Abstract
In Xenopus embryos, a dorsal–ventral patterning gradient is generated by diffusing Chordin/bone morphogenetic protein (BMP) complexes cleaved by BMP1/Tolloid metalloproteinases in the ventral side. We developed a new BMP1/Tolloid assay using a fluorogenic Chordin peptide substrate and identified an unexpected negative feedback loop for BMP4, in which BMP4 inhibits Tolloid enzyme activity noncompetitively. BMP4 binds directly to the CUB (Complement 1r/s, Uegf [a sea urchin embryonic protein] and BMP1) domains of BMP1 and Drosophila Tolloid with high affinity. Binding to CUB domains inhibits BMP4 signaling. These findings provide a molecular explanation for a long-standing genetical puzzle in which antimorphic Drosophila tolloid mutant alleles displayed anti-BMP effects. The extensive Drosophila genetics available supports the relevance of the interaction described here at endogenous physiological levels. Many extracellular proteins contain CUB domains; the binding of CUB domains to BMP4 suggests a possible general function in binding transforming growth factor-β (TGF-β) superfamily members. Mathematical modeling indicates that feedback inhibition by BMP ligands acts on the ventral side, while on the dorsal side the main regulator of BMP1/Tolloid enzymatic activity is the binding to its substrate, Chordin.
Keywords: BMP, BMP1, Chordin, CUB domain, Tolloid
Bone tissue can be induced in muscle or connective tissue by grafts of bone fragments or their alcohol extracts (Levander 1938). Urist (1965) discovered that this activity resided in the decalcified bone matrix and coined the term bone morphogenetic protein (BMP). BMP activity was purified using ectopic bone formation assays (Sampath and Reddi 1981). Once its components were cloned, the inducer preparation was found to contain growth factors of the transforming growth factor-β (TGF-β) superfamily designated BMP2 to BMP7 (Wozney et al. 1988; Celeste et al. 1990). An unexplained observation was that BMP1, the first bone-inducing protein identified, was a zinc metalloproteinase with homology with the Drosophila dorsal–ventral (D–V) patterning gene tolloid, but not with the BMP2–7 growth factors (Wozney et al. 1988).
In early animal development, D–V patterning is regulated by a gradient of BMP activity. This gradient is finely regulated by an extracellular network of secreted proteins such as Chordin (Chd), Twisted gastrulation (Tsg), and Crossveinless-2 (CV2) that have been conserved during evolution (Little and Mullins 2006; O'Connor et al. 2006; De Robertis 2008). A key switch in this pathway is provided by Tolloids, which cleave Chordin at two specific sites, releasing active BMP ligands from Chd/Tsg/BMP complexes (Marqués et al. 1997; Piccolo et al. 1997; De Robertis and Kuroda 2004). In addition to Chordin, tolloid-like metalloproteinases also digest the C-terminal peptide of fibrillar procollagens (hence, BMP1 is also known as procollagen C peptidase or BMP1/PCP), and other extracellular matrix proteins such as latent TGF-β-binding protein (LTBP), myostatin/GDF8 prodomain, lysyl oxidase, biglycan, and osteoglycin (Greenspan 2005; Hopkins et al. 2007). In the vertebrates, three Tolloid-like enzymes exist: Tolloid-like 1 (Tll-1, called Xolloid-related in Xenopus), Tolloid-like 2 (Tll-2, called Xolloid in Xenopus), and mammalian Tolloid (Dale et al. 2002; Hopkins et al. 2007). BMP1 is a shorter, alternatively spliced form of mammalian Tolloid. The BMP1 protein consists of the protease domain followed by CUB1, CUB2, epidermal growth factor (EGF), and CUB3 domains, whereas the other vertebrate Tolloids contain five CUB (Complement 1r/s, Uegf [a sea urchin embryonic protein] and BMP1) domains and two EGF domains, as is the case with Drosophila Tolloid (Fig. 1A).
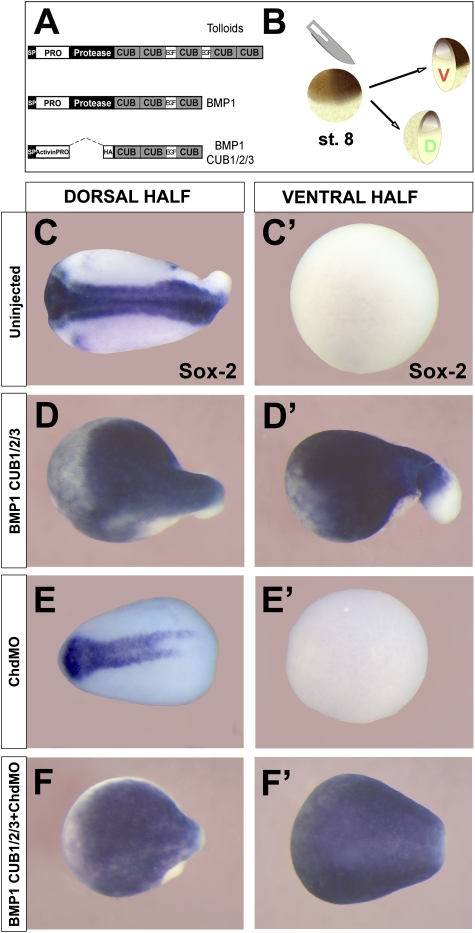
Figure 1.
BMP1 CUB domains dorsalize ventral half-embryos independently of Chordin. (A) Diagram of the generic Tolloid family metalloproteinase primary structure, BMP1, and the BMP1 CUB1/2/3 construct. (B) Xenopus laevis embryos were bisected into dorsal and ventral halves at late blastula/early gastrula stage using a surgical blade. (C,C′) Bisected wild-type embryos result in the dorsal half-embryo that self-regulates into a relatively normal embryo and a ventral belly piece lacking all neural structures marked by the pan-neural marker Sox2. (D,D′) BMP1 CUB1/2/3 mRNA (1 ng per embryo injected at the two-cell stage) dorsalizes the ventral half-embryo (n = 86, 96% positive for Sox2). (E,E′) Microinjection of morpholino oligos against Chordin (ChdMO) reduced neural structures in the dorsal half. (F,F′) The dorsalizing effect of BMP1 CUB1/2/3 mRNA in the ventral half-embryo is not affected by Chordin depletion (n = 82, 95% positive for Sox2).
CUB domains are protein modules containing four conserved cysteines, first described in the Complement 1r (C1r) and Complement 1s (C1s) proteins of the complement system. These proteins lent one initial to CUB domains, an acronym that stands for Complement 1r/s, Uegf (a sea urchin embryonic protein) and BMP1 (Bork and Beckmann 1993). CUB domains are widespread and have since been found in numerous extracellular proteins (http://www.expasy.org/prosite), but their biochemical function remains unknown.
Tolloids contain an astacin/metzincin protease domain of the type present in many enzymes that degrade extracellular matrix. Point mutations that inactivate the protease domain (Ferguson and Anderson 1992; Childs and O'Connor 1994; Finelli et al. 1994) generate antimorphic dominant-negative (DN) forms of tolloids (Piccolo et al. 1997; Wardle et al. 1999). One particularly potent construct was made by Blitz et al. (2000) by deleting the protease domain and expressing the C-terminal CUB/EGF region of BMP1 using an Activin proregion vector. Overexpression of this dominant-negative C-terminal BMP1 fragment (DN-BMP1) caused dorsalization of ventral mesoderm in Xenopus embryos, leading to the view that it acted by inhibiting endogenous BMP1 activity and stabilizing low levels of endogenous Chordin protein in the ventral region. The implication of this experiment was that Chordin might act as a short-range signal in Xenopus (Blitz et al. 2000). This presented a challenge for the D–V patterning field because Decapentaplegic/Short gastrulation (Sog/Dpp) complexes diffuse over long ranges in Drosophila (Shimmi et al. 2005; Wang and Ferguson 2005), and therefore Xenopus patterning would have had to function by a different molecular mechanism. However, recent work in Xenopus embryos has shown that BMPs diffuse from the dorsal to the ventral side of the embryo and that this long-range transport requires Chordin (Ben-Zvi et al. 2008; our unpublished observations). As shown below, we now find that ventral mesoderm can be dorsalized by the DN-BMP1 fragment in Chordin-depleted Xenopus embryos, suggesting a direct anti-BMP effect.
In Drosophila, tolloid (tld) was identified in the initial large-scale embryonic-lethal screens (Jürgens et al. 1984). An unusually rich allelic series of tld mutations exists that allowed, in a now classical study (Ferguson and Anderson 1992), the demonstration that Tolloid function is to increase the activity of the BMP homolog Dpp and to decrease the activity of the Chordin homolog Sog. Interestingly, about a third of the tld alleles were antimorphic mutations that enhanced the phenotype of weak dpp loss-of-function mutations (Ferguson and Anderson 1992; Childs and O'Connor 1994; Finelli et al. 1994).
Second site intragenic revertants of the antimorphic tld mutants were also isolated (Ferguson and Anderson 1992). Of five revertant mutations obtained, four mapped to the CUB domains and the other caused termination at the end of the protease domain (Childs and O'Connor 1994; Finelli et al. 1994). The molecular mechanism of the anti-BMP effect of tolloid antimorphic mutations has remained unexplained and was subsequently overshadowed by the discovery that the main pro-BMP activity of wild-type Tolloid is caused by the release of Dpp/BMP from inactive complexes when Sog/Chd is cleaved (Marqués et al. 1997; Piccolo et al. 1997). Similarly, the reason why a proteolytic enzyme such as BMP1 would copurify with BMP2–7 in bone-inducing fractions (Wozney et al. 1988) has remained a mystery to this day.
In the present study, we developed a quantitative assay for BMP1 activity using a novel fluorogenic chordin peptide substrate and used it to identify an unexpected inhibitory effect of BMP4 on BMP1 enzymatic activity. It was found that BMP4 is a noncompetitive enzyme inhibitor of BMP1 activity. Further analysis showed that BMP4 binds to isolated CUB domains of BMP1 with high affinity. In addition, the CUB domains of Drosophila Tolloid also bind BMP4 and BMP7 with dissociation constants of ∼20 nM. These findings open the door to the possibility that CUB domains may function in proteins as binding modules for members of the TGF-β superfamily of growth factors. A reaction-diffusion mathematical model (Turing 1952; Meinhardt and Gierer 2000) of Xenopus gastrulation indicated that the introduction of an enzymatic feedback step involving inhibition of BMP1/Tolloid by the BMP products released by its proteolytic reaction on Chordin may provide an extra layer of regulation in the ventral side of the embryo.
Results
The C-terminal BMP1 region dorsalizes independently of Chordin
We reinvestigated the effects of overexpressing DN-BMP1 consisting of the C terminus of BMP1 cloned in an Activin A expression vector (Fig. 1A; Blitz et al. 2000). We used bisection of Xenopus embryos at late blastula (Fig. 1B) as a sensitized method to gauge BMP signaling in the ventral half-embryos (Reversade and De Robertis 2005). We confirmed that DN-BMP1 (CUB1/2/3 plus EGF) strongly neuralized ventral half-embryos (Fig. 1, cf. C′ and D′). To test whether this anti-BMP phenotype was caused by stabilizing Chordin in the ventral half-embryo, we used antisense morpholinos (ChdMO) to deplete Chordin (Fig. 1E,E′). At the doses used, the morpholinos cause an almost complete depletion of Chordin protein in Xenopus embryos (Oelgeschläger et al. 2003). The DN-BMP1 C-terminal region was able to induce the neural marker Sox2 in ventral half-embryos even when Chordin was depleted (Fig. 1F′). This showed that the effects of DN-BMP1 are independent of Chordin. The dorsalizing effect was not caused by the stabilization of Chordin (via inhibition of endogenous Tolloids) and had to be due to some other molecular mechanism. This prompted us to re-examine the enzymology of BMP1/Tolloids.
A new fluorogenic assay for Tolloid Chordinase activity
Tolloid-like metalloproteinases are highly regulated during embryonic D–V patterning, and their activity is a rate-limiting step in the BMP signaling pathway. For example, Sizzled/Ogon is a secreted Frizzled-related protein (sFRP) that functions as a key competitive inhibitor of tolloid chordinase activity during D–V development (Lee et al. 2006; Muraoka et al. 2006). To better understand the kinetics and regulation of BMP/Tolloid activity, we developed a new enzymatic assay. Tolloids cleave Chordin at two discrete sites: one between the Cysteine-rich 1 (CR1) and CR2 domains, and the other between CR3 and CR4 (Marqués et al. 1997; Piccolo et al. 1997; Hopkins et al. 2007). We focused on the latter site because this is usually the first cleavage to take place and it is cut with high efficiency (Piccolo et al. 1997).
An octapeptide (Mca-SMQSDGAK-Dnp; hereafter referred to as Chordin peptide) was designed in accordance with the Tolloid target sequence on Xenopus Chordin protein (Fig. 2A). A fluorophore (7-methoxycoumarin-4-yl, Mca) and its quencher (2,4-dinitrophenyl, Dnp, conjugated to an additional Lys residue) flanked the Chordin sequence such that the cleavage of the Chordin peptide between the Ser and Asp residues resulted in emission of fluorescence from the Mca moiety at 405 nm. Chordin peptide was efficiently cleaved by BMP1 metalloproteinase (Fig. 2B). We previously used another fluorogenic substrate to measure Tolloid activity (Lee et al. 2006). However, this substrate was originally designed to test Caspase activity; a dedicated Chordin peptide substrate should report chordinase activity more faithfully. Of the Tolloid enzymes, we chose to use the commercially available recombinant human BMP1 (R&D Systems), because BMP1 is known to substitute for the effects of other Tolloids during embryonic development (Wardle et al. 1999; Jasuja et al. 2006).
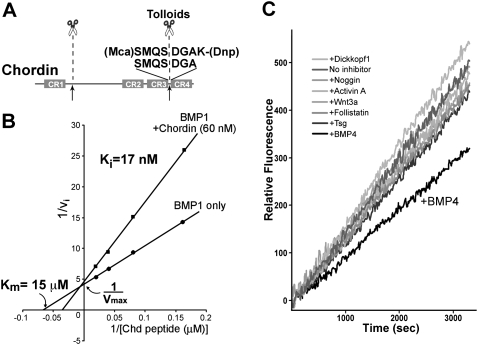
Figure 2.
A fluorometric assay for the Chordinase activity of Tolloids shows that BMP4 is a specific inhibitor of the reaction. (A) A new fluorogenic peptide substrate for Tolloids containing the sequence of the Xenopus Chordin cleavage site. (B) The Chd peptide substrate is efficiently digested by BMP1 enzyme, a reaction that is competitively inhibited by 60 nM full-length Chordin proteins. The Ki (17 nM) is similar to the Km of tolloids for full-length chordin (Lee et al. 2006). (C) BMP4, but not other regulators of D–V patterning, inhibits BMP1 metalloproteinase activity. All proteins were added at 120 nM final concentration.
A double-reciprocal Lineweaver-Burk plot showed that the Michaelis constant (Km) of BMP1 for Chordin peptide was 15 μM and, more importantly, that the addition of the natural substrate Chordin inhibited the peptide digestion with an inhibition constant (Ki) of 17 nM, very similar to the Km determined in a previous study on Tolloid cleavage using full-length Chordin protein as substrate (25 nM) (Lee et al. 2006). Moreover, the presence of Chordin shifted the apparent Km but did not affect the maximal velocity (Vmax), showing that the full-length Chordin protein acted as a competitive inhibitor of the reaction (Fig. 2B), competing with the Chordin peptide for the same active site on BMP1. We conclude that the new fluorogenic assay provides a quantitative readout for investigating the regulation of Tolloid/BMP1 activity.
BMP4 is a noncompetitive inhibitor of BMP1/Tolloids
We examined a panel of known extracellular proteins involved in D–V patterning for their ability to affect Tolloid/BMP1 activity. Surprisingly, we found that the BMP4 ligand specifically inhibited BMP1 enzyme (Fig. 2C). This was unexpected, since the main phenotypic effect of Tolloids is to enhance BMP signaling through cleavage of the BMP antagonist Chordin. Other proteins tested, such as Dickkopf1, Noggin, Activin A, Wnt3A, Follistatin, and Twisted gastrulation (Tsg), all applied at 120 nM, were without significant effects (Fig. 2C).
We next investigated the kinetics of inhibition of BMP1 by BMP4. Understanding the kinetic parameters of enzymes is important because they describe invariant physiochemical properties of the chemical reactions involved (Dixon and Webb 1979). Adding increasing amounts of recombinant BMP4 protein (R&D Systems) inhibited BMP1 activity in a concentration-dependent manner (Fig. 3A). BMP4 also inhibited the activity of the longer members of the tolloid-like metalloproteinase family (Fig. 1A), Xolloid-related and Xolloid, showing that the other tolloid enzymes were also subject to BMP4 regulation (data not shown). The linear quantitative fluorescent assay provided an accurate initial velocity measurement of BMP1 enzyme activity (Fig. 3A). These measurements were performed at different concentrations of substrate to characterize the inhibition of BMP1 enzyme by BMP4 (Fig. 3B). Lineweaver-Burk plots showing the inverse of initial velocity values plotted against the inverse of substrate concentration in the absence or presence of BMP4 showed that the inhibitor did not significantly alter the apparent affinity of BMP1 for the substrate (Km), but affected the maximal enzyme activity (Vmax) (Fig. 3C). This type of kinetics shows that BMP4 is a noncompetitive inhibitor of BMP1.
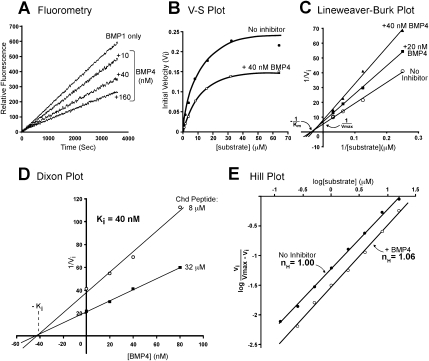
Figure 3.
BMP4 is a noncompetitive inhibitor of the BMP1 enzyme. (A) BMP4 inhibits BMP1 activity in a dose-dependent manner. (B) BMP4 inhibition cannot be competed by increasing amounts of substrate, as shown by the difference in maximal velocity with or without BMP4. This result is consistent with that expected of a noncompetitive enzyme inhibitor. (C) BMP4 affects the maximal velocity (Vmax), but not the Michaelis constant (Km) of the BMP1 enzyme. (D) Effects of different concentrations of inhibitor at two different substrate concentrations. The Dixon plot shows that BMP4 inhibited BMP1 with an inhibition constant (Ki) of 40 nM. (E) BMP4 does not exhibit cooperativity when inhibiting BMP1, indicating one-to-one binding is required for inhibition of catalytic activity.
The inhibition constant (Ki) of a noncompetitive inhibitor can be determined by varying the concentration of inhibitor at two fixed concentrations of substrate using the Dixon plot method (Dixon and Webb 1979). Because the two lines intersected at the abscissa, this confirmed the purely noncompetitive nature of the inhibition, and allowed us to determine a Ki of 40 nM (Fig. 3D). This is within physiological range (Hojima et al. 1985; Lee et al. 2006). We next asked whether the inhibition of the BMP1 enzyme by BMP4 displayed cooperativity. This was determined using a Hill plot (Fig. 3E). The slope, or Hill coefficient (nH), of this graph in the presence of inhibitor BMP4 was nH ≈ 1, suggesting that the BMP4-mediated inhibition is noncooperative.
We conclude from these results that BMP4 is a noncompetitive inhibitor of BMP1. In the early Xenopus embryo, this reaction would inhibit BMP1/tolloid enzymes when BMP4 is released by cleavage of Chordin, forming a classical metabolic negative feedback loop.
BMP1 CUB domains bind BMP4 with high affinity
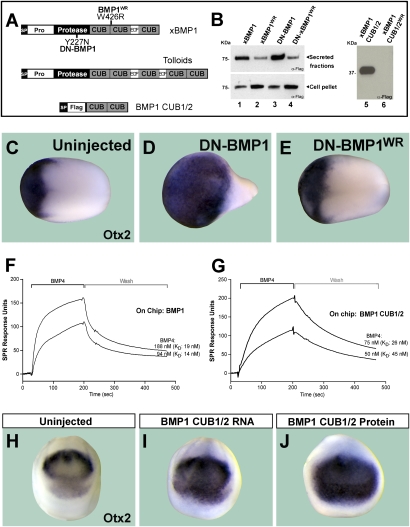
The noncompetitive kinetics of BMP4–BMP1 interaction suggested the existence of a direct binding between these two proteins. To investigate this, we generated Xenopus BMP1 constructs (Fig. 4A), expressed them in 293T cells, and affinity-purified the proteins. Since genetic analyses in Drosophila had identified four mutations in CUB domains that reverted the anti-Dpp effects of tolloid antimorphs (Childs and O'Connor 1994), we also tested whether the introduction of such mutations in its Xenopus homolog, BMP1 could prevent its interaction with BMP4. The first two CUB domains of Xenopus BMP1 and Drosophila tolloid are highly conserved, with 47% identity and 67% similarity (Supplemental Fig. 1). However, only one mutation in Drosophila tolloid (dTld W497R) corresponded to a residue conserved in Xenopus BMP1 (at position 426 in CUB2 domain of xBMP1). We therefore mutated this residue from Trp to Arg (W426R) in a wild-type xBMP1 (xBMP1WR), in a dominant-negative construct, DN-BMP1WR (generated by introducing a Tyr-to-Asn point mutation at position 227 in the protease domain) (Piccolo et al. 1997) and in a secreted construct comprising only the first and second CUB domains of BMP1 (BMP1 CUB1/2WR) (Fig. 4A). Anti-Flag Western blots of conditioned medium of 293T cells transfected with these constructs showed that this CUB2 point mutation reduced or prevented the secretion of xBMP1, DN-BMP1, and BMP1 CUB1/2 into the culture medium of transfected 293T cells (Fig. 4B, lanes 2,4,6). In Xenopus embryos, microinjection of DN-BMP1 mRNA caused dorsalization of the embryo (Fig. 4C,D, note expansion of the forebrain and mid-brain marker Otx2). Injection of DN-BMP1WR mRNA was without effect (Fig. 4E), presumably because it is poorly secreted.
Figure 4.
BMP1 CUB domains bind BMP4 growth factor. (A) Diagram of BMP1 and BMP1 CUB1/2 constructs and their mutations. (B) Mimicking an antimorph revertant mutant in BMP1 (DN-BMP1WR) or BMP1 CUB1/2 impairs protein secretion. (C–E) Overexpression of DN-BMP1 dorsalized embryos (note the enlarged head marked by Otx2), but the second site revertant mutation (DN-BMP1WR) had no effect. (F) BMP4 binds to full-length BMP1 on a BIAcore sensor chip. (G) Purified BMP1 CUB1 and CUB2 domains are sufficient for BMP4 binding. (H,I) Embryos injected with BMP1 CUB1/2 RNA were dorsalized, as indicated by the expanded Otx-2 expression domain (n = 83, 81% dorsalized). (J) Dorsalization (anti-BMP) effect was also observed after injection of BMP1 CUB1/2 purified protein into the blastula cavity (n = 85, 82% dorsalized).
To determine whether BMP4 and BMP1 bind to each other, we used commercial human BMP1 (R&D Systems) and affinity-purified wild-type xBMP1 CUB1/2 in surface plasmon resonance analyses (BIAcore). BMP1 was conjugated onto the sensor chip and changes in the refractive index caused by BMP4 supplied at a constant flow were measured (Fig. 4F). BMP4 was allowed to bind to BMP1 enzyme and the complexes were then washed with the same buffer without BMP4. The association and dissociation rates determined for different concentrations of BMP4 were then used to calculate an average equilibrium dissociation constant (KD) of ∼17 nM (Fig. 4F).
Having shown that recombinant human BMP1 bound directly to BMP4 with high affinity, we next mapped its binding site. The noncompetitive nature of the inhibition suggested that BMP4 would not bind directly to the active site in the enzyme. Since the sequence of BMP1 contains three CUB domains for which no function is known (Fig. 4A), we tested whether BMP1 fragments consisting only of CUB domains could bind BMP4. In surface plasmon resonance assays with BMP1 CUB1 and CUB2 domains (Fig. 4A, xBMP1 CUB1/2 lacking the EGF domain) conjugated onto the surface of the BIAcore chip, we observed high-affinity binding to BMP4 (average KD = 30 nM) (Fig. 4G). This interaction between CUB domains and BMP4 is within the physiological range and is comparable with that of the full-length BMP1 enzyme. In addition, CUB1/2 of xBMP1 bound to BMP7 immobilized on a chip (data not shown). Binding assays using proteins containing single CUB domains—BMP1 CUB1 or BMP1 CUB2—did not show strong affinity to BMP4 (data not shown), suggesting that multiple CUB domains may be required to generate a significant binding interface with BMP4.
To test whether BMP1 CUB1/2 had anti-BMP activity, we injected two-cell-stage Xenopus embryos with BMP1 CUB1/2 mRNA. We found that CUB1/2 lacking the EGF domain does indeed have dorsalizing (anti-BMP) activity in the Xenopus embryo (Fig. 4H,I). Microinjection of mRNA constructs expressing only CUB1 or CUB2 had some dorsalizing activity, but less than that of CUB1/2 mRNA (data not shown). Since BMP1 CUB1/2/3 (Fig. 1) is more dorsalizing than CUB1/2 or individual CUB mRNAs, the anti-BMP activity appears to be proportional to the number of CUB modules in each molecule. Anti-BMP phenotypes were also observed after microinjection of affinity-purified BMP1 CUB1/2 protein into the blastula cavity of Xenopus embryos (Fig. 4J). We conclude from these results that the enigmatic CUB domains of the BMP1 enzyme function as BMP4-binding modules.
Drosophila Tolloid (dTld) CUB domains bind BMP ligands
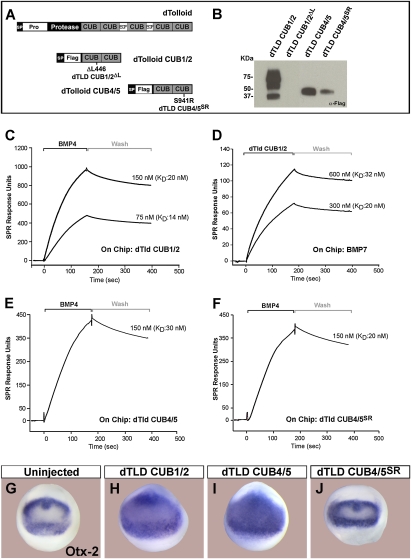
To test whether dTld also binds BMP growth factors, we generated CUB domain constructs of the dTld enzyme in 293T cells and affinity-purified them (Fig. 5A,B). In addition, we introduced in dTld CUB domain mutations for the remaining antimorph revertants sequenced by Childs and O'Connor (1994) for which we had not been able to identify homologous amino acids in Xenopus BMP1. The first mutation was a deletion of Leu 446 (ΔL446) in the CUB1 domain (dTld CUB1/2ΔL) (Fig. 5A). The other two mutations affected the same amino acid in the dTld CUB5 domain (Childs and O'Connor 1994). We therefore generated a dTld construct containing dTld CUB4/5 domains with a point mutation in CUB5 that changed Ser 941 into Arg (dTld CUB4/5SR). We found that deletion of Leu 446 blocked dTld CUB1/2 secretion in 293T cells (Fig. 5B). Similarly, introduction of the S914R mutation in CUB5 reduced secretion of CUB4/5 (Fig. 5B). However, this reduction was partial and we were able to purify small amounts of mutant dTldSR from the culture medium of 293T cells.
Figure 5.
dTld binds BMP4 and BMP7. (A) Diagram of dTld and the dTld CUB1/2 and CUB4/5 constructs. (B) dTld CUB1/2ΔL mutant is not secreted, and CUB4/5SR mutant has impaired secretion in transfected 293T cells. Multiple bands are presumably due to glycosylation. (C) dTld CUB1/2 binds BMP4 within the physiological range (KD ≈ 17 nM). (D) When BMP7 is placed on the sensor chip, dTld CUB1/2 protein is able to bind to it (KD ≈ 26 nM). (E,F) Both dTld CUB4/5 and CUB4/5SR bind BMP4 with similar KD. (G–J) dTld CUB1/2 (n = 81, 78% dorsalized) and CUB4/5 (n = 79, 77% dorsalized) have anti-BMP (dorsalizing) activity in a Xenopus assay, while dTld CUB4/5SR did not show any anti-BMP activity (n = 52).
Surface plasmon resonance analyses showed that, like their vertebrate counterparts, dTld CUB1/2 domains bound BMP4 in the physiological range, with a KD of 14–20 nM (Fig. 5C). The dTLD CUB1/2 protein also bound BMP7, with a KD between 20 and 32 nM (Fig. 5D). In a similar experiment with dTld CUB4/5 or CUB4/5SR cross-linked to the sensor chip, both proteins bound to BMP4 with dissociation constants in the 20–30 nM range (Fig. 5E,F). Thus, the dTld CUB S941R mutation affects the secretion but not the biochemical binding affinity of CUB4/5 to BMP4.
In microinjected Xenopus laevis embryos, dTld CUB1/2 or CUB4/5 mRNAs caused anti-BMP phenotypes, with expanded expression domain of Otx2 (Fig. 5G–I). However, dTld CUB4/5SR mRNA did not dorsalize the embryo (Fig. 5J), likely because of its poor secretion, since the mutation did not affect its ability to bind BMP4 (Fig. 5F).
We conclude that dTld can bind BMPs with high affinity and inhibit their activity in Xenopus assays. This molecular interaction may explain the anti-Dpp effects observed in Drosophila antimorphic tolloid mutants (Ferguson and Anderson 1992; Childs and O'Connor 1994). These experiments also suggest a molecular mechanism for the second site mutations in CUB domains that reverted the ability of Drosophila antimorphic tolloid mutations to enhance weak Dpp alleles (Ferguson and Anderson 1992; Childs and O'Connor 1994): They interfere with the secretion of dominant-negative mutant Tolloid protein, behaving essentially as null mutations.
Integrating negative feedback and embryonic patterning
A remarkable feature of vertebrate D–V patterning is that a key regulatory step is mediated by the activity of metalloproteinases that regulate the availability of Chordin and cause the release of active BMPs in the ventral side (Lee et al. 2006; Plouhinec and De Robertis 2009). The finding that BMP4 is a feedback inhibitor of the activity of BMP1/Tolloids raised the question of what effect such an interaction would have on embryonic patterning. To examine this, we constructed a mathematical model of D–V patterning based on reaction-diffusion equations (Turing 1952; Meinhardt and Gierer 2000; Plouhinec and De Robertis 2009).
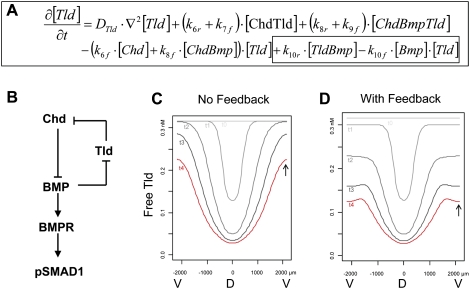
For simplicity, the model only incorporated the binding reactions among Tlds, Chd, BMPs, and BMP receptors (BMPRs), as well as the cleavage of Chd by Tld (Fig. 6B). As shown in Supplemental Figure 2, this requires a set of 14 different biochemical reactions. To correctly model the noncompetitive nature of the inhibition of Tlds by BMPs, it was necessary to include the formation of a transient enzyme–substrate complex of Tld and Chd that can be bound by the BMP4 inhibitor. This biochemical model was translated into a system of 14 interlinked partial differential equations, of which the one describing the evolution of free (active) Tolloid is shown in Figure 6A. The values of several parameters in those equations were chosen based on physicochemical measurements made in this study or previous work. For example, the affinity of BMP1/Tolloid for its substrate Chd was fixed at 17 nM (Fig. 2B), and the affinity of Tld for BMP4 was fixed at 14 nM (Fig. 4F). When no data were available, the unknown parameters were assigned estimated values spanning the physiologically relevant range (see the Materials and Methods; Supplemental Fig. 2).
Figure 6.
Mathematical modeling of the effects of the BMP negative feedback loop on BMP1/Tolloid (Tld) activity. (A) Partial differential equation describing the temporal evolution of free Tld concentration. The terms accounting for the inhibition of Tld by BMP (reaction 10 in Supplemental Fig. 1) are boxed. The reactions involving Tld and their corresponding parameters are described in Supplemental Figure 2. (B) Schematic diagram of the negative feedback loop between Tld, BMP, and Chd, which regulates the D–V gradient of pSmad1 activity. (C,D) Effect of BMP negative feedback on the temporal evolution of free Tld concentration profile. D and V indicate the position of the dorsal and ventral sides. Each concentration profile is labeled from t0 (0 h) to t4 (2 h) to indicate their evolution over time. The arrows indicate the final level of free Tld on the ventral side without (C) or with (D) BMP4 feedback. Note that the model predicts that free (active) Tld is low in the dorsal midline due to the large amounts of Chd substrate secreted by the dorsal side (Supplemental Fig. 3).
To determine the effect of the BMP4 inhibition on patterning, we examined two submodels: one with and one without BMP4–Tld interaction (Fig. 6B). We varied the unknown parameters and sorted the resulting simulations according to their ability to generate (1) a D–V pSmad gradient over time and (2) a different profile of Tolloid activity with or without inhibition by BMP4. Of the 48 different parameter sets tested, two-thirds generated a proper D–V gradient of BMP activity and, among those, one-half displayed a significant effect of BMP4 inhibition on free Tld (active enzyme) concentration profile and were analyzed further.
The first observation, which was unexpected, was that, independently of feedback, free Tld levels (indicating active enzyme) decreased over time on the dorsal side in parallel with the increase in Chordin levels (see Fig. 6C, curves t0–t4; Supplemental Fig. 3). Free Chd substrate present in the dorsal side saturates Tld enzyme by binding to it, decreasing its activity toward other substrates, such as the Chordin–BMP complex. The dorsal organizer produces massive amounts of Chordin in Xenopus (1.5 ng per embryo), reaching levels of at least 33 nM in the extracellular space if Chd were uniformly distributed in the extracellular space, undoubtedly reaching much higher levels on the dorsal side (Lee et al. 2006). Because the measured Km of tolloids for Chordin cleavage is 17–25 nM, this results in tolloids becoming fully engaged in Tld/Chd enzyme–substrate complexes on the dorsal side, so that the amount of active (free) Tld is greatly decreased in dorsal regions (Fig. 6C). Thus, Chd is the main inhibitor of Tld activity on the dorsal side.
The second observation, which was predictable from the biochemistry, was that when Tld activity with or without BMP feedback was compared, the inhibitory effect of BMP4 on Tld activity takes place mostly in the ventral side of the embryo. The effect of BMP4 feedback inhibition is to attenuate the levels of active (free) Tld ventrally (Fig. 6, cf. the position of the arrows in C and D).
From these mathematical modeling analyses, we conclude that the effect of BMP on the inhibition of Tld will be maximal on the ventral side of the Xenopus gastrula or in other regions of high BMP activity in the organism. Moreover, our modeling uncovered an unexpected role for Chordin as the major regulator of Tld activity on the dorsal side of the embryo. To our knowledge, this is the first time classical enzyme kinetics and reaction-diffusion mathematical modeling have been brought together in the analysis of cell–cell signaling.
Discussion
A new regulatory node in the Chordin/BMP pathway
The differentiation of cell types along the vertebrate D–V axis is regulated by an extracellular network of BMPs and their regulators, such as Chordin, BMP1/Tolloid, Tsg, and Crossveinless-2, in animals as diverse as Xenopus, Drosophila, zebrafish, amphioxus, hemichordates, and spiders (for review, see De Robertis 2008). In addition, in the vertebrates, additional extracellular BMP antagonists such as Noggin and Follistatin cooperate with the anti-BMP activity of Chordin (Khokha et al. 2005). The complexity of this biochemical pathway, shown in Figure 7, raises the question of why so many components and regulatory interactions are required to establish a simple gradient of BMP signaling through the transcription factors Smad1/5/8. One reason is that a stable gradient must be robustly maintained through many hours of development (from blastula until the end of gastrulation) at a time during which the three embryonic germ layers are undergoing massive morphogenetic movements (Tucker et al. 2008). In addition, the frog embryo must have the ability to adapt to changes in temperature in its environment.
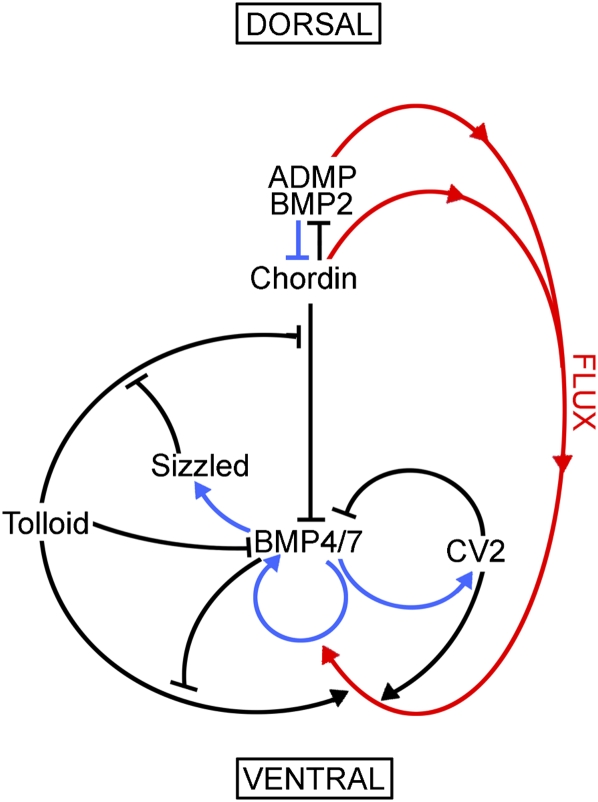
Figure 7.
Enzymatic regulation of Xenopus D–V patterning. This diagram depicts the extracellular network of biochemical protein–protein interactions that establishes the embryonic D–V axis through protein–protein interactions (black arrows), BMP-dependent transcription (blue arrows), and the flux of Chordin/BMP complexes from more dorsal regions to the ventral side, where it is bound by CV2 (red arrows). Tolloid and CV2 act as sinks for Chordin in the ventral side, where BMPs made in more dorsal regions can be released by Tolloid to reach peak signaling levels. The system is self-regulating because transcription of dorsal genes is repressed by BMP signals, while ventral genes are under the opposite transcriptional regulation (Reversade and De Robertis 2005; Lee et al. 2006; Ambrosio et al. 2008; Ben-Zvi et al. 2008). The two new reactions reported in this study are the inhibitory black arrows from BMP4/7 to Tolloid in the ventral side (enzyme activity inhibition) and from Tolloid to BMP4/7 (inhibition through binding and sequestration of the growth factor). Other important regulators of D–V patterning, such as Noggin and Follistatin (Khokha et al. 2005), are not included in this simplified model.
The patterning system must be resilient, given the self-regulating nature of development. When Xenopus embryos are cut in half, they will attempt to regenerate an embryo as perfect as possible, producing in some cases identical twins. This implies that cells in the dorsal and ventral poles of the early embryo communicate with each other, forming a self-regulating embryonic field. At a molecular level, these cell–cell communications can be explained by a pathway in which dorsal BMPs (ADMP and BMP2) and their antagonist, Chordin, are repressed at the transcriptional level by BMP signaling, while on the ventral side, BMP4/7 and CV2 are activated by the same signal, providing a self-regulating system (Reversade and De Robertis 2005; Ambrosio et al. 2008; Inomata et al. 2008). The key controlling element in this D–V conversation is provided by BMP1/Tolloid enzymes that degrade Chordin/BMP complexes releasing active BMP that are regulated by the Sizzled/Ogon-secreted competitive inhibitor (Lee et al. 2006; Muraoka et al. 2006). In this study, we introduced a novel regulatory node in the D–V patterning pathway, in which BMP4 serves as a feedback inhibitor of the BMP1 and Tolloid-related enzymes (Fig. 7).
A synthetic Chordin octapeptide spanning the C-terminal cleavage site that fluoresces when cleaved by Tolloids provided a quantitative enzymatic assay. This new assay was essential to the work, because both Chordin and Sog become better substrates for Tolloids when bound to BMP (Marqués et al. 1997; data not shown). It is therefore not possible to conduct a biochemical study on the digestion of full-length Chordin/Sog plus or minus BMP, because BMP affects both the substrate and the enzyme. The conformational change in the Chordin/Sog substrate would have precluded the discovery of the inhibition of enzyme activity by BMP4.
Inhibition of BMP1/tolloids by BMP4 was specific, because it was not observed with other proteins such as Activin A, Tsg, Follistatin, and Noggin. The kinetics followed those of a Michaelis-Menten noncompetitive inhibition. This meant that BMP4 affected the activity of the enzyme by binding to a site distinct from the catalytic center. We found that BMP4 binds directly to CUB domains with high affinity (Figs. 4, 5). The Ki or inhibition constant (concentration at which half of the enzyme is bound to the inhibitor) for BMP1 was in the 40 nM range, and in the 14–20 nM range when measured by direct binding. This is within physiological levels, since the Km (Michaelis constant or affinity of the enzyme for its substrate) of BMP/Tolloids for Chordin substrate was between 17 and 25 nM (Lee et al. 2006; this study), and of 96 nM for its BMP1/PCP activity (Hojima et al. 1985).
The ventral center of the Xenopus gastrula expresses a chordin-like protein called CV-2 that strongly binds Chordin/BMP complexes transported from more dorsal regions of the embryo (Ambrosio et al. 2008) and facilitates BMP signaling through its cognate receptors (Serpe et al. 2008) after cleavage of Chordin by BMP1/tolloids (Fig. 7). This suggests that in vivo free BMP is locally concentrated at sites of high CV2 and chordinase activity; it is in these regions that the negative feedback loop should be most effective. Not only will the BMP levels be highest, but also the Chordin levels will be lowest. The affinities of the interaction between BMP4/Tsg/Chordin and Tolloid may also be enhanced by the recently described Olfactomedin-related adaptor protein Ont-1, which brings together Chordin and tolloids (Inomata et al. 2008), and was not analyzed here.
The importance of the interaction between Tolloid and BMPs for developmental patterning in vivo is suggested by Drosophila genetics. A very large allelic series of tolloid mutants has been obtained that display a graded series of patterning defects along the D–V axis in Drosophila (Ferguson and Anderson 1992). This suggests that Tolloid provides a rate-limiting step during patterning. Therefore, any decrease in its activity caused by binding of BMPs would be expected to regulate the signaling gradient. The antimorphic tolloid mutations, which are proteolytically inactive but display anti-BMP effects, demonstrate that endogenous Tolloid enzyme is expressed at high enough levels to function antagonistically toward Dpp in vivo. Thus, at least in Drosophila, the interactions between Tolloid and BMPs discovered here function at physiological concentrations of D–V pathway components.
CUB domains of BMP1 and Drosophila Tolloid bind BMP4
There previously had been isolated reports showing that DN-BMP1/tolloids dorsalized Xenopus ventral mesoderm, which should lack Chordin (Wardle et al. 1999; Blitz et al. 2000). One possible interpretation for these results was the presence of a Chordin counterpart, such as CV2, expressed in the high-BMP regions of the embryo. However, we later found that CV2 is resistant to degradation by tolloids/BMP1 (Ambrosio et al. 2008). Instead, we now find that the anti-BMP effect of DN-tolloids, which can take place in Chordin-depleted embryos (Fig. 1), are due to the sequestration of BMP ligands through direct binding to CUB domains (Figs. 4, 5).
We initially hoped that the second site mutations described in Drosophila Tolloid CUB domains would point us to amino acid residues critical for Tolloid binding of BMP4. Instead, all second site mutations affected Tolloid/BMP1 protein secretion (Figs. 4, 5). These second site antimorphic revertants behave essentially as null mutations of tolloid because they are not secreted. It is likely that the original antimorphic mutants displayed anti-Dpp effects because they bound BMPs in the Drosophila embryo.
CUB domains are also required for enzymatic activity. In the case of BMP1/PCP, it has been shown that the procollagen substrate is not efficiently recognized when CUB2 of BMP1 is deleted (Hartigan et al. 2003). However, the protease domain plus CUB1 is sufficient for BMP1 chordinase activity (Petropoulou et al. 2005). In the case of Drosophila Tolloid, CUB4 and CUB5 are required to cleave Sog (Canty et al. 2006), and for Xolloid, CUB1 and CUB2 are required for recognition and cleavage of Chordin (Geach and Dale 2008). Thus, CUB domains in Tolloid/BMP1 have specific functions in substrate recognition. CUB domains are also required for secretion (Figs. 4B, 5B), in addition to serving as inhibitory BMP-binding sites. As an interaction between the BMP1 prodomain and BMP4 has also been reported (Jasuja et al. 2007), it should be noted that the prodomain was lacking in all the CUB domain constructs used in the present study.
CUB domains are present in many secreted or transmembrane proteins (Bork and Beckmann 1993), but their biochemical function remains unknown. The human genome contains 56 different loci encoding CUB domain-containing proteins (http://www.ensembl.org). Our finding that the CUB domains of BMP1 and Tolloid bind BMP4 suggests the exciting possibility that CUB domains may serve as binding modules for TGF-β superfamily ligands in other extracellular proteins as well. In the future it will be interesting to investigate, for example, the binding properties of the CUB domains found in Complement components C1r and C1s, which function in the opsonization of antigens. Another interesting protein is CUB domain-binding protein 1 (CDCP-1), a transmembrane receptor with three CUB domains that activates the Src tyrosine protein kinase and promotes metastases in human cancers (Siva et al. 2008); TGF-β also promotes metastases. Other CUB domain-containing proteins include membrane frizzled-related protein (MFRP), in which mutations in CUB domains cause nanophthalmos (Sundin et al. 2005); procollagen C-peptidase enhancer (PCPE), known as a potent enhancer of BMP1/PCP activity in procollagen processing (Takahara et al. 1994); the WNT transmembrane coreceptor Kremen (Mao et al. 2002); and many other extracellular or transmembrane proteins.
Integrating BMP4 inhibition into a reaction-diffusion model
We integrated the effects of enzymatic inhibition—in this case, noncompetitive inhibition by BMP4—into a reaction-diffusion model (Turing 1952; Meinhardt and Gierer 2000) to understand its effect on the BMP morphogen gradient of the early Xenopus embryo. This mathematical modeling predicted that Tld activity will be inhibited in ventral regions in which BMPs are present in high concentrations. An unexpected finding was that Chordin itself is a major regulator of BMP/Tolloid activity. At high concentrations, such as in the dorsal side of the frog gastrula and likely in the fly ventral blastoderm, Chd/Sog complexed with Tld is predicted to decrease the availability of free (active) BMP/Tolloid. This will inhibit degradation of Chordin–BMP complexes, preventing local BMP release and signaling, enabling the complex to diffuse further.
Conclusions and prospects
Our observations suggest that the Tolloid inhibition by BMP also takes place in fruit flies, which provide a system much more amenable to the visualization of gradients, and for which sophisticated mathematical modeling already exists (Eldar et al. 2002; Mizutani et al. 2005; Wang and Ferguson 2005; O'Connor et al. 2006; Umulis et al. 2006). In the future, it will be interesting to investigate whether CUB domains generally serve as BMP or TGF-β superfamily-binding modules. This approach has been productive in the case of the CR/vWFc domains of Chordin, which function as BMP-binding modules in many proteins (Larraín et al. 2000; De Robertis and Kuroda 2004; Zhang et al. 2007).
The present study suggests that the antimorphic revertant mutations discovered by Ferguson and Anderson (1992), and sequenced by Childs and O'Connor (1994) and Finelli et al. (1994), were based on direct Dpp–Tolloid associations and were indicators of a crucial step in the formation or maintenance of the self-adjusting D–V morphogen gradient. The findings in Drosophila and Xenopus also suggest that this extracellular negative feedback regulation was already present in the patterning system of Urbilateria, the last common ancestor of all bilateral animals that lived more than 535 million years ago (De Robertis 2008). Finally, the direct binding of BMP4 to BMP1 explains why highly purified bone-inducing protein preparations (Sampath and Reddi 1981; Wozney et al. 1988; Celeste et al. 1990) contained BMP1/Tolloid in addition to BMP2–7. It may be worthwhile to explore the value of BMP1 or its CUB domains as a delivery system for BMPs in therapeutic interventions, such as the repair of bone fractures.
Materials and methods
BMP1/dTolloid enzymatic assays
The fluorogenic substrate Mca-SMQSDGAK-Dnp was synthesized (Bachem) based on the sequence of the Tolloid cleavage site on Chordin. BMP1 CUB1/2 and Drosophila Tolloid CUB1/2 constructs were cloned into pCS2 by PCR and encoded amino acids 312–536 of BMP1 and 330–584 of Drosophila Tolloid with a signal peptide and Flag tag added to the N terminus (Piccolo et al. 1997). BMP1 or dTld CUB domain proteins were produced in human embryonic kidney 293T cells and affinity-purified using anti-Flag beads (Sigma). All other proteins used in this study, including recombinant human BMP1 and recombinant human BMP4, were purchased from R&D Systems unless specified otherwise. In vitro enzymatic digestion assays were performed usually with 25 μM fluorogenic Chordin peptide substrate in Xld Buffer (Piccolo et al. 1997) supplemented with 0.01% Brij 35. The enzymatic activity was measured on a fluorescent plate reader (excitation = 320 nm, emission = 405 nm). Initial velocities were determined from the rate of fluorescence increase over the first 60 min.
Enzyme kinetics
Double-reciprocal Lineweaver-Burk graphs were obtained by plotting the inverse of the Chd substrate peptide concentration against the inverse of the initial velocity (Vi) obtained from the fluorometric enzyme assays. Michaelis constant (Km), maximal velocity (Vmax), and inhibition constant (Ki) values were calculated as described (Dixon and Webb 1979). Dixon plots were generated by varying the inhibitor concentration and plotting it against the inverse of the initial velocity. Cooperative effects were examined with the Hill plot using Vmax and Vi values obtained from the Lineweaver-Burk plot.
Surface plasmon resonance analysis
Surface plasmon resonance measurements were performed on a BIAcore 3000 system. Proteins for conjugation were diluted to 10 μg/mL in 10 mM sodium acetate (pH 5.0) and immobilized on a carboxymethylated dextran (CM5) sensor chip using the amine coupling method up to a level exceeding 4000 response units. Binding and washes were performed in Xld Buffer. After each cycle, chip surfaces were regenerated by removing noncross-linked bound proteins with 10 mM HCl. Data were analyzed with BIAevaluation 4.1 software and curve-fitting was performed with the assumption of one-to-one binding.
Embryo manipulations
Bisection experiments were performed at early gastrula (stage 10) embryos by cutting two equal halves along the prospective D–V axis using a surgical blade (Reversade and De Robertis 2005). Bisected embryos were cultured in 0.3× Barth solution. For mRNA microinjection, 1 ng of X. laevis BMP1 or BMP1 CUB1/2/3 mRNA, or 2 ng of BMP1 or dTolloid CUB1/2, dTld CUB4/5, or their mutated mRNAs were used. xBMP1 CUB1/2, xBMP1 CUB1/2/3, and Drosophila Tolloid CUB1/2 and CUB4/5 were cloned in a pCS2 Activin proregion cassette vector (Piccolo et al. 1999). To inhibit endogenous Chordin synthesis, 16 ng of antisense morpholino oligos directed against each paralog were injected per X. laevis embryo (Oelgeschläger et al. 2003). Mutations in the X. laevis BMP1 or Drosophila Tolloid sequences (Y227N, W426R, ΔL446, and S941R) were introduced using the QuikChange II site-directed mutagenesis kit (Stratagene). Detailed procedures for mRNA synthesis and whole-mount in situ hybridization are available at http://www.hhmi.ucla.edu/derobertis/index.html.
Mathematical modeling of D–V patterning
Numerical simulations of the partial differential equations system were solved by the XMDS software (http://www.xmds.org) using the fourth/fifth-order adaptive Runge-Kutta method on a one-dimensional circle representing the frog embryonic D–V axis. Additional details on the 14 reactions modeled by these partial differential equations and reaction rates can be found in Supplemental Figure 2.
Acknowledgments
We thank Dr. A. Ambrosio for help with fluorogenic peptide design, D. Geissert and J. Greenan for technical assistance, members of our laboratory for critical readings of the manuscript, and the UCLA core facility led by R. Lehrer for help with the BIAcore analyses. This work was supported by the NIH (HD21502-23). E.M.D.R. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1839309.
Supplemental material is available at http://www.genesdev.org.
References
- Ambrosio AL, Taelman VF, Lee HX, Metzinger CA, Coffinier C, De Robertis EM. Crossveinless-2 is a BMP feedback inhibitor that binds Chordin/BMP to regulate Xenopus embryonic patterning. Dev Cell. 2008;15:248–260. doi: 10.1016/j.devcel.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zvi D, Shilo BZ, Fainsod A, Barkai N. Scaling of the BMP activation gradient in Xenopus embryos. Nature. 2008;453:1205–1211. doi: 10.1038/nature07059. [DOI] [PubMed] [Google Scholar]
- Blitz IL, Shimmi O, Wünnenberg-Stapleton K, O'Connor MB, Cho KW. Is chordin a long-range- or short-range-acting factor? Roles for BMP1-related metalloproteases in chordin and BMP4 autofeedback loop regulation. Dev Biol. 2000;223:120–138. doi: 10.1006/dbio.2000.9740. [DOI] [PubMed] [Google Scholar]
- Bork P, Beckmann G. The CUB domain. A widespread module in developmentally regulated proteins. J Mol Biol. 1993;231:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- Canty EG, Garrigue-Antar L, Kadler KE. A complete domain structure of Drosophila tolloid is required for cleavage of short gastrulation. J Biol Chem. 2006;281:13258–13267. doi: 10.1074/jbc.M510483200. [DOI] [PubMed] [Google Scholar]
- Celeste AJ, Iannazzi JA, Taylor RC, Hewick RM, Rosen V, Wang EA, Wozney JM. Identification of transforming growth factor β family members present in bone-inductive protein purified from bovine bone. Proc Natl Acad Sci. 1990;87:9843–9847. doi: 10.1073/pnas.87.24.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs S, O'Connor MB. Two domains of the tolloid protein contribute to its unusual genetic interaction with decapentaplegic. Dev Biol. 1994;162:209–220. doi: 10.1006/dbio.1994.1079. [DOI] [PubMed] [Google Scholar]
- Dale L, Evans W, Goodman SA. Xolloid-related: A novel BMP1/Tolloid-related metalloprotease is expressed during early Xenopus development. Mech Dev. 2002;119:177–190. doi: 10.1016/s0925-4773(02)00359-3. [DOI] [PubMed] [Google Scholar]
- De Robertis EM. Evo-Devo: Variations on ancestral themes. Cell. 2008;132:185–195. doi: 10.1016/j.cell.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis EM, Kuroda H. Dorsal–ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M, Webb EC. Enzymes. Academic; New York: 1979. [Google Scholar]
- Eldar A, Dorfman R, Weiss D, Ashe H, Shilo BZ, Barkai N. Robustness of the BMP morphogen gradient in Drosophila embryonic patterning. Nature. 2002;419:304–308. doi: 10.1038/nature01061. [DOI] [PubMed] [Google Scholar]
- Ferguson EL, Anderson KV. Localized enhancement and repression of the activity of the TGF-β family member, decapentaplegic, is necessary for dorsal-ventral pattern formation in the Drosophila embryo. Development. 1992;114:583–597. doi: 10.1242/dev.114.3.583. [DOI] [PubMed] [Google Scholar]
- Finelli AL, Bossie CA, Xie T, Padgett RW. Mutation analysis of the Drosophila tolloid gene, a human BMP-1 homolog. Development. 1994;120:861–870. doi: 10.1242/dev.120.4.861. [DOI] [PubMed] [Google Scholar]
- Geach TJ, Dale L. Molecular determinants of Xolloid action in vivo. J Biol Chem. 2008;283:27057–27063. doi: 10.1074/jbc.M804232200. [DOI] [PubMed] [Google Scholar]
- Greenspan DS. Biosynthetic processing of collagen molecules. Top Curr Chem. 2005;247:149–183. [Google Scholar]
- Hartigan N, Garrigue-Antar L, Kadler KE. Bone morphogenetic protein-1 (BMP-1). Identification of the minimal domain structure for procollagen C-proteinase activity. J Biol Chem. 2003;278:18045–18049. doi: 10.1074/jbc.M211448200. [DOI] [PubMed] [Google Scholar]
- Hojima Y, van der Rest M, Prockop DJ. Type I procollagen carboxyl-terminal proteinase from chick embryo tendons. J Biol Chem. 1985;260:15996–16003. [PubMed] [Google Scholar]
- Hopkins DR, Keles S, Greenspan DS. The bone morphogenetic protein 1/Tolloid-like metalloproteinases. Matrix Biol. 2007;26:508–523. doi: 10.1016/j.matbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata H, Haraguchi T, Sasai Y. Robust stability of the embryonic axial pattern requires a secreted scaffold for chordin degradation. Cell. 2008;134:854–865. doi: 10.1016/j.cell.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Jasuja R, Voss N, Ge G, Hoffman GG, Lyman-Gingerich J, Pelegri F, Greenspan DS. bmp1 and mini fin are functionally redundant in regulating formation of the zebrafish dorosoventral axis. Mech Dev. 2006;123:548–558. doi: 10.1016/j.mod.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Jasuja R, Ge G, Voss NG, Lyman-Gingerich J, Branam AM, Pelegri FJ, Greenspan DS. Bone morphogenetic protein 1 prodomain specifically binds and regulates signaling by bone morphogenetic proteins 2 and 4. J Biol Chem. 2007;282:9053–9062. doi: 10.1074/jbc.M610929200. [DOI] [PubMed] [Google Scholar]
- Jürgens G, Wieschaus E, Nüsslein-Volhard C, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. Rouxs Arch Dev Biol. 1984;193:283–293. doi: 10.1007/BF00848157. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Yeh J, Grammer TC, Harland RM. Depletion of three BMP antagonists from Spemann's organizer leads to a catastrophic loss of dorsal structures. Dev Cell. 2005;8:401–411. doi: 10.1016/j.devcel.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Larraín J, Bachiller D, Lu B, Agius E, Piccolo S, De Robertis EM. BMP-binding modules in Chordin: A model for signalling regulation in the extracellular space. Development. 2000;127:821–830. doi: 10.1242/dev.127.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HX, Ambrosio AL, Reversade B, De Robertis EM. Embryonic dorsal–ventral signaling: Secreted Frizzled-related proteins as inhibitors of Tolloid proteinases. Cell. 2006;124:147–159. doi: 10.1016/j.cell.2005.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levander G. A study of bone regeneration. Surg Gynecol Obstet. 1938;67:705–714. [Google Scholar]
- Little SC, Mullins MC. Extracellular modulation of BMP activity in patterning the dorsoventral axis. Birth Def Res. 2006;78:224–242. doi: 10.1002/bdrc.20079. [DOI] [PubMed] [Google Scholar]
- Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C, et al. Kremen proteins are Dickkopf receptors that regulate Wnt/β-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- Marqués G, Musacchio M, Shimell MJ, Wünnenberg-Stapleton K, Cho KWY, O'Connor MB. Production of DPP activity gradient in the early Drosophila embryo through the opposing actions of the SOG and TLD proteins. Cell. 1997;91:417–426. doi: 10.1016/s0092-8674(00)80425-0. [DOI] [PubMed] [Google Scholar]
- Meinhardt H, Gierer A. Pattern formation by local self-activation and lateral inhibition. Bioessays. 2000;22:753–760. doi: 10.1002/1521-1878(200008)22:8<753::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Mizutani CM, Nie Q, Wan FY, Zhang YT, Vilmos P, Sousa-Neves R, Bier E, Marsh JL, Lander AD. Formation of the BMP activity gradient in the Drosophila embryo. Dev Cell. 2005;8:915–924. doi: 10.1016/j.devcel.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka O, Shimizu T, Yabe T, Nojima H, Bae YK, Hashimoto H, Hibi M. Sizzled controls dorso–ventral polarity by repressing cleavage of the Chordin protein. Nat Cell Biol. 2006;8:329–338. doi: 10.1038/ncb1379. [DOI] [PubMed] [Google Scholar]
- O'Connor MB, Umulis D, Othmer HG, Blair SS. Shaping BMP morphogens gradients in the Drosophila embryo and pupal wing. Development. 2006;133:183–193. doi: 10.1242/dev.02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelgeschläger M, Kuroda H, Reversade B, De Robertis EM. Chordin is required for the Spemann organizer transplantation phenomenon in Xenopus embryos. Dev Cell. 2003;4:219–230. doi: 10.1016/s1534-5807(02)00404-5. [DOI] [PubMed] [Google Scholar]
- Petropoulou V, Garrigue-Antar L, Kadler KE. Identification of the minimal domain structure of bone morphogenetic protein-1 (BMP-1) for chordinase activity: Chordinase activity is not enhanced by procollagen C-proteinase enhancer-1 (PCPE-1) J Biol Chem. 2005;280:22616–22623. doi: 10.1074/jbc.M413468200. [DOI] [PubMed] [Google Scholar]
- Piccolo S, Agius E, Lu B, Goodman S, Dale L, De Robertis EM. Cleavage of Chordin by the Xolloid metalloprotease suggests a role for proteolytic processing in the regulation of Spemann organizer activity. Cell. 1997;91:407–416. doi: 10.1016/s0092-8674(00)80424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S, Agius E, Leyns L, Battacharyya S, Grunz H, Bouwmeester T, De Robertis EM. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature. 1999;397:707–710. doi: 10.1038/17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouhinec JL, De Robertis EM. Systems biology of the self-regulating morphogenetic gradient of the Xenopus gastrula. Cold Spring Harb Perspect Biol. 2009;1:a001701. doi: 10.1101/cshperspect.a001701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reversade B, De Robertis EM. Regulation of ADMP and BMP2/4/7 at opposite embryonic poles generates a self-regulating morphogen field. Cell. 2005;123:1147–1160. doi: 10.1016/j.cell.2005.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath TK, Reddi AH. Dissociative extraction and reconstitution of extracellular matrix components involved in local bone differentiation. Proc Natl Acad Sci. 1981;78:7599–7603. doi: 10.1073/pnas.78.12.7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpe M, Umulis D, Ralston A, Chen J, Olson DJ, Avanesov A, Othmer H, O'Connor MB, Blair SS. The BMP-binding protein Crossveinless 2 is a short-range, concentration-dependent, biphasic modulator of BMP signaling in Drosophila. Dev Cell. 2008;14:940–953. doi: 10.1016/j.devcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmi O, Umulis D, Othmer H, O'Connor MB. Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell. 2005;120:873–886. doi: 10.1016/j.cell.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siva AC, Wild MA, Kirkland RE, Nolan MJ, Lin B, Maruyama T, Yantiri F, Fredickson S, Bowdish KS, Xin H. Targeting CUB domain-containing protein 1 with a monoclonal antibody inhibits metastasis in a prostate cancer model. Cancer Res. 2008;68:3759–3766. doi: 10.1158/0008-5472.CAN-07-1657. [DOI] [PubMed] [Google Scholar]
- Sundin OH, Leppert GS, Silva ED, Yang JM, Dharmaraj S, Maumenee IH, Santos LC, Parsa CF, Traboulsi EI, Broman KW, et al. Extreme hyperopia is the result of null mutations in MFRP, which encodes a Frizzled-related protein. Proc Natl Acad Sci. 2005;102:9553–9558. doi: 10.1073/pnas.0501451102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara K, Kessler E, Biniaminov L, Brusel M, Eddy RL, Jani-Sait S, Shows TB, Greenspan DS. Type I procollagen COOH-terminal proteinase enhancer protein: Identification, primary structure, and chromosomal localization of the cognate human gene PCOLCE. J Biol Chem. 1994;269:26280–26285. [PubMed] [Google Scholar]
- Turing AM. The chemical basis of morphogenesis. Philos Trans R Soc Lond. 1952;237:37–72. doi: 10.1098/rstb.2014.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker JA, Mintzer KA, Mullins MC. The BMP signaling gradient patterns dorsoventral tissues in a temporally progressive manner along the anteroposterior axis. Dev Cell. 2008;14:108–119. doi: 10.1016/j.devcel.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umulis DM, Serpe M, O'Connor MB, Othmer HG. Robust, bistable patterning of the dorsal surface of the Drosophila embryo. Proc Natl Acad Sci. 2006;103:11613–11618. doi: 10.1073/pnas.0510398103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urist M. Bone: Formation by autoinduction. Science. 1965;150:893–899. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- Wang YC, Ferguson EL. Spatial bistability of Dpp–receptor interactions during Drosophila dorsal–ventral patterning. Nature. 2005;434:229–234. doi: 10.1038/nature03318. [DOI] [PubMed] [Google Scholar]
- Wardle FC, Welch JV, Dale L. Bone morphogenetic protein 1 regulates dorsal–ventral patterning in early Xenopus embryos by degrading chordin, a BMP4 antagonist. Mech Dev. 1999;86:75–85. doi: 10.1016/s0925-4773(99)00114-8. [DOI] [PubMed] [Google Scholar]
- Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, Wang EA. Novel regulators of bone formation: Molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- Zhang JL, Huang Y, Qiu LY, Nickel J, Sebald W. von Willebrand factor type C domain-containing proteins regulate bone morphogenetic protein signaling through different recognition mechanisms. J Biol Chem. 2007;282:20002–20014. doi: 10.1074/jbc.M700456200. [DOI] [PubMed] [Google Scholar]