Abstract
Physiological and premature aging are characterized by multiple defects in chromatin structure and accumulation of persistent DNA damage. Here we identify the NURD remodeling complex as a key modulator of these aging-associated chromatin defects. We demonstrate loss of several NURD components during premature and normal aging and we find aging-associated reduction of HDAC1 activity. Silencing of individual NURD subunits recapitulates some chromatin defects associated with aging and we provide evidence that structural chromatin defects precede DNA damage accumulation. These results outline a molecular mechanism for chromatin defects during aging.
A hallmark of normal and premature aging are global changes in chromatin including loss of heterochromatin structure, altered patterns of histone modifications 1–4, loss of key heterochromatin proteins 2, 3, and increased levels of persistent DNA damage 5, 6. To begin to elucidate the molecular mechanisms leading to these aging-related chromatin defects, we have taken advantage of the premature aging disorder Hutchinson-Gilford Progeria Syndrome (HGPS). This early childhood disease is caused by a recurrent denovo point mutation in exon 11 of the lamin A/C gene (LMNA), which encodes for the intermediate filament proteins lamin A and lamin C, two of the key architectural proteins of the cell nucleus 7. The disease-causing mutation in LMNA leads to the production of a mutant form of lamin A, referred to as progerin, which lacks 50 aa near the C-terminus, and acts in a dominant negative fashion causing multiple cellular defects of physiological and accelerated-aging 1, 2, 8, 9. In particular, HGPS cells exhibit several chromatin defects which are also characteristic for physiological aging including loss of heterochromatin structure, loss of methylation at H3K9 and H3K27, down-regulation of the heterochromatin protein HP1, and increased transcription of pericentromeric Satellite III repeats (SatIII) 2, 4. In addition, as normally aged cells, HGPS patient cells exhibit increased steady-state levels of DNA damage 3, 5.
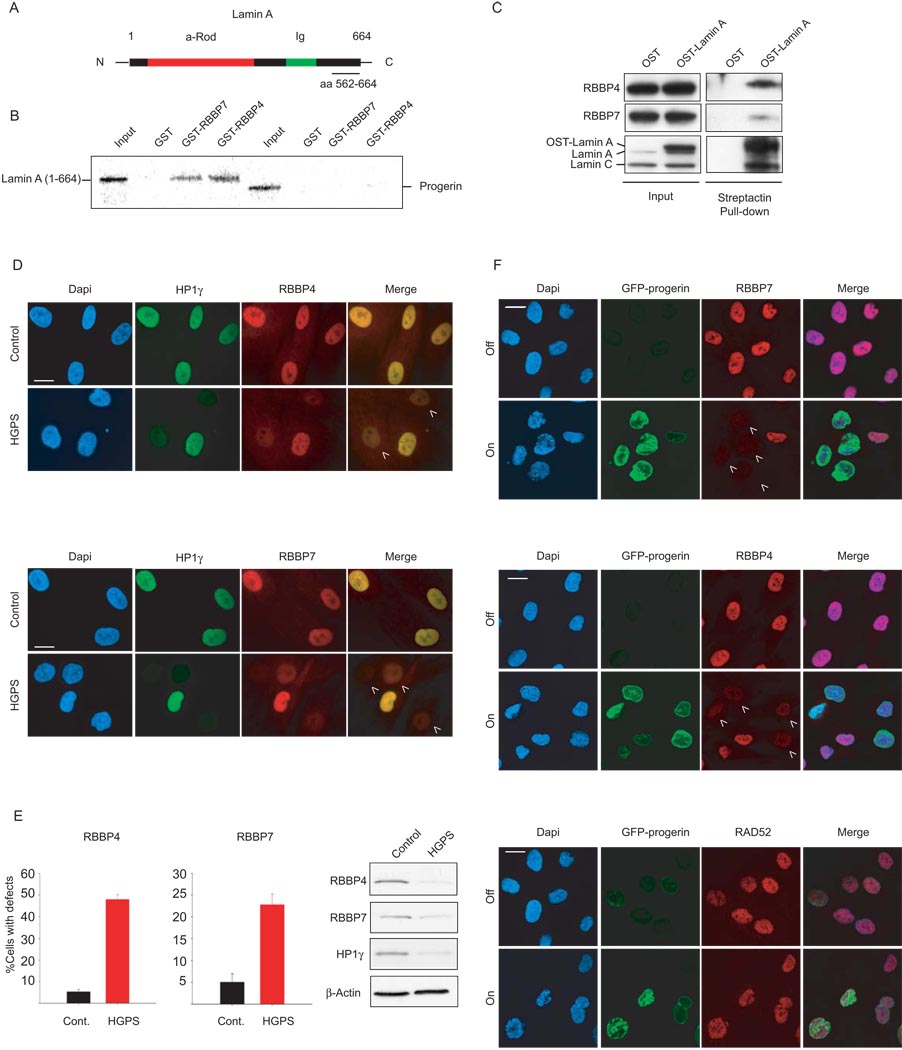
To identify the molecular basis of aging-associated chromatin defects, we performed a yeast two hybrid screen using a C-terminal region of lamin A (aa 562–664) which overlaps with the region deleted in progerin (Fig. 1A). We identified the WD40 domain chromatin protein RBBP4 as a lamin A interactor in four independent experiments. None of the other lamin A fragments tested (aa 1–118, aa 416–568, aa 604–657) interacted with RBBP4 by two-hybrid analysis (data not shown). The interaction between RBBP4 and the aa 562–664 fragment of lamin A was confirmed by GST-pull down (Supplementary Fig. 1A). RBBP4 and RBBP7 are evolutionary conserved histone binding proteins 10. They are shared subunits of several multi-protein complexes involved in the establishment of heterochromatin including the NUcleosome Remodeling and Deacetylase (NURD) complex 11 and the Polycomb PRC2 complex 12. RBBP4 is also the p48 subunit of the CAF-1 complex, which assembles chromatin upon DNA replication and DNA damage repair 13. In support of a physical interaction between lamin A and RBBP4/7, recombinant forms of both proteins bound in vitro to GST-lamin A (1–664) and to a lesser extend to a C-terminal fragment (residues 390 – 646) (Fig. 1B; Supplementary Fig. 1B). Importantly, neither protein bound to GST-progerin which lacks residues 607–657 of mature lamin A (Fig. 1B). The in vivo interaction of RBBP4, and more weakly of RBBP7, with lamin A was confirmed by immunoprecipitation of endogenous RBBP4/7 by overexpressed One-Strep (OST)-tagged lamin A (Fig. 1C). These results identify the two histone chaperone proteins RBBP4 and 7 as lamin A interactors.
Fig. 1. Loss of RBBP4 and RBBP7 in progerin expressing cells.
(A) Schematic representation of lamin A. The fragment used as a bait in the Y2H assay (aa 562–664) is underlined. (B) GST pulldown assays with recombinant GST-RBBP4/7 and in vitro transcribed and translated lamin A (aa 1–664) or progerin. (C) Crosslinked protein lysates were prepared from U2OS cells expressing One-Strep (OST)-tagged lamin A and then incubated with Streptactin beads. Input and pulled down material was probed by Western blot with antibodies against the indicated endogenous proteins. (D) Immunofluorescence staining on control and HGPS primary dermal fibroblasts with the indicated antibodies. DAPI, 4′, 6′-diamidino-2-phenylindole. Quantitative analysis of fluorescence intensity signal in control and HGPS patient cells. The percentages of cells with reduced protein level are indicated. N>200; values represent averages ± S.D from at least three experiments. Cells were analyzed around population doubling (PD) 25 in the RBBP4 experiments and around PD 20 in the RBBP7 experiments. Control cells were passage-matched to HGPS cells in each experiment. Western blot of cell lysates prepared from control and HGPS dermal fibroblasts. (E) Immunofluorescence with indicated antibodies in wt fibroblasts that were either induced (on) or not induced (off) to express a GFP-tagged version of progerin. Arrowheads indicate cells with reduced levels of heterochromatin proteins. Scale bars: 10 µm.
To ask whether RBBP4 and RBBP7 are involved in aging-associated chromatin defects, we probed their status in HGPS cells. Strikingly, a substantial fraction (48 +/− 2%) of HGPS patient cells showed reduced protein levels of RBBP4 when compared to passage- and age-matched controls from healthy donors by quantitative immunofluorescence microscopy as previously described (Fig. 1D, E)2, 3. The protein level of RBBP7 was similarly reduced in HGPS patient cells (Fig. 1D, E). As previously observed for other nuclear defects in HGPS cells, the extent of reduction was variable amongst cells in the population, but its extent was similar to that observed in HGPS for several other nuclear proteins including all isoforms of the heterochromatin protein HP1. Furthermore, loss of RBBP4/7 occurred in the same cells that exhibited lower levels of HP1 pointing to global chromatin defects in those cells (Fig. 1D) 2. Reduction of RBBP4 and RBBP7 in HGPS skin fibroblasts relative to control cells was confirmed by Western blot analysis on total cell lysates (Fig 1E). Similar observations were made in several independent primary HGPS patient cell lines (data not shown) and, as observed for HP1, the extent of RBBP4/7 reduction increased during cell passaging (data not shown; ref. 3, 4). RBBP4 and RBBP7 down-regulation occurred at the protein level since similar levels of mRNAs were present in HGPS and control fibroblasts as determined by RT-PCR (Supplementary Fig. 2A). Consistent with the reduction of RBBP4/7, the levels of the centromeric protein CENP-A was reduced in HGPS cells (Supplementary Fig. 2B, 2C), in agreement with earlier observations demonstrating loss of CENP-A after concomitant siRNA silencing of RBBP4 and RBBP7 14.
The loss of RBBP4 and RBBP7 was dependent on progerin. Upon controlled induction of a GFP-tagged version of progerin in immortalized wild-type skin fibroblasts using a tetracycline-dependent expression system 6, RBBP4 and RBBP7 were almost completely lost within 4 days when compared with the uninduced control (Fig 1F). The control protein RAD52 was not affected by GFP-progerin expression (Fig. 1F). Conversely, elimination of progerin pre-mRNA from HGPS skin fibroblasts by a previously characterized RNA morpholino approach2 rescued the levels of RBBP4 when compared to the same cells treated with a scrambled control oligonucleotide (Supplementary Fig. 2D). In addition, staining of HGPS primary fibroblasts with specific anti-progerin antibodies revealed that expression of progerin inversely correlated with RBBP4 levels in these cells (Supplementary Fig. 2E). Collectively, these results demonstrate loss of the histone chaperones RBBP4 and RBBP7 in HGPS cells in a progerin-dependent manner.
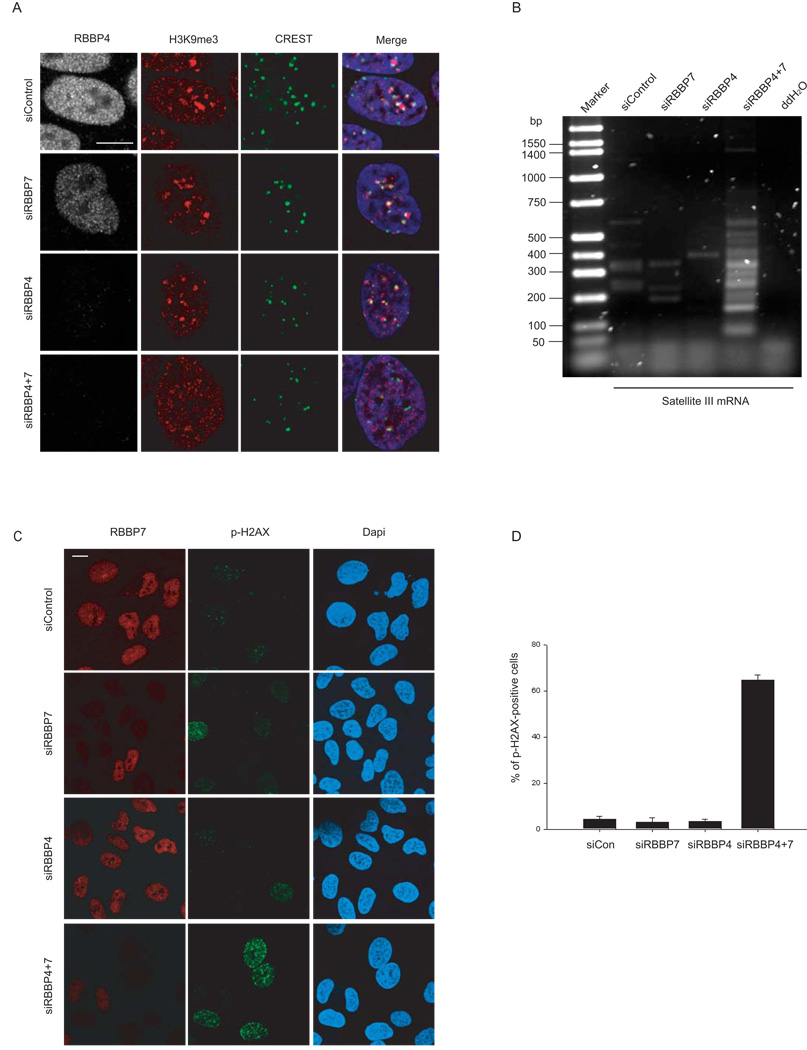
Given the role of RBBP4 and RBBP7 in various chromatin remodeling and assembly complexes, we hypothesized that their loss contributes to the changes in chromatin structure and elevated levels of DNA damage that occur during premature and normal aging. To directly test whether the absence of RBBP4/7 is sufficient to cause these defects, we analyzed the status of pericentromeric heterochromatin after siRNA-mediated knockdown of RBBP4 and RBBP7, either alone or in combination (Fig. 2). Efficient knockdown was confirmed by Western blotting (Supplementary Fig. 3A). As expected, cells treated with non-targeting control siRNA oligonucleotides displayed several bright foci of H3K9me3, corresponding to pericentromeric heterochromatin15, as confirmed by partial colocalization of these foci with CREST antibody which recognizes centromeric proteins (Fig. 2A). Upon simultaneous knock-down of RBBP4 and RBBP7 the focal accumulation of H3K9me3 was lost (Fig. 2A). Silencing of either RBBP4 or RBBP7 alone was not sufficient to cause heterochromatin alterations (Fig. 2A), most likely reflecting the functional overlap between these two highly similar proteins. In line with previous observations14 concomitant silencing of RBBP4 and RBBP7 led to loss of the centromeric protein CENP-A (Supplementary Fig. 3B). To further test whether RBBP4 and RBBP7 are involved in mediating the chromatin defects observed in HGPS cells, we probed the effect of loss of RBBP4 and RBBP7 on DNA SatIII repeat transcription by semi-quantitative RT-PCR (Fig. 2B). SatIII repeats are normally maintained in a transcriptionally silent state but are activated in HGPS cells, most likely as a result of heterochromatin disruption 4, 16. As observed in HGPS cells, simultaneous silencing of RBBP4 and RBBP7 caused a significant increase in SatIII transcription compared to control siRNA treated cells (Fig. 2B).
Fig. 2. Heterochromatin defects and increased DNA damage upon RBBP4 and RBBP7 silencing.
(A) HeLa cells were transfected with the indicated combinations of siRNA for 144 hrs and processed for immunofluorescence staining with the indicated antibodies. (B) Semi-quantitative, strand specific RT-PCR analysis of SatIII G-rich transcripts in cells knocked down for the indicated gene. (C) Detection of DNA damage in knock-down cells using α-phospho-H2AX antibody. (D) Quantitative analysis of the percentage of p-H2AX positive cells (defined as containing more than 8 foci per cell).Values represent averages ± S.D. from three experiments. N>200. Scale bars: 10 µm.
To test whether loss of RBBP4 and RBBP7 is also sufficient to induce the increased levels of DNA damage present in HGPS cells, we probed RBBP4 and RBBP7 depleted cells for the presence of phosphorylated histone H2AX, a marker of DNA damage 17. Similar to cells from HGPS patients and aged individuals, the percentage of cells containing multiple prominent phospho-H2AX foci increased from 5% in control cells to more than 60% in cells depleted of RBBP4 and RBBP7 (Fig. 2C, D) 3. The increase in endogenous DNA damage upon RBBP4/7 silencing was mirrored by an increased proportion of cells in S-phase as detected by FACS analysis (Supplementary Fig. 3C, D; P < 0.01 for Hela cells, P < 0.01 for U2OS cells), in agreement with the accumulation in S-phase of late-passage HGPS cells (Supplementary Fig. 3E, P < 0.01 ). While the percentage of cells with aberrant chromatin structure in RNAi-treated cells was significantly increased 72 h after RNAi knockdown when compared to the control (Supplementary Fig. 4A), DNA damage only became evident at 120h (Supplementary Fig. 4B), suggesting the occurrence of structural chromatin defects prior to DNA damage. Similar results were obtained after reduction of RBBP4/7 levels by induced expression of progerin (Supplementary Fig. 5).
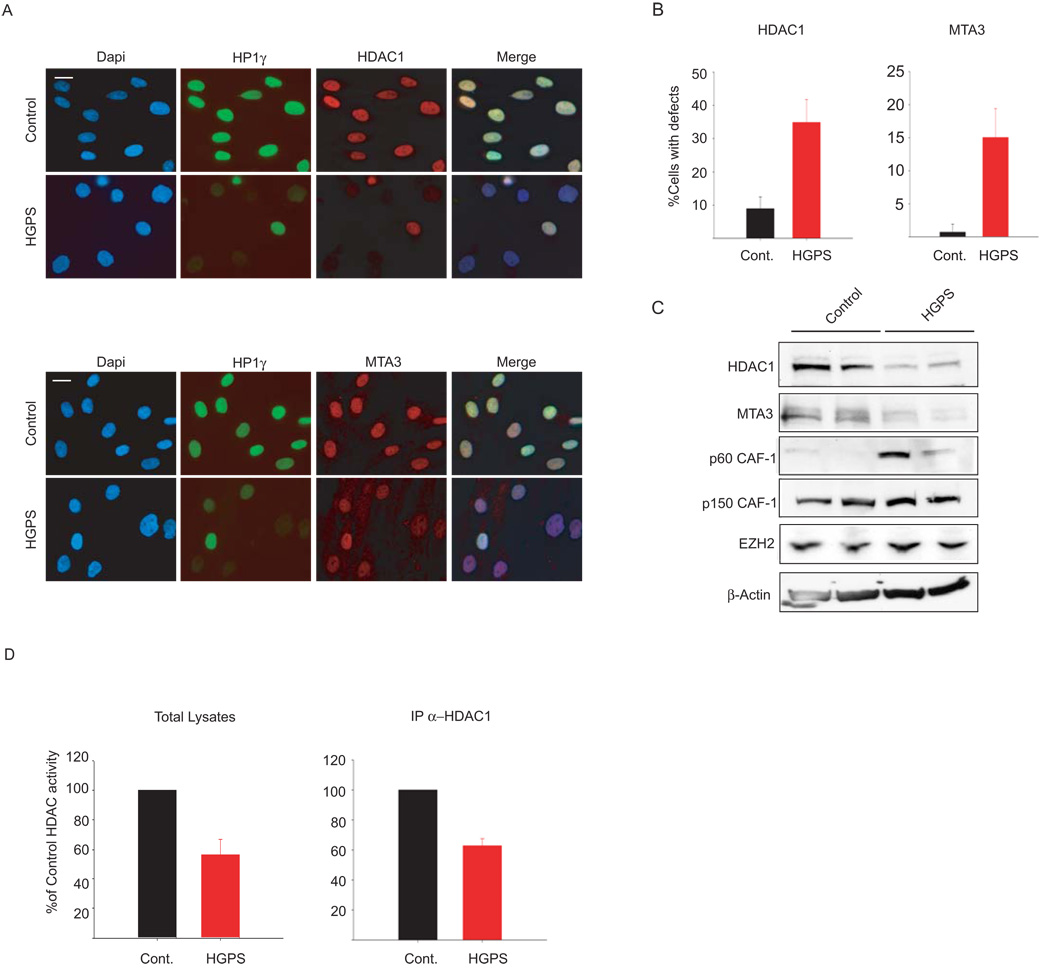
RBBP4 and RBBP7 are shared subunits of multiple protein complexes involved in chromatin metabolism, including the NURD complex and the PRC2 complex 11, 12. In addition, RBBP4 is an integral subunit of the CAF-1 complex 13. To test which of these complexes mediates the aging-associated chromatin defects caused by RBBP4 and RBBP7 loss, we tested the status of other subunits of the NURD, CAF-1 and PRC2 complexes in HGPS cells (Fig. 3). Similar to RBBP4 and RBBP7, two additional subunits of the NURD complex, MTA3 and HDAC1, were largely lost from HGPS cells as judged by quantitative single-cell microscopy and Western blotting (Fig. 3A–C). Consistent with a possible regulatory role of lamin A on the NURD complex, HDAC1 physically interacted with lamin A in vivo as demonstrated by its immunoprecipitation with OST-lamin A (Supplementary Fig. 6A). In contrast, protein levels of p150 and EZH2, the catalytic subunit of the CAF-1 and the PRC2 complexes, respectively, were unaffected in HGPS cells (Fig. 3C). The p60 subunit of the CAF-1 complex was up-regulated in HGPS primary fibroblasts (Fig. 3C), likely as a consequence of persistent DNA damage18. We conclude the NURD complex is lost from HGPS patient cells.
Fig. 3. Loss of NURD subunits in HGPS cells.
(A) Immunofluorescence staining of control and HGPS primary dermal fibroblasts with the indicated antibodies. (B) Quantitative analysis of the fluorescence intensity signal in control and HGPS cells. N>200; Values represent averages ± S.D. from three experiments. (C) Western blot with the indicated antibodies on total cell lysates prepared from dermal fibroblast from two unaffected individuals and two HGPS patients. (D) HDAC activity measured in total lysates or in HDAC1 immunoprecipitates prepared from primary fibroblasts. Values represent averages ± S.D. from three experiments. Scale bar: 10 µm.
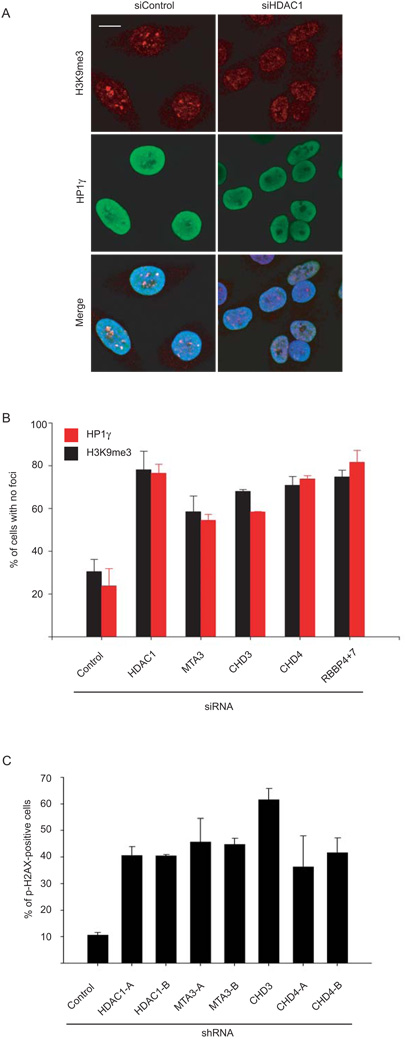
The loss of HDAC1 protein in HGPS cells pointed to the possibility that the observed chromatin defects are caused by loss of cellular HDAC1 deacetylase activity. To test this hypothesis we measured HDAC1 activity in HGPS patient cells. HDAC1 activity in total cell extracts and in HDAC1 immunoprecipitates prepared from HGPS patient cells was reduced by ~ 40% compared to matched control cells (Fig. 3D). To directly probe whether the NURD complex is responsible for the aging-associated chromatin defects, we individually knocked down the NURD subunits HDAC1, MTA3, CHD3 or CHD4 in Hela cells (Supplementary Fig. 6B). In a manner similar to RBBP4/7 knock down, silencing of any subunit increased the percentage of cells lacking H3K9me3 and HP1γ heterochromatin foci (Fig. 4A, B). Furthermore, shRNA mediated silencing of HDAC1, MTA3, CHD3 or CHD4 in primary human fibroblasts (Supplementary Fig. 6C, D) increased the percentage of cells containing phospho-H2AX positive foci to similar levels as observed in HGPS cells (Fig. 4C, D). These results demonstrate that HDAC1 activity is reduced in HGPS cells and that loss of NURD complex components is sufficient to recapitulate some aging-associated chromatin defects.
Fig. 4. Heterochromatin defects and increased DNA damage upon silencing of NURD subunits.
(A) HeLa cells were transfected with the indicated siRNA for 96 hrs and processed for immunofluorescence staining with the indicated antibodies. Scale bar: 10 µm. (B) Quantitation of the percentage of siRNA treated Hela cells in which H3K9me3 or HP1γ foci are absent. Values represent averages ± S.D. from two experiments. N>200. (C) Quantitation of the percentage of phospho-H2AX positive cells (defined as containing more than 8 foci per cell).Values represent averages ± S.D. from three experiments. N>200. Scale bar: 10 µm.
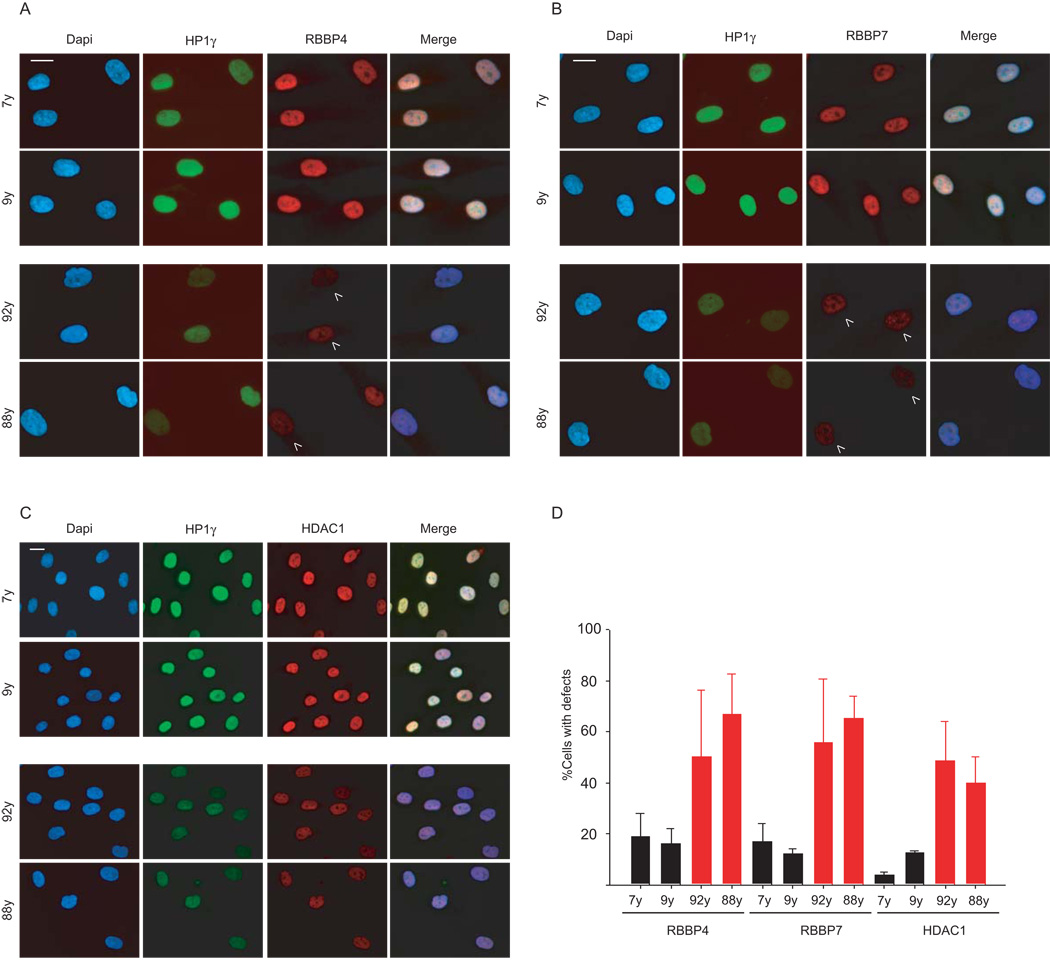
Since chromatin defects associated with HGPS also occur during physiological aging 3, we finally probed the status of the NURD subunits in cells from normally aged individuals. Quantitative immunofluorescence microscopy analysis on cells obtained from two young donors (7 and 9 years, respectively) and passage-matched primary fibroblasts from two old donors (92 and 88 years, respectively), revealed significant differences in RBBP4, RBBP7, and HDAC1 protein levels (Fig 5A–C). Cell populations from old individuals consistently showed a 4–5 fold increase in the percentage of cells with reduced protein levels of RBBP4, RBBP7 and HDAC1 (Fig. 5D). At the single cell level, the extent of reduction of NURD components strictly correlated with each other and with HP1γ levels, albeit less pronounced than that observed for HGPS cells as previously described 3. In contrast to HGPS cells, no loss of CENP-A was detected in normally aged cells. We conclude that loss of the RBBP4, RBBP7, and HDAC1 subunits of NURD is not limited to premature aging but is also a feature of physiologically aged cells.
Fig. 5. Loss of NURD subunits in physiological aging.
(A), (B) and (C) Immunofluorescence microscopy of passage-matched primary dermal fibroblasts from healthy young (7 yrs and 9 yrs) or old (92 yrs and 88 yrs) donors were fixed and stained with the indicated antibodies. (D) Quantitative analysis of the fluorescence intensity signal measured in dermal fibroblasts from young and old individuals. N>200; Values represent averages ± S.D. from three experiments. Arrowheads indicate cells with reduced levels of heterochromatin proteins. Scale bars: 10 µm.
Here we have identified the NURD chromatin remodeling complex as a mediator of aging-associated chromatin defects. We demonstrate reduction of several NURD subunits in cells from a progeroid syndrome and from normally aged individuals. Loss of any one of these components and reduction of HDAC1 activity is sufficient to recapitulate multiple aging-associated chromatin defects. The in vivo physical interaction of lamin A with RBBP4, RBBP7 and HDAC1 observed here, and previously suggested by biochemical fractionation 19, points towards a regulatory role for the nuclear lamina in the function of the NURD complex in normal cells.
We find that loss of RBBP4/7 is dependent on the presence of progerin and is an early event in the appearance of aging-associated chromatin defects. Upon induction of progerin or upon knock-down of RBBP4 and RBBP7, changes to heterochromatin structure occur and are followed by accumulation of DNA damage. We suggest that loss of critical chromatin components RBBP4/7 is an upstream event in the formation of aging-associated chromatin defects. Loss of RBBP4/7 compromises the establishment and maintenance of histone modifications and higher order chromatin structure, possibly making chromatin more susceptible to DNA damage. The increase in DNA damage might be a consequence of higher susceptibility of the affected chromatin to damage or alternatively due to impaired DNA replication 20. Interestingly, similar to the effects we observe upon silencing of RBBP4 and RBBP7, impairment of the H4K20 histone methyltransferase PR-Set7, which is necessary for proper H4K20 tri-methylation associated with heterochromatin, interferes with DNA replication, arrests cells in S-phase and causes increased levels of DNA damage 21, 22.
Our finding that multiple NURD components are lost in HGPS and normally aged cells suggests that the integrity of the NURD complex is compromised and its components are down-regulated during premature and physiological aging. Whether loss of these proteins occurs by activation of a specific protein degradation program or by interference with the normal homeostasis of the NURD protein complex remains to be determined. The involvement of a specific pathway is suggested by the fact that progerin affects the protein levels of several subunits of the NURD complex, but not of other RBBP4- or RBBP7-containing complexes, such as CAF-1 or PRC2. While we cannot rule out that the degradation mechanism of NURD components are distinct in HGPS cells and in normally aged cells, the fact that progerin is also expressed at low levels in cells from normally aged individuals is consistent with a conserved degradation mechanism 3. Overall, these results implicate declining NURD remodeling complex function in aging-associated chromatin defects and accumulation of DNA damage during aging.
Materials and Methods
Cell culture
Primary dermal fibroblast cell lines were obtained from the NIA and NIGMS collections of the Coriell Cell Repository (CCR), from the American Type Culture Collection (ATCC) or the Progeria Research Foundation (PRF). The cells from HGPS patients were AG01972 (Patient age 14 years, CCR, population doublings (PD) 20 at purchase), AG06297 (8 years, CCR, PD17), HGDAFN003 (2 years, PRF, PD7). Control cell lines were CRL-1474 (7 years, ATCC, PD19) GM00038 (9 years, CCR, PD17), AG05247 (88 years, CCR, PD15) and AG09602 (92 years, CCR, PD 12). Cells were used at PD’s 19–27. WI-38 primary human fetal fibroblasts (3 month gestation fetus, ATCC, PD8)were used at PD’s 10–18. All primary fibroblasts were grown in Minimum Essential Medium (MEM, Gibco) , 15% FCS, 2mM L-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin at 37 °C in 5% CO2. Growth and induction of immortalized, Tet-off human fibroblasts expressing GFP-Progerin was as previously described 6. U2OS cells stably expressing One-Strep-Lamin A were maintained in McCoy’s 5A medium (Gibco), 10% FCS, 2mM L-glutamine, 10 µg/ml blasticidin at 37 °C in 5% CO2.
Plasmids
Plasmids expressing GST -RBBP4/7 were a kind gift of B. Stillman (Cold Spring Harbor, NY)10. Vectors for in vitro transcription and translation of lamin A and progerin were constructed as follows: an EcoRI/BamHI fragment containing the entire CDS of lamin A (1–664) or progerin was excised from either pEGFP-Lamin A or pEGFP-Lamin Δ50 2, respectively, and cloned into the pCDNA3.1 (−) vector (Invitrogen) cut with the same enzymes. pGBKT7-lamin A (390–646) was a kind gift of T. Dittmer (NCI/NIH, Maryland). N-terminal One-Strep tag (IBA BioTagnology, Göttingen, Germany) oligonucleotides (Sigma, Zwijndrecht, Netherlands) were cloned into BamHI and EcoRI sites of pBabe puro (Plasmid 1764, Addgene). The retroviral pBabe puro One-Strep-Lamin A plasmid was generated by inserting lamin A from the pBabe puro HA-Lamin A vector23 into the SalI and EcoRI sites. In order to generate a lentiviral expression system a BamHI and EcoRI containing multiple cloning site was introduced in the pCDH1-MCS1-EF1-Puro vector (System BioSciences, Mountainview, California, USA), the puromycin cassette was replaced by a blasticidin cassette from the pLenti6/V5-GW/LacZ vector (Invitrogen, Carlsbad, California, USA) and the One-STrEP-tagged lamin A from the pBabe puro One-Strep-Lamin A vector was ligated into the BamHI and EcoRI restriction sites, thereby generating the lentiviral pCDHblast MCSNard OST-Lamin A vector. The Yeast Two Hybrid screen was performed as described24, 25
siRNA and shRNA constructs
siGenome SMART pool siRNA oligos against human RBBP4 and RBBP7 were purchased from Dharmacon-Thermo Scientific and transfected at 0 and 72h into Hela cells at 50 nM using Oligofectamine (Invitrogen) or Dharmafect1 (Dharmacon-Thermo), following the manufacturer instructions. RBBP4:
| Target 1: | 5′-GAUACUCGUUCAAACAAUA |
| Target2: | 5′-GAACUGCCUUUCUUUCAAU |
| Target3: | 5′-GGAUACUCGUUCAAACAAU |
| Target4: | 5′-AACAAUACUUCCAAACCAA |
| RBBP7: | |
| Target1: | 5′-GGAUAAGACCGUAGCUUUA |
| Target2: | 5′-CCACUGGUCUCCACAUAAU |
| Target3: | 5′-GGACACACUGCUAAGAUUU |
| Target4: | 5′-AAGUAAACCGUGCUCGUUA |
| HDAC1: | |
| Target1: | 5′-CUAAUGAGCUUCCAUACAA |
| Target2: | 5′-GAAAGUCUGUUACUACUAC |
| Target3: | 5′-GGACAUCGCUGUGAAUUGG |
| Target4: | 5′-CCGGUCAUGUCCAAAGUAA |
| MTA3: | |
| Target1: | 5′-GCAGAAACAUCAGUUGAAA |
| Target2: | 5′-UGACUAGCAUCAUUGAAUA |
| Target3: | 5′-CUUCAAUGACAUACGGCAA |
| Target4: | 5′-GUGCAACAGAAACGUCUAA |
| CHD3: | |
| Target1: | 5′-CGUAUGAGCUGAUCACCAU |
| Target2: | 5′-GAGGAGAAGUACUAUCGUU |
| Target3: | 5′-GAGGAGAAGUACUAUCGUU |
| Target4: | 5′-CAAGAUAGAUCAUAAGUUG |
| CHD4: | |
| Target1: | 5′-CCAAGGACCUGAAUGAUGA |
| Target2: | 5′-CAAAGGUGCUGCUGAUGUA |
| Target3: | 5′-GAAAGAGGCAUCUGUGAAA |
| Target4: | 5′-GAUGAUAUCCUCAAAUUUG |
GIPZ lentiviral vectors expressing shRNAmir sequences targeted against human HDAC1, MTA3, CHD3 and CHD4 were purchased from Open Biosystems-Thermo Scientific (Waltham, MA) and virus produced in 293FT packaging cells (Invitrogen) usingFUGENEHD (Roche), following the manufacturer’s instructions. Target WI38 fibroblasts (~ 50% confluent) were incubated for 8 hrs with a 1:1 mix of filtered viral supernatant and MEM+15%FBS and in the presence of 6 µg/ml Polybrene (Sigma-Aldrich). A second round of infection with fresh lentiviral vector-containing supernatant was performed after 24 hrs. 48–72 hrs after the first round of infection, puromycin (2 µg/ml) was added to the medium to select shRNAmir expressing cells. Experiments were performed 6 days after the first round of infection.
GIPZ non-targeting sequence: GIPZ non-targeting sequence: 5′-TCTCGCTTGGGCGAGAGTAAG
HDAC1-A: 5′-CCCGAATCCGCATGACTCATAA
HDAC1-B: 5′-AACCCATTCTTCCCGTTCTTAA
MTA3-A: 5′-CCCTAATATGCAGTGTAGATTA
MTA3-B: 5′-CCGGCCGTTTGTTGCTATTAAT
CHD3: 5′-AGGAATTACCACTATCTAGTAA
CHD4-A: 5′-CGCTGCTGACATCCTATGAATT
CHD4-B: 5′-CGCCCTCCAAGACAGAACTAAT
Antibodies
The following antibodies were used in immunofluorescence, Western blotting and immunoprecipitation: α-HP1γ mouse monoclonal (MAB3450, Chemicon), α-H3K9me3 rabbit polyclonal (4861, kindly provided by T. Jenuwein), α-RBBP4 mouse monoclonal (11G10, Abcam), α-RBBP4 rabbit polyclonal (ab1765, Abcam), α-RBBP7 rabbit polyclonal (ab3535, Abcam), α-RAD52 rabbit polyclonal (3425, Cell Signaling), α-p-H2AX mouse monoclonal (JBW301, Millipore), α-p-H2AX mouse monoclonal (2F3, Abcam), α-HDAC1 rabbit polyclonal (PA1-860, ABR), MTA3 rabbit polyclonal (A300-160A, Bethyl Laboratories), α-p150 CAF-1 mouse monoclonal SS1 and α-p60 CAF-1 mouse monoclonal SS53 (kind gifts of B. Stillman), α-EZH2 rabbit polyclonal (39103, Active Motif), α-CHD3 rabbit polyclonal (A301-219A, Bethyl Laboratories), α-CHD4 rabbit polyclonal (H-242, St. Cruz), α-CHD4 mouse monoclonal (91984, Genetex), α-Lamin A goat polyclonal (N-18, St. Cruz), α-progerin mouse monoclonal 13A4, (Alexis Biochemicals), α- centromeric proteins CREST human autoimmune serum (Antibodies Inc.), α-CENP-A mouse monoclonal (3–19, Abcam).
Immunofluorescence microscopy and image quantitation
Indirect immunofluorescence microscopy was performed as previously described 2.
Quantitative microscopy measurements were performed as previously described 2, 3. For single cell analysis, at least 200 cells per sample were counted in triplicate and the error bars in the histograms represent the standard deviation.
Western blotting
Cells were rinsed in PBS and lysed in either 1X Laemmli SDS-PAGE loading buffer or, alternatively, in a buffer containing 150 mM NaCl, 20 mM Hepes pH 7.4, 0.5% NP40, 1mM EDTA, 10 mM β-Glycero-phosphate, 1 mM NaF, 1 mM Na3VO4, Protease Inhibitors Cocktail SetIII (Calbiochem). Western blotting and immunodetection were performed as previously described 2.
Recombinant proteins expression and in vitro binding assays
GST-fusion proteins were expressed in E. coli BL21(DE3)pLysS (Promega) and affinity purified on glutathione Sepharose beads (GE Healthcare). For in vitro binding assays, GST fusion proteins (1–5 µg) immobilized on glutathione Sepharose beads were prewashed twice in NETN150 (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1mM EDTA, 0.5% NP40, 1 mM DTT), incubated 30 min at 4°C with Benzonase, washed once in NETN1000, blocked for 30 min in NETN150, 1 mg/ml BSA and then washed twice in NETN150. [35S]- labelled proteins were generated by in vitro transcription/translation using a T7TNTQuick-rabbit reticulocyte lysate system (Promega). Equal amounts of in vitro translated, [35S]-labelled proteins were incubated in a total volume of 50 µL of NETN150 with either GST or the appropriate GST fusion protein for 1 hr at 4°C in binding buffer. The beads were washed 6× with NETN150, boiled for 5 min in 1× SDS-PAGE Laemmli loading buffer, and resolved on a SDS-PAGE gel. The gel was dried and [35S]-labelled proteins were visualized using a Storm 860 (Molecular Dynamics).
RNA extraction and RT-PCR
RNA was extracted from cells using the RNeasy Mini Kit (QIAGEN) following the manufacturer instructions. Residual contaminating genomic DNA was eliminated by treating the RNA with Turbo DNA-free DNAse (Ambion). RBBP4 and RBBP7 mRNA levels in control and HGPS cells were measured by retrotranscribing 1 µg of RNA using random primers and the Mo-MulV RT enzyme included in the High Capacity Archive cDNA kit (Applied Biosystems) for 2 hr at 42 °C, after a denaturation step of 5 min at 65 °C. Equal volumes of cDNA were employed as template in a real-time Q-PCR reactions using iQ SYBR Green Supermix (Biorad) in a Biorad iCycler. Reaction conditions were: 3 min at 95 °C, 1 cycle; 20 sec at 95 °C, 30 sec at 56 °C, 41 cycles. Melting curves of the amplified product were generated to verify that a single amplicon was generated. Primers:
RBBP4–6-Fw (5′-TCTGTTTGGGTCAGTTGCTG-3′)
RBBP4–6-Rw (5′-AACTGAGTGGCTTGGTTTGG -3′)
RBBP7–6-Fw (5′-CTGGCCACTCAGCTGTTGTA -3′)
RBBP7–6-Rw (5′-AGGTGGTATTGGACCTGGTG -3′).
All the values were normalized to the internal control Cyclophilin A gene: Cycl-Fw (5′-GTCAACCCCACCGTGTTCTT-3′) and Cycl-Rw (5′-CTGCTGTCTTTGGGACCTTGT - 3′).
The G-rich strand of SatIII transcripts was retrotranscribed and PCR amplified as previously described 26.
HDAC1 immunoprecipitation and HDAC assays
CRL- 1474 and AG01972 primary fibroblasts were grown in 10 cm dishes to 70–90% confluency. Cells were rinsed in PBS and resuspended in Lysis buffer (300 mM NaCl, 50 mM Hepes pH 7.4, 0.5% NP40, 1 mM EDTA, 20% glycerol, 10 mM β-Glycero-phosphate, 1 mM NaF, 1 mM Na3VO4, Protease Inhibitors Cocktail SetIII (Calbiochem)). The cell lysate was incubated for 30 min at 4 °C on a rotating wheel and then centrifuged for 12 min at 12000 g. Total protein concentration of lysates was measured using the Bradford Assay reagent (Biorad). Equal quantities of protein lysate from different samples (1.5–2.0 mg) were diluted to 1 ml with Lysis buffer. Lysates were pre-cleared by incubation for 1hr at 4 °C with rabbit-preimmune serum and Protein A/G+ agarose (St. Cruz) and then incubated o/n at 4 °C with a α-HDAC1 rabbit polyclonal antibody. Immunocomplexes where captured by adding Protein A/G+ agarose. Beads were washed 5X in Lysis buffer and rinsed twice in HDAC assay buffer (see below). HDAC activity in total lysates and in HDAC1 immunoprecipitates was measured with the Fleur de Lys Fluorescent HDAC assay kit (Biomol) following the manufacturer instructions.
One-Strep-lamin A pulldown
U2OS cells stably expressing One-Strep-lamin A were grown in 150 mm dishes, fixed in formhaldeyde and lysed in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris pH 8.1). After extensive sonication, the SDS concentration in the lysate was lowered 10-fold by addition of SDS dilution buffer (0.01% SDS, 1.1 % Triton X-100, 1.2 mM EDTA, 17 mM Tris pH 8.1, 170 mM NaCl). Both SDS lysis buffer and SDS dilution buffer were supplemented with 2 mM ortohovanadate and protease inhibitors (Boehringer Mannheim, Indianapolis, USA). Undissolved fractions were discarded after a 10 minute centrifugation step at 4 degrees at 10, 000×g. Streptactin Matrix (IBA BioTagnology, Göttingen, Germany) prewashed in SDS dilution buffer was added and samples were incubated on a rotating wheel o/n at 4 °C. Precipitated protein complexes were washed extensively, and eluted from the beads by incubation at 95 °C in 1X Laemmli SDS-PAGE loading buffer.
Supplementary Material
Acknowledgements
We would like to thank P. Scaffidi and A. Marcello for critical reading of the manuscript and P. Scaffidi for sharing data and providing numerous suggestions during the course of the experiments. Fluorescence imaging was performed with the assistance of T. Karpova at the NCI Fluorescence Imaging Microscopy Facility. FACS cell cycle acquisition and analysis was performed with the help of K. McKinnon at the NCI FACS facility. We would also like to thank T. Jenuwein, B. Stillman and T. Dittmer for providing reagents. We thank H. Herrmann and E. Wanker for their support with the Y2H screening and subsequent confirmation studies. G.P. was partly supported by postdoctoral fellowships from the Telethon Italy Foundation and the Italian Association for Cancer Research (AIRC). K.H. and H.G. were supported by The Deutsche Forschungsgemeinschaft (DFG, SFB 577, projects A4 and Z2). This work was supported by an Intramural Research Program of the NIH, NCI, Center for Cancer Research.
References
- 1.Goldman RD, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nature Medicine. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science (New York, N.Y. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shumaker DK, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu B, et al. Genomic instability in laminopathy-based premature aging. Nature Medicine. 2005;11:780–785. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- 6.Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nature Cell Biology. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nature reviews. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- 8.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Sandre-Giovannoli A, et al. Lamin a truncation in Hutchinson-Gilford progeria. Science (New York, N.Y. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 10.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes & Development. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & Development. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi T, et al. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Molecular Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 16.Rizzi N, et al. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Molecular Biology of the Cell. 2004;15:543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. The Journal of Biological Chemistry. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 18.Nabatiyan A, Szuts D, Krude T. Induction of CAF-1 expression in response to DNA strand breaks in quiescent human cells. Molecular and Cellular Biology. 2006;26:1839–1849. doi: 10.1128/MCB.26.5.1839-1849.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. The Journal of Cell Biology. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng JC, Karpen GH. Epigenetic regulation of heterochromatic DNA stability. Current Opinion in Genetics & Development. 2008;18:204–211. doi: 10.1016/j.gde.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jorgensen S, et al. The histone methyltransferase SET8 is required for S-phase progression. The Journal of Cell Biology. 2007;179:1337–1345. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tardat M, Murr R, Herceg Z, Sardet C, Julien E. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. The Journal of Cell Biology. 2007;179:1413–1426. doi: 10.1083/jcb.200706179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Berlo JH, et al. A-type lamins are essential for TGF-beta1 induced PP2A to dephosphorylate transcription factors. Hum Mol Genet. 2005;14:2839–2849. doi: 10.1093/hmg/ddi316. [DOI] [PubMed] [Google Scholar]
- 24.Goehler H, et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Molecular Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 25.Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 26.Valgardsdottir R, et al. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Research. 2008;36:423–434. doi: 10.1093/nar/gkm1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.