Abstract
Transcription factors that play key roles in regulating embryonic stem (ES) cell state have been identified, but the chromatin regulators that help maintain ES cells are less well understood. A high-throughput shRNA screen was used to identify novel chromatin regulators that influence ES cell state. Loss of histone H3 Lys 9 (H3K9) methyltransferases, particularly SetDB1, had the most profound effects on ES cells. Chromatin immunoprecipitation (ChIP) coupled with massively parallel DNA sequencing (ChIP-Seq) and functional analysis revealed that SetDB1 and histone H3K9-methylated nucleosomes occupy and repress genes encoding developmental regulators. These SetDB1-occupied genes are a subset of the “bivalent” genes, which contain nucleosomes with H3K4me3 (H3K4 trimethylation) and H3K27me3 modifications catalyzed by Trithorax and Polycomb group proteins, respectively. These genes are subjected to repression by both Polycomb group proteins and SetDB1, and loss of either regulator can destabilize ES cell state.
Keywords: SetDB1, ES cell, H3K9 methylation, polycomb, repression, pluripotency
Embryonic stem (ES) cells provide an important model system to study the control of early development and hold significant potential for clinical therapies because of their distinctive capacity to both self-renew and differentiate into multiple lineages (Thomson et al. 1998; Reubinoff et al. 2000; Pera and Trounson 2004; Keller 2005; Hochedlinger and Jaenisch 2006; Yang and Smith 2007). Studies of key ES cell transcription factors—including Oct4, Sox2, Nanog, Ronin, Tcf3, and others—have led to models for ES cell transcriptional regulatory circuitry that can account for several important features of the gene expression program of these unique cells (Boyer et al. 2005; Loh et al. 2006; Chen et al. 2008; Cole et al. 2008; Dejosez et al. 2008; Jiang et al. 2008; Kim et al. 2008; Marson et al. 2008; Tam et al. 2008). These key transcriptional regulators occupy the promoters of actively transcribed genes encoding transcription factors, signaling components, and chromatin-modifying enzymes that promote ES cell self-renewal. They also occupy genes encoding a large set of developmental regulators that are silent in ES cells, but whose expression is associated with lineage commitment and cellular differentiation. Polycomb-repressive complexes (PRCs) co-occupy the genes encoding these developmental regulators to help maintain a silent transcriptional state in ES cells (Azuara et al. 2006; Boyer et al. 2006; Lee et al. 2006; Stock et al. 2007; van der Stoop et al. 2008).
The regulation of chromatin structure, which includes nucleosome remodeling and post-translational modification of histone proteins, is necessary for the establishment and maintenance of heritable gene expression patterns during development (Workman 2006; Dunn and Kingston 2007; Kouzarides 2007; Surani et al. 2007). Trithorax group (TrxG) and Polycomb group (PcG) protein complexes are key regulators of chromatin structure that are necessary for maintenance of the ES cell gene expression program and are required for segmental identity in the developing embryo (for review, see Schuettengruber et al. 2007). TrxG complexes catalyze histone H3 Lys 4 trimethylation (H3K4me3) in proximity to promoters of genes that experience transcription initiation by RNA polymerase II. Since most genes experience some level of transcription initiation, most promoters are occupied by nucleosomes with histone H3K4me3 (Guenther et al. 2007). The PcG protein complex PRC2 catalyzes H3K27me3, which contributes to repression of genes encoding key developmental regulators. Disruption of PcG function in ES cells leads to derepression of these developmental genes, alteration of the ES cell transcriptional program, and a loss of the ES cell state (Boyer et al. 2006; Lee et al. 2006; Pasini et al. 2007; Stock et al. 2007; Jaenisch and Young 2008; van der Stoop et al. 2008). In ES cells, genes encoding lineage-specific developmental regulators contain nucleosomes with both H3K4me3 and H3K27me3 modifications, and have thus been called “bivalent” (Bernstein et al. 2006). These genes apparently experience some level of transcription initiation, and thus TrxG-mediated histone H3K4 methylation, but are silenced by PcG complexes, which may occur through repression of transcription elongation (Stock et al. 2007).
Although many chromatin remodeling and modifying activities have been described, only a few are known to contribute to the control of ES cell state. We used a screening approach with a shRNA library to screen for additional chromatin regulators of ES cell state. Loss of histone H3K9 methyltransferases as well as PcG components was found to cause loss of ES cell state in this screen. Further study of the H3K9 methyltransferase SetDB1 revealed that it contributes to repression of a subset of genes encoding developmental regulators in ES cells. These results indicate that there is an additional layer of chromatin-mediated repression at these developmental genes, and that H3K9me3-modified nucleosomes are not exclusive to constitutive heterochromatin.
Results and Discussion
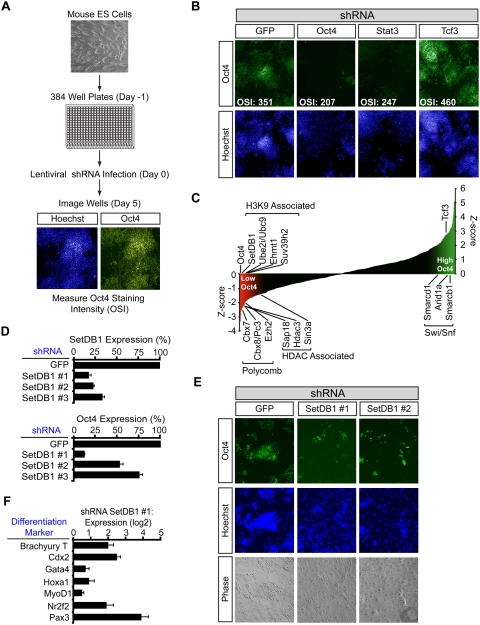
In order to identify chromatin regulators that may be involved in the establishment or maintenance of cell state, we screened murine ES (mES) cells with a shRNA library containing four to six hairpins directed against each of 197 chromatin regulators (Fig. 1A). ES cells were seeded into 384-well plates, and each well was infected with an individual lentiviral construct delivering a shRNA targeting a chromatin regulator. After selecting for stable integration of the shRNA construct, the cells were fixed, and the average Oct4 staining intensity of the cells in each well was measured. Control shRNAs targeting Oct4 and Stat3, positive regulators of pluripotency in mES cells (Nichols et al. 1998; Niwa et al. 1998; Hay et al. 2004), produced cells that lost Oct4 signal, as expected (Fig. 1B). In contrast, a control shRNA designed against Tcf3, a negative regulator of Oct4 and Nanog (Pereira et al. 2006; Cole et al. 2008; Tam et al. 2008), caused an increase in Oct4 staining. To normalize for plate variability, the average Oct4 staining intensity for the cells was used to calculate a Z-score for each well, taking into account values for negative control infections on each plate (see the Supplemental Material). The majority of the shRNAs targeting chromatin regulators produced Z-scores that were distributed between those observed for shRNAs targeting Oct4 and Tcf3, as expected (Fig. 1C; Supplemental Tables S1, S2).
Figure 1.
Identification of chromatin regulators of ES cell state. (A) Outline of the screening protocol. mES cells were seeded without a mouse embryonic fibroblast (MEF) feeder layer into 384-well plates. The following day, wells were infected with individual lentiviral shRNAs targeting chromatin regulators. Infections were done in quadruplicate on separate plates. Five days post-infection, cells were fixed and stained with Hoechst and for Oct4. Cells were identified based on the Hoechst staining, and the average Oct4 staining intensity was quantified using Cellomics software. (B) Representative images from control wells on a 384-well plate. (OSI) Average Oct4 staining intensity of the cells in the well. (C) Selected genes whose shRNAs have large effects on Oct4 staining intensity. Z-scores for Oct4 staining intensity were calculated as described in the Supplemental Material. (D) Validation with multiple SetDB1 shRNAs. mES cells were infected with the indicated shRNA, and Oct4 expression level and SetDB1 knockdown efficiency were evaluated by real-time quantitative PCR (qPCR). (E) SetDB1 knockdown causes reduced Oct4 protein levels and changes in colony morphology. mES cells were infected with the indicated shRNA and stained with Hoechst and for Oct4. (F) SetDB1 knockdown causes up-regulation of differentiation markers. mES cells were infected with SetDB1 shRNA #1, and the expression level of the indicated differentiation marker relative to a shRNA GFP control infection was quantified by real-time qPCR.
The chromatin regulators whose knockdowns reduced Oct4 levels nearly as profoundly as knockdown of Oct4 itself were of particular interest because many have been implicated in gene silencing, and many of these are associated with histone H3K9 and H3K27 methylation (Fig. 1C; Supplemental Table S1). The chromatin regulators associated with H3K9 methylation include SetDB1/ESET, Ube2i/Ubc9, Ehmt1, and Suv39h2. Those associated with H3K27 methylation include the Polycomb components Cbx7, Cbx8/Pc3, and Ezh2, and their identification in this screen is consistent with previous evidence implicating Polycomb in control of ES cell state (Schuettengruber et al. 2007). The loss of the histone deacetylase Hdac3 and the Sin3/HDAC complex components Sin3a and Sap18 had similar effects. The loss of certain chromatin regulators led to increased Oct4 levels, and these included three members of the SWI/SNF family (Smarcd1, Arid1a, and Smarcb1) (Fig. 1C). Thus, the results of this screen implicate multiple chromatin regulators in control of ES cell state.
One of the largest classes of chromatin regulators identified in the screen was associated with histone H3K9 methylation. From this class we selected the SetDB1 H3K9 methyltransferase (Schultz et al. 2002; Ayyanathan et al. 2003; Wang et al. 2003) for further detailed study because its loss produced the most profound effects on ES cell state (Fig. 1C; Supplemental Table S1; Supplemental Fig. S1) and SetDB1-null blastocysts have deficiencies in the inner cell mass (Dodge et al. 2004). Independent experiments demonstrated that multiple shRNAs targeting SetDB1 caused a loss in both SetDB1 and Oct4 expression levels and a loss of normal ES cell colony morphology (Fig. 1D,E; Supplemental Fig. S2). SetDB1 knockdown in a second mES cell line also resulted in a loss of Oct4 expression (Supplemental Fig. S2). Furthermore, SetDB1 knockdown led to increased expression of differentiation markers (Fig. 1F). These results confirm that SetDB1 contributes to the maintenance of ES cell state.
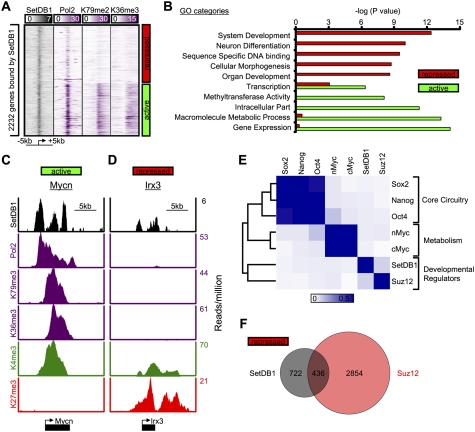
To gain insight into the role of SetDB1 in control of ES cell state, we used chromatin immunoprecipitation (ChIP) coupled with massively parallel DNA sequencing (ChIP-Seq) to identify genes that are occupied by this regulator. We used an antibody whose specificity was confirmed by ChIP and Western blot analyses (Supplemental Fig. S3). The ChIP-Seq results revealed that SetDB1 occupies the core promoter regions of 2232 genes (Supplemental Table S3). Approximately 48% of these showed evidence of active transcription (occupancy by RNA polymerase II and nucleosomes with methylated H3K79 and H3K36), whereas 52% appeared to be transcriptionally repressed (Fig. 2A). It was striking that the repressed genes were significantly enriched for regulators of development (Fig. 2B,D), while active genes were enriched for gene expression and metabolism (Fig. 2B,C). The association with repressed genes encoding developmental regulators led us to ask whether these genes were also occupied by PcG proteins. Indeed, SetDB1 and PRC subunit Suz12 are frequently colocalized in the ES cell genome; 38% of the repressed genes occupied by SetDB1 were also occupied by PcG subunit Suz12 (Fig. 2E,F). These results demonstrate that SetDB1 and PcG co-occupy a common set of repressed genes involved in developmental regulation, which may explain why both regulators were identified in the shRNA screen described above.
Figure 2.
SetDB1 occupies genes encoding developmental regulators in mES cells. (A) Binding density of regions surrounding the transcriptional start site (±5 kb) of 2232 genes bound by SetDB1. Genes were classified as active if they were enriched for RNA polymerase II, H3K79me2, and H3K36me3, and repressed if they lacked such evidence. (B) Gene Ontology (GO) analysis of genes occupied by SetDB1 that were categorized as active or repressed in mES cells. (C,D) SetDB1, RNA polymerase II, and histone H3K79me2, H3K36me3, H3K4me3, and H3K27me3 modification profiles for an actively transcribed gene (Mycn), an oncogene, and a repressed gene (Irx3), a homeodomain-containing transcription factor that is involved in spatially prepatterning the neural system. (E) Heat map showing colocalization frequency of SetDB1 and PRC subunit Suz12, together with other key ES cell transcriptional regulators. Colors in the heat map reflect the colocalization frequency of each pair of regulators (blue means more frequently colocalized). Regulators were clustered along both axes based on the similarity in their colocalization with other regulators. (F) Venn diagram representation of repressed genes occupied by SetDB1 and Suz12.
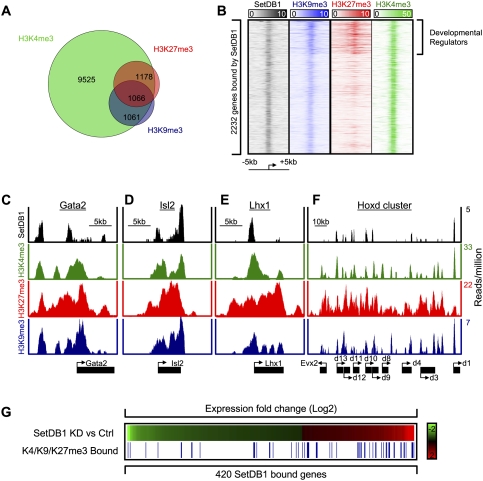
SetDB1 has been implicated in transcriptional repression through H3K9me3 (Schultz et al. 2002; Wang et al. 2003), so we used ChIP-Seq to identify genes occupied by nucleosomes containing H3K9me3. The specificity of the antibodies used in this experiment was confirmed by using competition assays with modified peptides (Supplemental Fig. S4). The results revealed that histone H3K9me3-modified nucleosomes occupy repetitive DNA elements located in constitutive heterochromatin, as described previously (Supplemental Tables S4, S5; Mikkelsen et al. 2007). The results also revealed that substantially more euchromatic genes are occupied by H3K9me3-modified nucleosomes than detected previously in ES cells (Supplemental Fig. S4; Supplemental Table S3; Mikkelsen et al. 2007). Essentially all of the 2127 euchromatic genes occupied by H3K9me3 were also occupied by histone H3K4me3 (Fig. 3A), consistent with evidence that most genes in ES cells have H3K4me3-modified nucleosomes (Kim et al. 2005; Guenther et al. 2007). About half of the genes occupied by H3K9me3 were also occupied by histone H3K27me3 (Fig. 3A). SetDB1 binding was detected at ∼22% of the genes occupied by histone H3K9me3 (Supplemental Fig. S5). The imperfect overlap between H3K9me3 and SetDB1 occupancy is not surprising because this H3K9 modification is catalyzed by other methyltransferases, and demethylation may occur at some sites. Approximately half of SetDB1-bound and H3K9me3-occupied genes showed a decrease in H3K9me3 following SetDB1 knockdown, suggesting that SetDB1 is in part responsible for H3K9me3 at this set of genes (Supplemental Fig. S6).
Figure 3.
Genes encoding developmental regulators are often occupied by SetDB1 and at least three histone modifications. (A) Venn diagram representation of genes occupied by nucleosomes modified with histone H3K4me3, H3K27me3, and H3K9me3. Approximately half of the genes that are occupied by both H3K4me3 and H3K27me3 are also occupied by H3K9me3. (B) Histone modifications in promoter-proximal regions of 2232 genes bound by SetDB1 surrounding the transcriptional start site (±5 kb). A set of developmental regulator genes are enriched for SetDB1 binding and nucleosomes containing H3K9me3, H3K27me3, and H3K4me3. (C–F) SetDB1 and histone H3K4me3, H3K27me3, and H3K9me3 occupancy profiles for Gata2, Isl2, Lhx1, and the Hoxd cluster. (G) Expression profiles showing changes in gene expression in SetDB1 knockdown cells relative to control shRNA cells. Red and green represent Log2 transformed probe signal levels. Genes occupied by histone H3K9me3, H3K27me3, and H3K4me3 are shown as blue lines. Genes up-regulated upon SetDB1 knockdown were 3.6-fold enriched (P < 10−14) for H3K4me3/H3K27me3/H3K9me3 occupancy (32 out of 164), while down-regulated genes were not enriched (16 out of 256).
Many of the 2232 genes bound by SetDB1 showed some evidence of H3K9me3 occupancy, as seen in the heat map in Fig 3B, but the highest density of histone H3K9me3 was found in the promoter regions of SetDB1-bound genes that are co-occupied by both H3K4me3 and H3K27me3, which frequently encode developmental regulators. Figure 3, C–F, shows how SetDB1 and histones H3K9me3, H3K27me3, and H3K4me3 occupy a subset of these developmental genes. It is evident from these and other gene tracks (Supplemental Fig. S7) that SetDB1 tends to occupy promoter regions. The methyltransferase was also found in regions that lack an annotated start site but have histone H3K4me3 and thus evidence suggesting transcription initiation (Fig. 3C,D). Histone H3K9me3 was found predominantly at promoters, but was also observed at lower levels throughout genes. We conclude that SetDB1 and histone H3K9me3 occupy a subset of the key developmental genes previously shown to be repressed by PcG proteins (Supplemental Table S3).
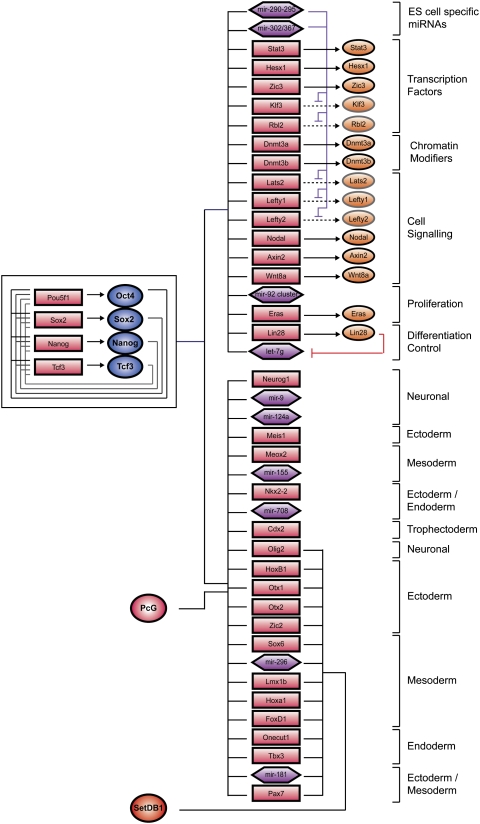
If SetDB1 and histone H3K9me3 contribute to repression of a set of genes encoding developmental regulators, loss of SetDB1 should lead to derepression of these genes. Indeed, knockdown of SetDB1 caused increased expression of a significant (P < 10−14) fraction of genes occupied by SetDB1 and enriched for nucleosomes containing H3K9me3, H3K4me3, and H3K27me3 (Fig. 3G). These results argue for the presence of an additional layer of repression mediated by SetDB1 and H3K9me3 at a subset of developmental regulators previously thought to be repressed only by PcG proteins (Fig. 4). These results also suggest that derepression of developmental regulators following loss of SetDB1 may be responsible, at least in part, for destabilizing ES cell state. However, it is possible that a function of SetDB1 at active genes (Fig. 2A; Wang et al. 2008; Loyola et al. 2009) might also be important for maintenance of ES cell state.
Figure 4.
Model for transcriptional regulation by SetDB1. A model of core ES cell regulatory circuitry is shown with a subset of genes occupied by the transcription factors and chromatin regulators depicted in the diagram. The interconnected autoregulatory loop is shown to the left. Active genes are shown at the top right, and inactive genes are shown at the bottom right. Transcription factors are represented by dark-blue circles, and PcG complexes and SetDB1 are indicated by a red circle. Gene promoters are represented by red rectangles, gene products are indicated by orange circles, and miRNA promoters are represented by purple hexagons.
The stability of cell state relies on the silencing of genes encoding regulators of other cell states. The ability to modify cell state holds great promise for regenerative medicine and makes it even more critical to understand how cell state is regulated (Jaenisch and Young 2008; Murry and Keller 2008). Chromatin regulators such as the PcG proteins contribute to the maintenance of cell type-specific gene expression states by establishing a repressed but poised transcriptional state at genes encoding developmental regulators (Bernstein et al. 2006; Boyer et al. 2006; Bracken et al. 2006; Lee et al. 2006; Schuettengruber et al. 2007). Negative control of these developmental regulators is essential for maintenance of a stable cell state because expression of transcription factors that produce cell type-specific gene expression programs can cause cells to acquire new states. Our results implicate histone H3K9 methyltransferases in maintenance of cell state, and show that one of these, SetDB1, functions by contributing to repression of many of the same developmental regulatory genes that are occupied by PcG proteins.
Materials and methods
A detailed description of all materials and methods can be found in the Supplemental Material.
Chromatin regulator shRNA screen
High-throughput RNAi screening was performed at the Broad Institute RNAi Platform (Moffat et al. 2006). mES cells were seeded at a density of 1500 cells per well in 384-well plates; wells were infected with an individual lentiviral shRNA construct targeting a chromatin regulator, treated with puromycin to select for stable integration of the shRNA, and cross-linked with 4% paraformaldehyde 5 d post-infection. Following staining with Hoechst and for Oct4, wells were imaged with an ArrayScan HCS Reader (Cellomics). Cells were identified with Cellomics software based on the Hoechst staining. After identification, the average Oct4 pixel intensity was quantified, and an average was calculated for all cells identified in the well.
Antibodies, ChIP, sequencing, and microarray analysis
Antibodies for H3K9me3 (Ab8898 [Abcam]; and Up07-442 [Upstate Biotechnologies]) and SetDB1 H-300 (sc-66884, Santa Cruz Biotechnologies) were used. Data for RNA polymerase II, H3K36me3, H3K79me2, H3K4me3, H3K27me3, nMyc, cMyc, Oct4, Sox2, Nanog, and TCF3 in mES cells were described previously (Mikkelsen et al. 2007; Chen et al. 2008; Marson et al. 2008; Seila et al. 2008). Expression analyses were carried out with Agilent DNA microarrays. ChIP-Seq experiments and analysis were performed as described previously (Guenther et al. 2008; Marson et al. 2008) in the Supplemental Material. All microarray and ChIP-Seq data from this study have been submitted to the GEO database (GSE18371).
Acknowledgments
We thank David Sabatini, Kathleen Ottina, and the RNAi Screening Platform at the Broad Institute, along with David Root, Serena Silver, Thomas Neiland, and Hanh Le for screening advice and technical support. We are grateful to James Evans and Victor Horodincu of the Whitehead-MIT Bioimaging Center and Sesha Tekur (Thermo Fisher) for imaging support and advice. We thank Jeong-Ah Kwon, Jennifer Love, Sumeet Gupta, and Thomas Volkert for help generating and analyzing ChIP-seq data. Immunofluorescence images were collected using the W.M. Keck Foundation Biological Imagining Facility at the Whitehead Institute and Whitehead-MIT Bioimaging Center. This work was supported by a Fellowship from the Canadian Institutes of Health Research (to S.B.), by an NIH NIGMS Post-doctoral Fellowship (to M.K.), and by NIH grant HG002668 (to R.Y.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1837309.
Supplemental material is available at http://www.genesdev.org.
References
- Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, Yamada Y, Tanaka K, Torigoe K, Rauscher FJ., III Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes & Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes & Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, Songyang Z, Thomson JA, Zwaka TP. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn RK, Kingston RE. Gene regulation in the postgenomic era: Technology takes the wheel. Mol Cell. 2007;28:708–714. doi: 10.1016/j.molcel.2007.11.022. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL, Croce CM, Nakamura T, Canaani E, Young RA. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes & Dev. 2008;22:3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay DC, Sutherland L, Clark J, Burdon T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells. 2004;22:225–235. doi: 10.1634/stemcells.22-2-225. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061–1067. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: Emergence of a new era in biology and medicine. Genes & Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Loyola A, Tagami H, Bonaldi T, Roche D, Quivy JP, Imhof A, Nakatani Y, Dent SY, Almouzni G. The HP1α–CAF1–SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009;10:769–775. doi: 10.1038/embor.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes & Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera MF, Trounson AO. Human embryonic stem cells: Prospects for development. Development. 2004;131:5515–5525. doi: 10.1242/dev.01451. [DOI] [PubMed] [Google Scholar]
- Pereira L, Yi F, Merrill BJ. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol Cell Biol. 2006;26:7479–7491. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: Somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., III SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Tam WL, Lim CY, Han J, Zhang J, Ang YS, Ng HH, Yang H, Lim B. T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells. 2008;26:2019–2031. doi: 10.1634/stemcells.2007-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- van der Stoop P, Boutsma EA, Hulsman D, Noback S, Heimerikx M, Kerkhoven RM, Voncken JW, Wessels LF, van Lohuizen M. Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS One. 2008;3:e2235. doi: 10.1371/journal.pone.0002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, Tempst P, Roeder RG, Zhang Y. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell. 2003;12:475–487. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman JL. Nucleosome displacement in transcription. Genes & Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- Yang X, Smith SL. ES cells derived from cloned embryos in monkey–a jump toward human therapeutic cloning. Cell Res. 2007;17:969–970. doi: 10.1038/cr.2007.105. [DOI] [PubMed] [Google Scholar]