as our current data surrounding knockdown of CBX7 may emphasize the dominant role for dense CpG-island DNA methylation over associated repressive chromatin states in maintaining heritable silencing.
We have also shown that over-expression of the PRC1 protein, CBX7, can lead to increased population doubling of EC cells as well as important epigenetic changes. The growth promotion observed is associated with suppression of genes frequently methylated in cancer including, CDH1, GATA4, sFRP4, and sFRP5. Additional gene expression profiling at the level of the whole genome suggests that a significant number of CBX7 target genes are both PcG marked and contain promoter CpG-islands. As stated, it has been previously postulated that promoters of genes susceptible to cancer specific DNA hypermethylation are enriched for PcG marked chromatin (3–5). We now suggest that such genes are also potential CBX7 targets.
We have further shown that CBX7 localization to target gene promoters results in assembly of a novel complex containing DNMTs. CBX7 mediated gene suppression requires DNMTs, as demonstrated in our study involving 5-aza-2'-deoxycytidine. These data provide the first clear link between PRC1 and DNA methylation machinery both by physical association as well as recruitment to target genes repressed by CBX7.
It is plausible that the initial gene suppression observed upon CBX7 overexpression may involve a non-catalytic function for DNMTs acting instead as transcriptional repressors. CBX7 mediated gene suppression is evident despite little increase in DNA methylation in CBX7 cells in the absence of ATRA treatment. In this regard, previous studies have demonstrated that all three DNMTs are capable of capable of repressing transcription (45–48) in a manner potentially separate from their ability to methylate CpGs (28, 45–48). However, the sustained presence of this DNMT containing complex at CBX7 target gene promoters may eventually allow for a modest induction or progression of promoter DNA methylation, particularly in genes that have low to no baseline methylation. This is more pronounced in ATRA treated cells. Work of others has shown that an antisense RNA causes DNA methylation of a TSG upon cell differentiation (49). Similarly, in our EC system, increased methylation and stable target gene suppression may confer partial resistance to differentiation in that CBX7 targets may be less susceptible to ATRA-induced gene expression. Such stable gene silencing may then promote a selective survival advantage, potentially giving rise to a population of EC cells that resembles adult cancer cells with more extensive aberrant CpG-island promoter DNA methylation. This type of evolution of promoter DNA hypermethylation may reflect a process by which a progenitor cell is rendered abnormal during transformation to an adult cancer cell.
In summary, we have provided a potential mechanism by which PRC1 may contribute to the maintenance of the stem-like state of pluripotent cells via stable repression of key target genes which would otherwise be maintained in a poised state available for signal transduction induced increases in gene expression. We now show that sustained expression and localization of the PRC1 protein, CBX7, in concert with key partner proteins, to target gene promoters in an embryonic tumor cell can blunt response to differentiation cues giving rise to a population of cells which model key epigenetic aspects of adult cancer. Together, our studies highlight a key potential role for the Pc component of the PcG silencing system in tumorigenesis through promotion of epigenetic changes in a progenitor-like cell (Fig. S5).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank J. Herman and B. Vogelstein for critical reading of this manuscript, and A. Ting for assistance in methylation analysis and Kathy Bender for manuscript submission. This work was supported by grants from the (ACS) American Cancer Society Fellowship Grant (HPM) and (NIH) National Institutes of Health CA116160 (SBB).
FUNDING
This work was supported by grants from the American Cancer Society (HPM) and National Institutes of Health (SBB).
REFERENCES
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 3.Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Widschwendter M, Fiegl H, Egle D, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 5.Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 6.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 9.Kirmizis A, Bartley SM, Kuzmichev A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vire E, Brenner C, Deplus R, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 11.McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res. 2007;67:5097–5102. doi: 10.1158/0008-5472.CAN-06-2029. [DOI] [PubMed] [Google Scholar]
- 12.Kondo Y, Shen L, Cheng AS, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 13.Paro R, Hogness DS. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci U S A. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gil J, Bernard D, Martinez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 15.Herman JG, Merlo A, Mao L, et al. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 16.Scott CL, Gil J, Hernando E, et al. Role of the chromobox protein CBX7 in lymphomagenesis. Proc Natl Acad Sci U S A. 2007;104:5389–5394. doi: 10.1073/pnas.0608721104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- 19.Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. ProcNatlAcadSci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bibikova M, Fan JB. GoldenGate assay for DNA methylation profiling. Methods Mol Biol. 2009;507:149–163. doi: 10.1007/978-1-59745-522-0_12. [DOI] [PubMed] [Google Scholar]
- 22.Rhee I, Bachman KE, Park BH, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 23.Easwaran HP, Schermelleh L, Leonhardt H, Cardoso MC. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 2004;5:1181–1186. doi: 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bestor TH. Cloning of a mammalian DNA methyltransferase. Gene. 1988;74:9–12. doi: 10.1016/0378-1119(88)90238-7. [DOI] [PubMed] [Google Scholar]
- 25.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 26.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 27.Bachman KE, Park BH, Rhee I, et al. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell. 2003;3:89–95. doi: 10.1016/s1535-6108(02)00234-9. [DOI] [PubMed] [Google Scholar]
- 28.Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 29.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- 31.Esteve PO, Chin HG, Smallwood A, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Epsztejn-Litman S, Feldman N, Abu-Remaileh M, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrews PW. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev Biol. 1984;103:285–293. doi: 10.1016/0012-1606(84)90316-6. [DOI] [PubMed] [Google Scholar]
- 34.Andrews PW, Gonczol E, Plotkin SA, Dignazio M, Oosterhuis JW. Differentiation of TERA-2 human embryonal carcinoma cells into neurons and HCMV permissive cells. Induction by agents other than retinoic acid. Differentiation. 1986;31:119–126. doi: 10.1111/j.1432-0436.1986.tb00392.x. [DOI] [PubMed] [Google Scholar]
- 35.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006 doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. JMolBiol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 39.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 40.Kondo Y, Shen L, Issa JP. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol. 2003;23:206–215. doi: 10.1128/MCB.23.1.206-215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen CT, Weisenberger DJ, Velicescu M, et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2'-deoxycytidine. Cancer Res. 2002;62:6456–6461. [PubMed] [Google Scholar]
- 42.Jacobsen CM, Narita N, Bielinska M, Syder AJ, Gordon JI, Wilson DB. Genetic mosaic analysis reveals that GATA-4 is required for proper differentiation of mouse gastric epithelium. Dev Biol. 2002;241:34–46. doi: 10.1006/dbio.2001.0424. [DOI] [PubMed] [Google Scholar]
- 43.Butler H, Ragoussis J. BeadArray-based genotyping. Methods Mol Biol. 2008;439:53–74. doi: 10.1007/978-1-59745-188-8_4. [DOI] [PubMed] [Google Scholar]
- 44.Zhang W, Glockner SC, Guo M, et al. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res. 2008;68:2764–2772. doi: 10.1158/0008-5472.CAN-07-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. NatGenet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 46.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. NatGenet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 47.Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. Embo J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 49.Yu W, Gius D, Onyango P, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.