Abstract
Recent observations highlight that the mammalian genome extensively communicates with itself via long-range chromatin interactions. The causal link between such chromatin cross-talk and epigenetic states is, however, poorly understood. We identify here a network of physically juxtaposed regions from the entire genome with the common denominator of being genomically imprinted. Moreover, CTCF-binding sites within the H19 imprinting control region (ICR) not only determine the physical proximity among imprinted domains, but also transvect allele-specific epigenetic states, identified by replication timing patterns, to interacting, nonallelic imprinted regions during germline development. We conclude that one locus can directly or indirectly pleiotropically influence epigenetic states of multiple regions on other chromosomes with which it interacts.
Keywords: Transvection, imprinting, epigenetics, replication timing, H19 ICR
The transvection principle generally describes epigenetic consequences from interactions between chromatin fibers of homologous as well as heterologous chromosomes (Tartof and Henikoff 1991; Duncan 2002). Originally discovered in Drosophila, evidence for allelic transvection—i.e., between homologous chromosomes—is scant in mice (Rassoulzadegan et al. 2002) and humans (Liu et al. 2008), whereas the more elusive nonallelic transvection principle (Duncan 2002) has not been documented in mammals. It is now widely accepted that distant chromosomal loci physically interact to influence the expressivity of the genome. Thus, multiple enhancers may converge on a single gene promoter (Deschenes et al. 2007; Cockerill 2008), and a single enhancer can loop to multiple promoters (Tsytsykova et al. 2007). Regulatory elements from neighboring domains or from other chromosomes may interact to generate chromatin structures poised for transcription (Osborne et al. 2004; Cai et al. 2006; Göndör and Ohlsson 2006; Lomvardas et al. 2006; Simonis et al. 2006; Zhao et al. 2006; Zhou et al. 2006) or repression (Ameres et al. 2005; Lanzuolo et al. 2007; Valenzuela et al. 2008). However, it is not known to what degree epigenetic states of higher-order chromatin conformations contribute to such chromatin cross-talk and, above all, how they are regulated by physical interactions between chromatin fibers (Göndör and Ohlsson 2009b). To explore these issues in some detail, we report here the genome-wide pattern of interactions with the H19 imprinting control region (ICR) using a previously described technique, termed 4C (circular chromosome conformation capture) (Zhao et al. 2006). This region was chosen for in-depth 4C analysis because its parental alleles are epigenetically differentially marked (Tremblay et al. 1997); it has unique features, including regulation of gene expression both in cis (Thorvaldsen et al. 1998; Kanduri et al. 2000; Kurukuti et al. 2006) and in trans (Ling et al. 2006; Zhao et al. 2006); and deletion of the maternal allele predisposes to colon cancer (Sakatani et al. 2005) and facilitates parthenogenesis in mice (Kono et al. 2004).
Results and Discussion
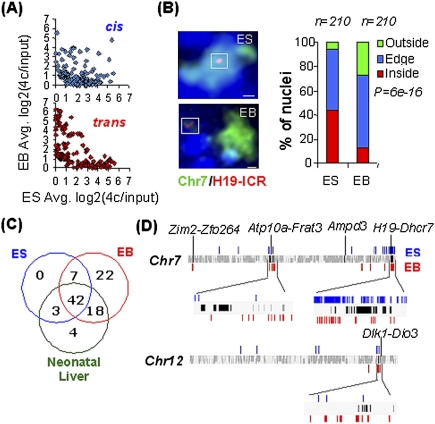
We previously performed a 4C analysis of H19 ICR-dependent networks to document epigenetic regulation of intra- and interchromosomal interactions (Zhao et al. 2006). The microarray analysis performed was limited to the 4C interactors identified by cloning and subsequent sequencing. To achieve a comprehensive genome-wide screen, 4C DNA samples of neonatal liver, neonatal brain, embryonic stem (ES), and derived embryoid body (EB) cells, all of mouse origin, were pooled and hybridized to tile path microarrays with a 100-base-pair (bp) resolution, covering the entire mouse genome. All sequences emerging as potential interactors were included in dedicated microarrays used to screen for patterns of long-range chromatin interactions during ES-to-EB cell differentiation. The scatter plots, displaying interaction frequencies in cis and trans, show a significant loss of intrachromosomal interactions when ES cells differentiate to EBs (P = 2.2956e-15) (Fig. 1A; Supplemental Fig. 1A). This may relate to the increased tendency of the bait to loop out from its chromosomal territory during ES cell differentiation (P = 6.015e-16) (Fig. 1B). The low biological variation in the ES and EB cell samples, each representing a pool of three independent samples (P = 0.8, P < 2.2e-16), is documented in the Supplemental Material (Supplemental Fig. 1A,B). Since EB cells represent all three germ layers, they are especially suited for identifying constitutive interactions present in many cell types. This strategy chiseled out a significant overrepresentation of known imprinted genes from 13 different chromosomes (false positive rate [FPR] = 6.5e-03). These interactions could also be confirmed in ES and neonatal liver cells, suggesting the widespread existence of physical networks of imprinted domains in several cell types independent of differentiation status (Fig. 1C) and expression levels of the involved loci (Supplemental Fig. 2). While the precise points of interaction can change during ES cell differentiation, they remained within the imprinted domains, as exemplified for four different imprinted clusters (Fig. 1D). Thus, overall epigenetic features associated with imprinted domains can be recognized by the H19 ICR in a diverse range of cell types.
Figure 1.
4C analysis of a genome-wide chromosomal network impinging in the H19 ICR. (A) Reprogramming of interactions during in vitro differentiation of ES cells. The scatter plots compare intra- and interchromosomal (cis and trans) interactions between ES and EB cells (P = 2.9e-15 for the difference in cis/trans interactions in ES and EB cells). (B) Difference in the location of the H19 ICR in relation to its chromosomal territory in ES cells and EBs. Chromosome 7 territory is shown in green, and Igf2/H19 is shown in red. Bar, 1 μm. (C) Significant overrepresentation of imprinted genes (out of 107 total) within 500 kb flanking the interactors (FPR = 6.5e-03, 104 randomizations). (D) Interactions at imprinted domains in ES cells and EBs identified at FDR = 0.05. Black bars represent the imprinted genes in the domain, while blue and red bars represent the location of interactors in ES and EB cells, respectively. Each domain is named as the first and last gene in the imprinted cluster shown. Magnified chromosomal regions are ∼3 Mb (Atp10a-Frat3), ∼2 Mb (H19-Dhrc7), and ∼1Mb (Dlk1-Dio3) in length. A detailed redundant list of interactors proximal to imprinted genes is given in Supplemental Table 4.
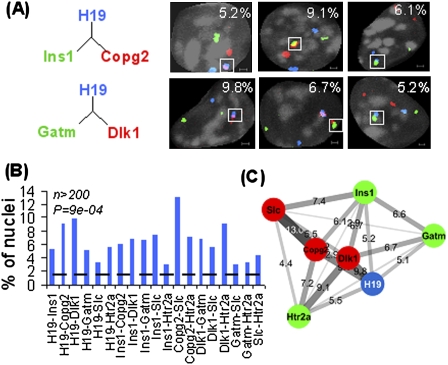
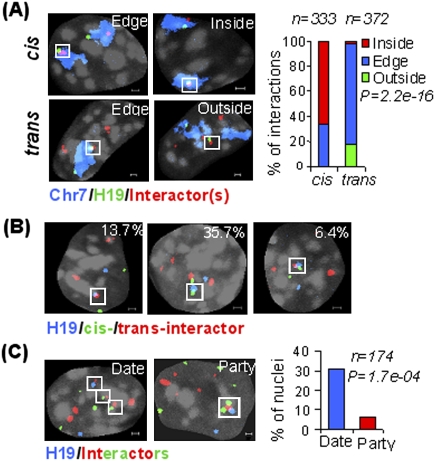
The extensive number of chromosomes physically impinging on the H19 ICR prompted an analysis of the dynamic pattern of interactions. As the 4C technique is a semiquantitative method at best for determining interaction frequencies (Göndör et al. 2008), we complemented this approach with DNA fluorescent in situ hybridization (FISH) analysis, which provides a more reliable indicator of interaction frequencies, although at a lower resolution. A maximum distance of 1 μm in three dimensions (3D) was used to define interacting loci in confocal microscopy analysis (Supplemental Fig. 3). Using this strategy, triple-color DNA FISH quantitative analysis of all-to-all interactions among imprinted loci from seven chromosomes in 3D (Supplemental Table 1) confirmed a widespread interaction network in ES cells (Fig. 2A–C). When we included a probe for chromosome 7 painting, we observed that cis interactions preferentially occurred inside the chromosome 7 territory containing the bait, while trans interactions mostly localized at the edge of the chromosome 7 territory (P = 2.2e-16) (Fig. 3A,B). To examine the prevalence of simultaneous interactions among imprinted regions, we mixed DNA FISH probes for all seven imprinted domains. Figure 3C shows that in 31% of the cases, imprinted genes interact in a pairwise manner that we term date interaction, and in 6% of the cases, more than two imprinted domains collide together to have party interaction. The lower than expected level of simultaneous interactions (6% observed vs. 16% expected, P = 1.72e-04) suggests that the interactions are highly dynamic, or that the juxtaposition of regions from several chromosomes is subject to restrictions in space. In this regard, we note that while the bulk of chromosome 7 is physically constrained, the distal region containing the bait is a highly mobile unit (Figs. 1B, 2A-C). These observations document that a particular locus in the genome can maintain extensive repertoires of cis and trans communications by its own dynamic movements.
Figure 2.
Quantitative in-situ analyses of trans chromosomal interactions. (A) Examples of nuclear staining of colocalized imprinted genes. Igf2/H19 is shown in blue, interactors are shown in red/green, and DAPI is shown in gray. All Z planes are merged together in these images; single-plane images are shown in Supplemental Fig. 6. Bar, 1 μm. (B) Frequencies of close proximity among imprinted loci. The dashed line shows the average interaction frequency for noninteracting controls. (C) Chromosomal network of imprinted loci. Length, thickness, and color gradient of each edge indicate corresponding frequency of close proximity.
Figure 3.
Dynamics of cis and trans interactions. (A) Preferred sites of cis and trans interactions as revealed by chromosome painting experiments. Chromosome 7 territory is shown in blue, Igf2/H19 is shown in green, cis/trans interactors are shown in red, and DAPI is shown in gray. (B) Frequency of cis (Atp10a in green) and trans (Dlk1 and Copg2 in red) interactions. (C) Date (pairwise) versus party (simultaneous) interactions. Igf2/H19 is shown in blue, while other trans imprinted loci are shown in red (Dlk1, Copg2, and Slc22a2) or green (Ins1, Gatm, and Htr2a). Bar, 1 μm.
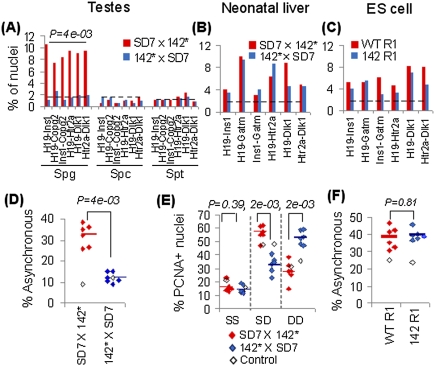
What happens to these chromosomal networks when the epigenetic states of imprinted domains are being reprogrammed? To answer this question, we turned our attention to male germline development. Supplemental Figure 4A shows that, in the mouse strains used in our studies, the paternal-specific methylation pattern of the maternal H19 ICR allele is acquired during spermatogonia-to-spermatocyte development concomitant with activation of BORIS, a CTCF paralog expressed primarily in spermatocytes (Loukinov et al. 2002). Moreover, the maternal H19 ICR allele is largely unmethylated while interacting with BORIS, as determined by bisulfite sequencing of BORIS chromatin-immunopurified DNA from adult mouse testis (Supplemental Fig. 4B). By these two criteria, the maternal-to-paternal switch in the epigenotype of the bait is linked with the spermatogonia–spermatocyte transition. This conclusion may reflect on the demonstration that the interactions between the Igf2/H19 domain and Copg2, Htr2a, Dlk1, and Ins1 could be observed in spermatogonia, but not in spermatocytes and round spermatids representing later stages of spermatogenesis when the reprogramming of the bait has been completed (Fig. 4A). We conclude that reprogramming the interactions between the imprinted bait and other imprinted domains overlaps with epigenetic reprogramming events during male germline development.
Figure 4.
Trans-regulation of epigenetic states by H19 ICR. (A) Frequencies of close proximity among imprinted domains in mouse spermatogonia (Spg), spermatocytes (Spc), and round spermatids (Spt). Cells were isolated from mice carrying a paternally (SD7 × 142*) or maternally (142* × SD7) inherited mutated H19 ICR allele defective in CTCF binding. The dotted line shows random control taken from noninteracting regions in our 4C screen. (B,C) Frequencies of close proximity in the neonatal liver of mouse crosses and ES cells (wild-type R1 and 142 R1 cells harboring a mutation in CTCF-binding sites within the maternally inherited H19 ICR allele; note that the 142 allele behaves the same as 142*, as illustrated in Supplemental Fig. 7). Dotted lines represents the noninteracting control. (D) Asynchronous replication timing pattern (percent SD) for spermatogonia cells from mouse crosses as above. Control is shown in open boxes and average values are shown in horizontal bars. (E) Replication timing pattern in PCNA-positive nuclei of neonatal liver. SS indicates two unreplicated/unseparated alleles, DD indicates two replicated and separated alleles, and SD represents one of each. (F) Asynchronous replication timing pattern in wild-type R1 and 142 R1 ES cells. More than 200 nuclei were counted for each instance in A–F. Genes analyzed are H19, Ins1, Copg2, Gatm, Htr2a, and Dlk1 in D, and H19, Ins1, Impact, Gatm, Htr2a, and Dlk1 in E and F.
To address the possibility that the “imprinted interactome” was directly or indirectly dependent on CTCF-binding sites within the H19 ICR, we analyzed the proximity between the Igf2/H19 and Gatm, Dlk1, Copg2, Htr2a, and Ins1 loci when a mutant form of H19 ICR (142* allele), unable to interact with CTCF (Pant et al. 2003), was inherited maternally. Figure 4A shows that the physical proximities between the imprinted domains and the mutated H19/Igf2 domain were absent in such spermatogonia, demonstrating that these interactions indeed are dependent on CTCF-binding sites during male germline development. This information prompted the question of whether the H19 ICR might also influence the interactions occurring between the other imprinted domains. To this end, we focused on the associations between Copg2 and Ins1 and Htr2 and Dlk1, interaction-pairs that typically excluded the Igf2/H19 locus when interacting with each other. Nonetheless, maternal inheritance of the 142* allele resulted in the loss of the Copg2/Ins1 and Htr2/Dlk1 associations (Fig. 4A), demonstrating that the wild-type maternal H19 ICR allele ascribes the interactions among other imprinted genes in pairwise manner in spermatogonia. We also conclude that the complete loss of interactions during male germline development reflects the inability of the paternal epigenotype to establish an “imprinted interactome.” Whether or not this means that the maternally inherited genome governs the physical network between imprinted chromatin fibers remains to be determined.
It is notable that the dependency of the physical closeness between imprinted domains on CTCF complexed to the maternal H19 ICR allele in spermatogonia is not paralleled in other cell types. Thus, the imprinted domains remained proximal to the Igf2/H19 domain when the mutant H19 ICR (142*) allele (Pant et al. 2003) was inherited from the mother in both neonatal liver and ES cells (Fig. 4B,C). These contrasting sets of data can be reconciled with the observation that the interaction network involves several contact points between entire imprinted domains and the H19 ICR in ES cells (Fig. 1D). By analogy, other regions within the imprinted domain that includes Igf2/H19 on the distal portion of chromosome 7 carrying maternal-specific epigenetic marks may be responsible for chromatin fiber interactions in somatic cells. This supposition is supported by the demonstration that the maternalization of the Igf2/H19 domain by a dysfunctional H19 ICR does not affect other parts of the entire imprinted domain on the distal part of chromosome 7 during female germline development (Cerrato et al. 2005).
To assess the functional outcome of these interaction patterns, we explored an epigenetic feature—i.e., asynchronous replication timing—commonly associated with imprinted genes (Simon et al. 1999). We documented previously that the replication timing of the maternally inherited Igf2/H19 domain switches from late to early in the absence of CTCF-binding sites within the H19 ICR (Bergstrom et al. 2007). It was thus possible that the H19 ICR might physically interact with other imprinted regions to influence their replication timing in cis and trans during a period when the bait is acquiring a maternal epigenotype. Due to the low amounts of spermatogonia available, replication timing was analyzed by DNA FISH. An as yet unreplicated allele for a certain gene is thus visualized by a single (S) dot, whereas replicated and separated alleles are displayed as doublets (D). The higher the proportion of SD cells, the more likely the examined genes replicate asynchronously. As this approach measures the endpoint of a replication process, it tends to exaggerate replication timing asynchrony estimates in unsynchronized cell populations to further enhance the sensitivity of the assay (Simon et al. 1999).
As expected for imprinted genes, each of the H19, Ins1, Cpog2, Gatm, Htr2a, and Dlk1 genes show a high proportion of SD signals—on average, 32%—in normal spermatogonia (Fig. 4D), indicative of asynchronous replication (Simon et al. 1999). Importantly, mutation of CTCF-binding sites on the maternal H19 ICR allele leads to a significant reduction, to <15% (P = 4e-03), in the number of spermatogonia with an SD signal at all examined imprinted loci (Fig. 4D). These data suggest that both parental alleles replicate synchronously in all of these instances. In contrast, a control region (Zbtb7c) that does not interact with the H19 ICR (dotted line in Fig. 4A) did not show any difference in its apparent replication timing pattern, irrespective of the (epi)genotype at the maternal H19 ICR (Fig. 4D). We conclude that CTCF-binding sites within the H19 ICR regulate the replication timing of the imprinted genes with which the H19 ICR interacts in cis and in trans during germline development. These data are consistent with the possibility that CTCF-binding sites within the maternal H19 ICR allele delay the reprogramming of the replication timing feature during male germline development. The interpretation that this inhibition to epigenetic reprogramming might normally be overcome by the activation of BORIS is supported by the demonstration that de novo methylation of the maternal H19 ICR allele is already initiated in the maternal allele interacting with BORIS (Supplemental Fig. 4B).
By extrapolation, CTCF complexed to the H19 ICR might both maintain and confer a maternal-specific late replication timing feature of the maternal and paternal alleles, respectively, during female germline development. Although the low number of oogonia in S phase precluded such an analysis, maternal, but not paternal, inheritance of the 142* allele led to a significant reduction of SD cells for all five imprinted genes in mouse neonatal liver (Fig. 4E). Moreover, this reduction in SD cells was completely paralleled by an increase in DD pattern among S-phase cells identified by PCNA staining, indicating that the late-replicating allele of the interacting imprinted gene replicates early in the absence of functional CTCF-binding sites within the H19 ICR in somatic cells. To establish whether this effect is germline-specific, we exploited the observation that the targeted allele in the 142 ES cell line (the ES cell clone used to generate the 142* mouse strain) was of maternal origin. Thus, bisulfite sequencing of the H19 ICR region in this ES cell clone revealed that the wild-type allele displayed a paternal epigenotype, largely methylated at its CpGs, while the mutated H19 ICR allele was largely unmethylated, indicating its maternal origin (Supplemental Fig. 5). Figure 4F shows that the asynchronous replication timing pattern was maintained for all imprinted domains, including the Igf2/H19 domain, despite the introduction of mutant CTCF-binding sites in the unmethylated H19 ICR allele. Thus, the CTCF-binding sites at the H19 ICR are essential for establishing the replication timing asynchrony during germline development, but not for its maintenance in cis and trans in ES cells.
We conclude that H19 ICR is a hub for the transvection of parent of origin-specific effects—i.e., replication timing patterns—to nonallelic loci on other chromosomes. Absence of any overrepresentation of CTCF-binding sites in the interacting sequences argues against cohesin being a major factor in stabilizing this network (data not shown). As these features require female germline transmission for their manifestation, defects in imprint resetting/acquisition during associated epigenetic reprogramming events may generate secondary epigenetic effects in the offspring via disruption of chromosomal networks. This scenario is underscored by the observation that epigenetic lesions in the H19 ICR can be linked with epigenetic lesions in other imprinted regions in pediatric cancer (Bliek et al. 2008). Interestingly, the only two regions known to prevent parthenogenesis, H19/Igf2 and Dlk1/Dio3 (Kawahara et al. 2007), interact in both the germline and somatic cells, and the replication timing pattern of Dlk1/Dio3 is regulated by the maternal H19 ICR allele, as shown here. The nonallelic transvection principle might also explain the spreading of imprinted states during mammalian radiation (Ohlsson et al. 2001). This notion is supported by the demonstration that the H19 ICR is the most ancient known ICR (Smits et al. 2008), and by the suggestion that replication timing asynchrony was an early epigenetic feature of imprinted domains (Cerrato et al. 2003) and was, perhaps, a vehicle to spread epigenetic features to replicons; i.e., clusters of genes (Göndör and Ohlsson 2009a). More generally, our observations provide a novel perspective on pleiotropic effects by epigenetic reprogramming events during development and disease potentially related to combinatorial repertoires of monoallelic expression of nonimprinted loci (Ohlsson 2007).
Materials and methods
ES cells and their differentiation into EBs
ES cell line R1 was cultured on a feeder layer of irradiated mouse fibroblast cell line STO, and in vitro differentiated into EBs during 12 d in the absence of leukemia inhibitory factor (LIF). Cells were directly harvested for 4C assays, as has been described (Zhao et al. 2006). For DNA FISH analysis, cells were attached to the chambered cover glass slide (Molecular Probes).
Testis and liver cell preparations
Testes were isolated from adult mice using the following strains and crosses: C57/Bl6, SD7 × 142*, and 142* × SD7 (in the order female × male). The 142* allele contains mutated CTCF-binding sites within the H19 ICR (Pant et al. 2003). Testes were macerated using scalpel blades in cold medium, and the resulting cell suspension was spotted on cover glass slides. Cells were covered in cold CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES, 0.5% Triton X-100, 1 mM EGTA at pH 6.8) for 10 min, and then in cold 4% paraformaldehyde (PFA) (pH 7–7.4) for 10 min. Freshly isolated neonatal liver cells (2 d post-partum) were dispersed in medium, and single-cell suspensions were spotted on cover slips. Cells were incubated for 30 min at 37°C to allow attachment, and fixed in 4% PFA for 10 min.
4C assay
The 4C assay was performed as described (see the Supplemental Material; Zhao et al. 2006; Göndör et al. 2008).
Three-dimensional DNA FISH
ES and EB cells were incubated on a chambered cover glass slide (Molecular Probes) for DNA FISH analysis, as has been described (see the Supplemental Material) (Zhao et al. 2006).
Immunofluorescence and immuno-FISH analysis
ES cell pluripotency markers were detected using the following antibodies: mouse antibody to SSEA-1 (MAB4301, Chemicon); goat antibody to Oct4 (ab27985, Abcam); rabbit antibody to Nanog (ab21603, Abcam). Immunodetection was combined with DNA FISH to ensure that the ES cells analyzed displayed markers for pluripotency. Anti-PCNA antibody (P8825, Sigma) was used to demarcate cells in S phase.
Methylation analysis
DNA methylation patterns of laser-mediated microdissection (see the Supplemental Material) of alkaline phosphatase-stained thin sections of adult mouse testis or chromatin-immunopurified DNA were performed using a previously described bisulfite protocol (Kerjean et al. 2001). Converted DNA was amplified by seminested PCR using the primers F1(2355), F2(2415), and R1(2748) (Tremblay et al. 1997). The purified PCR product was cloned into pGEMT-easy vector system (Promega) and sequenced using the Bigdye terminator cycle sequencing kit (Applied Biosystems).
Statistical analysis
NimbleGen tiling array data for 4C product was quantile-normalized across channels and further processed using the moving average method. Significantly enriched regions were identified at FDR = 0.05 using Bonferroni correction. Statistical tests for significance analysis were performed using R-package. A detailed list of statistical tests applied is given in Supplemental Table 3. The list of imprinted genes/domains was retrieved from https://atlas.genetics.kcl.ac.uk. The raw data for dedicated tiling array have been submitted to Gene Expression Omnibus (GEO) with the ID GSE14074.
Please see the Supplemental Material for a more detailed description of the methods.
Acknowledgments
Expert technical assistance from Anita Mattsson is gratefully acknowledged. This work was supported by the Swedish Science Research Council, the Swedish Cancer Research Foundation, the Swedish Pediatric Cancer Foundation, and the Lundberg Foundation, as well as HEROIC and ChILL (EU integrated projects).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.552109.
Supplemental material is available at http://www.genesdev.org.
References
- Ameres SL, Drueppel L, Pfleiderer K, Schmidt A, Hillen W, Berens C. Inducible DNA-loop formation blocks transcriptional activation by an SV40 enhancer. EMBO J. 2005;24:358–367. doi: 10.1038/sj.emboj.7600531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom R, Whitehead J, Kurukuti S, Ohlsson R. CTCF regulates asynchronous replication of the imprinted H19/Igf2 domain. Cell Cycle. 2007;6:450–454. doi: 10.4161/cc.6.4.3854. [DOI] [PubMed] [Google Scholar]
- Bliek J, Verde G, Callaway J, Maas SM, De Crescenzo A, Sparago A, Cerrato F, Russo S, Ferraiuolo S, Rinaldi MM, et al. Hypomethylation at multiple maternally methylated imprinted regions including PLAGL1 and GNAS loci in Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2008;17:611–619. doi: 10.1038/ejhg.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Cerrato F, Dean W, Davies K, Kagotani K, Mitsuya K, Okumura K, Riccio A, Reik W. Paternal imprints can be established on the maternal Igf2-H19 locus without altering replication timing of DNA. Hum Mol Genet. 2003;12:3123–3132. doi: 10.1093/hmg/ddg338. [DOI] [PubMed] [Google Scholar]
- Cerrato F, Sparago A, Di Matteo I, Zou X, Dean W, Sasaki H, Smith P, Genesio R, Bruggemann M, Reik W, et al. The two-domain hypothesis in Beckwith-Wiedemann syndrome: Autonomous imprinting of the telomeric domain of the distal chromosome 7 cluster. Hum Mol Genet. 2005;14:503–511. doi: 10.1093/hmg/ddi047. [DOI] [PubMed] [Google Scholar]
- Cockerill PN. NFAT is well placed to direct both enhancer looping and domain-wide models of enhancer function. Sci Signal. 2008;1:pe15. doi: 10.1126/stke.113pe15. [DOI] [PubMed] [Google Scholar]
- Deschenes J, Bourdeau V, White JH, Mader S. Regulation of GREB1 transcription by estrogen receptor α through a multipartite enhancer spread over 20 kb of upstream flanking sequences. J Biol Chem. 2007;282:17335–17339. doi: 10.1074/jbc.C700030200. [DOI] [PubMed] [Google Scholar]
- Duncan IW. Transvection effects in Drosophila. Annu Rev Genet. 2002;36:521–556. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- Göndör A, Ohlsson R. Transcription in the loop. Nat Genet. 2006;38:1229–1230. doi: 10.1038/ng1106-1229. [DOI] [PubMed] [Google Scholar]
- Göndör A, Ohlsson R. Replication timing and epigenetic reprogramming of gene expression: A two-way relationship? Nat Rev Genet. 2009a;10:269–276. doi: 10.1038/nrg2555. [DOI] [PubMed] [Google Scholar]
- Göndör A, Ohlsson R. Chromosome crosstalk in three dimensions. Nature. 2009b;461:212–217. doi: 10.1038/nature08453. [DOI] [PubMed] [Google Scholar]
- Göndör A, Rougier C, Ohlsson R. High-resolution circular chromosome conformation capture assay. Nat Protoc. 2008;3:303–313. doi: 10.1038/nprot.2007.540. [DOI] [PubMed] [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi C-F, Wolffe A, Ohlsson R, Lobanenkov A. Functional interaction of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- Kawahara M, Wu Q, Takahashi N, Morita S, Yamada K, Ito M, Ferguson-Smith AC, Kono T. High-frequency generation of viable mice from engineered bi-maternal embryos. Nat Biotechnol. 2007;25:1045–1050. doi: 10.1038/nbt1331. [DOI] [PubMed] [Google Scholar]
- Kerjean A, Vieillefond A, Thiounn N, Sibony M, Jeanpierre M, Jouannet P. Bisulfite genomic sequencing of microdissected cells. Nucleic Acids Res. 2001;29:E106. doi: 10.1093/nar/29.21.e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono T, Obata Y, Wu Q, Niwa K, Ono Y, Yamamoto Y, Park ES, Seo JS, Ogawa H. Birth of parthenogenetic mice that can develop to adulthood. Nature. 2004;428:860–864. doi: 10.1038/nature02402. [DOI] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Liu H, Huang J, Wang J, Jiang S, Bailey AS, Goldman DC, Welcker M, Bedell V, Slovak ML, Clurman B, et al. Transvection mediated by the translocated cyclin D1 locus in mantle cell lymphoma. J Exp Med. 2008;205:1843–1858. doi: 10.1084/jem.20072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, Chernukhin I, Mannan P, Larsson E, Kanduri C, Vostrov AA, et al. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc Natl Acad Sci. 2002;99:6806–6811. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R. Genetics. Widespread monoallelic expression. Science. 2007;318:1077–1078. doi: 10.1126/science.1150705. [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Paldi A, Graves JA. Did genomic imprinting and X chromosome inactivation arise from stochastic expression? Trends Genet. 2001;17:136–141. doi: 10.1016/s0168-9525(00)02211-3. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Pant V, Mariano P, Kanduri C, Mattsson A, Lobanenkov V, Heuchel R, Ohlsson R. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes & Dev. 2003;17:586–590. doi: 10.1101/gad.254903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M, Magliano M, Cuzin F. Transvection effects involving DNA methylation during meiosis in the mouse. EMBO J. 2002;21:440–450. doi: 10.1093/emboj/21.3.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakatani T, Kaneda A, Iacobuzio-Donahue CA, Carter MG, de Boom Witzel S, Okano H, Ko MS, Ohlsson R, Longo DL, Feinberg AP. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976–1978. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- Simon I, Tenzen T, Reubinoff BE, Hillman D, McCarrey JR, Cedar H. Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature. 1999;401:929–932. doi: 10.1038/44866. [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Smits G, Mungall AJ, Griffiths-Jones S, Smith P, Beury D, Matthews L, Rogers J, Pask AJ, Shaw G, VandeBerg JL, et al. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet. 2008;40:971–976. doi: 10.1038/ng.168. [DOI] [PubMed] [Google Scholar]
- Tartof KD, Henikoff S. Trans-sensing effects from Drosophila to humans. Cell. 1991;65:201–203. doi: 10.1016/0092-8674(91)90153-p. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes & Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay K, Duran K, Bartolomei M. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsytsykova AV, Rajsbaum R, Falvo JV, Ligeiro F, Neely SR, Goldfeld AE. Activation-dependent intrachromosomal interactions formed by the TNF gene promoter and two distal enhancers. Proc Natl Acad Sci. 2007;104:16850–16855. doi: 10.1073/pnas.0708210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L, Dhillon N, Dubey RN, Gartenberg MR, Kamakaka RT. Long-range communication between the silencers of HMR. Mol Cell Biol. 2008;28:1924–1935. doi: 10.1128/MCB.01647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- Zhou GL, Xin L, Song W, Di LJ, Liu G, Wu XS, Liu DP, Liang CC. Active chromatin hub of the mouse α-globin locus forms in a transcription factory of clustered housekeeping genes. Mol Cell Biol. 2006;26:5096–5105. doi: 10.1128/MCB.02454-05. [DOI] [PMC free article] [PubMed] [Google Scholar]