Abstract
The dosage compensation complex (DCC) in Drosophila globally increases transcription from the X chromosome in males to compensate for its monosomy. We discovered a male-specific conformation of the X chromosome that depends on the associations of high-affinity binding sites (HAS) of the DCC. The core DCC subunits MSL1–MSL2 are responsible for this male-specific organization. Contrary to emerging concepts, we found that neither DCC assembly nor the conformation of the male X chromosome are influenced by nuclear pore components. We propose that nuclear organization of HAS is central to the faithful distribution of the DCC along the X chromosome.
Keywords: Male-specific lethal; chromatin; transcription; nucleus, chromatin
Monitoring the relative localization of single loci and chromosomal domains within the nuclear space by high-resolution microscopy suggests a mutual influence between gene regulation and chromosome conformation as well as nuclear architecture (Sexton et al. 2007). Two general principles emerge. First, nuclear compartments with functional characteristics are defined by proximity to particular structures, such as the nuclear lamina or the nuclear pores. Chromosomal domains that are found associated with the lamina have a tendency to be repressed in humans and Drosophila (de Wit and van Steensel 2009). In contrast, optimal expression of some genes is enhanced by tethering to nuclear pores (Akhtar and Gasser 2007). A second organizational principle is the clustering of regulatory sequences in the nuclear interior in the form of bodies, clusters, or hubs, which are characterized by the accumulation of common protein interactors (Cavalli 2007; Fraser and Bickmore 2007). In combination, these peripheral and internal interactions partition chromosomes into loops and domains, restrict the mobility of these domains within the nucleus, and thus define a chromosome conformation. This organization bears the potential for coregulation of genes or chromosomal domains that are widely separated on the chromosome (de Wit and van Steensel 2009). In most cases, it is unclear whether the nonrandom chromosome conformation that can be observed by fluorescence in situ hybridization (FISH) microscopy and “chromosomal conformation capture” strategies (Simonis et al. 2007) is a cause or consequence of transcriptional coregulation of the participating genes.
Impressive examples for chromosome-wide transcriptional coregulation are provided by the dosage compensation phenomena in organisms with dimorphic sex chromosomes. Dosage compensation balances the relative expression between sex chromosomes and autosomes and adjusts the gene expression levels of monosomic sex chromosomes (like the X chromosomes in male mammals or Drosophila) to match the combined expression of the two female counterparts (Lucchesi et al. 2005). In mammals, one of the two X chromosomes in female nuclei is repressed by Polycomb proteins (Payer and Lee 2008). Active and inactive human female X chromosomes differ in shape and structure, the former having a larger surface-to-volume ratio (Eils et al. 1996; Rego et al. 2008). The inactive chromosome is nucleated by a kernel of constitutive and facultative heterochromatin harboring repetitive DNA elements, such as transposon-derived long interspersed sequences (LINE-1) (Clemson et al. 2006). During the process of inactivation, genes relocate into the inner Xist RNA domain, while genes that escape from silencing remain exposed to the transcription machinery in the outskirts of the chromosome territory (Chaumeil et al. 2006; Matarazzo et al. 2007; Rego et al. 2008). Clearly, the distinct conformation of the inactive X chromosome is an integral component of the chromosome-wide inactivation process.
In Drosophila, dosage compensation involves an approximately twofold activation of most genes on the single male X chromosome (Gelbart and Kuroda 2009). Fine-tuning of transcription is vital: Impaired dosage compensation is lethal for the males. This process is achieved by the dosage compensation complex (DCC; also termed male-specific lethal or MSL complex), which associates specifically with genes on the X chromosome and may regulate transcription at the level of elongation (Alekseyenko et al. 2006; Gilfillan et al. 2006). The DCC is formed by the assembly of five MSL proteins and two noncoding roX (RNA on the X) RNAs (Gelbart and Kuroda 2009). According to a prevalent model, the selective interaction of the DCC with the X chromosome involves several steps (Straub and Becker 2008; Gelbart and Kuroda 2009). The DCC is presumably assembled within the X chromosomal territory at the site of roX transcription (Kelley et al. 1999; Meller et al. 2000). The complex then associates with ∼150 “high-affinity sites” (HAS) (Straub et al. 2008), or “chromosomal entry sites” (Alekseyenko et al. 2008) that share a sequence motif required for DCC recruitment. However, it is currently not possible to predict a DCC-binding site considering just DNA sequence alone (Alekseyenko et al. 2008; Straub et al. 2008). Evidently, a crucial targeting principle is still missing (for discussion, see Straub and Becker 2008).
The HAS for DCC are clearly important for selective recognition of the X chromosome, but the majority of DCC binding occurs within the bodies of transcribed genes (Straub et al. 2005; Alekseyenko et al. 2006). The association of the DCC with transcribed chromatin is stabilized by preferential interaction of MSL3 with histone H3 methylated at Lys 36, a modification that is placed cotranscriptionally and marks active chromatin (Larschan et al. 2007; Bell et al. 2008). MSL3, along with the other RNA-binding DCC subunits MOF and MLE, appears to be particularly involved in “spreading” the DCC; i.e., its distribution from HAS to target genes (Gelbart and Kuroda 2009 and references therein). How the DCC is faithfully distributed from HAS to active genes is unknown. In contrast, the “core” subunits of the complex, MSL1 and MSL2, are specifically required for recognition of HAS.
To explore whether a higher-order chromosome organization may contribute to this process, we combined a two-color FISH, MSL2 immunostaining, and confocal microscopy to compare the three-dimensional (3D) localization of different HAS in female and male interphase nuclei. We first observed evidence for a male-specific chromosome conformation that is stably maintained through development and depends on the HAS interactors MSL1 and MSL2, but not on the “spreading” components MSL3 or MOF. These data suggest an involvement of X-chromosome conformation in the efficient and faithful targeting of the DCC.
Results and Discussion
A male-specific X-chromosome conformation
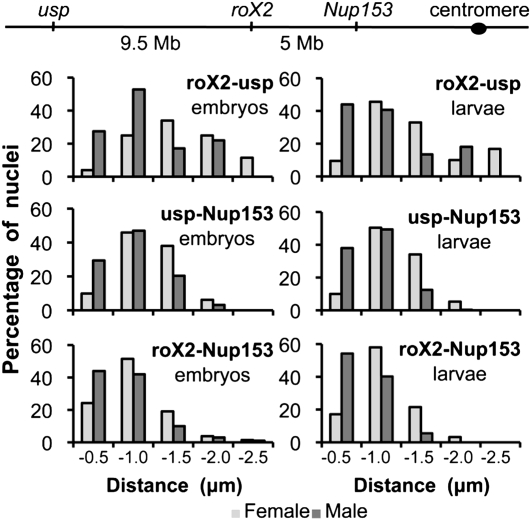
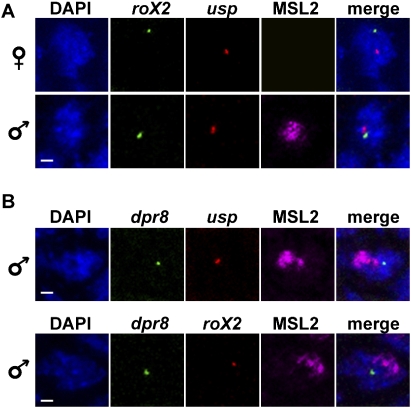
We assessed the conformation of the X chromosome by measuring the 3D distances between selected HAS for the DCC. We developed FISH probes specific for the ultraspiracle (usp) locus located at cytological position 2C, the roX2 locus located at 10C, and the Nup153 locus located at 14F (see Fig. 2, top [below]), containing HAS (Supplemental Fig. S1; Straub et al. 2008). The specificity of each probe for the selected locus was verified by FISH on polytene chromosomes of third instar larvae (Straub et al. 2008; data not shown). They were then used to determine the 3D distances between pairs of loci in various tissues and genetic conditions. Each two-color FISH experiment was followed by MSL2 immunostaining, which reveals the X-chromosomal territory and distinguishes male and female tissues. We selected interphase nuclei in the epidermal layer of wild-type (yw) embryos at stages 9–12, in which dosage compensation is effective. A calculation of the average nuclear radius, together with a comparison of the relative 3D distances of two autosomal loci (Supplemental Fig. S10), demonstrated the absence of general differences between male and female nuclei. We then compared the 3D distances between the different HAS located on the X chromosome. Due to the pairing between homologous chromosomes in Drosophila (Hiraoka et al. 1993), we observed only one FISH signal for each probe in the majority of female nuclei (for representative images, see Fig. 1; Supplemental Fig. S2). Analysis of male and female nuclei using the roX2 and usp probes revealed a continuum of distances with a striking difference between male and female nuclei. The significance of distance differences between the loci in the two sexes was evaluated by a two-tailed Mann-Whitney U-test. The two loci were significantly closer in males than in females (P-value < 2.3 × 10−6) (Fig. 2). In 27.5% of the male nuclei, usp and roX2 were within 0.5 μm, whereas this was only the case in 4% of the female nuclei (Supplemental Table S1). Visualization of the male X chromosome by MSL2 immunostaining showed that usp and roX2 resided within the MSL2 domain in 75% of the nuclei, and that the long-distance association between these two loci occurred within this domain (Fig. 1A).
Figure 2.
Long-distance associations between different HAS in male nuclei. A schematic representation of the probe localization on the X chromosome is presented at the top. Quantitative analysis of the distribution of 3D distances between the indicated pairs of loci measured in female (light gray) and male (dark gray) nuclei of embryos (left panel) or larvae (right panel) from the yw line. The bars show the percentage of nuclei with distances within the following ranges: 0–0.5 μm, 0.5–1 μm, 1–1.5 μm, 1.5–2 μm, and 2–2.5 μm. The average number of nuclei analyzed per condition was 220.
Figure 1.
Male-specific conformation of the dosage-compensated X chromosome. Long-range proximity between HAS for DCC binding in nuclei of whole-mount embryos. Pairs of chromosomal sites were visualized by two-color FISH in female or male embryos of the yw line (stages 9–12, wild type). DNA was stained with DAPI and the X-chromosome territory was painted with an antibody against MSL2 in male nuclei (there is no MSL2 in female nuclei). The figure shows single optical slices of representative images obtained for the roX2 and usp (A), for the dpr8 and usp (B, top), and for the dpr8 and roX2 (B, bottom) probes. A merge of the channels reveals the proximity of the HAS and their residence relative to the MSL2 territory in male nuclei. Bars, 1 μm.
The usp and Nup153 loci are physically separated by 14.5 Mb on the X chromosome. Strikingly, this increased linear distance was not reflected in space. The loci were found within 0.5 μm in 29.4% of the male nuclei (Fig. 2) and in only 9.9% of the female nuclei (P-value < 4.5 × 10−6). Accordingly, the roX2 and Nup153 loci were also significantly closer in males (in 44% of nuclei, the two loci are within 0.5 μm; P-value = 3.6 × 10−6). In this case, the physical proximity of these sites, separated by only 5 Mb, is reflected by a smaller average distance between them in female nuclei also (24.2% of the signals within 0.5 μm) (Fig. 2). Taken together, the data document long-range associations between three strong DCC-binding sites. The fact that this proximity was sex-specific shows that the X chromosome in male nuclei differs in conformation from the female X. This conformation may be brought about by the clustering of the HAS in space. Alternatively, a male-specific chromosome conformation may promote close contacts between HAS.
Maintenance of chromosome conformation during development
To determine whether the difference in chromosome conformation was maintained during development, we analyzed interphase nuclei in imaginal discs of third instar larvae. We confirmed that the three selected HAS are strikingly closer in male as compared with female nuclei (Fig. 2; Supplemental Fig. 2B). The percentage of nuclei showing 3D distances of <0.5 μm between the different HAS was even higher than in male embryonic nuclei (Supplemental Table S1), and HAS site pairing occurred within the MSL2 domain (Supplemental Fig. 2B). Apparently, the chromosome conformation, which appeared more stable than in embryos, is a feature of chromosomal organization that is independent of developmental stage.
X-chromosome sequences unbound by DCC do not show specific conformation
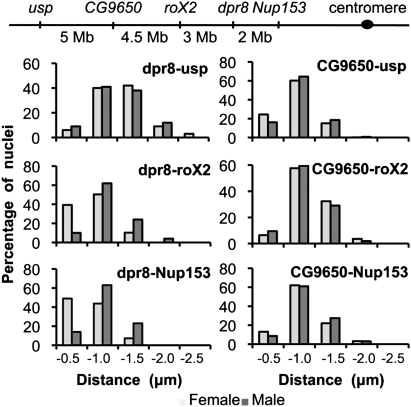
In order to determine whether the observed long-range associations were specific for DCC-binding sites, we developed two further probes that label X chromosomal sequences that do not bind DCC in male nuclei (see Supplemental Fig. S1 for a screen shot of the corresponding DCC profile). The probe located around the gene dpr8 at cytological position 12F resides in an area of ∼300 kb that is devoid of DCC. Two-color FISH on whole-mount embryos showed no significant distance difference between dpr8 and usp in female or male nuclei (Supplemental Fig. 3A). In contrast to the three HAS, the dpr8 locus was found outside of the territory in ∼70% of the nuclei (e.g., see Fig. 1B). Remarkably, the 3D distances between dpr8 and the roX2 or Nup153 loci, which are relatively close on the physical map of the chromosome (3 Mb and 2 Mb, respectively), were significantly shorter in female nuclei than in males (P-value < 2 × 10−6) (Supplemental Fig. S3B,C), as if X-chromosomal sequences not bound by DCC were driven out of the area of clustering into the periphery of the DCC territory. At later stages of development, the observations made for dpr8 were reproduced with even more striking values (Fig. 3; Supplemental Fig. S2C; see Supplemental Table S2 for P-values). The use of a fifth probe located around the CG9650 gene at cytological position 7A, which is 60 kb away from a HAS (Supplemental Fig. S1), confirmed that DCC-free sequences present on the X do not reside in the DCC territory and do not preferentially associate in male nuclei. The CG9650 locus does not particularly reside in the vicinity of the roX2, the Nup153, or the dpr8 loci in female or in male nuclei (Fig. 3; Supplemental Fig. 3D; see Supplemental Table 2 for P-values). This locus is relatively close to the usp locus in female nuclei, which mainly reflects the linear placement of the loci on the X chromosome. In male nuclei, the loci significantly separate (P = 2.5 × 10−4) (Fig. 3). In summary, the data show that HAS tend to cluster in male nuclei, but sites unbound by the DCC are more likely to be excluded from the core of the dosage-compensated chromosomal territory.
Figure 3.
X-chromosome sequences unbound by DCC do not show sex-specific association with HAS. The analysis was as in Figure 2.
DCC assembly, X-chromosome targeting, and X-chromosome conformation do not depend on nucleoporins Nup153 and Megator (Mtor)
Conceivably, a specific chromosome conformation may implicate interactions of MSL proteins with nuclear structures like the nuclear lamina or the pores (Akhtar and Gasser 2007). Recently, a study from the Akhtar laboratory (Mendjan et al. 2006) provided good candidates for such interactions. Mendjan et al. (2006) found the nuclear pore subunits Mtor and Nup153 in immunopurifications of MOF and MSL3. Upon ablation of the nucleoporins by RNAi, the DCC was observed to dissociate from the X chromosome in SL2 cell nuclei (Mendjan et al. 2006). In order to explore a possible role for Mtor and Nup153 on male-specific X-chromosome conformation, we used different inducible RNAi lines in which specific ablation of the nucleoporins can be induced at different developmental stages or in different tissues using the UAS-GAL4 system (Duffy 2002; Dietzl et al. 2007). We first analyzed the effect of a reduction of Mtor or Nup153 levels (Supplemental Fig. 4A) on MSL2 recruitment to the polytenic X chromosomes in salivary glands, using an appropriate GAL4 driver line (SGS3-GAL4). As a control, we used a driver line specific to the eye (ey-GAL4) that did not affect nucleoporin levels in the salivary glands. Double immunostaining using antibodies against Mtor or Nup153 and MSL2 confirmed the effective depletion of the nucleoporins on whole-mount salivary glands in the presence of SGS3-GAL4, and showed no effect on the intensity or integrity of DCC binding to the X chromosome (Supplemental Fig. 4B,C). This was confirmed on polytene chromosome spreads (Supplemental Fig. 4D). The depletion of the nucleoporins in interphase nuclei using the eye imaginal disc driver (ey-GAL4) frequently gave rise to eye phenotypes in either sex (Supplemental Fig. S5), yet it did not significantly affect the formation of the MSL2-stained X-chromosome territory in the eye imaginal discs (Supplemental Fig. 6). Since the effect observed by Mendjan et al. (2006) might depend on specific features of SL2 cells in culture, we treated SL2 cells with two different specific interfering RNAs (Supplemental Table S5) for 6 d. As for the larval tissues, the reduction of nucleoporin levels did not affect the integrity or intensity of the MSL2-stained chromosomal territory under those conditions (Supplemental Fig. 7). A possible explanation for the discrepancy between our results and the effects shown by Mendjan et al. (2006) could come from differences in the RNAi protocols. We used less dsRNA than Mendjan et al. (2006) (10 μg vs. 45 μg), and cells were analyzed after 6 d of RNAi instead of 9 d (the time employed by Mendjan et al. [2006]). We systematically observed a drop in cell numbers after 5 d (60%–70% of the NPC-depleted cells, compared with 85%–90% of untreated or GST-treated cells) and an increase in cell death starting after 7 d. This could reflect defects in mitosis progression as described for Mtor (Qi et al. 2004) and/or in nuclear import for Nup153 (Sabri et al. 2007). Clearly, our RNAi protocol was highly efficient. The effects of nucleoporin knockdown on the formation of the DCC territory under much harsher conditions are, in our opinion, indirect consequences of cell death.
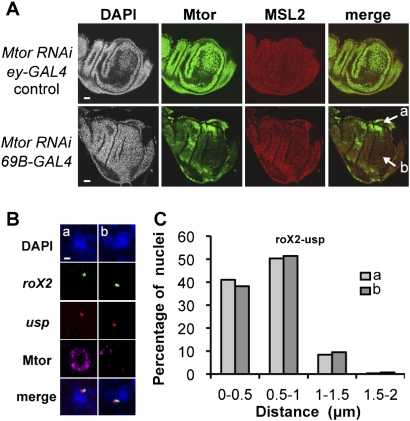
The fact that under our conditions of nucleoporin knockdown the MSL2 territory was still intact allowed us to test whether the presence of Mtor or Nup153 directly affects the specific X-chromosome conformation in males. We depleted Mtor in a large domain of the wing imaginal disc in the MtorRNAi-69BGAL4 line (Fig. 4A,B). We compared the 3D distances between the usp and the roX2 loci in areas of the disc where Mtor is present or absent (Fig. 4, areas a or b, respectively). The 3D distances were identical, independent of whether Mtor was present (Fig. 4C). We conclude that Mtor and Nup153 directly affect neither the integrity of the DCC domain on the X chromosome nor the male-specific chromosome conformation between the HAS.
Figure 4.
The male-specific X-chromosome conformation does not depend on nucleoporin Mtor. Mtor was ablated by RNAi in specific domains of wing imaginal discs. (A) Representative example of double immunostaining with Mtor (green) and MSL2 (red) antibodies in wing imaginal discs from male third instar larvae. Each panel corresponds to a projection of 10 Z-slices spaced by 0.5 μm. Bars, 20 μm. MtorRNAi-eyGAL4 and MtorRNAi-69BGAL4 correspond to transgenic lines where RNAi of Mtor expression was targeted to the eye disc (as a control) or the wing disc, respectively. 3D distances between the roX2 and usp sites were determined in male nuclei, which did (a) or did not (b, see arrows in merged panel) contain Mtor at their nuclear pores in the wing disc of the MtorRNAi-69BGAL4 larvae. (B) Two-color FISH with roX2 and usp probes followed by Mtor immunostaining in a wing disc of the kind shown in A. Single optical slices show representative nuclei residing in areas a and b of A. The presentation is as in Figure 1. (C) Quantitative analysis of 3D distances between the roX2 and usp loci nuclei taken from areas a or b. Two-hundred nuclei were analyzed from each area.
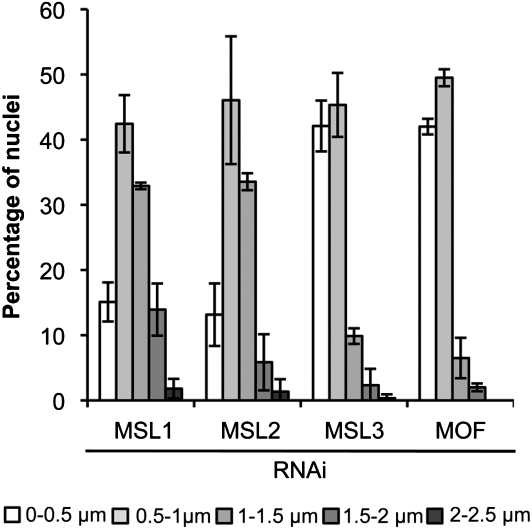
The male-specific X-chromosome conformation depends on MSL1–MSL2 but not on MSL3-MOF
In order to analyze whether DCC subunits affect the conformation of the male X chromosome, we depleted MSL1, MSL2, MSL3, or MOF by inducible RNAi. The efficiency of the specific ablation of those proteins using the SGS3-GAL4 driver was verified on polytene chromosomes. Knockdown of MSL1 prevented the binding of MSL2 and vice versa, in line with earlier genetic analyses (data not shown). Crossing homozygous flies of the 69B-GAL4 driver line with flies homozygous for the RNAi constructs directed against each DCC member, we obtained male third instar larvae where each of MSL1, MSL2, MSL3, or MOF were strongly depleted in the wing discs. The effectiveness of the knockdown was confirmed by strongly reduced numbers of adult male flies obtained from these crosses (Supplemental Table 3). We monitored the 3D distances between the usp and roX2 loci (Fig. 5) or between the usp and Nup153 loci (Supplemental Fig. S8) under those conditions and found that the absence of MSL1 or MSL2 strongly affected the association between these loci. Remarkably, the distance distribution in male MSL1RNAi or MSL2RNAi nuclei resembles that obtained in female nuclei for the same probes (Fig. 2). The sex-specific chromosome conformation therefore, depends on these core DCC subunits, in line with earlier observations that MSL1–MSL2 alone are capable of directing the DCC to HAS (Lyman et al. 1997; Kelley et al. 1999). Conversely, the efficient knockdown of MSL3 or MOF (Supplemental Fig S9) did not affect the male-specific proximity between the roX2 or the Nup153 and the usp loci (cf. Fig. 5, Supplemental Fig. S8 and Fig. 2). MSL3 and MOF are rather implicated in the spreading of the DCC from HAS to transcribed sequences, which constitute low-affinity binding sites for DCC (Larschan et al. 2007). The preferential interaction of MSL3 with the transcription-dependent H3K36 methylation mark promotes recruitment of the other DCC components to these low binding affinity sites. Acetylation of H4K16 mediated by MOF would facilitate the acquisition of opened chromatin structure, increasing the probability of a new event of transcription onto these novel DCC-binding sites.
Figure 5.
The male-specific X-chromosome conformation depends on MSL1 and MSL2. The quantitative analysis of 3D distances between the roX2 and usp loci in wing disc male nuclei was as in Figures 2 and 3, except that the larvae were analyzed in which levels of MSL1, MSL2, MSL3, or MOF were reduced by RNAi in the wing discs (69B-GAL4 driver). Error bars represent standard deviation from three independent experiments for each distance category. The average total number of nuclei analyzed per condition was 550.
Recognition of HAS by MSL1–2 is an early event in the multistep process of recruitment of DCC to the X chromosome. The sex-specific organization of HAS, which is mediated by MSL1–2, may thus contribute to assuring the efficient distribution of the complex. We envision the X chromosome to be structured in zones of decreasing DCC concentrations. DCC may be assembled at the site of roX RNA transcription within the X chromosome territory, containing HAS for MSL1–2 (Kelley et al. 1999). Sites of highest affinity for DCC may be clustered more internally, surrounded by layers of lower affinity, including active genes marked by H3K36 methylation. About 25% of X-chromosome sequences are bound by DCC (Alekseyenko et al. 2006; Gilfillan et al. 2006), and these constitute the nuclear domain stained by MSL2 antibody. X-chromosomal sequences that do not attract DCC—like intergenic regions, inactive genes, and genes that escape transcriptional dosage compensation—are interspersed with DCC-bound domains on the chromosome map. The fact that such a sequence (like dpr8 or CG9650) consistently resided outside of the MSL2 domain suggests that sequences not bound by the DCC may be arranged as an outer layer around the DCC-bound territory. Such an arrangement would be inverse to the one suggested for the inactive X chromosome on mammals (Chaumeil et al. 2006; Clemson et al. 2006).
We can only speculate about the mechanism responsible for the clustering of DCC HAS. We currently favor a “self-assembly” mechanism, based purely on concentrations of factors and interaction affinities. Accordingly, roX genes would initially establish a local focus of DCC enrichment within the X territory due to the assembly of the complex around the nascent roX transcript. HAS that encounter this focus during random chromosome movements will tend to dwell longer in its vicinity. Their presence will in turn lead to further concentration of DCC, establishing a positive feedback loop between DCC enrichment and the clustering of binding sites that will lead to a concentration gradient of DCC centered around the roX genes. Sites of lower affinity due to either the presence of degenerate sequence elements or the H3K36 methyl mark-associated with active transcription will be bound by DCC and incorporated into the domain (Dahlsveen et al. 2006; Larschan et al. 2007), whereas loci without DCC-binding sites will tend to be excluded. Because the recognition of HAS relies mainly on MSL1–2, knockdown of MSL3 or MOF does not affect their clustering. However, loss of MSL3-MOF is also lethal for males, which illustrates the importance of distributing the DCC on to the bulk of binding sites at transcribed genes. Further 3D structuring of the chromosome may then depend on transcription; for example, by cohabitation of dosage-compensated transcription units in transcription factories (Fraser and Bickmore 2007). However, unlike the situation in mammals where the female X chromosomes show extreme differences in expression, the male and female X chromosomes of Drosophila differ in their activity state by up to only twofold. The example of the dosage-compensated X chromosome thus serves as a prominent example for the intimate relationship between chromosome conformation and coordinate regulation of a large number of genes.
Materials and methods
Fly stocks and handling
Flies were raised on standard corn meal/yeast extract medium. Inducible RNAi and GAL4 driver lines were obtained from VDRC (Vienna) and the Bloomington Stock Center, respectively. They are described in the Supplemental Material.
FISH-I on whole-mount tissues
Each FISH probe consisted of a pool of six to seven PCR fragments (1.2–1.5 kb). Each amplicon was spaced by ∼2 kb, to cover ∼20 kb for each locus. Probe preparation and two-color FISH followed by immunostaining on whole-mount tissues were as described (Bantignies et al. 2003). A detailed protocol is presented in the Supplemental Material.
Double immunostaining on imaginal discs of third instar larvae
Imaginal discs of third instar larvae were fixed in 4% formaldehyde in PBT (PBS, 0.1% Tween20) for 20 min at room temperature. After several washes in PBT, the specimens were blocked in PBS/0.3% Triton/10% NGS (BS) for 2 h at room temperature and incubated overnight at 4°C with the primary antibodies. Larval tissues were washed in PBS/0.3% Triton, blocked 1 h in BS, and incubated sequentially for 1 h with different secondary antibodies diluted in BS (see the Supplemental Material for details). After, several washes in PBS/0.3% Triton, DNA was counterstained with DAPI and separated imaginal discs were mounted on slides in Prolong Antifade Medium (Molecular Probes).
Microscopy and image analysis
Series of confocal sections through whole-mount embryos or imaginal discs were collected with a Leica SP5 microscope equipped with a Plan/Apo 63× 1.4 NA oil immersion objective. For each optical section, images (512 × 512 pixels) were collected sequentially for three to four channels. RGB stacks were reconstructed with the three-channels function in ImageJ (after chromatic shift correction) and used as raw data files for 3D FISH distance analysis. Detailed 3D distance calculation and statistical analysis are described in the Supplemental Material.
Acknowledgments
We thank members of the laboratory for critical readings of the manuscript, Irina Solovej and Thomas Cremer for an introduction into the microscope and for microscope time, A. Akhtar for nucleoporin antibodies, and T. Straub for advice on probe selection. This work was supported by the Deutsche Forschungsgemeinschaft through SFB TR 5. C.G was supported by an EMBO long-term fellowship.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.539509.
Supplemental material is available at http://www.genesdev.org.
References
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes & Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F, Grimaud C, Lavrov S, Gabut M, Cavalli G. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes & Dev. 2003;17:2406–2420. doi: 10.1101/gad.269503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Conrad T, Kind J, Wirbelauer C, Akhtar A, Schubeler D. Transcription-coupled methylation of histone H3 at lysine 36 regulates dosage compensation by enhancing recruitment of the MSL complex in Drosophila melanogaster. Mol Cell Biol. 2008;28:3401–3409. doi: 10.1128/MCB.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G. Chromosome kissing. Curr Opin Genet Dev. 2007;17:443–450. doi: 10.1016/j.gde.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes & Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlsveen IK, Gilfillan GD, Shelest VI, Lamm R, Becker PB. Targeting determinants of dosage compensation in Drosophila. PLoS Genet. 2006;2:e5. doi: 10.1371/journal.pgen.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, van Steensel B. Chromatin domains in higher eukaryotes: Insights from genome-wide mapping studies. Chromosoma. 2009;118:25–36. doi: 10.1007/s00412-008-0186-0. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Duffy JB. GAL4 system in Drosophila: A fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Eils R, Dietzel S, Bertin E, Schrock E, Speicher MR, Ried T, Robert-Nicoud M, Cremer C, Cremer T. Three-dimensional reconstruction of painted human interphase chromosomes: Active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J Cell Biol. 1996;135:1427–1440. doi: 10.1083/jcb.135.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Gelbart ME, Kuroda MI. Drosophila dosage compensation: A complex voyage to the X chromosome. Development. 2009;136:1399–1410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan GD, Straub T, de Wit E, Greil F, Lamm R, van Steensel B, Becker PB. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes & Dev. 2006;20:858–870. doi: 10.1101/gad.1399406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF, Parmelee SJ, Rykowski MC, Agard DA, Sedat JW. The onset of homologous chromosome pairing during Drosophila melanogaster embryogenesis. J Cell Biol. 1993;120:591–600. doi: 10.1083/jcb.120.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- Lyman LM, Copps K, Rastelli L, Kelley RL, Kuroda MI. Drosophila male-specific lethal-2 protein: Structure/function analysis and dependence on MSL-1 for chromosome association. Genetics. 1997;147:1743–1753. doi: 10.1093/genetics/147.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarazzo MR, Boyle S, D'Esposito M, Bickmore WA. Chromosome territory reorganization in a human disease with altered DNA methylation. Proc Natl Acad Sci. 2007;104:16546–16551. doi: 10.1073/pnas.0702924104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller VH, Gordadze PR, Park Y, Chu X, Stuckenholz C, Kelley RL, Kuroda MI. Ordered assembly of roX RNAs into MSL complexes on the dosage-compensated X chromosome in Drosophila. Curr Biol. 2000;10:136–143. doi: 10.1016/s0960-9822(00)00311-0. [DOI] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: How mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Qi H, Rath U, Wang D, Xu YZ, Ding Y, Zhang W, Blacketer MJ, Paddy MR, Girton J, Johansen J, et al. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol Biol Cell. 2004;15:4854–4865. doi: 10.1091/mbc.E04-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rego A, Sinclair PB, Tao W, Kireev I, Belmont AS. The facultative heterochromatin of the inactive X chromosome has a distinctive condensed ultrastructure. J Cell Sci. 2008;121:1119–1127. doi: 10.1242/jcs.026104. [DOI] [PubMed] [Google Scholar]
- Sabri N, Roth P, Xylourgidis N, Sadeghifar F, Adler J, Samakovlis C. Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport. J Cell Biol. 2007;178:557–565. doi: 10.1083/jcb.200612135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nat Struct Mol Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- Straub T, Becker PB. DNA sequence and the organization of chromosomal domains. Curr Opin Genet Dev. 2008;18:175–180. doi: 10.1016/j.gde.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Straub T, Gilfillan GD, Maier VK, Becker PB. The Drosophila MSL complex activates the transcription of target genes. Genes & Dev. 2005;19:2284–2288. doi: 10.1101/gad.1343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 2008;4:e1000302. doi: 10.1371/journal.pgen.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]