Abstract
The histone H3 Lys 9 (H3K9) methyltransferase Eset is an epigenetic regulator critical for the development of the inner cell mass (ICM). Although ICM-derived embryonic stem (ES) cells are normally unable to contribute to the trophectoderm (TE) in blastocysts, we find that depletion of Eset by shRNAs leads to differentiation with the formation of trophoblast-like cells and induction of trophoblast-associated gene expression. Using chromatin immmunoprecipitation (ChIP) and sequencing (ChIP-seq) analyses, we identified Eset target genes with Eset-dependent H3K9 trimethylation. We confirmed that genes that are preferentially expressed in the TE (Tcfap2a and Cdx2) are bound and repressed by Eset. Single-cell PCR analysis shows that the expression of Cdx2 and Tcfap2a is also induced in Eset-depleted morula cells. Importantly, Eset-depleted cells can incorporate into the TE of a blastocyst and, subsequently, placental tissues. Coimmunoprecipitation and ChIP assays further demonstrate that Eset interacts with Oct4, which in turn recruits Eset to silence these trophoblast-associated genes. Our results suggest that Eset restricts the extraembryonic trophoblast lineage potential of pluripotent cells and links an epigenetic regulator to key cell fate decision through a pluripotency factor.
Keywords: Oct4, embryonic stem cells, histone methylation, pluripotency, transcriptional repression, trophoblast
Preimplantation embryonic development is characterized by a progressive restriction in developmental potentials (Loebel et al. 2003; Rossant 2004). The initial totipotent blastomeres will undergo the first cell fate segregation with the formation of trophectoderm (TE) and inner cell mass (ICM), which develop into the extraembryonic and embryonic lineages, respectively (Loebel et al. 2003; Rossant 2004). Embryonic stem (ES) cells derived from the ICM are pluripotent (Smith 2001; Rossant 2008), whereas trophoblast stem (TS) cells derived from the TE are multipotent (Tanaka et al. 1998). When introduced back into the blastocysts, ES cells are unable to differentiate into extraembryonic tissues derived from the TE, but they can give rise to all embryonic tissues (Beddington and Robertson 1989).
Lineage-specific transcription factors are required for ICM and TE cell fate determination (Ralston and Rossant 2005). The POU domain transcription factor Oct4 (encoded by the Pou5f1 gene) is highly expressed in ES cells (Scholer et al. 1990; Palmieri et al. 1994). It is expressed throughout the early embryo until the blastocyst stage, when its expression becomes restricted to the ICM. Pou5f1 mutant mice embryos die around the time of implantation. Although the mutant blastocysts appear to possess a morphologically normal ICM, the cells within the ICM actually differentiate into trophoblast cells and not cells of the embryonic lineages (Nichols et al. 1998). This switch from embryonic to extraembryonic cell fate can also be recapitulated in ES cells. Reducing the expression of Pou5f1 by half induces ES cells to differentiate into trophoblasts (Niwa et al. 2000).
The Cdx2 gene encodes a caudal-related transcription factor that is essential for the specification of the TE fate and development of the TE. Cdx2-null mouse embryos die prior to implantation with Pou5f1 aberrantly expressed in the TE (Strumpf et al. 2005). In the absence of Cdx2, the mutant blastocysts fail to express markers of TE differentiation. In ES cells, depletion of Oct4 induces Cdx2 expression through the release of its direct repression of Cdx2 (Niwa et al. 2005). Conversely, ectopic expression of Cdx2 interferes with the transcriptional activator function of Oct4 through binding at the Pou5f1 promoter (Niwa et al. 2005). Hence, Cdx2 and Oct4 are implicated in reciprocal repression of each other's function to specify the first lineage segregation of the TE and the ICM.
Besides transcription factors, epigenetic mechanisms are also required for the restriction of extraembryonic trophoblast lineage potential in ES cells (Surani et al. 2007). Hence, it is of interest to investigate the role of epigenetic regulators in modulating the embryonic and extraembryonic fate of ES cells. Eset (also known as Setdb1) represses gene expression through catalyzing the methylation of mono- and dimethylated states of histone H3 Lys 9 residue to form H3K9me2 and H3K9me3, respectively (Yang et al. 2002; Wang et al. 2003). These marks are generally associated with transcriptional silencing and are bound by corepressors such as HP1 (Kouzarides 2002; Lachner and Jenuwein 2002). Disruption of Eset by gene targeting results in peri-implantation lethality (Dodge et al. 2004). Eset-null blastocysts show defective ICM outgrowth, and ES cells cannot be derived from these blastocysts. Thus, we reasoned that Eset may play an important role in ES cell biology.
In this study, we show that depletion of Eset by RNAi induces ES cells to differentiate. Genome-wide location analysis of Eset reveals that Eset targets genes involved in trophoblast lineage specification and differentiation. We confirmed that genes that are preferentially expressed in the TE (Tcfap2a and Cdx2) are bound and repressed by Eset in ES cells. At these genes, the recruitment of Eset is dependent on Oct4. Eset-depleted ES cells can incorporate into the TE of a blastocyst. Thus, the pluripotency factor Oct4 partners with an epigenetic regulator, Eset, to restrict trophoblast lineage potential in ES cells.
Results
Depletion of Eset induces ES cell differentiation
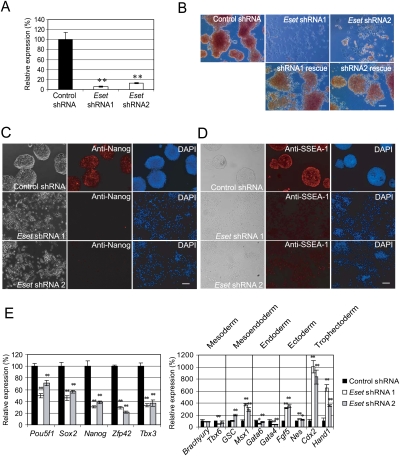
To study the roles of Eset in ES cells, we depleted its expression using RNAi. Two shRNA constructs targeting different regions of the Eset transcript were used to establish the knockdown effects. Both constructs were effective in reducing the RNA and protein (Fig. 1A; Supplemental Fig. S1). Strikingly, the colony morphology of the Eset knockdown ES cells was lost, indicating differentiation of the cells. The common properties of ES cells, alkaline phosphatase activity, and presence of Nanog and SSEA-1 were also reduced upon knockdown of Eset transcripts, strongly indicative of differentiation (Fig. 1B–D). Importantly, we were able to rescue the morphology phenotype by coexpression of RNAi-immune Eset cDNAs for both Eset shRNAs, indicating that the knockdown effects are specific to Eset (Fig. 1B; Supplemental Fig. S2). To confirm cellular differentiation, we measured the transcripts of ES cell-associated genes and genes induced upon differentiation. Pou5f1, Sox2, Nanog, Zfp42, and Tbx3 were reduced while Msx1, Fgf5, Cdx2, and Hand1 were induced (Fig. 1E). The induction of TE markers Cdx2 and Hand1 is consistent with ES cells differentiating into trophoblast-like cells (Fig. 1E). Some of the differentiated cells showed trophoblast giant cell morphology, with dramatically expanded cytoplasm and nuclei (Supplemental Fig. S3). To probe into other genes whose expression was affected after Eset depletion, cDNA microarray experiments were performed to capture the gene expression changes upon Eset knockdown. The level of transcripts coding for self-renewal regulators such as Klf4, Esrrb, and Tcl1 was reduced, while trophoblast lineage-associated genes such as Cdx2, Tcfap2a, Fgfr2, Plf, and Mmp9 were coordinately up-regulated (Supplemental Fig. S4; Niwa et al. 2005; Strumpf et al. 2005; Winger et al. 2006) . Although the data support the notion that Eset depletion leads to the formation of trophoblast-like cells, it is also likely that cell types of other lineages are formed (Fig. 1E, right panel) as marker genes for mesendoderm and ectoderm lineages are also up-regulated. To further characterize the Eset-depleted ES cells, we analyzed their ability to form colonies in a replating assay. Transfected cells were dissociated with trypsin and replated to allow the cells to expand into colonies. Depletion of Eset reduced the number of ES cell colony-forming units (CFUs) by threefold to 17-fold, as compared with the control knockdown (Supplemental Fig. S5). Taken together, our results indicate that Eset is critical for the maintenance of ES cell properties.
Figure 1.
Eset is required for the maintenance of ES cells. (A) Depletion of Eset by RNAi. Level of Eset transcripts after knockdown was measured by real-time PCR analysis. Data are represented as mean ± SD; n = 3. (**) P < 0.005. (B) Eset knockdown leads to ES cell differentiation. The ES cells were transfected with plasmids expressing control Luc shRNA, Eset shRNA1, or Eset shRNA2. In control shRNA transfected cells, colony morphology typical for undifferentiated ES cells was maintained. Flattened fibroblast-like cells were formed after Eset depletion. The cells were selected with puromycin for 5 d before alkaline phosphatase assay was performed to detect undifferentiated ES cells. Cotransfection of RNAi-immune Eset rescued the differentiation phenotype induced by the Eset shRNAs. Bar, 100 μm. (C,D) Immunofluorescence staining with Nanog or SSEA-1 antibody. Bar, 100 μm. (E) Real-time PCR analysis of ES cell-associated gene expression (left panel) and lineage-specific marker gene expression (right panel) in Eset and control knockdown cells. Data are represented as mean ± SD; n = 3. (*) P < 0.05; (**) P < 0.005.
Genomic targets of Eset identified by chromatin immunoprecipitation (ChIP) and sequencing (ChIP-seq) analysis
Eset is involved in euchromatic gene silencing, and interacts with a number of proteins associated with transcriptional repression (Ayyanathan et al. 2003; Mulligan et al. 2008). To identify the genes repressed by Eset, we mapped the Eset-binding sites in mouse ES cells using a ChIP-based approach. Chromatin extracts prepared from ES cells were subjected to ChIP using an anti-Eset antibody to enrich for Eset-bound chromatin. Western blot and immunofluorescence assays confirmed that our antibody is specific toward Eset (Supplemental Figs. S1, S6A). The ChIP-enriched DNA was subjected to massive parallel short tag-based sequencing (Barski et al. 2007; Johnson et al. 2007; Mikkelsen et al. 2007; Robertson et al. 2007; Chen et al. 2008). The uniquely mapped tags were then used to reconstruct a whole-genome-binding profile of Eset. We validated 20 loci and showed that the ChIP signal is reduced upon Eset depletion (Supplemental Fig. S6B).
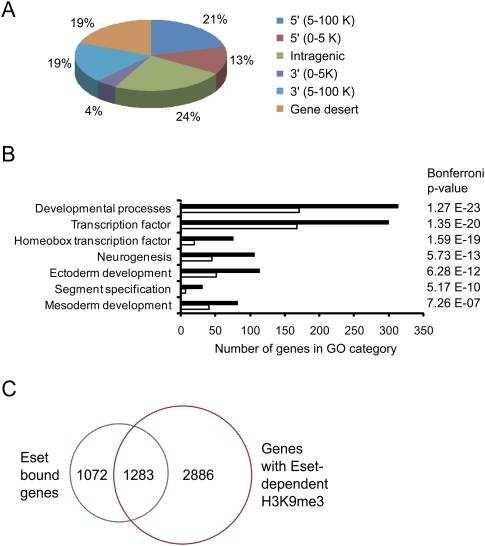
Eset occupancy can be found at intragenic and intergenic regions (Fig. 2A). For the 2355 Eset-bound genes (Supplemental Table 1), we analyzed the enrichment of Gene Ontology (GO) categories classified under the PANTHER database (Mi et al. 2005). Several GO categories are significantly overrepresented. For example, Eset preferentially binds to genes involved in developmental processes, neurogenesis, ectoderm development, mesoderm development, and segment specification (Fig. 2B). Genes encoding for transcription factors, especially homeobox transcription factors, are also overrepresented in the Eset-bound gene list. Many homeobox transcription factors are known to regulate tissue-specific genes and play important roles in development (Shashikant et al. 1991). By analyzing publicly available expression data sets for ES cells, we found that Eset-bound target genes generally show lower expression than non-Eset-bound genes (Supplemental Fig. S7). This is consistent with the role of Eset as a transcriptional repressor.
Figure 2.
Genome-wide mapping of Eset in ES cells using ChIP-seq technology. (A) Distribution of Eset-binding sites. Locations of Eset-binding sites relative to the nearest Refseq genes. The percentages of binding sites at the respective locations are shown. (B) GO analysis of Eset-bound genes. Black bars represent the number of Eset-bound genes (observed) in the respective GO category. White bars represent the number of genes expected by chance. (C) Intersection of Eset-bound genes and genes that show Eset-dependent H3K9me3.
We postulate that Eset could suppress gene expression through the methylation of H3K9. To this end, we set out to identify the genomic regions that show Eset-dependent H3K9 trimethylation (H3K9me3). Chromatin extracts were prepared from ES cells transfected with control or Eset shRNA constructs. These extracts were subjected to ChIP using anti-H3K9me3 antibodies to enrich for nucleosomes with H3K9me3. By comparing these two ChIP-seq data sets, we identified genomic regions that show reduced H3K9me3 after Eset depletion (Supplemental Table 2). Fifty-five percent (1283) of the Eset-bound genes have reduced H3K9me3 after Eset knockdown, indicating that Eset is mediating H3K9me3 of these genes (Fig. 2C). Eset harbors H3K9me2 methyltransferase activity; however, our attempt to perform a ChIP-seq experiment was unsuccessful because the anti-H3K9me2 antibody is less efficient in enriching for H3K9me2.
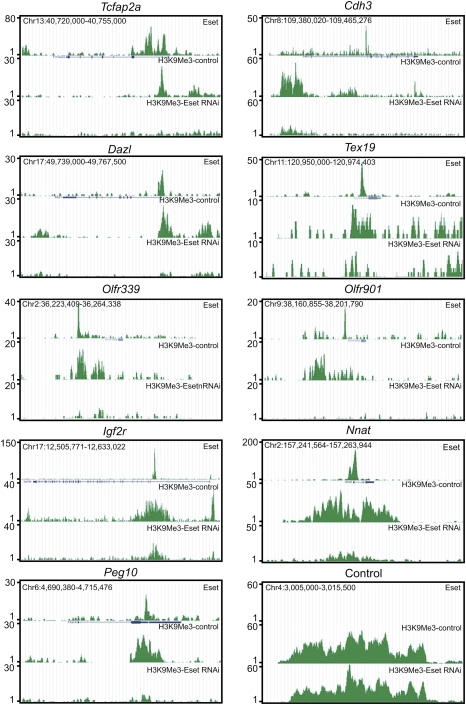
Eset occupancy and Eset-dependent H3K9me3 were observed at several genes expressed in the trophoblast lineages: Tcfap2a and Cdh3 (also known as placental cadherin) (Fig. 3). Other examples of Eset-bound genes involved in trophoblast development are Cdx2, Id2, and Eomes, but they do not show Eset-dependent H3K9me3. The binding of Eset to these genes suggests a functional link between this transcriptional repressor and trophoblast lineage regulation in ES cells (see later sections for further evidence).
Figure 3.
Eset and H3K9me3 profiles of genes. Eset and H3K9me3 profiles on the trophoblast lineage genes Tcfap2a and Cdh3, genes associated with germ cells Dazl and Tex19, olfactory receptor genes Olfr339 and Olfr901, and imprinted genes Igf2r, Nnat, and Peg10.
Interestingly, Eset binds to Dazl and Tex19.1 (Fig. 3). The expression of both genes is restricted to germ cells and pluripotent stem cells (Kuntz et al. 2008). Disruption of Dazl leads to a reduction in the number of primordial germ cells (Haston et al. 2009). The same study also shows that differentiation of mouse ES cells to germ cells is dependent on Dazl. Male Tex19.1-null mice exhibit impaired spermatogenesis (Ollinger et al. 2008). Our data suggest that Eset down-regulates these genes in ES cells.
In ES cells, imprinted loci are marked with H3K9me3, but not H3K27me3 (Mikkelsen et al. 2007). Of the 20 known and putative imprinted loci, we found Eset binding to 15 of them (Fig. 3; Supplemental Table 3). Among these Eset-bound imprinted genes, Peg10, H19, and Grb10 were induced upon Eset knockdown (Supplemental Fig. S8). Furthermore, eight of these genes show Eset-dependent H3K9me3, suggesting that they are targets of Eset. The silenced olfactory receptor genes (Olfr339 and Olfr901) are also targets of Eset (Fig. 3).
In summary, our genome-wide analysis of Eset-binding analysis uncovers novel putative Eset targets, and these data depict the interactions between a transcriptional repressor and the genome of ES cells.
Eset regulates the expression of trophoblast-associated genes Tcfap2a and Cdx2
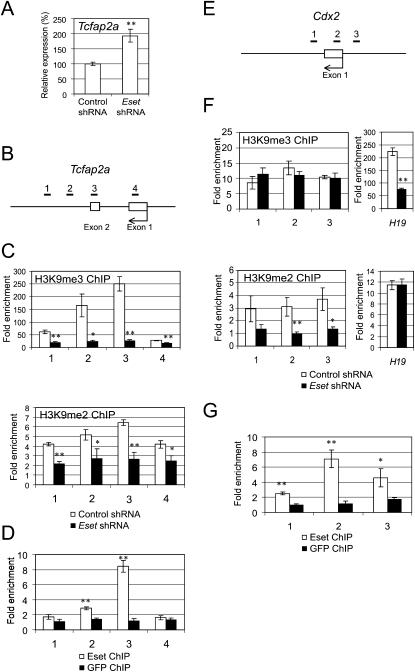
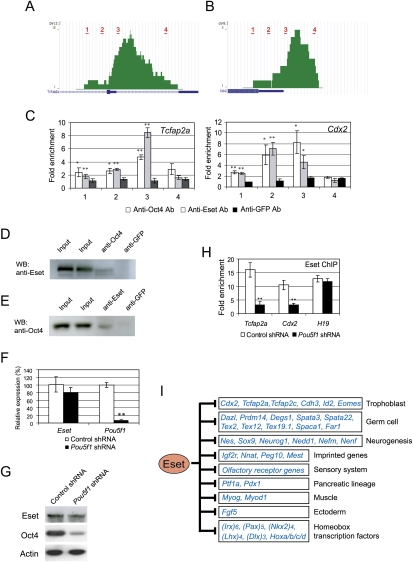
Next, we focused on dissecting the molecular basis of how Eset suppresses the expression of the trophoblast-associated genes. Tcfap2a, a member of the activating protein 2 (AP-2) transcription factor family, is preferentially expressed in the TE of the blastocyst (Winger et al. 2006). Tcfap2c, another member of the AP-2 family, is expressed throughout the preimplantation period and becomes restricted to the extraembryonic lineages at the time of implantation. Tcfap2c knockout studies revealed that it is essential in the extraembryonic lineages for early post-implantation development (Auman et al. 2002). Double knockout of Tcfap2a and Tcfap2c showed a more severe phenotype than individual knockout, indicating that these genes play redundant roles during the peri-implantation stage, presumably by affecting the trophoblast cell lineage (Winger et al. 2006). We confirmed that Tcfap2a expression is induced after Eset depletion (Fig. 4A; Supplemental Fig. S9). Depletion of Eset led to a reduction of H3K9me3 over the exon 2 region of Tcfap2a (Fig. 4B,C). This is consistent with our ChIP-seq results showing Eset-dependent H3K9me3 at this region. Eset also harbors H3K9me2 methyltransferase activity; however, as mentioned previously, we were unable to successfully enrich H3K9me2, as the enrichment is not efficient enough to subject to ChIP-seq analysis. To circumvent this, we performed a more sensitive ChIP-qPCR assay to quantitate the change in H3K9me2 and found that the H3K9me2 level is also reduced after Eset depletion (Fig. 4C, bottom panel). Hence, Eset regulates both H3K9me3 and H3K9me2 at Tcfap2a. Consistent with our ChIP-seq results (Fig. 3), we confirmed Eset occupancy at Tcfap2a (Fig. 4D). Our ChIP-seq data also show that Tcfap2c is bound by Eset. Similar to Tcfap2a, the level of Tcfap2c was also induced upon Eset knockdown, and the H3K9me3 and H3K9me2 levels were dependent on Eset (Supplemental Fig. S10). These results indicated that Tcfap2c is also a target of Eset.
Figure 4.
Eset regulates H3K9 methylation at Tcfap2a and Cdx2 loci. (A) Real-time PCR analysis of Tcfap2a expression after knockdown using Eset shRNA construct. Control shRNA targets luciferase sequence. The levels of the transcripts were normalized against control Luc shRNA. (B) Scheme of the amplicons (black bars labeled 1–4) used to analyze ChIP-enriched fragments over the Tcfap2a. (C) Eset regulates H3K9me3 and H3K9me2 levels of Tcfap2a. ChIP assays were performed using anti-H3K9me3 (top panel) or anti-H3K9me2 (bottom panel) antibodies with extracts prepared from Eset knockdown (black bars) and control knockdown (white bars) cells. (D) Eset binds to Tcfcp2a. ChIP assays were performed with Eset or control GFP antibodies, and DNA samples were measured by real-time PCR using primers targeting the amplicons shown in B. (E) Scheme of the amplicons (black bars labeled 1–3) used to analyze ChIP-enriched fragments over the Cdx2. (F) Eset regulates H3K9me2 level of Cdx2. ChIP assays were performed using anti-H3K9me3 (top panel) or anti-H3K9me2 (bottom panel) antibodies with extracts prepared from Eset knockdown (black bars) and control knockdown (white bars) cells. H3K9me3 at Cdx2 was not affected by Eset depletion, but H3K9me3 at H19 showed Eset-dependent H3K9me3. H3K9me2 level at Cdx2 was significantly reduced, but H3K9me2 level at the control H19 locus was not altered by Eset depletion. (G) Eset binds to Cdx2. ChIP assays were performed with Eset or GFP antibodies, and DNA samples were measured by real-time PCR using primers targeting the amplicons shown in E. All the data are represented as mean ± SD; n = 3. (*) P < 0.05; (**) P < 0.005.
Cdx2 encodes a caudal-related transcription factor expressed in the TE of blastocysts, and is essential for normal TE development (Strumpf et al. 2005). Cdx2 is bound by Eset (Supplemental Fig. S11), and its mRNA level was significantly increased after Eset knockdown (Fig. 1E; Supplemental Fig. S9) in ES cells. However, the H3K9me3 signal is not altered in Eset-depleted ES cells as compared with control ES cell at the loci (Supplemental Fig. S11). To explore this observation, we carried out a ChIP-qPCR assay with a series of primer pairs (Fig. 4E). With H3K9me3 ChIP, we confirmed our ChIP-seq data showing that there is no significant change in H3K9me3 level at the Cdx2 promoter after Eset knockdown, while the H19 locus showed Eset-dependent trimethylation (Fig. 4F). However, ChIP using an anti-H3K9me2 antibody showed a reduction of H3K9me2 after Eset depletion (Fig. 4F), indicating that Eset regulates H3K9me2, but not H3K9me3 at this site. As a control, we showed that the level of H3K9me2 at H19 locus was not altered by Eset depletion (Fig. 4F). The binding of Eset at Cdx2 is also confirmed by ChIP-qPCR (Fig. 4G). Hence, Tcfap2a and Cdx2 are direct targets of Eset.
Eset-depleted morula cells show a higher level of Tcfap2a and Cdx2 expression
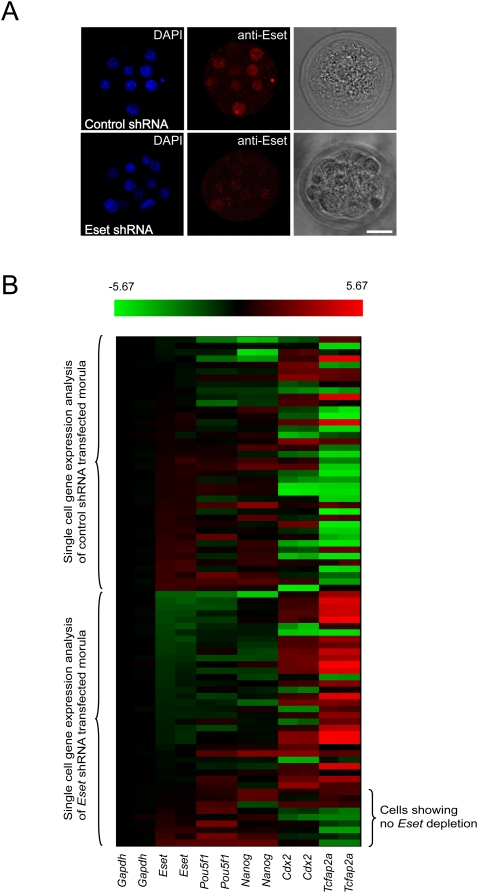
Eset is expressed during early embryogenesis, as measured by TaqMan real-time PCR assays (Supplemental Fig. S12A). The expression of Eset is ubiquitous at the blastocyst stage, and is expressed at the same level in both the ICM and TE (Supplemental Fig. S12B). These results are consistent with a previous study that measured reporter gene expression from the targeted Eset allele (Dodge et al. 2004). To address the role of Eset during early embryogenesis, we transfected constructs expressing Eset shRNA or control shRNA into both blastomeres of two-cell-stage embryos, and cultured these embryos until the morula stage. Immunofluorescence staining with Eset antibody confirms that Eset expression is significantly depleted (Fig. 5A). The blastocysts developed from Eset knockdown morulae showed defective ICM outgrowth, and this is consistent with the knockout phenotype of Eset-null blastocysts (Supplemental Fig. S13; Dodge et al. 2004). When we analyzed the whole morulae developed from Eset shRNA-transfected cells, we observed only a modest knockdown effect due to the depletion of Eset (data not shown). We reasoned that this may be due to incomplete knockdown of Eset in some of the cells of the morulae, making it difficult to detect robust changes at the whole-morula level. To get around this problem, we performed single-cell expression analysis (Jedrusik et al. 2008) on the Eset knockdown morulae. Single-cell expression analysis of the Eset knockdown morulae showed that the Eset level of some cells is not down-regulated, suggesting an explanation as to why it is difficult to observe robust changes in whole morulae. Overall, we found that Eset shRNA-treated morula cells show an elevated level of Tcfap2a and Cdx2 as compared with the control morula cells (Fig. 5B). Hence, these data suggest that Eset represses Tcfap2a and Cdx2 expression during early embryogenesis.
Figure 5.
Eset depletion induces TE-specific transcription factor expression in morula cells. (A) Eset expression is reduced in the Eset shRNA transfected morula as compared with control shRNA transfected morula. Bar, 20 μm. (B) Expression of ICM and TE lineage-specific transcription factors in control shRNA- or Eset shRNA-tranduced morula cells by single-cell qRT–PCR analysis. The heat map displays the mean centered cycling threshold (Ct) values after being normalized with Gapdh. The expression of ICM-specific genes Nanog and Pou5f1 was reduced, but TE-specific genes Cdx2 and Tcfap2a were induced in the Eset shRNA-transduced morula cell as compared with the control. Red indicates increased expression compared with control, whereas green means decreased expression.
Depletion of Eset at the two-cell stage leads to preferential incorporation of the cells into TE
As the depletion of Eset in two-cell embryos resulted in the up-regulation of Tcfap2a and Cdx2 at the morula stage (Fig. 5), we next addressed whether this will lead to preferential incorporation of Eset-depleted cells in the TE. To this end, we used an embryo aggregation assay described by Wakayama and colleagues (Kishigami et al. 2006). Eset shRNA or control shRNA constructs were transfected into two-cell embryos (Supplemental Fig. S14A). At the four-cell stage, nontreated and transfected embryos were aggregated and allowed to develop to the blastocyst stage in microwells. As the shRNA constructs harbor a GFP reporter, we were able to identify the transfected cells by following GFP fluorescence.
As expected, we found that the control shRNA-transduced totipotent cells can incorporate in both the ICM and TE. Interestingly, Eset shRNA-transduced cells were preferentially found in the TE (Supplemental Fig. S14B). From three independent experiments, 24 out of 32 blastocysts derived from Eset shRNA-transduced embryos show GFP expression only in TE; only eight blastocysts show both ICM and TE incorporation (Supplemental Fig. S14C). This compares with the control experiments, where all 34 blastocysts of control shRNA-tranduced embryo aggregates show GFP in both the ICM and TE. These results show that depletion of Eset at the two-cell stage leads to preferential incorporation of the cells into the TE.
Eset-depleted ES cells incorporate into the TE
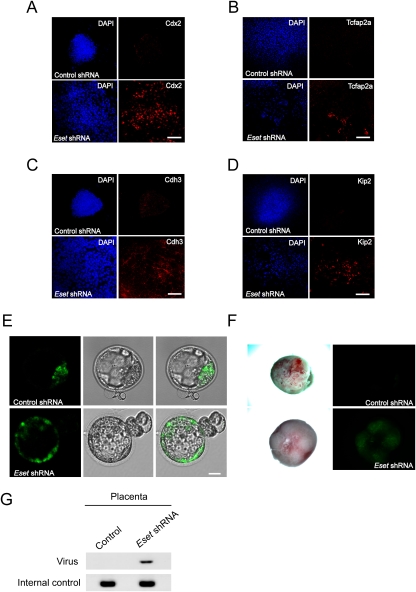
To further substantiate our finding that Eset depletion leads to the formation of trophoblast-like cells, we transduced ES cells with lentivirus expressing Eset shRNA or control shRNA. As we observed cell death after Eset knockdown (Fig. 1B, Supplemental Fig. S5), it is likely that the differentiated cells could not proliferate in culture medium for the propagation of ES cells. Therefore, after lentivirus transduction and isolation of GFP-positive cells by FACS, we maintained the cells under TS cell culture medium to increase the likelihood of the trophoblast-like cells surviving (Tanaka et al. 1998). After 5 d of culture in TS cell growth condition on mouse embryonic fibroblasts (MEF), we stained the cells for Cdx2, Tcfap2a, and another TE marker, Cdh3 (also known as placental cadherin) (Niwa et al. 2005). We could detect cells with Cdx2, Tcfap2a, and Cdh3 protein (Fig. 6A–C). After four more days in the same TS cell culture media, we stained the cells for Kip2, a trophoblast cell marker (Kalantry et al. 2006). Expression of Kip2 was detected in Eset-depleted cells (Fig. 6D). No Cdx2, Tcfap2a, Cdh3, or Kip2-positive cells were detected in ES cell lines transduced with control shRNA (Fig. 6A–D). Furthermore, we detected cells with trophoblast giant cell morphology (Supplemental Fig. S15A) and transcripts coding for trophoblast giant cell markers (Pl1, Pl2, and Plf) (Supplemental Fig. S15B). This indicates that a mixture of undifferentiated and differentiated trophoblast cells is present in the culture.
Figure 6.
Eset-depleted cells incorporate into TE. Immunofluorescence staining of Eset and control knockdown cells with anti-Cdx2 (A), anti-Tcfap2a (B), anti-Cdh3 (C), and anti-Kip2 (D) antibodies. ES cells were transduced with lentivirus expressing shRNA (Eset shRNA or control Luc shRNA) and GFP. Three days post-infection, FACS was used to isolate GFP-positive cells. The cells were then cultured in TS cell medium for 5–9 d before immunostaining. The cells were also stained with DAPI to locate the nuclei. Bars, 100 μm. (E) Eset or control knockdown cells were injected into four- to eight-cell-stage embryos. The embryos were then allowed to develop into blastocysts and were visualized with fluorescence microscopy to detect GFP-positive cells. For Eset knockdown cells, 25 out of 25 blastocysts showed TE incorporation. However, only one out of 22 blastocysts showed TE incorporation for the control knockdown cells. Bars, 20 μm. (F) Chimeric placental tissues were obtained from the Eset shRNA knockdown ES cells, but not from the control shRNA knockdown ES cells. GFP fluorescence images of the placental tissues were captured by fluorescence microscopy. (G) Genotyping of the placental tissues shown in E.
The induction of TS cell marker Cdx2 (Figs. 1E, 6A; Supplemental Fig. S3) upon Eset depletion suggests the presence of trophoblast progenitor cells. Hence, we compared the expression profile of Eset-depleted cells with that of TS cells (Supplemental Fig. S16). Among 159 genes preferentially expressed in TS cells, 104 of them were up-regulated, as revealed by our Eset knockdown microarray analysis. It is of interest to note that Gata3, which has been shown recently to be specifically expressed in the TE of blastocysts, is induced upon Eset depletion in ES cells (Home et al. 2009).
We were, however, unable to derive TS cells from Eset-depleted ES cells. The induction of trophoblast giant cell markers Pl1, Pl2, Plf, and Kip2 is indicative of the presence of differentiated trophoblasts (Fig. 6D; Supplemental Fig. S15). It is possible that Eset is required to maintain TS cells, and with long-term depletion of Eset in our experiments, the progenitor cells may differentiate. To test this hypothesis, we infected TS cells with lentiviruses expressing control or Eset shRNA. TS cell markers (Eomes and Cdx2) were reduced, and both the intermediate diploid trophoblast marker (Tpbpa) and trophoblast giant cell markers (Pl1, Pl2, and Plf) were induced (Supplemental Fig. S17A). Cells with giant cell-like morphology (large nuclei and expanded cytoplasm) and Kip2 expression were obtained in and Eset knockdown experiment but not in a control RNAi experiment (Supplemental Fig. S17B). This is consistent with the presence of differentiated trophoblast giant cells among the cells that originated from Eset-depleted ES cells cultured in TS cell medium (Supplemental Fig. S15). Overall, these data suggest that Eset may also be required to maintain the undifferentiated state of TS cells and potentially explain our futile attempts to derive TS cells from Eset-depleted ES cells. However, we cannot exclude the possibility that Eset-depleted ES cells give rise to trophoblasts without transiting through a TS cell stage.
To determine whether the Eset-depleted cells give rise to functional trophoblasts, the Eset shRNA or control shRNA-transduced cells were microinjected into four- to eight-cell-stage embryos to assay for incorporation into ICM and TE. As the lentiviruses carried a GFP expression cassette, we were able to trace the transduced cells by green fluorescence. Remarkably, Eset shRNA-transduced cells were incorporated into the TE for all 25 injected blastocysts. Although we could also detect GFP-positive cells in the ICM, this may be due to inadequate knockdown of Eset resulting in the appropriate maintenance of embryonic cells contributing to ICM (Fig. 6E). For control shRNA cells, only one out of 22 blastocysts showed TE incorporation. To test whether the Eset shRNA-transduced cells can contribute to placental tissues, these chimeric blastocysts were transferred to pseudopregnant foster mothers. GFP imaging, GFP staining, and genotyping results support the presence of transduced cells in the placental tissues (Fig. 6F,G; Supplemental Fig. S18). Overall, the results indicate that Eset depletion leads to a change in cell fate to form TE.
Oct4 recruits Eset to trophoblast-associated genes
As Oct4 is known to restrict TE fate in ES cells (Nichols et al. 1998; Niwa et al. 2000), we examined the binding profile of Oct4 at Tcfap2a and Cdx2 (Chen et al. 2008). The data show that Oct4 binds in close proximity to Eset-bound regions at these genes (Fig. 7A,B). The cobinding of Oct4 and Eset was further confirmed by ChIP-qPCR assay (Fig. 7C). Furthermore, we also analyzed the overlap of Eset sites with our previously generated Oct4, Sox2, Nanog, and Suz12 ChIP-seq data sets (Chen et al. 2008). Eset shows statistically significant overlap with Oct4 and Suz12 data sets. However, Eset is not significantly cobound with Sox2 and Nanog (Supplemental Fig. S19). The induction of trophoblast-associated genes and cobinding of Oct4 and Eset prompted us to test the hypothesis that Oct4 could interact with Eset. To this end, we performed coimmunoprecipitation experiments using ES cell extracts. Oct4 was associated with Eset using Oct4 and Eset antibodies (Fig. 7D,E). To demonstrate that Oct4 is required for Eset recruitment in living ES cells, we down-regulated Pou5f1 by RNAi and examined Eset occupancy by ChIP. At the 1-d time point after shRNA-mediated Pou5f1 knockdown, we did not observe significant reduction in the Eset transcript and protein levels (Fig. 7F,G). However, Eset binding at Tcfap2a and Cdx2 was reduced dramatically, while Eset occupancy at H19 locus was not affected (Fig. 7H).
Figure 7.
Oct4 recruits Eset to Tcfap2a and Cdx2. (A) Screen shots of the genome browser showing Oct4 ChIP-seq profiles at Tcfap2a locus. Red bars show the locations of amplicons used for ChIP-qPCR assay. (B) Screen shots of the genome browser showing Oct4 ChIP-seq profiles at the Cdx2 locus. Red bars show the locations of amplicons used for the ChIP-qPCR assay. (C) ChIP assays were performed using anti-Oct4, anti-Eset, or anti-GFP antibodies. Quantitative real-time PCR analyses of ChIP samples were carried out using primer pairs for Tcfap2a and Cdx2 loci. The locations of the amplicons are shown in A and B. Fold enrichment represents the abundance of enriched DNA fragments over a control region. GFP ChIP served as mock ChIP. Data are presented as the mean ± SD; n = 3. (*) P < 0.05; (**) P < 0.005. (D) Eset interacts with Oct4 in ES cells. Anti-Oct4 or anti-GFP antibodies were used for immunoprecipitation and associated proteins were analyzed by Western blotting using an anti-Eset antibody. (E) Reverse coimmunoprecipitation experiment showing that Oct4 is present in anti-Eset immunoprecipitation samples but not control anti-GFP immunoprecipitation samples. (F) Analysis of Eset and Pou5f1 transcripts after Pou5f1 knockdown. A construct expressing Pou5f1 shRNA was transfected into ES cells for 1 d, and cells were harvested for real-time PCR analysis of the transcripts. Data are represented as mean ± SD; n = 3. (**) P < 0.005. (G) Analysis of Eset and Oct4 after Pou5f1 knockdown. A construct expressing Pou5f1 shRNA was transfected into ES cells for 1 d and cells were harvested for Western blot analysis for Oct4, Eset, and Actin. (H) Eset occupancy at Tcfap2a, Cdx2, and H19 in Pou5f1 knockdown and control ES cells was analyzed by ChIP-qPCR. Data are represented as mean ± SD; n = 3. (**) P < 0.005. (I) A model to depict different classes of genes repressed by Eset.
In TS cells, Oct4 is absent but Cdx2, Tcfap2a, and Eset are expressed (Supplemental Fig. S20A). Hence, it is of interest to examine Eset occupancy and H3K9 methylations at Tcfap2a and Cdx2 in TS cells. Our ChIP experiments revealed that Eset is not associated with these genes, and they showed only background levels of H3K9me3 and H3K9me2 (Supplemental Fig. S20B).
Taken together, we conclude that Oct4 recruits Eset to mediate H3K9 methylation and transcriptional repression of Cdx2 and Tcfap2a in ES cells.
Discussion
Restriction of trophoblast lineage potential in ES cells
There are only a few circumstances when ES cells can be directed toward the TE fate. First, the removal of Oct4 triggers differentiation of ES cells into TE (Nichols et al. 1998; Niwa et al. 2000). Hence, Oct4 is implicated as a gatekeeper with a function to override the TE fate (Pesce and Scholer 2001). Second, ectopic expression of Cdx2 can induce ES cell to differentiate into the trophoblast lineage (Niwa et al. 2005; Tolkunova et al. 2006). Elevated expression of Cdx2 suppresses the expression of Pou5f1 through the direct binding to an Oct-Sox element and possibly interferes with the activation function of Oct4 at this site (Niwa et al. 2005). On the other hand, Oct4 is known to bind to the Cdx2 gene and repress its expression (Boyer et al. 2005; Niwa et al. 2005; Loh et al. 2006). A reciprocal inhibition model has been proposed to explain the mutual exclusion of Oct4 and Cdx2 to specify extraembryonic and embryonic cell fate in the ES cells and blastocysts. Third, activation of Ras can also direct ES cells to switch to a trophoblast fate (Lu et al. 2008). It is not clear, however, how the Ras–MAPK signaling influences the key transcriptional regulators to bring about the switch in cell fate. Fourth, it has been shown recently that Dnmt1-null ES cells differentiate into trophoblast lineage under TS cell culture conditions (Ng et al. 2008). In the absence of DNA methylation, Elf5 becomes preferentially induced under culture conditions that promote trophoblast formation. The up-regulation of transcription factor Elf5 in turn activates Cdx2. Hence, DNA methylation of the Elf5 promoter serves as a locking mechanism to prevent precocious induction of Cdx2.
In this study, our results demonstrate that Eset is important in restricting extraembryonic trophoblast lineage potential in ES cells. Recent studies, along with ours, show that Cdx2 is subjected to both genetic and epigenetic regulation, and these repressive mechanisms are required to maintain pluripotency of mouse ES cells (Boyer et al. 2005; Niwa et al. 2005; Loh et al. 2006). In addition to Cdx2, Eset also negatively regulates other genes, such as Tcfap2a and Tcfap2c. Both genes are implicated in the regulation of trophoblast cell lineage during early post-implantation development (Winger et al. 2006). Hence, Eset is likely to restrict extraembryonic trophoblast lineage potential in ES cells through repressing multiple genes involved in trophoblast lineage development. Eset is unlikely to function through Elf5 (Ng et al. 2008), as Elf5 is not induced and its promoter remains methylated upon Eset depletion (data not shown). Although we show evidence to link Eset to the regulation of extraembryonic lineage, our study does not exclude the possibility that Eset also regulates differentiation to other lineages. For example, genes associated with other lineages (Msx1 and Fgf5) are induced upon Eset down-regulation.
The roles of H3K9 methyltransferases during development and in ES cells
Several of the H3K9 methyltransferase genes (Suv39h1, Suv39h2, GLP, G9a, and Eset) have been disrupted in mouse embryos and ES cells. Suv39h1 and its closely related protein, Suv39h2, catalyze the formation of H3K9 trimethylation. Suv39h1 and Suv39h2 double-mutant embryos show a defect after embryonic day 12.5 (E12.5) (Peters et al. 2001; Lehnertz et al. 2003). Suv39h-null ES cells can be derived and propagated in culture. Hence, Suv39h methyltransferases are not required to maintain ES cell identity. They are, however, required to direct H3K9me3 at pericentric repeats (Lehnertz et al. 2003).
G9a regulates H3K9 mono- and dimethylation at the euchromatin, and G9a-null embryos die at E9.5 (Tachibana et al. 2002). GLP is a structurally and functionally related protein of G9a, and these two proteins form a stoichiometric heteromeric complex (Tachibana et al. 2005). Similar to G9a, GLP-null embryos die at E9.5 (Tachibana et al. 2005). Both G9a and GLP-null ES cells are viable and can be maintained like wild-type ES cells. As a result of reduction in H3K9 mono- and dimethylation at the euchromatic regions in these mutant ES cells, HP1 localization at euchromatin sites is affected. Hypomethylation also leads to derepression of Mage-a genes in these mutant ES cells. It is also of interest to note that G9a-null ES cells are defective in growth under conditions that promote differentiation (Tachibana et al. 2002). These results suggest that G9a plays a more important role in differentiated cells than in pluripotent cells. Indeed, G9a has been shown to mediate H3K9 methylation of Pou5f1 when ES cells are induced to differentiate (Feldman et al. 2006). The heterochromatinization of the Pou5f1 promoter is a key mechanism to providing tight silencing of this pluripotency gene in non-ES cells.
Among these histone H3K9 methyltransferases, only Eset has been shown to play a critical role at an earlier stage of development. Homozygous deletion of Eset results in peri-implantation lethality between E3.5 and E5.5 (Dodge et al. 2004). Eset-null blastocysts show defective ICM outgrowth, and ES cells cannot be derived from these blastocysts. As we show in this study, Eset is unique among the H3K9 methyltransferases in that it is required to maintain ES cell identity and control lineage decision. Hence, the different members of the H3K9 methyltransferases play different roles during development and in ES cells.
Interface between genetic and epigenetic networks
Using a ChIP-seq approach, we mapped the whole-genome Eset-binding profiles in ES cells. Apart from genes involved in the trophoblast lineage, it is also striking that genes implicated in germ cell biology and imprinted genes are bound by Eset (Fig. 7I). Similar to the case of polycomb target genes, many of the Eset-bound genes encode for developmentally regulated transcription factors (Boyer et al. 2006; Lee et al. 2006). The transcriptionally silenced Eset target genes may also be poised for activation in the right cell types.
We and others previously mapped the regulatory networks governed by the core ES cell transcription factors Oct4, Sox2, and Nanog (Boyer et al. 2005; Chen et al. 2008; Kim et al. 2008). Co-occupancy by this transcription factor trio is one of the key features of the ES cell transcriptional regulatory network. A proportion of these sites are associated with the polycomb repressor complexes that mediate H3K27 methylation (Lee et al. 2006; Endoh et al. 2008). Our genome-wide Eset location analysis also reveals that Eset cobinds a small fraction of the Oct4 sites. These cobound sites are points of integration of the Eset and Oct4 regulatory networks. This interface can be explained by the biochemical interactions between Eset and Oct4. Importantly, we demonstrate that the recruitment of Eset to several target genes is also dependent on Oct4. At other non-Oct4 cobound sites, the recruitment of Eset is presumably mediated through other sequence-specific DNA-binding transcription factors. We showed previously that Oct4 preferentially up-regulates histone demethylases that remove repressive H3K9 methylations (Loh et al. 2006). These histone demethylases are involved in preventing the accumulation of repressive H3K9 methylations at genes required for self-renewal of ES cells. The present study provides a distinct mechanism for how Oct4 interacts with a histone H3K9 methylase to maintain pluripotency.
We also observed a statistically significant overlap between Eset- and Suz12-bound sites. This suggests that the two histone methyltransferase complexes may function cooperatively to silence certain target genes through H3K9 and H3K27 methylations. The two repressive mechanisms may be required to provide multiple layers of gene silencing in ES cells.
In conclusion, our results demonstrate that Eset is important in restricting extraembryonic trophoblast lineage potential in ES cells, and implicate Eset as a novel epigenetic regulator with a key role in determining the first lineage segregation. Our findings also indicate that Oct4 uses an epigenetic regulator to coordinately silence genes involved in TE maintenance or development through modulating the repressive H3K9 methylation. This represents a novel mechanism to suppress extraembryonic trophoblast lineage potential by a key pluripotency transcription factor.
Materials and methods
Cell culture and transfection
Feeder-free E14 mouse ES cell culture and transfection were maintained as described previously (Loh et al. 2007), except 0.8 μg/mL puromycin (Sigma) was added to the medium for selection after transfection. Cells were maintained for 5 d prior to harvesting. The rescue assay was conducted the same as described (Jiang et al. 2008), except 2 μg of shRNA construct were cotransfected with 2 μg of pCAG-RNAi-immune Eset constructs, and cells were maintained in normal medium supplemented with 0.8 μg/mL puromycin and 1 μg/mL hygromycin.
RNAi assay
shRNA constructs were designed as described previously (Jiang et al. 2008). Two shRNA constructs for Eset were designed to target 19-base-pair (bp) transcript-specific regions. The sequences targeted by the shRNAs are as follows: Eset shRNA1, GAACCTATGTTTAGTATGA; Eset shRNA2, GTGGAAGTCTCGAGTTGAA. The control shRNA sequence was GATGAAATGGGTAAGTACA, which targets the luciferase gene. Total RNA was extracted with Trizol (Invitrogen) and purified with an RNeasy mini-kit (Qiagen). Reverse transcription was performed with 1 μg of total RNA using the SuperScript II kit (Invitrogen). Real-time PCR analysis was performed by using the ABI Prism 7900HT machine (Applied Biosystems) with the SYBR Green mixture. For each primer, only one correct size band was formed. All experiments were repeated at least three times with different batches of ES cells. The final results were normalized against β-actin expression.
Expression DNA microarray analysis
mRNAs derived from Eset shRNA1 and control Luc shRNA-treated ES cells were reverse-transcribed, labeled, and analyzed using the Illumina microarray platform (Sentrix Mouse-6 Expression BeadChip version 1.1). Arrays were processed as per the manufacturer's instructions. Three biological repeats of the profiles were used to generate statistically significant gene lists. Significance analysis of microarrays (SAM) was used to select differentially expressed genes. The thresholds set for the differently expressed genes were a >1.5-fold change and a q-value <0.05. Data for the microarray experiments can be found under GEO accession number GSE17439.
ES cell replating assay
After 3 d of puromycin selection, shRNA-transfected ES cells were trypsinized and resuspended in medium. Ten-thousand cells were plated onto gelatin-coated 60-mm plates for colony formation assay. After 5 d, emerging colonies were stained for alkaline phosphatase activity to characterize ES colonies and differentiated cell colonies.
Generation of Eset antibodies
cDNA encoding the amino acids 401∼613 of Eset was cloned into pET42b (Novagen) for the production of His-tagged fusion proteins. The recombinant proteins were produced in Escherichia coli (BL21) after IPTG induction and purified with Ni-NTA-sepharose (Qiagen) columns. The purified antigens were then used to immunize rabbits for polyclonal antibody production.
ChIP assay
ChIP assay was carried out as described previously (Jiang et al. 2008). Briefly, the cells were cross-linked with 1% (w/v) formaldehyde for 10 min at room temperature, and formaldehyde was then inactivated by the addition of 125 mM glycine. Chromatin extracts containing DNA fragments with an average size of 500 bp were immunoprecipitated using anti-Oct4 (sc-8628, Santa Cruz Biotechnologies), anti-H3K9Me2 (ab1220, Abcam), anti-H3K9Me3 (ab8898, Abcam), anti-Eset Ab1 (antibody against amino acids 401∼613 of mouse Eset raised in rabbit), or anti-GFP (sc-9996, Santa Cruz Biotechnologies) antibodies. The ChIP-enriched DNA was then decross-linked and analyzed by real-time PCR using the ABI PRISM 7900 sequence detection system and SYBR Green master mix. Relative occupancy values (also known as fold enrichments) were calculated by determining the immunoprecipitation efficiency (ratios of the amount of immunoprecipitated DNA to that of the input sample) and were normalized to the level observed at control regions, which was defined as 1.0. The coordinates for the control region 1 was chr6: 123,352,993–123,353,158 (mm5 genome build), and the control region 2 was chr5: 140,388,587–140,388,755. For all the primers used, each gave a single product of the right size.
Analysis of ChIP-seq data sets
Peak calling based on the Eset ChIP-seq data (11,159,322 uniquely mapped tags) was performed using MACS (Zhang et al. 2008) with a P-value cutoff of 1e-12. This resulted in 4633 peaks. The control RNAi H3K9me3 ChIP-seq library contained 12,920,863 uniquely mapped tags and the Eset RNAi H3K9me3 ChIP-seq library contained 10,391,825 uniquely mapped tags. We used the CCAT software (H Xu, in prep.) to find regions that were significantly depleted in H3K9me3 after Eset RNAi. CCAT was run in “region mode,” which detects changes over broader genomic regions, rather than the localized peaks detected as most peak callers do. We ordered the list of Eset-dependent H3K9me3-marked regions by fold change between control and Eset RNAi (corrected for sequencing depth). At the threshold of 2.5-fold change, we obtained 10,798 Eset-dependent H3K9me3 regions. The Eset-binding sites or H3K9me3 regions were assigned to each gene if they occurred within 50 kb from the transcription start site. As each gene may have multiple Eset-binding sites or H3K9me3 regions, the total number of genes associated with the binding site or histone modification is less than the number of identified sites or Eset-dependent H3K9me3. A total of 2355 genes are bound by Eset, and 4169 genes contain Eset-dependent H3K9me3 regions. We analyzed the enrichment of GO categories using the PANTHER database (Gene Expression tools). We determined the over- or underrepresentation of PANTHER classification categories using binomial statistics with Bonferroni correction for each molecular function, biological process, or pathway term in PANTHER. Data for the ChIP-seq experiments can be found under GEO accession number GSE17642.
Cobinding analysis
We assessed the overlap of the 4633 Eset peaks with other transcription factors (Oct4, Sox2, Nanog, and Suz12) in mouse ES cells by intersecting the peak list with data from a previous study (Chen et al. 2008). We allowed up to 200 bp between the borders of two peaks. Instead of assessing overrepresentation by comparing the observed overlaps with overlaps with random regions, we used a control library generated from sequencing input DNA. This was done to partially correct for possible bias due to uneven fragmentation and read-mapping. We used a nonstringent P-value threshold (1e-3) in MACS to call peaks, and then randomly selected 40,000 of these peaks. Overlaps to these regions were used as a baseline control for each transcription factor. The observed number of overlaps between the transcription factor and Eset was then compared with this baseline, and statistical significance of Eset enrichment for each transcription factor was calculated by using Fisher's exact test.
Generation of Eset knockdown cells from ES cells
Eset shRNA or control shRNA oligos were cloned into the EcoRI and ClaI sites of the pLVTH lentivirus vector (Plasmid 12262, Addgene). This plasmid harbors the GFP reporter, and GFP can be used to trace the infected cells. Lentivirus expressing Eset shRNA or control shRNA was generated and used to infect ES cells. GFP-positive cells were sorted 3 d after lentivirus infection with BD FACSAria cell sorter (BD Bioscience).
Immunofluorescence microscopy
Cells were fixed in 4% paraformaldehyde and permeabilized with 0.5% Triton X-100, followed by blocking with 1% BSA in PBS. The samples were then stained with anti-Cdh3 antibody (Clone56C1, Neomarkers) at 1:200, anti-Cdx2 antibody (CDX2-88, BioGenex) at 1:200, anti-Tcfap2a (SC-184, Santa Cruz Biotechnologies) at 1:100, P57Kip2 (RB-1637-P, Neomarker), anti-Nanog (RCAB0002P-F, Cosmo Bio) at 1:50, or anti-SSEA-1 (SC-21702. Santa Cruz Biotechnologies) at 1:200. The primary antibodies were detected with the appropriate secondary antibodies conjugated with Alexa Fluor 546 (Molecular Probes). Images were captured with a confocal microscope (inverted LSM 5 DUO system, Zeiss).
TE incorporation and embryo chimera assays
Forty-seven four- to eight-cell-stage embryos were collected from FVB female mice after mating. Eight to 10 Eset shRNA-treated GFP-positive ES cells or control shRNA-treated GFP-positive ES cells were injected into the embryos by Piezo Micro Manipulator (PMAS-CT150, PMM) under a fluorescent microscope (Olympus). The embryos were then cultured in KSOM medium until the blastocyst stage. To localize the incorporation of the GFP-positive cells within the embryos, the fluorescence images were captured with a confocal microscope (inverted LSM 5 DUO system; Zeiss). Some blastocysts were further transferred to the uterus of a pseudopregnant female to check the incorporation of GFP-positive cells.
Gene expression profiling of preimplantation embryos and dissected TE and ICM cells
TE cells and ICM cells were dissected manually from the E4.5 embryo under a dissection microscope using a glass knife. Total RNA was extracted from individual embryos or dissected tissues using the PicoPure RNA isolation kit (Arcturus Bioscience). The entire RNA preparation was used for cDNA synthesis for 2 h at 37°C using the high-capacity cDNA archive kit (Applied Biosystems). An eighth of each cDNA was preamplified for genes of interest by 16 cycles of amplification (each cycle: 15 sec at 95°C and 4 min at 60°C) using the TaqMan PreAmp Master Mix Kit (Applied Biosystems). The preamplified products were diluted 10-fold before analysis. Real-time reactions were performed in technical duplicate with master mix (Applied Biosystems) in a 48.48 Dynamic Array on a BioMark System (Fluidigm). Threshold cycle (Ct) values were calculated from the system's software (BioMark Real-time PCR Analysis).
Single-cell gene expression analysis
Single-cell gene expression analysis was carried out as described in Jedrusik et al. (2008). The zona pellucidae of the two-cell-stage embryos was removed by brief exposure to acid Tyrode's solution and then transfected with Eset shRNA or control shRNA overnight using Lipfectamine 2000 in M2 medium. Thereafter, the embryos were transferred to fresh KSOM medium and cultured until morula stage. The morula was then treated with trypsin-EDTA for 10 min at 37°C and further dissociated into single cells by mouth pipetting. Single cells were transferred to a PCR tube by a mouth pipette for reverse transcription and preamplification by using CellDirect one-step qRT–PCR kit (SKU no.11754-100, Invitrogen) with TaqMan primers as described (Diehn et al. 2009). The resulting amplified cDNA from each cell was diluted five times and loaded to sample inlets of a biomark chip with TaqMan q-PCR mix (Applied Biosystem). Primer assays were inserted into the assay inlet with a duplicate. The chip was then loaded for 1 h in a chip loader (Nanoflex, Fluidigm), and then transferred to the thermocycle (Biomark, Fluidigm) for fluorescent quantification. The results presented were obtained from three independent experiments. The Ct value of each reading was first normalized with Gapdh to get ΔCt. The ΔΔCt between ΔCt of individual assays and the mean ΔCt of all cells in the same assay was further plotted as a heat map with Mayday software.
Embryo aggregation assay
The two-cell-stage embryos with the zone pellucidae removed were transfected with Eset shRNA or control shRNA overnight using Lipfectamine 2000 in M2 medium. Untreated wild-type embryos were cultured in KSOM medium for aggregation. On the second day, the four-cell-stage treated embryos were selected for aggregation. The untreated embryos, which were also four-cell stage, were then removed from the zona pellucidae. Two untreated embryos and one transfected embryo were placed in KSOM medium in one small well of a plastic dish for aggregation (Kishigami et al. 2006). Twenty-four hours to 36 h after aggregation, the aggregates gave rise to a “giant” blastocyst. The blastocysts were then imaged with a confocal microscope (inverted LSM 5 DUO system, Zeiss) to localize the GFP-positive cells.
Coimmunoprecipitation assays
Immunoprecipitation assays were performed from whole-cell lysates of ES cells transfected with overexpression plasmids. Anti-GFP (Sc-8334 and Sc 5384, Santa Cruz Biotechnologies), anti-Eset, and anti-Oct4 (Sc-8628, Santa Cruz Biotechnologies) antibodies were used to pull down the protein complexes. Immunoprecipitated complexes bound by the above antibody were washed extensively with 0.3% digitonin wash buffer (50 mM Tris-HCl at pH 8, 150 mM NaCl, 1 mM EDTA, 0.3% digitonin, 10% glycerol plus Roche protease inhibitor cocktail). The interacting protein bands are resolved with 10% SDS-PAGE gel and transferred to the PVDF membrane, followed by detection with an appropriate primary antibody, an HRP-conjugated second antibody, and an ECL reagent.
Acknowledgments
We are grateful to Satoshi Tanaka for TS cells, and Ching-Aeng Lim, Kuee-Theng Kuay, Xiangling Ng, Fang Fang, Petra Kraus, and Junliang Tay for technical assistance. We thank Tara Huber, Jia-Hui Ng, Edwin Cheung, Thomas Lufkin, Masafumi Muratani, and Andrew Hutchins for critical comments on the manuscript and insightful discussion. We thank the Genome Technology and Biology group for sequencing. We thank Azim Surani and Rick Young for communicating unpublished results. We are grateful to the Biomedical Research Council (BMRC), Agency for Science, Technology, and Research (A*STAR) and Singapore Stem Cell Consortium for funding.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1831909.
Supplemental material is available at http://www.genesdev.org.
References
- Auman HJ, Nottoli T, Lakiza O, Winger Q, Donaldson S, Williams T. Transcription factor AP-2γ is essential in the extra-embryonic lineages for early postimplantation development. Development. 2002;129:2733–2747. doi: 10.1242/dev.129.11.2733. [DOI] [PubMed] [Google Scholar]
- Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, Yamada Y, Tanaka K, Torigoe K, Rauscher FJ., 3rd Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes & Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beddington RS, Robertson EJ. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–737. doi: 10.1242/dev.105.4.733. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O, Toyoda T, Otte AP, Okano M, Brockdorff N, Vidal M, et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Haston KM, Tung JY, Reijo Pera RA. Dazl functions in maintenance of pluripotency and genetic and epigenetic programs of differentiation in mouse primordial germ cells in vivo and in vitro. PLoS One. 2009;4:e5654. doi: 10.1371/journal.pone.0005654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Home P, Ray S, Dutta D, Bronshteyn I, Larson M, Paul S. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem. 2009 doi: 10.1074/jbc.M109.016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik A, Parfitt DE, Guo G, Skamagki M, Grabarek JB, Johnson MH, Robson P, Zernicka-Goetz M. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes & Dev. 2008;22:2692–2706. doi: 10.1101/gad.486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T. The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol. 2006;8:195–202. doi: 10.1038/ncb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishigami S, Wakayama S, Thuan NV, Ohta H, Mizutani E, Hikichi T, Bui HT, Balbach S, Ogura A, Boiani M, et al. Production of cloned mice by somatic cell nuclear transfer. Nat Protoc. 2006;1:125–138. doi: 10.1038/nprot.2006.21. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Kuntz S, Kieffer E, Bianchetti L, Lamoureux N, Fuhrmann G, Viville S. Tex19, a mammalian-specific protein with a restricted expression in pluripotent stem cells and germ line. Stem Cells. 2008;26:734–744. doi: 10.1634/stemcells.2007-0772. [DOI] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Loebel DA, Watson CM, De Young RA, Tam PP. Lineage choice and differentiation in mouse embryos and embryonic stem cells. Dev Biol. 2003;264:1–14. doi: 10.1016/s0012-1606(03)00390-7. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes & Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CW, Yabuuchi A, Chen L, Viswanathan S, Kim K, Daley GQ. Ras-MAPK signaling promotes trophectoderm formation from embryonic stem cells and mouse embryos. Nat Genet. 2008;40:921–926. doi: 10.1038/ng.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan P, Westbrook TF, Ottinger M, Pavlova N, Chang B, Macia E, Shi YJ, Barretina J, Liu J, Howley PM, et al. CDYL bridges REST and histone methyltransferases for gene repression and suppression of cellular transformation. Mol Cell. 2008;32:718–726. doi: 10.1016/j.molcel.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, Hemberger M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol. 2008;10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Ollinger R, Childs AJ, Burgess HM, Speed RM, Lundegaard PR, Reynolds N, Gray NK, Cooke HJ, Adams IR. Deletion of the pluripotency-associated Tex19.1 gene causes activation of endogenous retroviruses and defective spermatogenesis in mice. PLoS Genet. 2008;4:e1000199. doi: 10.1371/journal.pgen.1000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri SL, Peter W, Hess H, Scholer HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- Pesce M, Scholer HR. Oct-4: Gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Ralston A, Rossant J. Genetic regulation of stem cell origins in the mouse embryo. Clin Genet. 2005;68:106–112. doi: 10.1111/j.1399-0004.2005.00478.x. [DOI] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Rossant J. Lineage development and polar asymmetries in the peri-implantation mouse blastocyst. Semin Cell Dev Biol. 2004;15:573–581. doi: 10.1016/j.semcdb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Rossant J. Stem cells and early lineage development. Cell. 2008;132:527–531. doi: 10.1016/j.cell.2008.01.039. [DOI] [PubMed] [Google Scholar]
- Scholer HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- Shashikant CS, Utset MF, Violette SM, Wise TL, Einat P, Einat M, Pendleton JW, Schughart K, Ruddle FH. Homeobox genes in mouse development. Crit Rev Eukaryot Gene Expr. 1991;1:207–245. [PubMed] [Google Scholar]
- Smith AG. Embryo-derived stem cells: Of mice and men. Annu Rev Cell Dev Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes & Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes & Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Tolkunova E, Cavaleri F, Eckardt S, Reinbold R, Christenson LK, Scholer HR, Tomilin A. The caudal-related protein cdx2 promotes trophoblast differentiation of mouse embryonic stem cells. Stem Cells. 2006;24:139–144. doi: 10.1634/stemcells.2005-0240. [DOI] [PubMed] [Google Scholar]
- Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, Tempst P, Roeder RG, Zhang Y. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell. 2003;12:475–487. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Winger Q, Huang J, Auman HJ, Lewandoski M, Williams T. Analysis of transcription factor AP-2 expression and function during mouse preimplantation development. Biol Reprod. 2006;75:324–333. doi: 10.1095/biolreprod.106.052407. [DOI] [PubMed] [Google Scholar]
- Yang L, Xia L, Wu DY, Wang H, Chansky HA, Schubach WH, Hickstein DD, Zhang Y. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene. 2002;21:148–152. doi: 10.1038/sj.onc.1204998. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]