Abstract
Mammalian mRNAs lose and acquire proteins throughout their life span while undergoing processing, transport, translation, and decay. How translation affects messenger RNA (mRNA)–protein interactions is largely unknown. The pioneer round of translation uses newly synthesized mRNA that is bound by cap-binding protein 80 (CBP80)–CBP20 (also known as the cap-binding complex [CBC]) at the cap, poly(A)-binding protein N1 (PABPN1) and PABPC1 at the poly(A) tail, and, provided biogenesis involves pre-mRNA splicing, exon junction complexes (EJCs) at exon–exon junctions. Subsequent rounds of translation engage mRNA that is bound by eukaryotic translation initiation factor 4E (eIF4E) at the cap and PABPC1 at the poly(A) tail, but that lacks detectable EJCs and PABPN1. Using the level of intracellular iron to regulate the translation of specific mRNAs, we show that translation promotes not only removal of EJC constituents, including the eIF4AIII anchor, but also replacement of PABPN1 by PABPC1. Remarkably, translation does not affect replacement of CBC by eIF4E. Instead, replacement of CBC by eIF4E is promoted by importin β (IMPβ): Inhibiting the binding of IMPβ to the complex of CBC–IMPα at an mRNA cap using the IMPα IBB (IMPβ-binding) domain or a RAN variant increases the amount of CBC-bound mRNA and decreases the amount of eIF4E-bound mRNA. Our studies uncover a previously unappreciated role for IMPβ and a novel paradigm for how newly synthesized messenger ribonucleoproteins (mRNPs) are matured.
Keywords: Iron response element, EJC, CBP80–CBP20, PABP, IMPα, IMPβ, RAN
In mammalian cells, two different messenger ribonucleoproteins (mRNPs) serve as templates for protein synthesis, each for distinct purposes. Newly synthesized mRNP initially undergoes a pioneer round of translation that generally ensures the quality of gene expression. In particular, nonsense-mediated mRNA decay (NMD), which largely functions as a quality control mechanism by eliminating messenger RNAs (mRNAs) that prematurely terminate translation, occurs as a consequence of nonsense codon recognition during a pioneer round of translation (for recent reviews, see Chang et al. 2007; Isken and Maquat 2007; Mühlemann et al. 2008; Shyu et al. 2008; Silva and Romão 2009). In contrast, subsequent steady-state rounds of translation do not detectably support NMD, but instead provide for the bulk of cellular protein synthesis (Lejeune et al. 2002; Chiu et al. 2004; Matsuda et al. 2007; Woeller et al. 2008). Accordingly, steady-state rounds of translation are often targeted by mechanisms that conditionally regulate gene expression in response to changes in physiological circumstances, as exemplified by the inhibitory effects of cellular stress brought about by prolonged hypoxia or serum starvation (e.g., see Flynn and Proud 1995; Raught et al. 2000; Gingras et al. 2001; Koritzinsky et al. 2006; Oh et al. 2007a,b; Sonenberg 2008 and references therein).
Pioneer translation initiation complexes are physically distinct from steady-state translation initiation complexes. One distinguishing feature is the cap-binding proteins (CBPs). Whereas pioneer translation initiation complexes are associated with the CBP heterodimer CBP80–CBP20, which is also called the cap-binding complex (CBC), steady-state translation initiation complexes are bound at their caps by eukaryotic translation initiation factor 4E (eIF4E) (Ishigaki et al. 2001; Lejeune et al. 2002; Hosoda et al. 2005; Sonenberg 2008). Additionally, pioneer translation initiation complexes, unlike steady-state translation initiation complexes, contain the largely nuclear poly(A)-binding protein N1 (PABPN1); if they derived from pre-mRNA splicing, then they would also consist of exon junction complexes (EJCs) of proteins (Lejeune et al. 2002; Hosoda et al. 2006; Kashima et al. 2006). These distinguishing features contribute to the specialized functions of each initiation complex. For example, the preferential inactivation of steady-state translation initiation relative to pioneer translation initiation during prolonged hypoxia or serum starvation is due to the activation of 4E-BP1 (Koritzinsky et al. 2006), which inhibits the translation of eIF4E-bound mRNA but not CBC-bound mRNA (Chiu et al. 2004; Matsuda et al. 2007; Woeller et al. 2008), and to the redistribution of eIF4E to stress granules (Oh et al. 2007a,b). In contrast, the pioneer round of translation, as measured by the efficiency of NMD, occurs during prolonged hypoxia or serum starvation, presumably to permit the surveillance of newly synthesized mRNAs that will later undergo steady-state rounds of translation when stress is removed (Oh et al. 2007a,b).
Despite important differences, both translation complexes share a number of factors that actively direct protein synthesis and include PABPC1, eIF2α, eIF3, eIF4G, eukaryotic release factor 1 (eRF1), and eRF3 (McKendrick et al. 2001; Chiu et al. 2004; Lejeune et al. 2004; Hosoda et al. 2006; Kashima et al. 2006; Isken et al. 2008). Furthermore, both complexes ultimately assemble into polysomes (Stephenson and Maquat 1996; Chiu et al. 2004; Sato et al. 2008 and references therein). The finding that CBC-bound mRNAs associate with smaller polysomes than their eIF4E-bound counterparts suggests that the replacement of CBC by eIF4E occurs on polysomes, consistent with data indicating that CBC-bound mRNA is exported to the cytoplasm (Visa et al. 1996; Chiu et al. 2004; Cheng et al. 2006), where all rounds of translation are thought to occur (Sato et al. 2008). Moreover, the NMD factors UPF2 and UPF3X, which are also EJC constituents (Lykke-Andersen et al. 2000, 2001; Kim et al. 2001; Lejeune et al. 2002; Chamieh et al. 2008), like CBP80 and PABPN1, are associated in polysome profiles with fewer ribosomes than either eIF4E or PABPC1 (Chiu et al. 2004). Studies indicate that the Y14 EJC constituent also cosediments in polysome profiles with fewer ribosomes than PABPC1 (Dostie and Dreyfuss 2002).
Little is known about how a pioneer translation initiation complex is converted to a steady-state translation initiation complex. One possible activator of mRNP remodeling could be the pioneer round of translation itself. In fact, two lines of evidence suggest that the removal of the EJC component Y14 from newly synthesized mRNP involves translation (Dostie and Dreyfuss 2002). First, mRNA generated by splicing in cell-free extracts coimmunoprecipitates with Y14 after incubation in translationally active extracts, provided the 5′-untranslated region (UTR) harbors a hairpin structure that blocks translation. Second, spliced mRNA produced from a transiently introduced intron-containing plasmid in HEK293T cells coimmunoprecipitates with Y14 after purification from cytoplasmic lysates, provided the 5′-UTR harbors a hairpin structure that blocks translation. It is likely but not proven that other EJC constituents are also removed by translating ribosomes.
Here, we aim to determine how the process of translation affects remodeling of pioneer translation initiation complexes to steady-state translation initiation complexes. Results indicate that translation not only augments the removal of EJC constituents, including the EJC anchor eIF4AIII, but also enhances the replacement of PABPN1 by PABPC1. Remarkably, however, translation does not promote the replacement of CBC by eIF4E. Instead, we provide evidence that binding of the nuclear transport receptor importin β (IMPβ) to CBP80-bound IMPα at the cap of newly synthesized mRNA promotes the replacement of CBC by eIF4E. We demonstrate that IMPα coimmunoprecipitates with the CBC-containing 5′ end of newly synthesized mRNA in a way that is augmented when the interaction of IMPα with IMPβ is inhibited. Furthermore, inhibiting the interaction of IMPα with IMPβ increases the coimmunoprecipitation (co-IP) of CBP80 with mRNA and concomitantly decreases the co-IP of eIF4E with mRNA. We discuss the significance of this previously unappreciated mechanism of karyopherin-mediated mRNP remodeling.
Results
Translation promotes the removal of EJC constituents UPF3X and eIF4AIII from newly synthesized mRNA but does not promote the replacement of CBC by eIF4E
The translocation of newly synthesized mRNPs through nuclear pore complexes involves many changes in protein:mRNA and protein:protein interactions. As a rule, specific proteins either dissociate from or become associated with mRNAs during the process of mRNA export from the nucleus to the cytoplasm (e.g., see Tran et al. 2007 and references therein). Nevertheless, essentially nothing is known about how the pioneer round of translation contributes to the remodeling of newly synthesized mRNPs. To assess the role of translation in the replacement and/or loss of particular mRNP proteins that constitute the pioneer translation initiation complex, we initially used human β-globin (Gl) mRNA, since information is available on its association with a number of established mRNP proteins (e.g., see Schaal and Maniatis 1999; Huang and Steitz 2001; Ishigaki et al. 2001; Lejeune et al. 2002; Chiu et al. 2004; Sato et al. 2008).
We first aimed to determine if the replacement of CBC by eIF4E during mRNP biogenesis in HeLa cells involves translation. CBP80 and CBP20 associate with cap structures as a heterodimer rather than individually: CBP20, like eIF4E, sandwiches the 7-methylguanine of the cap structure between two amino acid side chains (Marcotrigiano et al. 1997; Matsuo et al. 1997; Niedzwiecka et al. 2002; Tomoo et al. 2002), and its binding to an mRNA cap requires CBP80 (Izaurralde et al. 1994; Mazza et al. 2001, 2002; Calero et al. 2002). HeLa cells were generated that stably express pIRE-Gl Norm or pIRE-Gl Ter, each of which encodes Gl mRNA harboring in its 5′-UTR the iron response element (IRE) that typifies the 5′-UTR of ferritin heavy chain (referred to here simply as ferritin) mRNA (Fig. 1A; Gray and Hentze 1994; Thermann et al. 1998). pIRE-Gl Norm lacks a premature termination codon (PTC), whereas pIRE-Gl Ter harbors a PTC at position 39 so as to encode mRNA that is targeted for NMD (Gray and Hentze 1994; Thermann et al. 1998; Zhang et al. 1998). Iron regulatory proteins (IRPs) fail to bind the IRE in the presence of intracellular iron, whereas IRPs bind the IRE and block IRE-containing mRNA translation in the absence of intracellular iron by inhibiting formation of the 43S preinitiation complex (Klausner et al. 1993; Gray and Hentze 1994; Hentze and Kühn 1996; Muckenthaler et al. 1998). Since NMD targets PTC-containing Gl mRNA and does not affect the level of PTC-containing Gl pre-mRNA (Zhang et al. 1998), stably transfected clonal cell lines that express an equal amount of Gl Norm or Gl Ter pre-mRNA were chosen for further use.
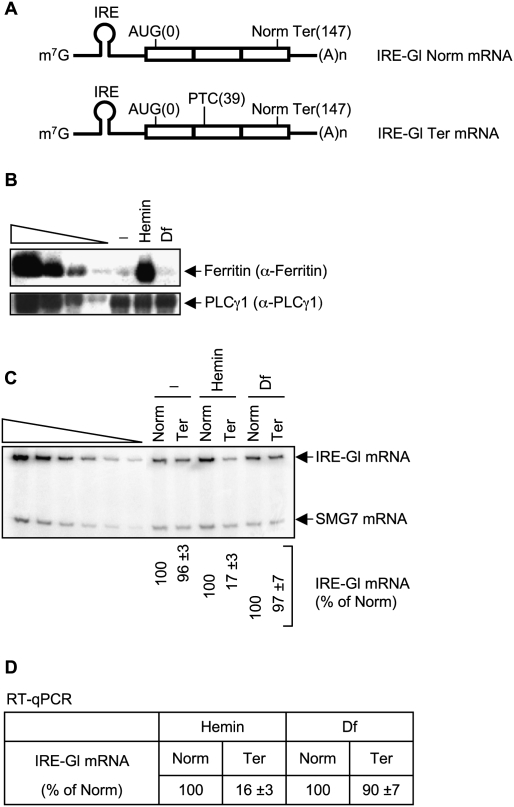
Figure 1.
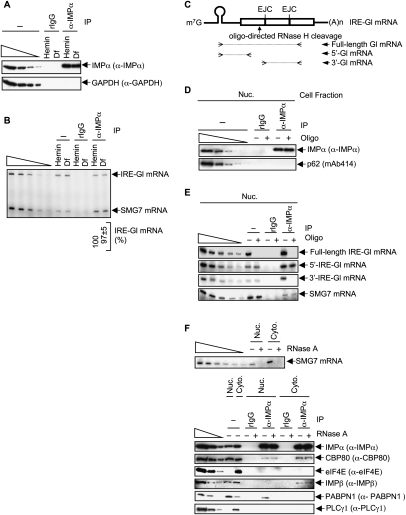
Evidence that hemin promotes and Df inhibits the translation of IRE-Gl mRNA. (A) Structures of IRE-Gl mRNA, either lacking a PTC (Norm) or containing a PTC at position 39 (Ter). Horizontal lines represent Gl 5′-UTRs and 3′-UTRs, and boxes represent each of the three Gl exons joined by splicing-generated exon–exon junctions. (m7G) cap structure; (IRE) iron response element; [AUG(0)] translation initiation codon; [Norm Ter(147)] normal termination codon; [(A)n] poly(A) tail. Lysates were generated from HeLa cells that stably express IRE-Gl mRNA, either Norm or Ter. (B) Western blotting using anti-ferritin (α-ferritin) or anti-PLCγ1 in the absence (−) or presence of hemin or Df. Serial dilutions of protein in the left four lanes demonstrate that the Western blot analysis is semiquantitative. (C) Using RT–sqPCR, the level of IRE-Gl mRNA, either Norm or Ter, was normalized to the level of cellular SMG7 mRNA, where the normalized level of IRE-Gl Norm mRNA in the presence of either hemin or Df is defined as 100. Serial dilutions of RNA in the left six lanes demonstrate that the RT–PCR analysis is semiquantitative. (D) cDNA that was generated using RT as in C was amplified using qPCR. The level of IRE-Gl mRNA, either Norm or Ter, was normalized to the level of β-actin mRNA. Normalized values were then calculated as a percentage of the normalized value of IRE-Gl Norm mRNA in the presence of either hemin or Df, which is defined as 100%. Results derive from three independently performed experiments and did not vary by more than the amounts shown.
Cell lines were left untreated or were treated with either 50 μM hemin, which is a source of iron and therefore allows for the translation of IRE-Gl mRNA, or 50 μM deferoxamine mesylate (Df), which chelates iron and consequently inhibits the translation of IRE-Gl mRNA. Protein and RNA in total cell lysates were analyzed by Western blotting and RT–PCR, respectively. The level of cellular ferritin provided a measure of IRE function. Ferritin was abundant in the presence of hemin but essentially undetectable with Df treatment or no treatment (Fig. 1B, where the level of PLCγ1 controls for variations in protein loading), as would be predicted since hemin promotes ferritin mRNA translation, whereas Df or no treatment inhibits ferritin mRNA translation. RT coupled to either semiquantitative PCR (sqPCR) or real-time quantitative PCR (qPCR) revealed that hemin also promoted IRE-Gl NMD so that the level of IRE-Gl Ter mRNA was reduced to ∼17% of Norm, whereas Df or no treatment inhibited IRE-Gl NMD so that the level of IRE-Gl Ter mRNA was ∼96%–97% of Norm (Fig. 1C,D, where the level of endogenous SMG7 mRNA or β-actin mRNA controlled for variations in RNA loading, respectively). These results are consistent with an earlier report that an IRP-mediated block in the translation of IRE-Gl Ter mRNA abrogates NMD (Thermann et al. 1998).
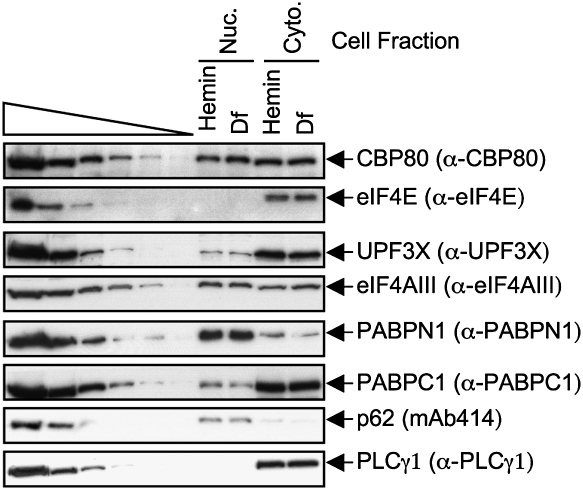
To examine mRNP, lysates of HeLa cells that express IRE-Gl Norm mRNA (hereafter called IRE-Gl mRNA) were immunoprecipitated using (1) anti-CBP80, (2) anti-eIF4E, or, to control for nonspecific IP, (3) normal rabbit serum (NRS). Additionally, in proof-of-principle experiments, lysates of cells that express IRE-Gl mRNA were also immunoprecipitated using anti-UPF3X or anti-eIF4AIII, each of which reacts with an EJC constituent. In HeLa cells, as in other mammalian cells, the CBC constituents CBP80 and CBP20 are largely nuclear proteins, whereas eIF4E is largely cytoplasmic (e.g., see Lejbkowicz et al. 1992; Wilson et al. 1999; McKendrick et al. 2001), and, importantly, this intracellular distribution was comparable in hemin-treated and Df-treated cells (Fig. 2, where p62 or PLCγ1 control for, respectively, nuclear contamination of the cytoplasmic fraction or cytoplasmic contamination of the nuclear fraction). UPF3X and eIF4AIII shuttle between the nucleus and the cytoplasm according to heterokaryon assays (Lykke-Andersen et al. 2000; Serin et al. 2001; Shibuya et al. 2004). Consistent with previous results using untreated cells (Ferraiuolo et al. 2004; our unpublished data for UPF3X), Western blotting demonstrated that both UPF3X and eIF4AIII are readily detected in both nuclear and cytoplasmic fractions (Fig. 2). Furthermore, the intracellular distribution of each protein appeared to be unaffected by hemin or Df treatment (Fig. 2).
Figure 2.
Nuclear and cytoplasmic distributions of mRNP proteins in HeLa cells. Western blot analysis using nuclear and cytoplasmic HeLa cell fractions and the specified antibody (α). The cellular distributions of p62, with which mAb414 primarily reacts, and PLCγ1 demonstrate that the cytoplasmic fraction is free of detectable nuclear contamination and vice versa. Serial dilutions of protein in the left six lanes demonstrate that the Western blot analysis is semiquantitative. Results are derived from two independently performed experiments.
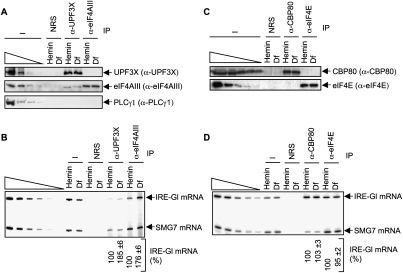
Western blotting revealed that, as expected, anti-UPF3X immunoprecipitated UPF3X and eIF4AIII but not detectably PLCγ1 (Fig. 3A). Also as expected, anti-eIF4AIII immunoprecipitated eIF4AIII and not detectably PLCγ1 (our inability to detect UPF3X may reflect that only a fraction of cellular eIF4AIII is present in EJCs, or that anti-eIF4AIII physically precludes the co-IP of UPF3X) (Fig. 3A). RT–sqPCR revealed an ∼1.9-fold to ∼1.8-fold increase in the amount of IRE-Gl mRNA that was immunoprecipitated by, respectively, anti-UPF3X or anti-eIF4AIII from Df-treated cells relative to hemin-treated cells (Fig. 3B), which was confirmed using RT–qPCR (data not shown). These results are consistent with data demonstrating that translation promotes the removal of the Y14 EJC constituent from mRNA (Dostie and Dreyfuss 2002). Considering that eIF4AIII forms the EJC anchor (Ballut et al. 2005; Tange et al. 2005; Andersen et al. 2006; Bono et al. 2006), these data also suggest that translation—presumably the pioneer round of translation—promotes the removal of the entire EJC from mRNA. Notably, analyses of cytoplasmic lysates that, unlike total cell lysates, contain a significant fraction of EJC-bound mRNA that is accessible to the translational machinery revealed a larger (∼3.7-fold) increase in the amount of EJC-bound IRE-Gl mRNA in Df-treated cells relative to hemin-treated cells (Supplemental Fig. S1).
Figure 3.
Translation decreases the co-IP of the UPF3X and eIF4AIII EJC constituents with IRE-Gl mRNA but does not affect the co-IP of CBP80 or eIF4E with IRE-Gl mRNA. (A) Lysates from hemin-treated or Df-treated HeLa cells that stably express IRE-Gl Norm mRNA, referred to here as IRE-Gl mRNA, were immunoprecipitated using the specified antibody (α) or, as a control for nonspecific IP, NRS. (B) RT–sqPCR analysis of IRE-Gl mRNA and SMG7 mRNA before or after the IP shown in A, where the normalized level of IRE-Gl mRNA in the presence of hemin is defined as 100. (C) Western blotting before and after IP using NRS, anti-CBP80, or anti-eIF4E. (D) As in B, except the samples from the IP shown in C were used. Results are representative of three independently performed experiments.
In contrast to IPs involving anti-UPF3X or anti-eIF4AIII, IPs involving anti-CBP80 or anti-eIF4E (Fig. 3C) did not reveal a significant difference in the amount of IRE-Gl mRNA that was immunoprecipitated from Df-treated cells compared with hemin-treated cells, as measured using either RT–sqPCR (Fig. 3D) or RT–qPCR (data not shown). Since a block in IRE-Gl mRNA translation does not result in an accumulation of CBC-bound IRE-Gl mRNA or a decrease in the amount of eIF4E-bound IRE-Gl mRNA, it follows that the pioneer round of translation is not required for the replacement of CBP80 by eIF4E at the cap of IRE-Gl mRNA. Importantly, similar conclusions can be drawn from studies of cellular ferritin mRNA (Supplemental Fig. S2), which is the source of the IRE in IRE-Gl mRNA. Notably, our ability to detect eIF4E-bound mRNA in the absence of translation (i.e., in Df-treated cells that, prior to treatment, were not translating IRE-containing mRNAs) (Supplemental Fig. S2) provides additional evidence that translation is not required for the generation of eIF4E-bound mRNA from CBC-bound mRNA.
It may be that a Df-mediated block in IRE-Gl mRNA translation inhibits IRE-Gl mRNA export to the cytoplasm, given that a cycloheximide-mediated block in translation was shown to slow the rate of mRNA export to the cytoplasm (Muralidhar and Johnson 1988). However, cell fractionation studies revealed that Df treatment had no appreciable effect on the relative levels of nuclear and cytoplasmic IRE-Gl mRNA (Supplemental Fig. S3).
Translation promotes the replacement of PABPN1 by PABPC1 on newly synthesized mRNA
PABPN1 is a mostly nuclear protein that also copurifies with the cytoplasm (Hosoda et al. 2006), and its cellular distribution was not detectably influenced by hemin or Df treatment (Fig. 2). PABPN1, which coimmunoprecipitates with CBP80 (Ishigaki et al. 2001), begins associating with newly synthesized transcripts prior to splicing and remains associated with newly synthesized mRNA that has undergone NMD and, thus, the pioneer round of translation (Hosoda et al. 2006). PABPC1 is mostly cytoplasmic but also nuclear (Hosoda et al. 2006), and its cellular distribution was not altered by hemin or Df treatment (Fig. 2). Like PABPN1, PABPC1 coimmunoprecipitates with CBP80 (Chiu et al. 2004) and begins associating with newly synthesized transcripts prior to splicing (Hosoda et al. 2006). Furthermore, PABPC1 appears to activate pioneer rounds of translation similarly to how it activates steady-state rounds of translation, since Paip2, which inhibits the interaction of PABPC1 with poly(A), inhibits NMD (Chiu et al. 2004).
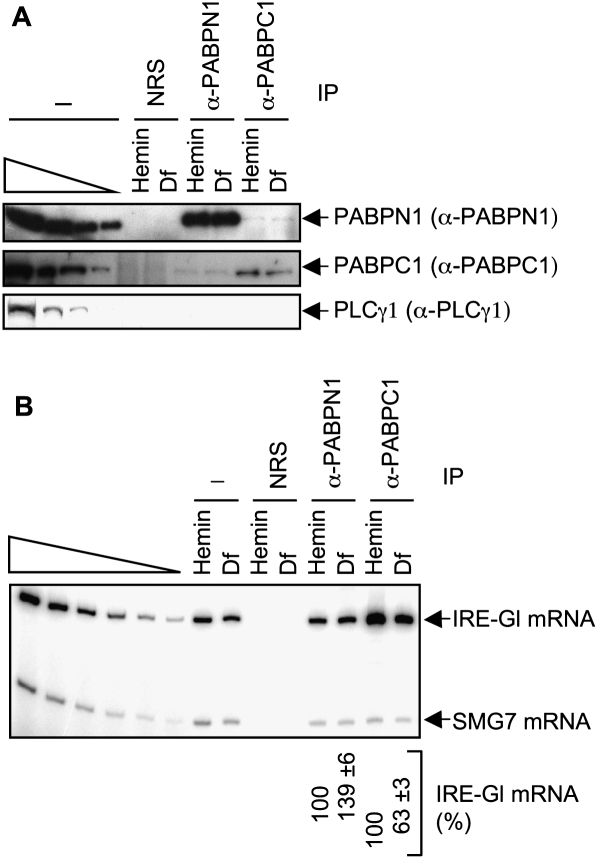
To determine if translation influences the association of either PABPN1 or PAPBC1 with mRNA, lysates of HeLa cells expressing IRE-Gl mRNA were immunoprecipitated using (1) anti-PABPN1, (2) anti-PABPC1, or, to control for nonspecific IP, (3) NRS. Western blotting demonstrated the specificity of each IP (Fig. 4A). Remarkably, RT–sqPCR revealed an ∼1.4-fold increase and an ∼1.6-fold decrease in the amount of IRE-Gl mRNA that was immunoprecipitated by, respectively, anti-PABPN1 and anti-PABPC1 in Df-treated cells compared with hemin-treated cells (Fig. 4B; data not shown for RT–qPCR). Similar results were obtained by assaying ferritin mRNA (Supplemental Fig. S2). Furthermore, there was an ∼2.5-fold increase in the amount of PABPN1-bound IRE-Gl mRNA and an ∼2.4-fold decrease in the amount of PABPC1-bound IRE-Gl mRNA in cytoplasmic lysates from Df-treated cells relative to cytoplasmic lysates from hemin-treated cells (Supplemental Fig. S1). The larger effects observed using cytoplasmic lysates compared with total cell lysates (Fig. 4B) is consistent with the idea that newly synthesized mRNA is susceptible to translation in the cytoplasm but not the nucleus. We conclude that translation augments the removal of PABPN1 and/or increases the binding of PABPC1 to mRNA, which results in the replacement of PABPN1 by PABPC1.
Figure 4.
Translation decreases the co-IP of PABPN1 and increases the co-IP of PABPC1 with IRE-Gl mRNA. Lysates from hemin-treated or Df-treated HeLa cells that stably express IRE-Gl mRNA were immunoprecipitated using the specified antibody (α). (A) Western blotting before (−) and after IP. (B) RT–sqPCR analysis of RNA before and after IP. Results are representative of three independently performed experiments.
Evidence that IMPα associates with the 5′ end of newly synthesized mRNA via CBP80
Data indicate that CBC, which binds to newly synthesized transcripts within nuclei, is a constituent of the pioneer translation initiation complex that is replaced by eIF4E in the cytoplasm. In vitro binding studies of CBC and eIF4E to mono-, di-, and tetranucleotide 5′-cap analogs indicate that CBC binds to cap structures with higher affinity than eIF4E (Worch et al. 2005). Therefore, the replacement of CBC by eIF4E at an mRNA cap appears to involve transitioning from a more tightly bound complex to a more loosely bound complex. If translation does not promote the replacement of CBC by eIF4E, what does?
IMPα is an adaptor protein that generally binds the nuclear localization signal (NLS) of cytoplasmic cargo that is destined for import into nuclei (for review, see Cook et al. 2007; Goldfarb 2009). Görlich et al. (1996b) found that IMPα in the nuclei of Xenopus laevis oocytes and the Saccharomyces cerevisiae IMPα ortholog Srp1p, which is largely nuclear, interact with CBC that is bound to small nuclear RNA (snRNA) in a cap-dependent manner. Unlike most IMPα–NLS complexes, the interaction of IMPα with the bipartite NLS of CBP80 is remarkably resistant to high salt concentrations and is not dissociated within nuclei (Lewis and Izaurralde 1997). IMPα was shown to coimmunoprecipitate with U1ΔSm snRNA, which has a monomethylated cap because it is mutated so as to be retained within nuclei. Furthermore, IMPβ was demonstrated to dissociate Srp1p–CBC from an in vitro synthesized cap structure (Görlich et al. 1996b) and, thus, presumably but not certainly in vivo.
The direct binding of the karyopherin IMPβ to IMPα in the cytoplasm generally docks an IMPα–NLS complex at the nuclear pore and mediates its import into nuclei, after which the direct binding of nuclear RAN–GTP to IMPβ releases import substrate and IMPα into the nucleoplasm (Cook et al. 2007). It follows that IMPβ binding to IMPα–CBC at the caps of pioneer translation initiation complexes in the cytoplasm could mediate the dissociation of CBC from newly exported mRNAs independently of translation, which could in turn augment the binding of eIF4E to the resulting unoccupied 5′ caps.
To begin to test this hypothesis, we assayed for the co-IP of IMPα or, as a control for nonspecific IP, rabbit IgG (rIgG) with IRE-Gl mRNA in hemin-treated or Df-treated HeLa cells that stably express IRE-Gl mRNA. Western blotting demonstrated the specificity of the anti-IMPα IP (Fig. 5A). RT–sqPCR revealed that, indeed, IRE-Gl mRNA as well as SMG7 mRNA were immunoprecipitated using anti-IMPα in a way that was unaffected by the intracellular iron level (Fig. 5B). To determine if the IP of IRE-Gl mRNA using anti-IMPα is due to IMPα binding to the cap-containing mRNA 5′ end, as our hypothesis would predict, nuclear lysates, which are rich in newly synthesized mRNA, were incubated with RNase H and an antisense DNA oligonucleotide prior to IP using anti-IMPα or rIgG. The antisense DNA oligonucleotide directs the cleavage of IRE-Gl mRNA within exon 1, resulting in a cap-containing 5′-end cleavage product of 138 nucleotides (nt) and an EJC- and poly(A) tail-containing 3′-end cleavage product of 526 nt, excluding the poly(A) tail (Fig. 5C).
Figure 5.
IMPα stably associates with CBP80 at the cap of newly synthesized IRE-Gl mRNA. (A) Western blot analysis of lysates from hemin-treated or Df-treated HeLa cells that stably express IRE-Gl mRNA, either before (−) or after IP using rIgG or anti-IMPα (α-IMPα). (B) RT–sqPCR of IRE-Gl and SMG7 mRNAs from the same lysates used in A. (C) Diagram of IRE-Gl mRNA, where boxes denote exons, the hairpin structure denotes the IRE, and the solid arrow specifies the position of oligonucleotide (oligo)-directed RNase H cleavage within exon 1. Open arrowheads represent primers used to generate the RT–sqPCR product of full-length Gl mRNA or its 5′ or 3′ region. (D) Western blot analysis of nuclear lysates from untreated HeLa cells stably expressing IRE-Gl mRNA, either before (−) or after (+) oligonucleotide-directed RNase H cleavage followed by IP using rIgG or anti-IMPα. (E) RT–sqPCR analysis of IRE-Gl mRNA from nuclear lysates analyzed in D, except that lysates prior to IP were also analyzed to demonstrate that the oligonucleotide-directed cleavage of IRE-Gl mRNA was complete. (F) RT–PCR analysis of SMG7 mRNA (top) and Western blot analysis of the specified proteins (bottom) from nuclear and cytoplasmic lysates that were (+) or were not (−) treated with RNase A and subsequently subjected to IP using rIgG or anti-IMPα. Results are representative of two or three independently performed experiments.
Western blotting confirmed the specificity of the anti-IMPα IP (Fig. 5D). RT–sqPCR of samples prior to IP demonstrated that oligonucleotide-directed RNase H cleavage was complete, as evidenced by the lack of detectable full-length IRE-Gl mRNA (Fig. 5E). IMPα was found to coimmunoprecipitate with the cap-containing 5′-cleavage product and not the EJC- and poly(A)-containing 3′-cleavage product (Fig. 5E), which is consistent with data indicating that the association of IMPα with CBP80 is atypical of other mRNP-binding proteins that contain an NLS. Supporting the idea that IMPα coimmunoprecipitates with the 5′-cleavage product via binding to CBP80, IMPα coimmunoprecipitated with CBP80 in both nuclear and cytoplasmic fractions (Fig. 5F). In contrast, IMPα did not coimmunoprecipitate with eIF4E (Fig. 5F), which lacks an NLS. Notably, the co-IP of IMPα and CBP80, like the co-IP of IMPα and IMPβ, was insensitive to RNase A treatment (Fig. 5F), in keeping with the interactions being direct (Görlich et al. 1996b). The co-IP of IMPα and CBP80 was detected in both nuclear and cytoplasmic fractions (Fig. 5F), suggesting that IMPα could associate with cap-bound CBP80 within nuclei. In fact, our findings that IMPα coimmunoprecipitates with nuclear IRE-Gl pre-mRNA (Supplemental Fig. S4A) as well as 28S and 18S rRNAs (Supplemental Fig. S4B), and associates with monosomes and small polysomes, as does CBP80 (Supplemental Fig. S4C), indicate that IMPα indeed begins to interact with cap-bound CBP80 in the nucleus prior to splicing and remains bound in the cytoplasm during the pioneer round of translation.
We conclude that IMPα stably associates with the 5′ end of IRE-Gl mRNA via CBP80 and not via eIF4E. In support of this, and consistent with data showing that IMPα does not coimmunoprecipitate with the mRNA poly(A) tail (Fig. 5E), the co-IP of IMPα with PABPN1 in the nuclear fraction was sensitive to RNase A treatment (Fig. 5F, where the co-IP of IMPα with PABPN1 in the cytoplasmic fraction was undetectable).
Evidence that IMPβ promotes the replacement of CBC by eIF4E at an mRNA cap
We next tested if inhibiting the interaction of IMPα with IMPβ by transiently expressing the IMPβ-binding domain (IBB) of IMPα affects the replacement of CBC by eIF4E on newly synthesized mRNA. IBB is a 41-amino-acid arginine-rich region of IMPα that binds directly to IMPβ (Görlich et al. 1996a; Palmeri and Malim 1999). Initially, we chose to assay cellular MYC (c-MYC) and c-FOS mRNAs rather than IRE-Gl mRNA, since the shorter-than-average half-lives of c-MYC and c-FOS mRNAs (e.g., ∼7–15 min in cultured mammalian cells) (Shyu et al. 1989; Kabnick and Housman 1988) would ensure that a significant percentage of each mRNA in the cytoplasmic fraction consists of newly synthesized mRNA.
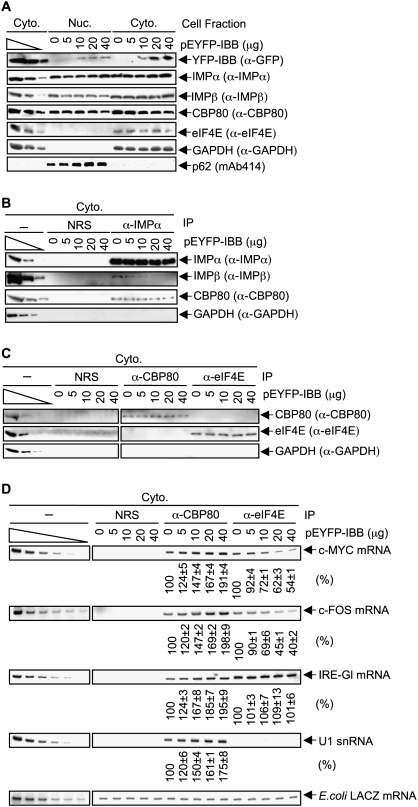
HeLa cells were transfected with 0, 5, 10, 20, or 40 μg of pEYFP-IBB (Li and Zheng 2004). Western blotting of nuclear and cytoplasmic lysates revealed that increasing amounts of epitope-tagged IBB were produced with increasing amounts of IBB expression vector without affecting the cellular abundance of either IMPα or IMPβ (Fig. 6A, where the level of GAPDH controlled for variations in cytoplasmic protein loading). Notably, IBB expression did not result in a detectable accumulation of either IMPα or IMPα-bound CBP80 in the cytoplasm (Fig. 6A,B). This is likely because there exists an IMPβ-independent pathway by which IMPα and, presumably as a consequence, IMPα-bound CBP80 can migrate from the cytoplasm into nuclei (Miyamoto et al. 2002), although the relative activities of the IMPβ-dependent and IMPβ-independent pathways of IMPα-mediated nuclear import remain to be determined. Nevertheless, increasing amounts of IBB did inhibit the co-IP of IMPα with IMPβ (Fig. 6B), which was the purpose of expressing IBB in these experiments.
Figure 6.
Inhibiting the interaction of IMPα with IMPβ by expressing the IBB region of IMPα augments the co-IP of CBP80 with c-MYC mRNA, c-FOS mRNA, IRE-Gl mRNA, and U1 snRNA, and concomitantly decreases the co-IP of eIF4E with c-MYC and c-FOS mRNAs. (A) HeLa cells (1 × 107) were transiently transfected with pEYFP-IBB (0, 5, 10, 20, or 40 μg) and, when needed, pUC19 to bring the total amount of transfecting DNA to 40 μg. Nuclear and cytoplasmic lysates were subjected to Western blot analysis using the specified antibody (α). The distributions of p62 and GAPDH demonstrate that cellular fractions are free of detectable cross-contamination. (B) Cytoplasmic lysates from HeLa cells that had been transfected as in A were analyzed by Western blotting either before (−) or after IP using NRS or anti-IMPα. (C) Cytoplasmic lysates generated as in A were subjected to Western blotting before or after IP using NRS, anti-CBP80, or anti-eIF4E. (D) RT–sqPCR analysis of c-MYC mRNA, c-FOS mRNA, IRE-Gl mRNA, or U1 snRNA in the same IPs of cytoplasmic lysates that were analyzed in C. RNA was purified before or after IP. Samples after IP were spiked with a small amount of E. coli RNA, and RT–sqPCR measurements of the level of E. coli LACZ mRNA were used to control for variations in RNA recovery. (The left six lanes of LACZ mRNA do not derive from dilutions of samples before IP but from dilutions of E. coli RNA.) For each IP, the level of c-MYC, c-FOS, IRE-Gl mRNA, or U1 snRNA in cells lacking IBB was defined as 100. Using quantitations of 28S rRNA to control for variations in RNA recovery and loading, IBB expression was found to have no effect on the cytoplasmic level of any of the three mRNAs tested (data not shown). Results are representative of two independently performed experiments.
Next, cytoplasmic lysates from cells expressing increasing amounts of epitope-tagged IBB were immunoprecipitated using anti-CBP80, anti-eIF4E, or, as a control for nonspecific IP, NRS. Variations in the level of IBB expression did not change the amounts of CBP80 or eIF4E that were immunoprecipitated using the respective antibody (Fig. 6C). RT–sqPCR demonstrated that expressing increasing amounts of YFP-IBB indeed resulted in the co-IP of increasing amounts of c-MYC and c-FOS mRNAs and U1 snRNA with CBP80 (Fig. 6D). These increases were ∼1.9-fold, ∼2.0-fold, and ∼1.8-fold, respectively, when cells were transfected with 40 μg of pEYFP-IBB (Fig. 6D). These findings are consistent with IMPβ dissociating IMPα and CBP80 from newly synthesized c-MYC and c-FOS mRNAs and U1 snRNA. Furthermore, expressing increasing amounts of YFP-IBB resulted in the co-IP of decreasing amounts of c-MYC and c-FOS mRNAs with eIF4E (Fig. 6D), as would be expected if eIF4E binding to mRNA caps were promoted by the IMPβ-mediated dissociation of CBP80 from mRNA caps. U1 snRNA, which harbors a trimethylated cap in the cytoplasm, did not detectably coimmunoprecipitate with eIF4E (Fig. 6D), as was evident from several observations: (1) Cellular U snRNPs appear to be largely free of sequestering CBPs, based on their quantitative IP using anti-trimethyl G (Bringmann et al. 1983); (2) eIF4E binds trimethylated caps 10-fold less efficiently than it binds monomethylated caps (Wieczorek et al. 1999); and (3) snurportin binds the trimethylated cap of cytoplasmic U snRNAs (Will and Lührmann 2001) so as to mediate together with IMPβ snRNA import into nuclei to function in pre-mRNA splicing (Rollenhagen and Pante 2006; Cook et al. 2007).
For the sake of completeness, the effect of YFP-IBB on the co-IP of either CBP80 or eIF4E with IRE-Gl mRNA was also measured. While expressing increasing amounts of YFP-IBB resulted in the co-IP of increasing amounts of IRE-Gl mRNA with CBP80 (Fig. 6D), our failure to detect an effect of YFP-IBB on the co-IP of eIF4E with IRE-Gl mRNA (Fig. 6D) verifies that pre-existing levels of cytoplasmic eIF4E-bound IRE-Gl mRNA mute the ability to measure the decreased acquisition of eIF4E by newly synthesized IRE-Gl mRNA. We conclude that the binding of IMPβ to IMPα–CBC at the 5′ caps of newly synthesized mRNAs contributes to mRNP remodeling by promoting the removal of CBC and, concomitantly, its replacement by eIF4E.
As an alternative way to assess the effect of IMPβ on the remodeling of newly synthesized mRNP, we aimed to inhibit the binding of IMPβ to IMPα by transiently expressing RAN(Q69L), which is a variant of the RAS-related small guanine nucleotide-binding protein RAN that binds but cannot hydrolyze GTP (Bischoff et al. 1994; Klebe et al. 1995). Normally, the complex of RAN–GTP and IMPβ is transported from nuclei to the cytoplasm, where the GTPase-activating protein RAN-GAP stimulates the hydrolysis of the GTP bound to RAN; this, in turn, frees IMPβ to bind to IMPα (Cook et al. 2007).
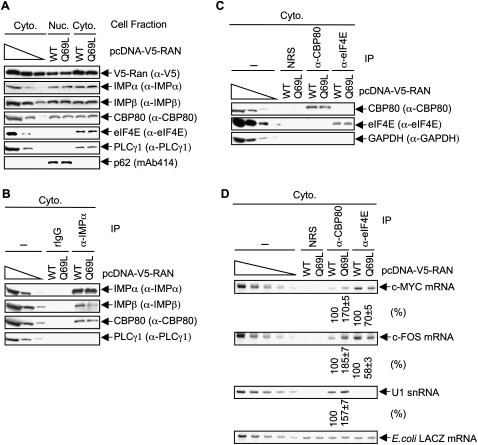
HeLa cells were transiently transfected with pcDNA3.1D/V5-His-TOPO- RAN(Q69L) [hereafter called pcDNA-V5-RAN(Q69L)] or, as a control, pcDNA-V5-RAN(WT), which produces wild-type (WT) V5-RAN. Western blotting of nuclear and cytoplasmic fractions revealed that neither V5-RAN(Q69L) nor V5-RAN(WT) affected the cellular abundance of IMPα or IMPβ (Fig. 7A). Furthermore, V5-RAN(Q69L) did not result in a detectable accumulation of either IMPα or IMPα-bound CBP80 in the cytoplasm (Fig. 7A,B) even though it inhibited the co-IP of IMPα with IMPβ (Fig. 7B), consistent with results obtained using YFP-IBB (Fig. 6). Additionally, V5-RAN(Q69L) did not change the amounts of CBP80 or eIF4E that were immunoprecipitated using anti-CBP80 or anti-eIF4E, respectively (Fig. 7C). However, RT–sqPCR demonstrated that V5-RAN(Q69L) increased the co-IP of c-MYC and c-FOS mRNAs and U1 snRNA with CBP80 by ∼1.7-fold, ∼1.9-fold, and ∼1.6-fold, respectively (Fig. 7D), consistent with IMPβ dissociating IMPα and CBP80 from each newly synthesized transcript. Furthermore, V5-RAN(Q69L) decreased the co-IP of c-MYC and c-FOS mRNAs with eIF4E by ∼1.4-fold and ∼1.7-fold, respectively (Fig. 7D). These findings reinforce the idea that the binding of IMPβ to IMPα–CBC at the 5′ caps of newly synthesized mRNAs contributes to mRNP remodeling by promoting the removal of CBC and, concomitantly, its replacement by eIF4E.
Figure 7.
Inhibiting the interaction of IMPα with IMPβ by expressing RAN(Q69L) augments the co-IP of CBP80 with c-MYC, c-FOS, IRE-Gl mRNAs, and U1 snRNA, and concomitantly decreases the co-IP of eIF4E with c-MYC and c-FOS mRNAs. (A) HeLa cells (1 × 107) were transiently transfected with pcDNA-RAN(WT) or pcDNA-RAN(Q69L) (30 μg). Nuclear and cytoplasmic lysates were subjected to Western blot analysis using the specified antibody (α). The distributions of p62 and PLCγ1 demonstrate that cellular fractions are free of detectable cross-contamination. (B) As in Figure 6B, except PLCγ1 was used to control for IP specificity. (C) As in Figure 6C. (D) As in Figure 6D, except IRE-Gl mRNA was not measured. Notably, it has been shown that RANQ69L expression, like IBB expression (Fig. 6), does not alter the cytoplasmic level of mRNA (Clouse et al. 2001). Results are representative of two independently performed experiments.
Discussion
Proper remodeling of newly synthesized mRNP is essential to the fidelity of gene expression. Remodeling occurs within nuclei, on both sides of the nuclear pore complex during the process of mRNA export from the nucleus to the cytoplasm, and in the cytoplasm (e.g., see Tran et al. 2007 and references therein). Results reported here provide evidence that remodeling in the cytoplasm can occur as a consequence of mRNA associating with translationally active ribosomes during the pioneer round of translation or with IMPβ independently of translation. The pioneer round of translation promotes EJC removal and the replacement of PABPN1 by PABPC1 (Fig. 3, 4; Supplemental Figs. S1, S2). This finding for EJCs is consistent with the rule that termination codons situated more than ∼50–55 nt upstream of a splicing-generated exon–exon junction generally trigger NMD (Nagy and Maquat 1998): If a termination codon resides less than ∼50–55 nt upstream of an exon–exon junction or downstream from the junction, then the translating ribosome is thought to physically displace the EJC (Lejeune and Maquat 2005). The removal of EJCs by translating ribosomes is also consistent with a report from Gehring et al. (2009) that appeared while this paper was under review. Ribosome-bound PYM as well as overexpressed PYM were found to promote the removal of EJCs from spliced mRNA, implicating the involvement of translation-dependent and, possibly, translation-independent mechanisms, depending on how much of cytoplasmic PYM is not associated with ribosomes and can bind EJCs. How translation promotes the replacement of PABPN1 by PABPC1 is less certain. PABPC1 is known to interact directly with eRF3, which is thought to bind mRNA during the process of translation termination and, in theory, could facilitate the binding of PABPC1 to translationally active mRNA (Hoshino et al. 1999). Notably, the region of PABPC1 that interacts with eRF3 is not conserved in PABPN1 (Kühn and Wahle 2004).
Strikingly, the pioneer round of translation is of no apparent consequence to the replacement of CBC by eIF4E (Fig. 3; Supplemental Fig. S2). In this regard, it is notable that the CBC is encountered only by the scanning 40S ribosomal subunit rather than by the translationally active 80S ribosome, as are EJCs that are displaced by the pioneer round of translation. Instead of depending on translation, we find that the exchange of CBPs is promoted by binding of the nuclear transport receptor IMPβ to the adaptor IMPα that is associated with the NLS of CBP80 on newly synthesized CBC-bound mRNA. First, IMPα detectably coimmunoprecipitates with CBP80 but not eIF4E, and the interaction of IMPα with CBP80 is resistant to RNase A treatment (Fig. 5). Second, IMPα coimmunoprecipitates with newly synthesized mRNP—specifically, with the 5′-cap-containing fragment, and not with the significantly larger 3′-EJC- and poly(A)-containing fragment (Fig. 5). Third, inhibiting the interaction of IMPα with IMPβ by expressing either the IBB of IMPα or a variant of RAN that cannot hydrolyze GTP increases the co-IP of CBP80 with mRNA and, as a result, decreases the co-IP of eIF4E with mRNA (Figs. 6, 7). We conclude that the replacement of CBC by eIF4E is regulated by the RAN–GTP/GDP gradient that promotes the binding of IMPβ to IMPα in the cytoplasm and the dissociation of IMPβ from IMPα in the nucleus (Fig. 8; Cook et al. 2007). Consistent with this conclusion, while this paper was under review, Dias et al. (2009) reported the structure of an IMPα–CBC complex. Using this structure, a model of the IMPβ–IMPα–CBC interaction, and biochemical and mutagenesis data, they formulated that IMPβ binds not only the IBB of IMPα but also the C-terminal region of CBP20, so as to weaken the affinity of CBP20 for capped RNA and promote the release of CBC.
Figure 8.
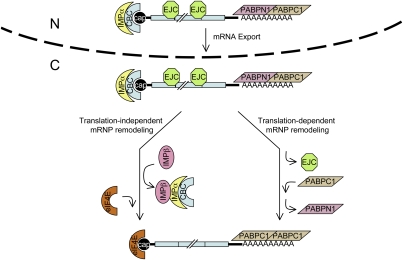
Schematic of translation-induced remodeling and translation-independent, IMPβ-induced remodeling of the pioneer translation initiation complex. The pioneer translation initiation complex is exported from nuclei (N) to the cytoplasm (C) bound by (1) CBC at the 5′-cap; (2) EJCs, which contain eIF4AIII and NMD factors that include UPF3X situated upstream of exon–exon junctions, provided the mRNA underwent splicing; (3) PABPN1 and PABPC1 at the poly(A) tail; and (4) other initiation factors that are not shown, including eIF4G and eIF3. Notably, IMPα associates with CBC via the NLS of CBP80, and the analysis of unspliced pre-mRNA indicates that IMPα begins associating with CBC-bound transcripts prior to their splicing (Supplemental Fig. S4), possibly concomitantly with CBC binding to nascent transcripts. Data indicate that the pioneer round of translation removes the EJCs (Fig. 3; Supplemental Fig. S2) and promotes the replacement of PABPN1 by PABPC1 (Fig. 4; Supplemental Figs. S1, S2) but is inconsequential to the association of IMPα with the CBC (Fig. 5) or the replacement of CBC by eIF4E (Fig. 3). Instead, our findings suggest that the interaction of IMPβ with IMPα promotes the replacement of CBC by eIF4E (Figs. 6, 7), which depends on the RAN–GTP-mediated delivery of nuclear IMPβ to the cytoplasm and, subsequently, promotion of IMPβ binding to IMPα–CBC by the GTPase-activating protein RAN-GAP. The replacement of CBC by eIF4E is followed by the trafficking of IMPβ–IMPα–CBC to nuclei. Once in the nucleus, IMPβ dissociates from IMPα–CBC. This frees IMPα–CBC to bind to the 5′ cap of newly made transcripts so as to function once again in the nuclear export and cytoplasmic remodeling of mRNP.
As implied above, our results from the experiments that employed the site-directed cleavage of IRE-Gl mRNA followed by IP with anti-IMPα indicate that we detect IMPα interacting with CBP80 but not with other mRNA-bound proteins. This may be attributable to the combination of IMPα (1) entering into an atypically strong interaction with CBP80 (Lewis and Izaurralde 1997) and (2) lacking an affinity for other proteins that bind mRNA. Among the proteins that typify newly synthesized mRNA, PABPN1 (Calado et al. 2000), SR proteins (see below), and hnRNPA1 (Pollard et al. 1996; Bonifaci et al. 1997) are recycled back to the nucleus by a transport mechanism that apparently does not involve IMPα.
In theory, the replacement of CBC by eIF4E at an mRNA 5′ cap could be augmented not only by IMPβ binding to IMPα–CBC at the cap but also by mass action, since the level of eIF4E relative to CBC is higher in the cytoplasm than in nuclei (Fig. 2). Additionally, it has been suggested (but not demonstrated) that replacement could be mediated by specific interactions among eIF4E, CBC, and eIF4G (Fortes et al. 2000; McKendrick et al. 2001) that decrease the affinity of CBC for the cap. However, whereas the binding of eIF4G to eIF4E does strengthen the interaction between eIF4E and the mRNA cap (Haghighat and Sonenberg 1997; Gross et al. 2003), there is currently no reason to think that eIF4G could not also stabilize the interaction between CBC and the cap considering that eIF4G appears to be a functional constituent of the pioneer translation initiation complex; i.e., eIF4G is present on mRNA prior to the binding of eIF4E (Lejeune et al. 2004).
Conceivably, mechanisms of protein trafficking other than those mediated by IMPβ–IMPα interactions could also be involved in remodeling newly synthesized mRNP to steady-state mRNP. As one of a number of possibilities, the IMPβ family member transportin-SR, which directly imports serine–arginine-rich (SR) splicing factors into nuclei after they are phosphorylated in the cytoplasm (Lai et al. 2000, 2001), may mediate the removal of SR proteins from newly synthesized mRNA. Alternatively, or additionally, phosphorylation per se may promote the removal of SR proteins from newly synthesized mRNA. The findings that SF2/ASF detectably coimmunoprecipitates with CBP80 but not eIF4E (9G8 was not tested) (Sato et al. 2008) and that both 9G8 and SF2/ASF are present in fractions containing monosomes and small polysomes rather than large polysomes (Sanford et al. 2004, 2005; Swartz et al. 2007) suggest that each is released from mRNA after the pioneer round of translation initiates. Thus, transportin-SR, possibly together with the pioneer round of translation, could promote the release of 9G8 and SF2/ASF from mRNA.
It is now clear that IMPβ, which was once thought to exclusively mediate the nuclear import of IMPα–NLS cargo, functions not only in adaptor-independent protein import but also in a number of roles during interphase and mitosis (for review, see Harel and Forbes 2004). As examples, IMPβ functions during interphase as a microtubule motor adaptor that carries certain viruses to nuclei, as a chaperone for positively charged and newly made nuclear proteins, and in signaling axonal damage to the neuronal cell body. IMPβ also functions during mitosis after nuclear breakdown in centrosome cohesion as well as the assembly of spindles, nuclear membranes, and nuclear pores. Our studies reveal yet another role for IMPβ in mRNP remodeling, and they portend that this role may extend to other members of the IMPβ superfamily of nuclear transport receptors.
There exists one other example by which a karyopherin may, but does not certainly, mediate the remodeling of newly synthesized mRNP. The example derives from studies of the karyopherin KAP104 in S. cerevisiae. It remains controversial if KAP104 is directly responsible for stripping the mRNA-binding protein NAB2 off newly synthesized mRNA, or if the DEAD-box protein DBP5 does the stripping and KAP104 merely recycles NAB2 back to the nucleus (for review, see Goldfarb 2009). However, a report that appeared while we were writing up our work provides data consistent with the idea that KAP104 mediates the dissociation of not only NAB2 but also NAB4/HRP1 from mRNA at the yeast bud tip and neck so as to promote the localized transition of translationally repressed mRNA to a translationally competent state (van den Bogaart et al. 2009). Future studies from a number of laboratories will undoubtedly test these and additional members of the karyopherin family of proteins for the potential to function in one or more of the many steps that comprise mRNP remodeling.
Materials and methods
Cell culture and transfections
HeLa cells were cultured in DMEM (GIBCO-BRL) with 10% fetal bovine serum (GIBCO-BRL). For stable transfections, HeLa cells (1.4 × 106 per 60-mm dish) were transfected with 10 μg of the specified pIRE-Gl derivative (Thermann et al. 1998) and 1 μg of pCI-neo using Lipofectamine Plus (Invitrogen). Two days after transfection, the medium was supplemented with 500 μg/mL geneticin (GIBCO-BRL). Individual geneticin-resistant colonies were isolated, and those expressing comparable levels of IRE-Gl pre-mRNA were identified and expanded for analysis. Cells were treated with 50 μM hemin (Sigma) or 50 μM Df (Sigma) for 18 h prior to harvesting. Alternatively, cells were transiently transfected with the specified plasmids using Lipofectamine 2000 (Invitrogen). Total cell or nuclear and cytoplasmic lysates were prepared as described previously (Sato et al. 2008).
Oligonucleotide-directed RNase H cleavage of mRNP
Nuclear extract from 2 × 107 HeLa cells was incubated with 0.1 U/mL RNase H (New England BioLabs) and 3 mM MgCl2 in the absence or presence of 0.25 mg/mL 5′-CACGTTCACCTT-3′ for 30 min at 30°C. This oligonucleotide anneals to and directs the cleavage of IRE-Gl mRNA at 30–41 nt upstream of the exon 1–exon 2 junction.
IPs
Samples for the analysis of protein and RNA were generated before and after IP, and IPs were performed as reported (Ishigaki et al. 2001).
Western blotting
Antibodies consisted of mAb414 (BABCO), anti-PLCγ1 (Santa Cruz Biotechnologies), anti-ferritin (Abcam), anti-UPF3X (Santa Cruz Biotechnologies), anti-eIF4AIII (Li et al. 1999), anti-CBP80 (Izaurralde et al. 1994), anti-eIF4E (Santa Cruz Biotechnologies), anti-PABPN1 (a gift from David Bear), anti-PABPC1 (Kuyumcu-Martinez et al. 2004), anti-IMPα (Bethyl Laboratories), anti-IMPβ (Bethyl Laboratories), anti-GFP (Santa Cruz Biotechnologies), anti-GAPDH (Santa Cruz Biotechnologies), and anti-V5 (Invitrogen).
RT coupled to sqPCR
cDNA synthesis using SuperScript II reverse transcriptase (Invitrogen) and PCR using mRNA-specific primers (IDT) were performed as described (Sato et al. 2008). cDNA of IRE-Gl or ferritin mRNA was amplified using the primer pair 5′-CCTGCTTCAACAGTGCTTGG-3′ (sense) and 5′-CCGAAGCACTTTCTTGCC-3′ (antisense) or 5′-CCATGTCTTACTACTTTGACCGC-3′ (sense) and 5′-GTTGGTCACGTGGTCACCC-3′ (antisense), respectively. cDNA of SMG7 mRNA was analyzed using a previously reported primer pair (Kim et al. 2005). cDNA for c-MYC or c-FOS mRNA was amplified using 5′-GCAAGACTCCAGCGCCTTCT-3′ (sense) and 5′-ACCTTTTGCCAGGAGCCTGC-3′ (antisense) or 5′-AAGGTGGAACAGTTATCTCC-3′ (sense) and 5′-CTAGTTGGTCTGTCTCCGCTTGG-3′ (antisense), respectively. cDNA of U1 snRNA was amplified using the primer pair 5′-ATACTTACCTGGCAGGG-3′ (sense) and 5′-AAGCGCGAACGCAGTCCCCC-3′ (antisense). cDNA of Escherichia coli LACZ mRNA was amplified using the primer pair 5′-CGCGAGGCGGTTTTCTCC-3′ (sense) and 5′-GCCTGAATGGCGAATGGCG-3′ (antisense). Primers used to amplify cDNA of the oligonucleotide-directed and RNase H-catalyzed 5′-cleavage product of IRE-Gl mRNA were the same sense primer that was used to amplify full-length IRE-Gl mRNA and 5′-CACAGGGCAGTAACGG-3′ (antisense). cDNA of the corresponding 3′-cleavage product was amplified using 5′-GCTGCTGGTGGTCTACCC-3′ (sense) and the same antisense primer that was used to amplify full-length IRE-Gl mRNA. The simultaneous analysis of serial dilutions of RNA ensured that RT-sqPCR analyses were quantitative. RT–sqPCR products were quantitated using Typhoon PhosphorImaging (Molecular Dynamics).
RT coupled to qPCR
RNA was converted to cDNA by random priming as described above for RT coupled to sqPCR. Specific cDNAs were analyzed by qPCR, which was performed at the University of Rochester Functional Genomics Center using an ABI 7900 PCR machine. TaqMan probes (ABI) consisted of human HBB (hemoglobin, β; assay ID Hs00747223-gl) to detect IRE-Gl mRNA, either Norm or Ter, and human ACTB (β actin; part no. 4326315E) to detect β-actin mRNA. All samples were analyzed in triplicate, and the levels of test mRNAs were normalized to the level of β-actin mRNA. Relative mRNA levels were determined from CT values according to the ΔΔCT method (Applied Biosystems).
Acknowledgments
We thank Fabrice Lejeune for generating stable HeLa cell transfectants, Andreas Kulozik and Matthias Hentze for IRE-Gl plasmids, Richard Cerione for pcDNA-V5-RAN(WT) and pcDNA-V5-RAN(Q69L), Nahum Sonenberg for anti-eIF4AIII, Elisa Izaurralde for anti-CBP80, David Bear for anti-PABPN1, Richard Lloyd for anti-PABPC1, and Hoi-Yeung Li and Yixian Zheng for pEYFP-IBB. We also thank Mutsuhito Ohno and David Goldfarb for helpful advice, and Olaf Isken for comments on the manuscript. This work was supported by NIH R01 GM 059614 to L.E.M and a post-doctoral fellowship from the Japan Society for the Promotion of Science to H.S.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1817109.
Supplemental material is available at http://www.genesdev.org.
References
- Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Séraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Klebe K, Kretschmer J, Wittinghofer W, Ponstingl H. Ran GAP1 induces GTPas activity of nuclear Ras-related Ran. Proc Natl Acad Sci. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin β2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Bringmann P, Rinke J, Appel B, Reuter R, Lührmann R. Purification of snRNPs U1, U2, U4, U5 and U6 with 2,2,7-trimethylguanosine-specific antibody and definition of their constituent proteins reacting with anti-Sm and anti-(U1)RNP antisera. EMBO J. 1983;2:1129–1135. doi: 10.1002/j.1460-2075.1983.tb01557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A, Kutay U, Kühn U, Wahle E, Carmo-Fonseca M. Deciphering the cellular pathway for transport of poly(A)-binding protein II. RNA. 2000;6:245–256. doi: 10.1017/s1355838200991908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero G, Wilson KF, Ly T, Rios-Steiner JL, Clardy JC, Cerione RA. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat Struct Biol. 2002;9:912–917. doi: 10.1038/nsb874. [DOI] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Chiu SY, Lejeune F, Ranganathan AC, Maquat LE. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes & Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse NC, Luo MJ, Zhou Z, Reed R. A Ran-independent pathway for export of spliced mRNA. Nat Cell Biol. 2001;3:97–99. doi: 10.1038/35050625. [DOI] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Dias SMG, Wilsoon KF, Rojas KS, Ambrosio AIB, Cerione RA. The molecular basis for the regulation of the cap-binding complex by the importins. Nat Struct Mol Biol. 2009;16:930–937. doi: 10.1038/nsmb.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol. 2002;12:1060–1067. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo MA, Lee CS, Ler LW, Hsu JL, Costa-Mattioli M, Luo MJ, Reed R, Sonenberg N. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc Natl Acad Sci. 2004;101:4118–4123. doi: 10.1073/pnas.0400933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn A, Proud CG. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J Biol Chem. 1995;270:21684–21688. doi: 10.1074/jbc.270.37.21684. [DOI] [PubMed] [Google Scholar]
- Fortes P, Inada T, Preiss T, Hentze MW, Mattaj IW, Sachs AB. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol Cell. 2000;6:191–196. [PubMed] [Google Scholar]
- Gehring NE, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009;137:536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes & Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb DS. How to grow a bud: An importin acts in asymmetric division. Nat Cell Biol. 2009;11:243–245. doi: 10.1038/ncb0309-243. [DOI] [PubMed] [Google Scholar]
- Görlich D, Henklein P, Laskey RA, Hartmann E. A 41 amino acid motif in importin-α confers binding to importin-β and hence transit into the nucleus. EMBO J. 1996a;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey RA, Mattaj IW, Izaurralde E. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996b;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- Gray NK, Hentze MW. Regulation of protein synthesis by mRNA structure. Mol Biol Rep. 1994;19:195–200. doi: 10.1007/BF00986961. [DOI] [PubMed] [Google Scholar]
- Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- Haghighat A, Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J Biol Chem. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- Harel A, Forbes DJ. Importin β: Conducting a much larger cellular symphony. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Kühn LC. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc Natl Acad Sci. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- Hosoda N, Kim YK, Lejeune F, Maquat LE. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- Hosoda N, Lejeune F, Maquat LE. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol Cell Biol. 2006;26:3085–3097. doi: 10.1128/MCB.26.8.3085-3097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes & Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- Kabnick KS, Housman DE. Determinants that contribute to cytoplasmic stability of human c-fos and β-globin mRNAs are located at several sites in each mRNA. Mol Cell Biol. 1988;8:3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes & Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science. 2001;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- Klausner RD, Rouault TA, Harford JB. Regulating the fate of mRNA: The control of cellular iron metabolism. Cell. 1993;72:19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- Klebe C, Bischoff FR, Ponstingl H, Wittinghofer A. Interaction of nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochem. 1995;34:639–647. doi: 10.1021/bi00002a031. [DOI] [PubMed] [Google Scholar]
- Koritzinsky M, Magagnin MG, van den Beucken T, Seigneuric R, Savelkouls K, Dostie J, Pyronnet S, Kaufman RJ, Weppler SA, Voncken JW, et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 2006;25:1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn U, Wahle E. Structure and function of poly(A) binding proteins. Biochim Biophys Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez M, Belliot G, Sosnovtsev SV, Chang KO, Green KY, Lloyd RE. Calicivirus 3C-like proteinase inhibits cellular translation by cleavage of poly(A)-binding protein. J Virol. 2004;78:8172–8182. doi: 10.1128/JVI.78.15.8172-8182.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY. A human importin-β family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J Biol Chem. 2000;275:7950–7957. doi: 10.1074/jbc.275.11.7950. [DOI] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Tarn WY. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc Natl Acad Sci. 2001;98:10154–10159. doi: 10.1073/pnas.181354098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejbkowicz F, Goyer C, Darveau A, Neron S, Lemieux R, Sonenberg N. A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc Natl Acad Sci. 1992;89:9612–9616. doi: 10.1073/pnas.89.20.9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Ranganathan AC, Maquat LE. eIF4G is required for the pioneer round of translation in mammalian cells. Nat Struct Mol Biol. 2004;11:992–1000. doi: 10.1038/nsmb824. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur J Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- Li HY, Zheng Y. Phosphorylation of RCC1 in mitosis is essential for producing a high RanGTP concentration on chromosomes and for spindle assembly in mammalian cells. Genes & Dev. 2004;18:512–527. doi: 10.1101/gad.1177304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Imataka H, Morino S, Rogers GW, Jr, Richter-Cook NJ, Merrick WC, Sonenberg N. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol Cell Biol. 1999;19:7336–7346. doi: 10.1128/mcb.19.11.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- Matsuda D, Hosoda N, Kim YK, Maquat LE. Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nat Struct Mol Biol. 2007;14:974–979. doi: 10.1038/nsmb1297. [DOI] [PubMed] [Google Scholar]
- Matsuo H, Li H, McGuire AM, Fletcher CM, Gingras AC, Sonenberg N, Wagner G. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol. 1997;4:717–724. doi: 10.1038/nsb0997-717. [DOI] [PubMed] [Google Scholar]
- Mazza C, Ohno M, Segref A, Mattaj IW, Cusack S. Crystal structure of the human nuclear cap binding complex. Mol Cell. 2001;8:383–396. doi: 10.1016/s1097-2765(01)00299-4. [DOI] [PubMed] [Google Scholar]
- Mazza C, Segref A, Mattaj IW, Cusack S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21:5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKendrick L, Thompson E, Ferreira J, Morley SJ, Lewis JD. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol Cell Biol. 2001;21:3632–3641. doi: 10.1128/MCB.21.11.3632-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Hieda M, Harreman MT, Fukumoto M, Saiwaki T, Hodel AE, Corbett AH, Yoneda Y. Importin α can migrate into the nucleus in an importin β- and Ran-independent manner. EMBO J. 2002;21:5833–5842. doi: 10.1093/emboj/cdf569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenthaler M, Gray NK, Hentze MW. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol Cell. 1998;2:383–388. doi: 10.1016/s1097-2765(00)80282-8. [DOI] [PubMed] [Google Scholar]
- Mühlemann O, Eberle AB, Stalder L, Zamudio Orozco R. Recognition and elimination of nonsense mRNA. Biochim Biophys Acta. 2008;1779:538–549. doi: 10.1016/j.bbagrm.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Muralidhar MG, Johnson LF. Delayed processing/export of messenger RNA under conditions of reduced protein synthesis. J Cell Physiol. 1988;135:115–121. doi: 10.1002/jcp.1041350116. [DOI] [PubMed] [Google Scholar]
- Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Niedzwiecka A, Stepinski J, Darzynkiewicz E, Sonenberg N, Stolarski R. Positive heat capacity change upon specific binding of translation initiation factor eIF4E to mRNA 5′ cap. Biochemistry. 2002;41:12140–12148. doi: 10.1021/bi0258142. [DOI] [PubMed] [Google Scholar]
- Oh N, Kim KM, Cho H, Choe J, Kim YK. Pioneer round of translation occurs during serum starvation. Biochem Biophys Res Commun. 2007a;362:145–151. doi: 10.1016/j.bbrc.2007.07.169. [DOI] [PubMed] [Google Scholar]
- Oh N, Kim KM, Choe J, Kim YK. Pioneer round of translation mediated by nuclear cap-binding proteins CBP80/20 occurs during prolonged hypoxia. FEBS Lett. 2007b;581:5158–5164. doi: 10.1016/j.febslet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Palmeri D, Malim MH. Importin β can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin α. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Raught B, Gingras AC, Gygi SP, Imataka H, Morino S, Gradi A, Aebersold R, Sonenberg N. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 2000;19:434–444. doi: 10.1093/emboj/19.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollenhagen C, Pante N. Nuclear import of spliceosomal snRNPs. Can J Physiol Pharmacol. 2006;84:367–376. doi: 10.1139/y05-101. [DOI] [PubMed] [Google Scholar]
- Sanford JR, Gray NK, Beckmann K, Cáceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes & Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Ellis JD, Cazalla D, Cáceres JF. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc Natl Acad Sci. 2005;102:15042–15047. doi: 10.1073/pnas.0507827102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Hosoda N, Maquat LE. Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol Cell. 2008;29:255–262. doi: 10.1016/j.molcel.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Schaal TD, Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol Cell Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4) Mol Cell Biol. 2001;21:209–223. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- Shyu AB, Greenberg ME, Belasco JE. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes & Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: To translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AL, Romão L. The mammalian nonsense-mediated mRNA decay pathway: To decay or not to decay! Which players make the decision? FEBS Lett. 2009;583:499–505. doi: 10.1016/j.febslet.2008.12.058. [DOI] [PubMed] [Google Scholar]
- Sonenberg N. eIF4E, the mRNA cap-binding protein: From basic discovery to translational research. Biochem Cell Biol. 2008;86:178–183. doi: 10.1139/O08-034. [DOI] [PubMed] [Google Scholar]
- Stephenson LS, Maquat LE. Cytoplasmic mRNA for human triosephosphate isomerase is immune to nonsense-mediated decay despite forming polysomes. Biochimie. 1996;78:1043–1047. doi: 10.1016/s0300-9084(97)86728-4. [DOI] [PubMed] [Google Scholar]
- Swartz JE, Bor YC, Misawa Y, Rekosh D, Hammarskjöld ML. The shuttling SR protein 9G8 plays a role in translation of unspliced mRNA containing a constitutive transport element. J Biol Chem. 2007;282:19844–19853. doi: 10.1074/jbc.M701660200. [DOI] [PubMed] [Google Scholar]
- Tange TO, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, Ishida T, Taniguchi T, Hasegawa H, Terashima A, et al. Crystal structures of 7-methylguanosine 5′-triphosphate (m(7)GTP)- and P(1)-7-methylguanosine-P(3)-adenosine-5′,5′-triphosphate (m(7)GpppA)-bound human full-length eukaryotic initiation factor 4E: Biological importance of the C-terminal flexible region. Biochem J. 2002;362:539–544. doi: 10.1042/0264-6021:3620539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- van den Bogaart G, Meinema AC, Krasnikov V, Veenhoff LM, Poolman B. Nuclear transport factor directs localization of protein synthesis during mitosis. Nat Cell Biol. 2009;11:350–356. doi: 10.1038/ncb1844. [DOI] [PubMed] [Google Scholar]
- Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj IW. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek Z, Niedzwiecka-Kornas A, Chlebicka L, Jankowska M, Kiraga K, Stepinski J, Dadlez M, Drabent R, Darzynkiewicz E, Stolarski R. Fluorescence studies on association of human translation initiation factor eIF4E with mRNA cap-analogues. Z Naturforsch [C] 1999;54:278–284. doi: 10.1515/znc-1999-3-420. [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Wilson KF, Fortes P, Singh US, Ohno M, Mattaj IW, Cerione RA. The nuclear cap-binding complex is a novel target of growth factor receptor-coupled signal transduction. J Biol Chem. 1999;274:4166–4173. doi: 10.1074/jbc.274.7.4166. [DOI] [PubMed] [Google Scholar]
- Woeller CF, Gaspari M, Isken O, Maquat LE. NMD resulting from encephalomyocarditis virus IRES-directed translation initiation seems to be restricted to CBP80/20-bound mRNA. EMBO Rep. 2008;9:446–451. doi: 10.1038/embor.2008.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worch R, Niedzwiecka A, Stepinski J, Mazza C, Jankowska-Anyszka M, Darzynkiewicz E, Cusack S, Stolarski R. Specificity of recognition of mRNA 5′ cap by human nuclear cap-binding complex. RNA. 2005;11:1355–1363. doi: 10.1261/rna.2850705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun X, Qian Y, Maquat LE. Intron function in the nonsense-mediated decay of β-globin mRNA: Indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]