Abstract
Sterol regulatory element-binding proteins (SREBPs) are a subfamily of basic helix–loop–helix leucine zipper (bHLH-LZ) transcription factors that are conserved from fungi to humans and are defined by two key features: a signature tyrosine residue in the DNA-binding domain, and a membrane-tethering domain that is a target for regulated proteolysis. Recent studies including genome-wide and model organism approaches indicate SREBPs coordinate cellular lipid metabolism with other cellular physiologic processes. These functions are broadly related as cellular adaptation to environmental changes ranging from nutrient fluctuations to toxin exposure. This review integrates classic features of the SREBP pathway with newer information regarding the regulation and sensing mechanisms that serve to assimilate different cellular physiologic processes for optimal function and growth.
Keywords: INSIG, SCAP, SREBPs, cell–environment, nutrient fluctuation
Sterol regulatory element-binding proteins (SREBPs) define a distinct subclass of basic helix–loop–helix (bHLH) proteins
The HLH family of eukaryotic transcription factors was first identified through the cloning and analysis of the proteins E12 and E47, which dimerize to bind immunoglobulin enhancers (Murre et al. 1989). This functional protein domain was quickly recognized as a common structural motif that is present in many eukaryotic transcription factors (Murre et al. 1994). Many members of the HLH family also have a basic domain that extends outward from the N-terminal helix, and several also contain a leucine zipper motif on the C-terminal side that participates in dimerization (Simionato et al. 2007).
The basic domain makes sequence-specific contacts with palindromic recognition sites in DNA that have a core composition of 5′-CANNTG-3′. bHLH domain proteins typically bind a specific version of this “E-box” element with high affinity. Sequence-specific DNA recognition by individual family members is mediated through preferences for specific nucleotides in the middle and flanking positions immediately outside of the core (Blackwell et al. 1990; Murre et al. 1994).
The SREBPs comprise a subfamily of bHLH leucine zipper (bHLH-LZ) proteins that were initially identified as transcription factors for key genes of lipid metabolism and adipocyte differentiation (Tontonoz et al. 1993; Yokoyama et al. 1993). There are three mammalian SREBP isoforms: SREBP-1a and SREBP-1c are encoded from a single gene owing to differential promoter usage and alternative splicing, and SREBP-2 is encoded by a different gene (Osborne 2000). Based on a sequence alignment of a limited set of mammalian bHLH-LZ proteins, SREBPs were unique in having a tyrosine residue at a position that corresponded to an arginine in all of the other bHLH-LZ proteins that were compared (Kim et al. 1995). Studies also demonstrated that this tyrosine allowed SREBPs to bind not only E-box inverted repeats but also direct repeat variants called the sterol regulatory element (SRE), 5′-ATCACCCCAC-3′ (Kim et al. 1995), which was first identified in the promoter for the human LDL (low-density lipoprotein) receptor through mutational analysis to be a high-affinity SREBP-binding site (Dawson et al. 1988; Briggs et al. 1993). In comparing the crystal structure of an SREBP-1 homodimer bound to a DNA fragment containing the LDL receptor SRE to the crystal structure of a heterodimer containing the bHLH-LZ domains from Myc and Max bound to a canonical E-box element, it was determined that the signature tyrosine residue provided structural flexibility to the SREBPs, allowing the basic domain to fold into a slightly different conformation that was critical for high-affinity contacts with the LDL receptor promoter SRE (Parraga et al. 1998).
A second defining feature of SREBPs is the large C-terminal half of the protein that contains two closely spaced membrane-spanning helices that line up in the endoplasmic reticulum (ER) membrane in a hairpin configuration. This juxtaposes the N-terminal domain containing the transcriptional activation and DNA-binding domains with the C-terminal regulatory domain on the cytoplasmic side of the membrane (Brown and Goldstein 1999). In SREBPs from yeast to humans, the C-terminal domain interacts with the SREBP cleavage-activating protein (SCAP), which is an escort protein that carries SREBPs from the ER to the Golgi apparatus in response to metabolic regulatory cues (Hughes et al. 2005). Once in the Golgi, the SREBPs are released from the membrane through a sequential two-step proteolytic process discussed in more detail below.
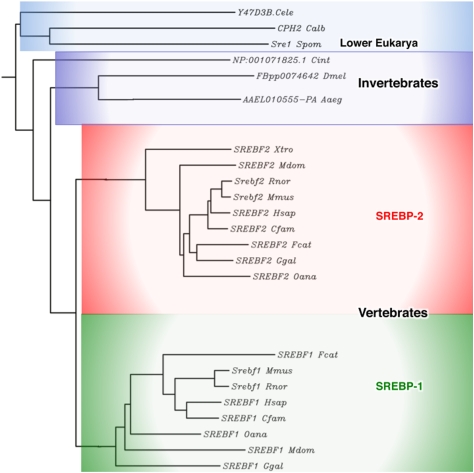
Using the signature tyrosine and membrane attachment domain as key identifiers, a phylogenetic analysis demonstrates that there are predicted SREBP homologs across the eukarya (Fig. 1; Table 1). There is a single SREBP-related gene in fungi, Caenorhabditis elegans, and all invertebrates. A simple interpretation of the analysis suggests that a single gene duplication event occurred in vertebrates after separating from invertebrates during evolution. This is interesting, because in mammals, SREBP-2 preferentially activates genes of cholesterol metabolism, whereas SREBP-1 preferentially activates genes of fatty acid metabolism, and most if not all invertebrates are cholesterol auxotrophs because they lack key enzymes of the sterol biosynthetic pathway (Karlson 1970). Because of the preferential roles for mammalian SREBP-1 and SREBP-2 in fatty acid and cholesterol metabolism, respectively (Horton et al. 2002), it is tempting to speculate that the gene duplication occurred at a time when it was necessary to independently regulate fatty acid and cholesterol biosynthesis and metabolism.
Figure 1.
SREBP evolution. An evolutionary tree was constructed for the SREBP homologs from different species. Only a subset of species within each major category is shown for simplicity. (Cele) Caenorhabditis elegans; (Calb) Candida albicans; (Spom) Schizosaccharomyces pombe; (Cint) Ciona intestinalis; (Dme) Drosophila melanogaster; (Aaeg) Aedes aegypti; (Xtro) Xenopus tropicalis; (Mdom) Musca domesticus; (Rnor) Rattus norvegicus; (Mmus) Mus musculus; (Hsap) Homo sapiens; (Cfam) Canis familiaris; (Fcat) Felis catus; (Ggal) Gallus gallus; (Oana) Ornithorhynchus anatinus.
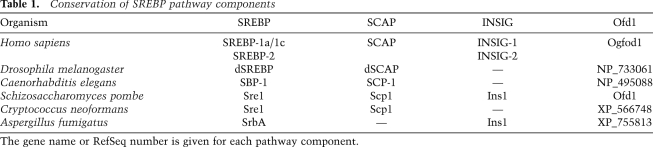
Table 1.
Conservation of SREBP pathway components
The gene name or RefSeq number is given for each pathway component.
Regulation of SREBP proteolytic maturation
Whether the SCAP–SREBP complex remains in the ER or is recognized as a target by the COPII trafficking machinery for movement to the Golgi is a highly regulated process. In mammals, ER exit is controlled by metabolite-regulated binding of SCAP to the INSIG proteins (Yang et al. 2002). There are two mammalian INSIG proteins encoded by separate genes (Yabe et al. 2002), and their expression is regulated differently: INSIG-1 is an SREBP target gene, and INSIG-2 is repressed by insulin in the liver (Horton et al. 2003a; Yabe et al. 2003). INSIGs have overlapping functions, and most biochemical studies have focused on INSIG-1.
INSIGs are resident ER membrane proteins that interact directly with SCAP and prevent the SCAP–SREBP complex from entering the COPII-dependent ER–Golgi trafficking pathway (Fig. 2; Sun et al. 2007). The SCAP–INSIG association is regulated by the binding of regulatory sterols directly to INSIG or SCAP (Radhakrishnan et al. 2007; Sun et al. 2007). The polytopic membrane region of SCAP contains a conserved sterol-sensing domain that is also found in other proteins involved in cholesterol regulation and signaling, such as HMG CoA reductase, NPC-1, and the hedgehog receptor Patched (Chang et al. 2006). Cholesterol binds directly to mammalian SCAP and NPC-1 (Ohgami et al. 2004; Radhakrishnan et al. 2004; Infante et al. 2008), and key metabolites modulate the binding of reductase and SCAP to the INSIG proteins (Sever et al. 2003; Radhakrishnan et al. 2007).
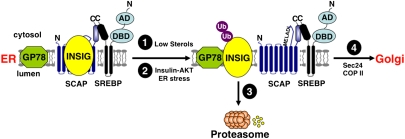
Figure 2.
Multiple pathways for SREBP regulation in the ER. SCAP is a polytopic ER membrane protein (blue), and its cytoplasmic C terminus interacts with the C terminus of the two-pass ER precursor form of SREBP (black). The bHLH-LZ DNA-binding domain (DBD) and N-terminal activation domain (AD) of SREBP are also shown. When bound to cholesterol, SCAP interacts with INSIG. This association keeps the SCAP–SREBP complex in the ER membrane. SREBP–SCAP movement to the Golgi apparatus is regulated by at least two pathways: (1) When cholesterol levels are depleted, a conformational change occurs in SCAP, decreasing its interaction with INSIG and exposing the MELADL COPII targeting signal. (2) Insulin–AKT and ER stress also result in INSIG dissociating from SCAP, and SCAP–SREBP moves to the Golgi presumably through the same pathway. (3) Once free of SCAP, INSIG interacts with the membrane-bound E3 ubiquitin ligase GP78 and, following ubiquitination, INSIG is degraded by the proteasome. (4) Once free of INSIG, SCAP escorts SREBP to the Golgi apparatus through interaction with the Sec24 subunit of COPII. The fate of SREBPs in the Golgi are discussed in the text.
When cholesterol levels in the ER fall below a critical threshold, the conformation of SCAP changes to no longer bind INSIG (Fig. 2, pathway 1). This unmasks a critical interaction motif of SCAP with the core sequence MELADL, which interacts with the Sec24 subunit of the COPII trafficking complex and facilitates movement of the SCAP–SREBP cargo to the Golgi apparatus (Sun et al. 2007). While it is difficult to quantify ER cholesterol relative to total cellular cholesterol, a recent study showed that cholesterol ranges from <1% to 12% of total ER lipid. Transport of SCAP–SREBP is blocked at ER cholesterol concentrations ≥5% of total ER lipid, indicating that this is the critical threshold for regulation (Radhakrishnan et al. 2008). Two distinct Golgi membrane-bound proteases, site-1 protease (S1P) and S2P, then cleave the precursor molecule (Goldstein et al. 2006). S1P first clips the large membrane-tethered immature SREBP in half by cleaving at the C-terminal side of a leucine residue in the RXXL motif located within the lumenal loop (Duncan et al. 1997). Once this cleavage has occurred, the N-terminal pro-SREBP is cleaved within the membrane-spanning domain by S2P (Duncan et al. 1998). The activity of S2P is an example of regulated intramembrane proteolysis (RIP), an evolutionarily conserved process from bacteria to human that is responsible for converting a membrane-bound precursor into a soluble active form in response to physiologic signals (Brown et al. 2000). S2P releases the N-terminal soluble SREBP transcription factor containing both its bHLH-LZ and transcriptional activation domains, which is then efficiently targeted to the nucleus through a pathway requiring interaction with importin-β (Nagoshi and Yoneda 2001).
After its sterol-dependent dissociation from SCAP, INSIG-1 interacts with the membrane-bound E3 ubiquitin ligase GP78, resulting in INSIG-1 ubiquitination and proteasomal degradation (Fig. 2; Lee et al. 2006). The rapid degradation of INSIG after dissociation from SCAP makes the subsequent retention of SCAP–SREBP dependent on new synthesis of INSIG, and is therefore critical to the dynamic nature of the feedback regulatory mechanism. INSIG-1 is an SREBP target gene, and the levels of INSIG would rise similarly to other SREBP targets and in concert with cellular cholesterol, providing a mechanism to re-establish the SCAP–INSIG association and limit SREBP maturation as cholesterol levels return above the critical threshold level (Gong et al. 2006).
This elaborate pathway has been defined mainly through the analysis of mammalian SREBPs. However, the molecular components have been identified in many other distantly related species (Table 1). Although the key proteins are conserved, the SREBP pathway in the fission yeast system responds to sterol levels as a secondary response to limited oxygen supply (Hughes et al. 2005), whereas in Drosophila and probably other invertebrates that are cholesterol auxotrophs (Karlson 1970), SREBP maturation does not respond to sterol concentrations, but rather to phosphatidylethanolamine (PE) (Dobrosotskaya et al. 2002). An INSIG homolog has not been found in Drosophila (Rawson 2003) despite the fact that processing of the Drosophila SREBP homolog is regulated by sterols when expressed in mammalian cells (Rosenfeld and Osborne 1998). Details of the regulatory mechanism for the trafficking of the SCAP/SREBP complex in Drosophila and how PE is involved remain to be determined.
SREBP regulation by insulin
In addition to regulation by sterols, increased expression and aberrant regulation of SREBP-1c are associated with diabetes and fatty liver (Shimomura et al. 1999b), and enhanced nuclear accumulation of SREBP-1c in the liver by insulin signaling has been known for several years (Horton et al. 1998; Kim et al. 1998; Shimomura et al. 1999a). However, because the amounts of SREBP-1c mRNA and precursor protein were also increased by insulin, whether there was an active effect of insulin on the maturation of SREBP was not clear. More recent studies suggest insulin enhances the hepatic processing of SREBP-1c (Hegarty et al. 2005). Insulin signaling activates AKT, which has been reported to be directly involved in the movement of SREBP–SCAP from the ER to Golgi (Fig. 2; Du et al. 2006; Yellaturu et al. 2009). The precursor SREBP-1c protein is also a substrate for AKT phosphorylation in vitro (Yellaturu et al. 2009). Insulin also activates protein kinase C λ (PKCλ), which has been associated with defective liver lipid metabolism through changes in SREBP-1c gene expression (Matsumoto et al. 2003; Taniguchi et al. 2006), and more recent studies indicate that PKCλ activation of SREBP-1c is associated with obesity, diabetes, and hyperlipidemia in animal models (Sajan et al. 2009a,b).
The effects of AKT on SREBP-1 function are complex, as there appear to be both rapamycin-sensitive (mTORC1-dependent) and rapamycin-insensitive effects of AKT on the level of mature nuclear SREBP-1 protein (Porstmann et al. 2008). One of the rapamycin-insensitive effects is likely through the direct action of AKT on SREBP-1 maturation. However, AKT probably also stabilizes the nuclear form of SREBP-1 through inhibition of GSK3 (Cross et al. 1995), which can phosphorylate nuclear SREBP-1 at two closely spaced residues close to its C terminus (Sundqvist et al. 2005). Once phosphorylated by GSK3, SREBP-1 interacts with the E3 ubiquitin ligase Fbw7, resulting in ubiquitination and degradation by the proteasome.
SREBPs and the ER stress response
ER stress results from the sensing of misfolded proteins in its lumen (Ron and Walter 2007) and is associated with aberrant cellular lipid accumulation (Rutkowski et al. 2008; Lee and Glimcher 2009). Because SREBPs activate genes of lipid biosynthesis and are maintained as precursors in the ER membrane, it was reasonable to hypothesize that enhanced SREBP processing due to ER stress might explain at least part of the mechanism for lipid overload. The first experimental observation that hinted at such a mechanism was that processing of the ER membrane-bound precursor form of activating transcription factor 6 (ATF6), an ER stress-related transcription factor, required the identical Golgi-located S1P and S2P proteases involved in SREBP maturation (Ye et al. 2000). In the ensuing years, several reports have documented an association between ER stress and aberrant cellular lipid control (Werstuck et al. 2001; Ji et al. 2006; Lee et al. 2008), and the involvement of SREBP in the process has been confirmed (Werstuck et al. 2001; Lee and Ye 2004). In fact, the induction of ER stress by either hypotonic conditions or select chemical inducers decreases levels of INSIG-1, providing a mechanism for the activation of SREBP (Fig. 2, pathway 2; Lee and Ye 2004).
Additional studies have revealed a role for the ER stress-related PERK kinase in regulating SREBP maturation. Lipogenesis and SREBP-1 processing were significantly reduced in mammary tissue or mouse embryo fibroblasts (MEFs) that lack PERK due to gene disruption (Bobrovnikova-Marjon et al. 2008). PERK phosphorylates and inactivates eIF2α to inhibit general protein synthesis during the response to ER stress to limit the accumulation of misfolded proteins (Harding et al. 1999). This would also acutely reduce levels of proteins that turn over relatively rapidly, such as INSIG-1. Bobrovnikova-Marjon et al. (2008) suggested that, in the absence of PERK, INSIG-1 levels would be relatively higher, resulting in greater retention of the SCAP–SREBP complex in the ER, thus limiting SREBP maturation. Therefore, it is noteworthy that studies of sterol regulation suggest that INSIG-1 is relatively stable when complexed with SCAP (Lee et al. 2006). It would be interesting to determine whether PERK itself or another component of the ER stress response additionally regulates SCAP–INSIG interaction, which would have an even more significant effect on the level of INSIG-1. In another study, overexpression of a soluble form of PERK in cultured cells resulted in a decrease in SREBP processing, indicating that the relationship between PERK and SREBP maturation may be complex (Harding et al. 2005).
As mentioned above, ATF6 maturation requires the S1P and S2P that modify SREBPs. Additionally, ER retention of ATF6 is mediated through interaction with the ER stress-related chaperone GRP78 (Shen et al. 2002). A recent report demonstrated that overexpression of GRP78 in the livers of ob/ob mice decreased steatosis and inhibited SREBP-1 processing in the liver and primary hepatocytes treated with both insulin and ER stress inducers (Kammoun et al. 2009). These results are intriguing and suggest a more intimate relationship between ER stress and SREBPs than recognized previously. It will be interesting to determine how GRP78 interacts with the other components of the SREBP maturation pathway and whether physiologic changes in GRP78 modulate SREBP activity.
The relationship between ER stress and lipogenesis extends beyond SREBPs, as a liver-specific knockout of the key ER stress regulator XBP-1 results in a decrease in expression of a subset of lipogenic genes and lower levels of plasma lipids due to a decrease in hepatic triglyceride secretion (Lee et al. 2008). Importantly, these effects appear to be separate from the ER stress response and effects on protein secretion, indicating that XBP-1 functions as a molecular bridge between the ER stress response and hepatic lipid production, which further suggests a close association of these two seemingly disparate processes.
Regulation of SREBP gene transcription
In addition to the multicomponent pathway for regulated processing of SREBPs, there is also evidence for regulation of SREBP gene transcription as a mechanism to alter SREBP levels. The SREBP-1a promoter is relatively simple and weak and is composed of multiple Sp1-binding elements proximal to the transcription start site (Zhang et al. 2005). There is an NF-kB-binding site in the promoter, but there is no evidence yet to indicate that SREBP-1a is regulated through an NF-kB-dependent pathway (Zhang et al. 2005). In contrast, the SREBP-1c promoter is highly regulated by both insulin and oxysterols. While the mechanism for insulin regulation is presently unclear, the signaling pathway is physiologically significant and is a major contributing factor to SREBP-1c's role in lipogenesis. Insulin also has an effect on SREBP-1 processing, as discussed above. Oxysterol regulation of SREBP-1c is mediated by a liver X receptor (LXR)-dependent pathway (Repa et al. 2000). In fact, steatosis and hypertriglyceridemia associated with the oral delivery of synthetic LXR agonists is a consequence of elevated expression of SREBP-1c and other lipogenic target genes by pharmacologic signaling through LXR (Joseph et al. 2002).
The 5′-flanking sequence of the SREBP-2 gene has binding sites for SREBP and thyroid hormone receptor (TR), and there is evidence for regulation by both mechanisms (Sato et al. 1996; Shin and Osborne 2003). The autoregulation implies that the basal level of SREBP-2 in the ER is low and not sufficient to meet the cholesterol needs of the cell under prolonged sterol deficiency. The association of hypercholesterolemia with low thyroid hormone levels has been known for decades (Mason et al. 1930), and the connection was traced to a decrease in expression of functional LDL receptors secondary to thyroid hormone deficiency (Ness et al. 1990). Because the LDL receptor gene is an SREBP-2 target, the regulation of SREBP-2 by thyroid hormone provides a mechanism to link hypercholesterolemia with hypothyroidism (Shin and Osborne 2003).
Target gene activation by SREBPs
Nuclear SREBPs activate target genes through binding to defined SREs in promoters of target genes. The first identified SREBP-regulated genes were those encoding the LDL receptor and HMG CoA synthase. Nearly identical 10-base-pair (bp) elements were first defined by sterol-dependent promoter reporter gene assays (Dawson et al. 1988; Smith et al. 1988), and then by direct DNA binding studies with purified SREBP in vitro (Briggs et al. 1993; Wang et al. 1993). As additional SREBP target genes were identified and their DNA-binding sites were characterized by functional mutagenesis, it was somewhat surprising that not one well-defined target site contained the palindromic E-box element typical for bHLH-LZ proteins (Osborne 1995). Instead, all of the interaction sites were related to the direct repeat SRE (Athanikar and Osborne 1998).
The physiological basis for this conundrum was investigated through converting the SRE of the LDL receptor promoter into the E-box that is recognized with high affinity by SREBP in vitro (Athanikar and Osborne 1998). While in vitro binding was maintained at a similar affinity in the E-box-converted promoter, gene expression was constitutively high relative to the native promoter and insensitive to a standard sterol depletion protocol known to alter SREBP nuclear accumulation. The explanation for this result is based on the two unique properties of SREBPs mentioned above: the key tyrosine residue in the DNA-binding domain, and the sterol-sensitive membrane-anchoring domain. By converting the LDL receptor promoter into an E-box, the resulting promoter became a target for other constitutively nuclear bHLH-LZ proteins that activate transcription through E-box binding and are not subject to sterol-regulated nuclear accumulation.
In addition to defining the direct repeat SRE, functional studies on a small number of SREBP target promoters indicated that an additional more generic transcription factor that bound in close proximity was essential for SREBPs to stimulate target gene expression (Sanchez et al. 1995; Ericsson et al. 1996; Guan et al. 1997; Dooley et al. 1999; Magaña et al. 2000). NF-Y/CBF, CREB/ATF, and Sp1 have all been identified as SREBP coregulatory factors in different promoters. SREBPs interact directly with all three proteins in the absence of DNA, and the interaction domains of both partners are required for the concerted activation of transcription from target promoters. Interestingly, this coregulatory model for SREBP activation has been supported by recent promoter-wide chromatin immunoprecipitation (ChIP)–chip and genome-wide ChIP-seq interrogation of SREBP-1 binding. In these studies, motifs corresponding to NF-Y (Reed et al. 2008) and Sp1 (Reed et al. 2008; Seo et al. 2009) were preferentially coenriched close to genomic target sites for SREBP-1 in both HepG2 cells (Reed et al. 2008) and the mouse liver (Seo et al. 2009).
Roles for SREBPs beyond core lipid metabolic processes
As discussed above, SREBP maturation is stimulated by both lipid signals as well as by other physiologic cues. Thus, it is likely that SREBPs connect fundamental aspects of lipid metabolism with other physiologic responses, and several studies support this hypothesis. In some of these cases, SREBP-binding sites were localized in regulatory regions of specific genes such as caspase 2 (Logette et al. 2005) and p21/WAF (Inoue et al. 2005). In other instances, SREBP activation was shown to be important for other cellular activities, such as phagocytosis (Castoreno et al. 2005) and the defensive response to pore-forming bacterial α-toxin (Gurcel et al. 2006). In both of these instances, SREBP target genes in lipid metabolism were activated presumably to keep up with the demand for new membrane mass required in each response. However, whether SREBPs activated additional genes that are more specifically related to phagocytosis or toxin response was not addressed. SREBP-1 has also been directly implicated in controlling cell size through AKT signaling that is at least partially dependent on the mTORC1 pathway (Porstmann et al. 2008). Taken together, the lipid and nutrient regulation of SREBPs underscores their fundamental involvement in other physiologic responses broadly related to cell–environment interactions.
More comprehensive genome-wide studies also support a role for SREBPs beyond lipid metabolism. Horton et al. (2003a) hypothesized that SREBP target genes should (1) exhibit elevated levels of expression when SREBP-1 or SREBP-2 was overexpressed in livers of transgenic mice, and (2) show reduced expression when nuclear translocation of SREBPs was blocked by inactivation of SCAP. Candidate SREBP target genes were then identified through a comparative genome-wide microarray approach for hepatic gene expression from the corresponding transgenic or knockout mice. Most genes involved in cholesterol biosynthesis and several genes of lipogenesis, including glucose breakdown and fatty acid and triglyceride biosynthesis, exhibited the pattern of expression expected for SREBP target genes. However, several other genes not directly related to lipid metabolism were also identified as potential SREBP target genes in this study.
In a more recent investigation designed to identify sites for SREBP binding across the entire genome, a ChIP-seq approach was used with an SREBP-1 antibody and hepatic chromatin prepared from fasted/refed mice, conditions known to increase nuclear SREBP-1c abundance (Seo et al. 2009). An alignment of the sequences contained in the 427 SREBP-1-binding peaks (false discovery rate of 0.002) revealed a new enriched motif. This new element is a variant of the SRE direct repeat sequence and corresponds to an orphan motif that was identified by a purely computational approach as being enriched across mammalian species. However, the corresponding transcription factor for this motif was unknown (Xie et al. 2005). This ChIP-seq study suggested that the missing transcription factor is SREBP-1. When the genes associated with the SREBP-1-binding peaks in this study were categorized by Gene Ontology (GO), enriched clusters involved in carbohydrate and lipid metabolism were identified as expected. However, several other GO categories exhibited a high degree of enrichment, including protein metabolism and trafficking, apoptosis, cell structure, and proliferation/differentiation.
Support for the involvement of SREBP-1 in cellular processes related to these additional GO categories is suggested by several observations. As discussed above, SREBPs undergo a regulated itinerary from the ER to the Golgi followed by proteolysis (Brown and Goldstein 1999). SREBPs are also associated with apoptosis (Wang et al. 1996; Logette et al. 2005; Gibot et al. 2009), and SREBP-1 is regulated by phosphorylation in a cell cycle-dependent fashion (Bengoechea-Alonso et al. 2005). The activation of p21/WAF by SREBP-1 also provides a direct link with cell cycle and growth control (Inoue et al. 2005). Additionally, SREBP-1 is downstream from TORC1 signaling, where it has been proposed to be responsible for the activation of lipid metabolic genes during cellular growth (Porstmann et al. 2008). Because TORC1 signaling also induces other anabolic processes required for cell growth, it is possible that SREBP-1 might contribute to the activation of these processes as well.
An additional study used a ChIP–chip hybridization protocol and promoter tiling arrays to map SREBP-1-binding sites in chromatin from cultures of insulin-treated human hepatoma HepG2 cells. In contrast to the ChIP-seq experiments, this study identified a classic bHLH E-box site enriched in promoters that scored positive for binding SREBP-1 (Reed et al. 2008). Differences between these two global approaches may be due to the fact that SREBP-1a is the major isoform in cultured cell lines but SREBP-1c is 10 times more abundant than SREBP-1a in the liver (Shimomura et al. 1997). However, as noted above, experimental evidence indicates that the direct repeat recognition by SREBPs is central to their physiological function, suggesting that the E-box sites may not correspond to physiologically important SREBP-binding sites.
Additional microarray-based and high-throughput cDNA expression/computational filtering approaches have been used to identify cellular processes potentially regulated by SREBPs (Rome et al. 2008; Bartz et al. 2009; Chatterjee et al. 2009). As with the genome-wide ChIP studies mentioned above, how successful these global approaches ultimately prove to be at revealing new roles for SREBPs in physiology awaits further in-depth investigation. Thus far, a ChIP–chip approach provided the basis for a study that demonstrated that cholesterol-dependent activation of SREBP-2 in the small intestine regulates the expression and activity of at least a handful of bitter taste receptor G-protein-coupled receptors (T2Rs) (Jeon et al. 2008). Jeon et al. (2008) hypothesized that this pathway is an important second line of defense after taste aversion to prevent the absorption of bitter-tasting and potentially toxic components from the diet that are overrepresented in cholesterol-poor plant-based foods. In another recent genome-wide study (Bartz et al. 2009), TMEM97, a protein identified as an SREBP target in the stringent microarray approach of Horton et al. (2003a) was shown to be associated with the intracellular cholesterol regulatory protein NPC-1 in endolysosomal compartments, where it may influence intracellular cholesterol trafficking.
Transcriptional activation by SREBPs
The only identified transcriptional activation domain in SREBPs is located at their extreme N termini (Sato et al. 1994). The alternative mRNAs that specify either SREBP-1a or SREBP-1c encode distinct N-terminal domains with drastically different potencies for transcriptional activation (Shimano et al. 1997). Alternative promoter usage and differential splicing have been observed for many transcription factor genes, and in several instances this alters the structure of an activation domain. Most similar to SREBP-1, alternative promoters that result in N-terminal heterogeneity are common in the nuclear receptor family where different versions of an N-terminal constitutive activation domain (AF1) are produced (Zhu et al. 1995; Oberste-Berghaus et al. 2000; Huber et al. 2002). However, in most cases, the real physiological significance of these variants has not been fully explored because most studies have focused on the nuclear receptor AF2 activation domain, which is located close to the ligand-binding domain at the C terminus. The interest in AF2 is because its structure is subject to agonist/antagonist-induced conformational change that modulate coactivator/corepressor recruitment and function in response to appropriate ligand-dependent signaling pathways (Glass and Rosenfeld 2000).
The single SREBP-1a activation domain contains residues that are key to interaction with the non-DNA-binding transcriptional coactivators CBP/p300 and the mediator complex (MED) (Oliner et al. 1996; Ericsson and Edwards 1998; Nåår et al. 1998). CBP has intrinsic histone acetyltransferase activity as well as multiple independent protein–protein interaction domains (Goodman and Smolik 2000). SREBP-1a interacts in vitro with both the N-terminal C/H1 and KIX domains of CBP, and when each of these isolated domains was overexpressed in cultured cells, both peptide fragments blocked the ability of SREBP-1a to stimulate gene transcription (Oliner et al. 1996; Ericsson and Edwards 1998).
While the dominant-negative approach can be useful for identifying sites of interaction between CBP and a critical region required for coactivator recruitment in transcription factors like SREBP-1, such studies do not directly address whether the isolated interactions with the KIX or C/H1 domain are important for activation of transcription in the context of full-length CBP/p300. To address this issue, point mutations were engineered into conserved residues of the SREBP-1a activation domain, and the mutant proteins were tested for both interaction with the isolated KIX and C/H1 domains in vitro and stimulation of transcription in a cultured cell transfection assay (Toth et al. 2004). All mutations that decreased SREBP-dependent gene transcription also decreased interaction with the C/H1 region, whereas only a subset of mutations that decreased interaction with the isolated KIX domain decreased transcriptional activation by SREBP-1a. These results suggested that interaction with the C/H1 domain of CBP is important for SREBP-1a activation. However, they do not rule out the possibility that there is a distinct physiological setting or subset of target genes where the KIX–SREBP interaction is also important.
MED—also referred to in the literature as DRIP (vitamin D receptor-interacting protein), TRAP (thyroid receptor-activating protein), or ARC (activator-recruiting complex)—is a large multisubunit complex that is required for RNA polymerase II transcription across eukarya (Malik and Roeder 2000). Individual MED subunits interact with different DNA-bound transcription factors to increase gene expression, and SREBP-1a has been shown to interact with both the MED14 (Toth et al. 2004) and MED15 (Yang et al. 2006) subunits. The Med14 interaction is interesting because MED14 was first described in Saccharomyces cerevisiae as Rgr1, an essential gene for nutrient metabolism, and a null allele resulted in a failure to store carbohydrate (Sakai et al. 1990). The SREBP-1 homolog in C. elegans also interacts with MED15, and knockdown studies suggest both MED15 and SREBP-1 are important for lipid regulation in C. elegans (Yang et al. 2006). Interestingly, many of the key SREBP-1 residues required for interaction with CBP and MED14 are present in SREBP-1a but not in SREBP-1c (Toth et al. 2004). In fact, SREBP-1a-specific amino acids that are required for interaction with CBP and MED14 are conserved across vertebrates, but there is limited primary sequence homology between the vertebrate and C. elegans N termini. The fact that these critical residues are not present in SREBP-1c explains why it is a relatively poor activator of transcription relative to SREBP-1a. Many of the conserved amino acids that interact with CBP and MED14 are also present in SREBP-2, which has a similar potency for gene activation as SREBP-1a.
Most eukaryotic transcription factors bind DNA as homodimers or heterodimers and, in the case of all homodimers and many heterodimeric combinations, each monomer contains a separate transcriptional activation domain. When two potent activation domains are present in the dimer, it is difficult to determine whether both domains actively participate in recruiting coactivators to stimulate gene expression or whether one monomer contributes anything in addition to orienting the dimer for specific DNA recognition. This is particularly relevant to the SREBPs, where, in addition to homotypic interactions, there is the potential to form SREBP-1a–SREBP-1c or SREBP-2–SREBP-1c heterodimers as well.
To address this issue, all homodimeric and heterodimeric combinations were analyzed separately by covalently linking the individual monomeric coding units together using an unstructured peptide linker (Datta and Osborne 2005). These covalent dimeric constructs preferentially form intramolecular dimers that bind DNA in vitro similar to the corresponding proteins expressed as monomers (Neuhold and Wold 1993; Datta and Osborne 2005). In analyzing the activation of several SREBP-responsive promoters, the homodimeric constructs stimulated gene expression to similar levels as the corresponding monomers, with SREBP-1c being a very weak activator (Datta and Osborne 2005). Similar to other reports, SREBP-1c stimulated promoter activity at least 10-fold less compared with the other two SREBPs when expressed at equal protein levels. However, when the SREBP-1a–SREBP-1c or SREBP-1c–SREBP-2 heterodimers were expressed, the level of promoter activation was intermediate between the high values for the SREBP-1a–SREBP-1a or SREBP-2–SREBP-2 homodimers and the low level for the SREBP-1c–SREBP-1c dimer. Indeed, there was no difference when a mutant version that deleted the complete SREBP-1 activation domain was substituted for SREBP-1c in the heterodimeric fusion with either SREBP-1a or SREBP-2. These results indicate that the activation domain from each monomeric partner functions to stimulate gene transcription, and that, in the heterodimeric form, the SREBP-1c activation domain is functionally silent.
The exact role for each SREBP monomer in the recruitment and function of coactivators remains to be determined. However, these observations have significant functional implications, as they suggest that SREBP-1c could be viewed as an in vivo modulator of the activity of SREBP-1a or SREBP-2. Therefore, induction of SREBP-1c expression in tissues where its basal level is low compared with the other SREBPs may dampen the overall cellular SREBP activity. In fact, there is support for such a model from targeted overexpression studies in mice. When SREBP-1a was overexpressed in adipose tissue, there was significant adipocyte hypertrophy with increased secretion of fatty acids, as expected for a lipid synthetic transcription factor (Horton et al. 2003b). On the other hand, similar overexpression of SREBP-1c resulted in severe loss of adipose tissue mass accompanied with insulin resistance and diabetes, which is reminiscent of congenital lipodystrophy in humans (Shimomura et al. 1998). These observations provide strong evidence that the two SREBP-1 isoforms may indeed have opposing effects on gene expression and tissue function in vivo under certain circumstances.
SREBP in model organisms
Model organism research into the function of SREBP has revealed important similarities and differences between these systems and the mammalian SREBP pathway. In the cholesterol auxotrophs Drosophila melanogaster and C. elegans, SREBP controls fatty acid synthesis and functions to maintain fat supply. In fungi, SREBP is a hypoxic transcription factor that controls adaptation to low oxygen growth. Work in pathogenic fungi identifies SREBP as a candidate antifungal drug target for the treatment of opportunistic fungal infections.
SREBP in flies
The first nonmammalian SREBP homolog was identified in D. melanogaster, dSREBP/HLH106, which was curious because fruit flies cannot synthesize cholesterol and therefore are natural cholesterol auxotrophs (Theopold et al. 1996; Rosenfeld and Osborne 1998). D. melanogaster encodes homologs of SREBP, SCAP, S1P, and S2P, but lacks an Insig homolog (Table 1). Detailed mechanistic studies into the regulation of dSREBP using cultured insect cells revealed that dSREBP is proteolytically activated in response to levels of the glycerophospholipid PE (Dobrosotskaya et al. 2002). Under conditions of low PE, dSREBP up-regulates enzymes required for fatty acid and phospholipid synthesis, such as acetyl CoA synthase, acetyl CoA carboxylase, fatty acid synthase, and phosphocholine cytidylyltransferase (Dobrosotskaya et al. 2002; Seegmiller et al. 2002). Consistent with this, cells deficient in dSREBP have reduced levels of fatty acid and phospholipid synthesis. Regulation of dSREBP proteolysis by phospholipids rather than cholesterol as in mammals suggests that dSCAP has the capacity to sense different classes of lipids in the feedback control of lipid homeostasis, but the metabolite interaction mechanism for SCAP sensing needs further exploration.
Studies of dSREBP function in the whole animal demonstrate that dSREBP is required for larval development (Kunte et al. 2006). Flies lacking dSREBP are fatty acid auxotrophs due to defects in fatty acid synthesis, and development can be rescued by addition of fatty acids to the diet. Supplementation with soy lipid extract repressed dSREBP activation in larvae, consistent with the cultured cell data that dSREBP is regulated by phospholipids. The mechanism of dSREBP proteolysis mirrors that of mammalian cells, with dSCAP facilitating ER-to-Golgi transport and interaction of dSREBP with homologs of S1P and S2P. Interestingly, studies of dS2P-null flies revealed an alternative pathway for dSREBP activation that requires the caspase Drice and permits larval development in the absence of normal Site-2 protease cleavage (Amarneh et al. 2009; Matthews et al. 2009). These data indicate that dSREBP functions in the D. melanogaster fat body and gut to promote fatty acid synthesis essential for development.
The function of dSREBP in fatty acid synthesis is also closely linked to viral pathogenesis in D. melanogaster. A genome-wide RNAi screen identified dSREBP and enzymes of fatty acid biosynthesis as essential for viral replication in cultured insect cells (Cherry et al. 2006). dSREBP is required for Golgi maintenance and for formation of a cytoplasmic vesicular compartment required for viral replication. Interestingly, experiments using a regulated dSREBP allele indicate that dSREBP is not required for viability of adult flies, but that dSREBP is required for viral replication in adult animals. These results highlight functions of SREBP beyond regulation of lipid homeostasis and provide a link to viral pathogenesis.
SREBP in worms
Targeted genetic and RNAi studies identified the C. elegans SREBP (SBP-1/LPD-1) as required for fat accumulation (Ashrafi et al. 2003; McKay et al. 2003). Animals lacking SBP-1 fail to accumulate fat stores in the intestine, and gene expression analyses indicate that SBP-1 is required for expression of lipogenic genes, such as fatty acid synthase, acetyl CoA carboxylase, and stearoyl CoA desaturase (McKay et al. 2003; Yang et al. 2006). In addition to its role in straight-chain fatty acid synthesis, SBP-1 is required for monomethyl branched-chain fatty acid (BCFA) synthesis through the control of two long-chain fatty acid elongases, elo-5 and elo-6, and the very-long-chain acyl CoA synthetase acs-1 (Kniazeva et al. 2004, 2008). BCFA synthesis is essential for post-embryonic development and growth control in worms (Kniazeva et al. 2008), and BCFAs signal through a nutrient-sensing pathway that functions independently from the insulin receptor/DAF-2 pathway. Thus, SBP-1 plays an important role in C. elegans developmental regulation by promoting production of BCFA synthesis.
As in mammals, worm SREBP cooperates with transcriptional coregulators to control gene expression. SBP-1 functions with the transcription mediator MDT-15/ARC105 and the CBP/p300 acetyltransferase CBP-1 to control straight-chain fatty acid homeostasis and BCFA homeostasis, respectively (Taubert et al. 2006; Yang et al. 2006; Kniazeva et al. 2008). Like D. melanogaster, C. elegans is a sterol auxotroph and lacks an Insig homolog, raising the possibility that SBP-1 may also be regulated by a nonsterol lipid. Indeed, SBP-1-dependent activation of acs-1 is negatively feedback-regulated by the BCFA end product C17ISO. C. elegans contains a SCAP homolog (Table 1), but whether proteolysis of SBP-1 is regulated and whether fatty acids directly control SBP-1 activity are unknown.
SREBP in fungi
The full extent of SREBP evolutionary conservation was recognized with the discovery of a functional SREBP pathway in the fission yeast Schizosaccharomyces pombe. Sequence database searches revealed homologs of SREBP, SCAP, and INSIG, called Sre1, Scp1, and Ins1, respectively (Table 1; Hughes et al. 2005). As in mammalian cells, Sre1–Scp1 form a complex, and Sre1 is proteolytically cleaved and activated in response to cellular depletion of the fungal sterol ergosterol (Hughes et al. 2005, 2007). However, unlike in mammalian cells, sterol regulation of Sre1 does not require the INSIG homolog (Hughes et al. 2005). Rather, Ins1 negatively regulates activity of the sterol pathway enzyme HMG CoA reductase, a second function for INSIG in mammalian cells (Goldstein et al. 2006; Burg et al. 2008). While Sre1–Scp1 responds to pharmacological inhibition of sterol synthesis, the physiological trigger for Sre1 activation is hypoxia (Hughes et al. 2005). Ergosterol synthesis is highly oxygen-consumptive, requiring 12 molecules of dioxygen (Rosenfeld and Beauvoit 2003). Under low oxygen, ergosterol synthesis is inhibited, stimulating Sre1 proteolysis (Fig. 3A). Thus, Sre1–Scp1 sense sterol synthesis as an indirect measure of environmental oxygen. While some fungi contain a functional Site-2 protease (discussed below), fission yeast lack a recognizable S2P, suggesting that perhaps, as in D. melanogaster, an alternative mechanism exists for Sre1 cleavage.
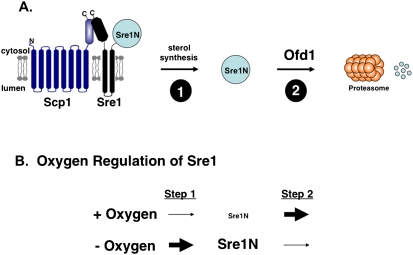
Figure 3.
Oxygen regulates the active, cleaved Sre1N transcription factor by two mechanisms. (A, step 1) Oxygen-dependent ergosterol synthesis inhibits Sre1 proteolysis and production of Sre1N. (Step 2) Oxygen positively regulates Ofd1, which accelerates the proteasomal degradation of Sre1N. (B) In the presence of oxygen, Sre1 cleavage is low, and Ofd1-dependent degradation is high. Under low oxygen, these two mechanisms are reversed, and Sre1N accumulates.
Genome-wide gene expression studies revealed that Sre1 is principally a transcriptional activator that up-regulates genes required for adaptation to hypoxia, including several oxygen-dependent enzymes in biosynthetic pathways for ergosterol, heme, and sphingolipids (Todd et al. 2006). Consistent with the requirement for hypoxic gene expression, cells lacking Sre1 or Scp1 fail to grow under low oxygen. Sre1 does not control expression of glycolytic or respiratory genes, indicating that other hypoxic transcription factors exist in S. pombe. Promoter analysis of the sre1+ gene revealed two DNA-binding sites that resembled the SRE found in the human LDL receptor promoter, and expression of Sre1 mRNA, like mammalian SREBP-2, is autoregulated in a positive feedback loop (Todd et al. 2006; Sehgal et al. 2007).
The importance of Sre1 for hypoxic adaptation is underscored by the presence of a second oxygen-dependent regulatory mechanism that controls Sre1 activity. The prolyl 4-hydroxylase domain protein Ofd1 controls activity of the soluble N-terminal cleavage product of Sre1, called Sre1N. In the presence of oxygen, Ofd1 accelerates degradation of Sre1N by the proteasome (Fig. 3A; Hughes and Espenshade 2008). Unlike the prolyl hydroxylases that regulate degradation of the mammalian hypoxia-inducible factor HIF-1, the hydroxylase domain of Ofd1 is not required for Sre1N degradation (Dann and Bruick 2005). Rather, the N-terminal hydroxylase domain functions as an oxygen sensor to regulate activity of a C-terminal degradation domain (CTDD) (Hughes and Espenshade 2008). Activity of the Ofd1 CTDD is regulated by the oxygen-dependent direct binding of the inhibitor Nro1 (Lee et al. 2009). Under low oxygen, Nro1 binds Ofd1, which leads to stabilization of Sre1N and increased target gene expression. Thus, oxygen acts at two points to regulate Sre1 activity (Fig. 3B). First, oxygen is required for sterol synthesis, which regulates Sre1 proteolysis. Second, Sre1N stability is regulated by oxygen through Ofd1. Together, these two oxygen-sensing mechanisms function as switches to rapidly up-regulate Sre1-dependent transcription under low oxygen, allowing adaptation to hypoxia and continued cell growth (Fig. 3B).
The function of SREBP as a hypoxic transcription factor is conserved across distantly related fungi. Studies of the opportunistic pathogen Cryptococcus neoformans demonstrated that Sre1 is a hypoxic transcription factor that activates expression of genes required for ergosterol synthesis and iron acquisition (Chang et al. 2007; Chun et al. 2007). While it appears that S. pombe lacks a Site-2 protease, C. neoformans has a Site-2 protease that is required for Sre1 activation (Chun et al. 2007; Bien et al. 2009). Importantly, Sre1 is required for virulence and disease progression in two different mouse models of cryptococcosis, indicating that Sre1 is essential for adaptation to the host environment. Notably, cells lacking Sre1 are defective for ergosterol synthesis and are therefore hypersensitive to antifungal azole drugs that target ergosterol synthesis. These findings suggest that inhibitors of Sre1 activity may be effective antifungals when used in combination with current therapies.
The requirement of SREBP for fungal pathogenesis was confirmed recently in an unrelated opportunistic pathogen, Aspergillus fumigatus. As in C. neoformans, SREBP was required for hypoxic growth, regulation of sterol biosynthetic enzymes, resistance to azole drugs, and fungal virulence (Willger et al. 2008). Of note, A. fumigatus lacks a SCAP homolog but codes for Ofd1 (Table 1), suggesting that oxygen regulation of SREBP may be modular in fungi. An SREBP homolog is also required for hyphal growth in Candida albicans (Lane et al. 2001), a process required for pathogenesis, and the rice blast Magnaporthe grisea also contains an SREBP homolog, raising the possibility that SREBP pathway inhibitors could be effective antifungals in a number of disease settings.
Oxygen regulation of metazoan SREBPs
Work from fungi clearly establishes SREBP as a hypoxic transcription factor. However, whether oxygen controls SREBP activity in other systems is unclear. One report in C. elegans indicates that low oxygen up-regulates SBP-1, but the mechanism by which this occurs is not known (Taghibiglou et al. 2009). The ability of fission yeast cells to sense ergosterol synthesis as a measure of environmental oxygen is coupled to the fact that this yeast does not import exogenous sterols. The ability of mammalian SREBP–SCAP to respond to oxygen is therefore complicated because mammalian cells receive cholesterol from both synthetic as well as endocytic receptor-mediated pathways. However, an Ofd1-like mechanism could control SREBP activity in response to oxygen, independent of sterol-regulated proteolysis. Further studies of mammalian SREBP and its nonlipogenic functions may reveal links to oxygen control.
Closing remarks
The physiologic role for the SREBP pathway was initially defined in the regulation of core pathways of lipid metabolism, and there is overwhelming support for this association. However, studies in several organisms provide substantial evidence for a wider role for SREBPs in the response to external signals. The variety of metabolic and environmental inputs to the SREBP pathway, such as insulin and ER stress, highlights the centrality of lipogenesis in cell function and survival. Work in model organisms revealed a requirement for SREBP in the hypoxic response and for adaptation of fungal pathogens to the host environment. Interestingly, SREBP is also required for host cell response to bacterial toxins, placing these unique transcription factors on both sides of a host–pathogen encounter.
Acknowledgments
We thank Xiaohui Xie for assistance with the evolutionary analysis of SREBPs. We also wish to acknowledge the contributions of Michael S. Brown and Joseph L. Goldstein to the field and for their mentorship, which continues to influence how we approach scientific investigation. The work in our laboratories is supported by NIH grants HL48044 (to T.O.), DK71021 (to T.O.), and HL077588 (to P.E.). P.E. is an Established Investigator of the American Heart Association and a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1854309.
References
- Amarneh B, Matthews KA, Rawson RB. Activation of sterol regulatory element-binding protein by the caspase Drice in Drosophila larvae. J Biol Chem. 2009;284:9674–9682. doi: 10.1074/jbc.M900346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Athanikar JN, Osborne TF. Specificity in cholesterol regulation of gene expression by coevolution of sterol regulatory DNA element and its binding protein. Proc Natl Acad Sci. 1998;95:4935–4940. doi: 10.1073/pnas.95.9.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz F, Kern L, Erz D, Zhu M, Gilbert D, Meinhof T, Wirkner U, Erfle H, Muckenthaler M, Pepperkok R, et al. Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 2009;10:63–75. doi: 10.1016/j.cmet.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Bengoechea-Alonso MT, Punga T, Ericsson J. Hyperphosphorylation regulates the activity of SREBP1 during mitosis. Proc Natl Acad Sci. 2005;102:11681–11686. doi: 10.1073/pnas.0501494102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien CM, Chang YC, Nes WD, Kwon-Chung KJ, Espenshade PJ. Cryptococcus neoformans Site-2 protease is required for virulence and survival in the presence of azole drugs. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.06895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Bobrovnikova-Marjon E, Hatzivassiliou G, Grigoriadou C, Romero M, Cavener DR, Thompson CB, Diehl JA. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proc Natl Acad Sci. 2008;105:16314–16319. doi: 10.1073/pnas.0808517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MR, Yokoyama C, Wang X, Brown MS, Goldstein JL. Nuclear protein that binds sterol regulatory element of low density receptor promoter I: Identification of the protein and delineation of its target nucleotide sequence. J Biol Chem. 1993;268:14490–14496. [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Burg JS, Powell DW, Chai R, Hughes AL, Link AJ, Espenshade PJ. Insig regulates HMG-CoA reductase by controlling enzyme phosphorylation in fission yeast. Cell Metab. 2008;8:522–531. doi: 10.1016/j.cmet.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoreno AB, Wang Y, Stockinger W, Jarzylo LA, Du H, Pagnon JC, Shieh EC, Nohturfft A. Transcriptional regulation of phagocytosis-induced membrane biogenesis by sterol regulatory element binding proteins. Proc Natl Acad Sci. 2005;102:13129–13134. doi: 10.1073/pnas.0506716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TY, Chang CC, Ohgami N, Yamauchi Y. Cholesterol sensing, trafficking, and esterification. Annu Rev Cell Dev Biol. 2006;22:129–157. doi: 10.1146/annurev.cellbio.22.010305.104656. [DOI] [PubMed] [Google Scholar]
- Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol Microbiol. 2007;64:614–629. doi: 10.1111/j.1365-2958.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Szustakowski JD, Nanguneri NR, Mickanin C, Labow MA, Nohturfft A, Dev KK, Sivasankaran R. Identification of novel genes and pathways regulating SREBP transcriptional activity. PLoS One. 2009;4:e5197. doi: 10.1371/journal.pone.0005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S, Kunte A, Wang H, Coyne C, Rawson RB, Perrimon N. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2006;2:e102. doi: 10.1371/journal.ppat.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun CD, Liu OW, Madhani HD. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 2007;3:e22. doi: 10.1371/journal.ppat.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Dann CE, III, Bruick RK. Dioxygenases as O2-dependent regulators of the hypoxic response pathway. Biochem Biophys Res Commun. 2005;338:639–647. doi: 10.1016/j.bbrc.2005.08.140. [DOI] [PubMed] [Google Scholar]
- Datta S, Osborne TF. Activation domains from both monomers contribute to transcriptional stimulation by SREBP dimers. J Biol Chem. 2005;280:3338–3345. doi: 10.1074/jbc.M411222200. [DOI] [PubMed] [Google Scholar]
- Dawson PA, Hofmann SL, Van Der Westhuyzen DR, Sudhof TC, Brown MS, Goldstein JL. Sterol-dependent repression of low density lipoprotein receptor promoter mediated by 16-basepair sequence adjacent to binding site for transcription factor Sp1. J Biol Chem. 1988;263:3372–3379. [PubMed] [Google Scholar]
- Dobrosotskaya IY, Seegmiller AC, Brown MS, Goldstein JL, Rawson RB. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science. 2002;296:879–883. doi: 10.1126/science.1071124. [DOI] [PubMed] [Google Scholar]
- Dooley KA, Bennett MK, Osborne TF. A critical role for CREB as a co-activator in sterol regulated transcription of HMG CoA synthase promoter. J Biol Chem. 1999;274:5285–5291. doi: 10.1074/jbc.274.9.5285. [DOI] [PubMed] [Google Scholar]
- Du X, Kristiana I, Wong J, Brown AJ. Involvement of Akt in ER-to-Golgi transport of SCAP/SREBP: A link between a key cell proliferative pathway and membrane synthesis. Mol Biol Cell. 2006;17:2735–2745. doi: 10.1091/mbc.E05-11-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan EA, Brown MS, Goldstein JL, Sakai J. Cleavage site for sterol-regulated protease localized to a Leu–Ser bond in the lumenal loop of sterol regulatory element binding protein-2. J Biol Chem. 1997;272:12778–12785. doi: 10.1074/jbc.272.19.12778. [DOI] [PubMed] [Google Scholar]
- Duncan E, Dave UP, Sakai J, Goldstein JL, Brown MS. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J Biol Chem. 1998;273:17801–17809. doi: 10.1074/jbc.273.28.17801. [DOI] [PubMed] [Google Scholar]
- Ericsson J, Edwards PA. CBP is required for sterol-regulated and sterol regulatory element-binding protein-regulated transcription. J Biol Chem. 1998;273:17865–17870. doi: 10.1074/jbc.273.28.17865. [DOI] [PubMed] [Google Scholar]
- Ericsson J, Jackson SM, Edwards PA. Synergistic binding of sterol regulatory element-binding protein and NF-Y to farnesyl diphosphate synthase promoter is critical for sterol-regulated expression of the gene. J Biol Chem. 1996;271:24359–24364. doi: 10.1074/jbc.271.40.24359. [DOI] [PubMed] [Google Scholar]
- Gibot L, Follet J, Metges JP, Auvray P, Simon B, Corcos L, Le Jossic-Corcos C. Human caspase-7 is positively controlled by SREBP-1 and SREBP-2. Biochem J. 2009;420:473–483. doi: 10.1042/BJ20082057. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear recpetors. Genes & Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Gong Y, Lee JN, Lee PC, Goldstein JL, Brown MS, Ye J. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell Metab. 2006;3:15–24. doi: 10.1016/j.cmet.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes & Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Guan G, Dai P-H, Osborne TF, Kim JB, Shechter I. Multiple regulatory elements regulate transcription of the human squalene synthase gene. J Biol Chem. 1997;272:10295–10302. doi: 10.1074/jbc.272.15.10295. [DOI] [PubMed] [Google Scholar]
- Gurcel L, Abrami L, Girardin S, Tschopp J, van der Goot FG. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell. 2006;126:1135–1145. doi: 10.1016/j.cell.2006.07.033. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Khersonsky S, Marciniak S, Scheuner D, Kaufman RJ, Javitt N, Chang YT, Ron D. Bioactive small molecules reveal antagonism between the integrated stress response and sterol-regulated gene expression. Cell Metab. 2005;2:361–371. doi: 10.1016/j.cmet.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty BD, Bobard A, Hainault I, Ferre P, Bossard P, Foufelle F. Distinct roles of insulin and liver X receptor in the induction and cleavage of sterol regulatory element-binding protein-1c. Proc Natl Acad Sci. 2005;102:791–796. doi: 10.1073/pnas.0405067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci. 2003a;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shimomura I, Ikemoto S, Bashmakov Y, Hammer RE. Overexpression of sterol regulatory element-binding protein-1a in mouse adipose tissue produces adipocyte hypertrophy, increased fatty acid secretion, and fatty liver. J Biol Chem. 2003b;278:36652–36660. doi: 10.1074/jbc.M306540200. [DOI] [PubMed] [Google Scholar]
- Huber RM, Murphy K, Miao B, Link JR, Cunningham MR, Rupar MJ, Gunyuzlu PL, Haws TF, Kassam A, Powell F, et al. Generation of multiple farnesoid-X-receptor isoforms through the use of alternative promoters. Gene. 2002;290:35–43. doi: 10.1016/s0378-1119(02)00557-7. [DOI] [PubMed] [Google Scholar]
- Hughes BT, Espenshade PJ. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 2008;27:1491–1501. doi: 10.1038/emboj.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Lee CY, Bien CM, Espenshade PJ. 4-Methyl sterols regulate fission yeast SREBP-Scap under low oxygen and cell stress. J Biol Chem. 2007;282:24388–24396. doi: 10.1074/jbc.M701326200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante RE, Abi-Mosleh L, Radhakrishnan A, Dale JD, Brown MS, Goldstein JL. Purified NPC1 protein. I. Binding of cholesterol and oxysterols to a 1278-amino acid membrane protein. J Biol Chem. 2008;283:1052–1063. doi: 10.1074/jbc.M707943200. [DOI] [PubMed] [Google Scholar]
- Inoue N, Shimano H, Nakakuki M, Matsuzaka T, Nakagawa Y, Yamamoto T, Sato R, Takahashi A, Sone H, Yahagi N, et al. Lipid synthetic transcription factor SREBP-1a activates p21WAF1/CIP1, a universal cyclin-dependent kinase inhibitor. Mol Cell Biol. 2005;25:8938–8947. doi: 10.1128/MCB.25.20.8938-8947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon TI, Zhu B, Larson JL, Osborne TF. SREBP-2 regulates gut peptide secretion through intestinal bitter taste receptor signaling in mice. J Clin Invest. 2008;118:3693–3700. doi: 10.1172/JCI36461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C, Chan C, Kaplowitz N. Predominant role of sterol response element binding proteins (SREBP) lipogenic pathways in hepatic steatosis in the murine intragastric ethanol feeding model. J Hepatol. 2006;45:717–724. doi: 10.1016/j.jhep.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Joseph SB, Laffitte BA, Patel PH, Watson MA, Matsukuma KE, Walczak R, Collins JL, Osborne TF, Tontonoz P. Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J Biol Chem. 2002;277:11019–11025. doi: 10.1074/jbc.M111041200. [DOI] [PubMed] [Google Scholar]
- Kammoun HL, Chabanon H, Hainault I, Luquet S, Magnan C, Koike T, Ferre P, Foufelle F. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest. 2009;119:1201–1215. doi: 10.1172/JCI37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlson P. Terpenoids in insects. Biochem Soc Symp. 1970;29:145–156. [PubMed] [Google Scholar]
- Kim JB, Spotts GD, Halvorsen Y-D, Shih H-M, Ellenberger T, Towle HC, Spiegelman BM. Dual DNA binding specificity of ADD1/SREBP-1 controlled by a single amino acid in the basic helix–loop–helix domain. Mol Cell Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Sarraf P, Wright M, Yao KM, Mueller E, Solanes G, Lowell BB, Speigelman BM. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP-1c. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeva M, Crawford QT, Seiber M, Wang CY, Han M. Monomethyl branched-chain fatty acids play an essential role in Caenorhabditis elegans development. PLoS Biol. 2004;2:E257. doi: 10.1371/journal.pbio.0020257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeva M, Euler T, Han M. A branched-chain fatty acid is involved in post-embryonic growth control in parallel to the insulin receptor pathway and its biosynthesis is feedback-regulated in C. elegans. Genes & Dev. 2008;22:2102–2110. doi: 10.1101/gad.1692008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunte AS, Matthews KA, Rawson RB. Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell Metab. 2006;3:439–448. doi: 10.1016/j.cmet.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Lane S, Zhou S, Pan T, Dai Q, Liu H. The basic helix–loop–helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via TEC1. Mol Cell Biol. 2001;21:6418–6428. doi: 10.1128/MCB.21.19.6418-6428.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Glimcher LH. Intersection of the unfolded protein response and hepatic lipid metabolism. Cell Mol Life Sci. 2009;66:2835–2850. doi: 10.1007/s00018-009-0049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JN, Ye J. Proteolytic activation of sterol regulatory element-binding protein induced by cellular stress through depletion of Insig-1. J Biol Chem. 2004;279:45257–45265. doi: 10.1074/jbc.M408235200. [DOI] [PubMed] [Google Scholar]
- Lee JN, Song B, DeBose-Boyd RA, Ye J. Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. J Biol Chem. 2006;281:39308–39315. doi: 10.1074/jbc.M608999200. [DOI] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Stewart EV, Hughes BT, Espenshade PJ. Oxygen-dependent binding of Nro1 to the prolyl hydroxylase Ofd1 regulates SREBP degradation in yeast. EMBO J. 2009;28:135–143. doi: 10.1038/emboj.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logette E, Solary E, Corcos L. Identification of a functional DNA binding site for the SREBP-1c transcription factor in the first intron of the human caspase-2 gene. Biochim Biophys Acta. 2005;1738:1–5. doi: 10.1016/j.bbalip.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Magaña MM, Koo S-H, Towle HC, Osborne TF. Different sterol regulatory element binding protein -1 Isoforms utilize distinct co-regulatory factors to activate the promoter for fatty acid synthase. J Biol Chem. 2000;275:4726–4733. doi: 10.1074/jbc.275.7.4726. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- Mason RL, Hunt HM, Hurxthal L. Blood cholesterol values in hyperthyroidism and hypothyroidism. N Engl J Med. 1930;203:1273–1278. [Google Scholar]
- Matsumoto M, Ogawa W, Akimoto K, Inoue H, Miyake K, Furukawa K, Hayashi Y, Iguchi H, Matsuki Y, Hiramatsu R, et al. PKCλ in liver mediates insulin-induced SREBP-1c expression and determines both hepatic lipid content and overall insulin sensitivity. J Clin Invest. 2003;112:935–944. doi: 10.1172/JCI18816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews KA, Kunte AS, Tambe-Ebot E, Rawson RB. Alternative processing of sterol regulatory element binding protein during larval development in Drosophila melanogaster. Genetics. 2009;181:119–128. doi: 10.1534/genetics.108.093450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay RM, McKay JP, Avery L, Graff JM. C. elegans: A model for exploring the genetics of fat storage. Dev Cell. 2003;4:131–142. doi: 10.1016/s1534-5807(02)00411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, Schonleber McCaw P, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding protein, daughterless MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME, Matthews JR, Quong MW, Rivera RR, Stuiver MH. Structure and function of helix–loop–helix proteins. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- Nåår AM, Beaurang PA, Robinson KM, Oliner JD, Avizonis D, Scheek S, Zwicker J, Kadonaga JT, Tjian R. Chromatin, TAFs and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes & Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Yoneda Y. Dimerization of sterol regulatory element-binding protein 2 via the helix–loop–helix-leucine zipper domain is a prerequisite for its nuclear localization mediated by importin β. Mol Cell Biol. 2001;21:2779–2789. doi: 10.1128/MCB.21.8.2779-2789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness GC, Pendleton LC, Li YC, Chiang JYL. Effect of thyroid hormone on hepatic cholesterol 7ahydroxylase, LDL receptor, HMG CoA reductase, farnesyl pyrophosphate synthetase and apolipoprotein A-1 mRNA levels in hypophysectomized rats. Biochem Biophys Res Commun. 1990;173:1150–1156. doi: 10.1016/0006-291x(90)91568-d. [DOI] [PubMed] [Google Scholar]
- Neuhold LA, Wold B. HLH forced dimers: Tethering MyoD to E47 generates a dominant positive myogenic factor insulated from negative regulation by Id. Cell. 1993;74:1033–1042. doi: 10.1016/0092-8674(93)90725-6. [DOI] [PubMed] [Google Scholar]
- Oberste-Berghaus C, Zanger K, Hashimoto K, Cohen RN, Hollenberg AN, Wondisford FE. Thyroid hormone-independent interaction between the thyroid hormone receptor β2 amino terminus and coactivators. J Biol Chem. 2000;275:1787–1792. doi: 10.1074/jbc.275.3.1787. [DOI] [PubMed] [Google Scholar]
- Ohgami N, Ko DC, Thomas M, Scott MP, Chang CC, Chang TY. Binding between the Niemann-Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc Natl Acad Sci. 2004;101:12473–12478. doi: 10.1073/pnas.0405255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliner JD, Andresen JM, Hansen SK, Zhou S, Tjian R. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes & Dev. 1996;10:2903–2911. doi: 10.1101/gad.10.22.2903. [DOI] [PubMed] [Google Scholar]
- Osborne TF. Transcriptional control mechanisms in the regulation of cholesterol balance. Crit Rev Eukaryot Gene Expr. 1995;5:317–335. doi: 10.1615/critreveukargeneexpr.v5.i3-4.50. [DOI] [PubMed] [Google Scholar]
- Osborne TF. Sterol regulatory element binding proteins (SREBPs): Key regulators of nutritional homeostasis and insulin action. J Biol Chem. 2000;275:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- Parraga A, Bellsolell L, Ferre-D'Amare AR, Burley SK. Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 Å resolution. Structure. 1998;6:661–672. doi: 10.1016/s0969-2126(98)00067-7. [DOI] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Sun LP, Kwon HJ, Brown MS, Goldstein JL. Direct binding of cholesterol to the purified membrane region of SCAP: Mechanism for a sterol-sensing domain. Mol Cell. 2004;15:259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Oxysterols block transport by binding to Insig. Proc Natl Acad Sci. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: A delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson RB. The SREBP pathway—Insights from Insigs and insects. Nat Rev Mol Cell Biol. 2003;4:631–640. doi: 10.1038/nrm1174. [DOI] [PubMed] [Google Scholar]
- Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4:e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes & Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rome S, Lecomte V, Meugnier E, Rieusset J, Debard C, Euthine V, Vidal H, Lefai E. Microarray analyses of SREBP-1a and SREBP-1c target genes identify new regulatory pathways in muscle. Physiol Genomics. 2008;34:327–337. doi: 10.1152/physiolgenomics.90211.2008. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rosenfeld E, Beauvoit B. Role of the non-respiratory pathways in the utilization of molecular oxygen by Saccharomyces cerevisiae. Yeast. 2003;20:1115–1144. doi: 10.1002/yea.1026. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JM, Osborne TF. HLH106, a Drosophila sterol regulatory element-binding protein in a natural cholesterol auxotroph. J Biol Chem. 1998;273:16112–16121. doi: 10.1074/jbc.273.26.16112. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajan MP, Standaert ML, Nimal S, Varanasi U, Pastoor T, Mastorides S, Braun U, Leitges M, Farese RV. The critical role of atypical protein kinase C in activating hepatic SREBP-1c and NFκB in obesity. J Lipid Res. 2009a;50:1133–1145. doi: 10.1194/jlr.M800520-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajan MP, Standaert ML, Rivas J, Miura A, Kanoh Y, Soto J, Taniguchi CM, Kahn CR, Farese RV. Role of atypical protein kinase C in activation of sterol regulatory element binding protein-1c and nuclear factor κ B (NFκB) in liver of rodents used as a model of diabetes, and relationships to hyperlipidaemia and insulin resistance. Diabetologia. 2009b;52:1197–1207. doi: 10.1007/s00125-009-1336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Shimizu Y, Kondou S, Chibazakura T, Hishinuma F. Structure and molecular analysis of RGR1, a gene required for glucose repression of Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4130–4138. doi: 10.1128/mcb.10.8.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez HB, Yieh L, Osborne TF. Cooperation by sterol regulatory element-binding protein and Sp1 in sterol regulation of low density lipoprotein receptor gene. J Biol Chem. 1995;270:1161–1169. doi: 10.1074/jbc.270.3.1161. [DOI] [PubMed] [Google Scholar]
- Sato R, Yang J, Wang X, Evans MJ, Ho YK, Goldstein JL, Brown MS. Assignment of the membrane attachment, DNA binding, and transcriptional activation domains of sterol regulatory element-binding protein-1 (SREBP-1) J Biol Chem. 1994;209:17267–17273. [PubMed] [Google Scholar]
- Sato R, Inoue J, Kawabe Y, Kodama T, Takano T, Maeda M. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J Biol Chem. 1996;271:26461–26464. doi: 10.1074/jbc.271.43.26461. [DOI] [PubMed] [Google Scholar]
- Seegmiller AC, Dobrosotskaya I, Goldstein JL, Ho YK, Brown MS, Rawson RB. The SREBP pathway in Drosophila: Regulation by palmitate, not sterols. Dev Cell. 2002;2:229–238. doi: 10.1016/s1534-5807(01)00119-8. [DOI] [PubMed] [Google Scholar]
- Sehgal A, Lee CY, Espenshade PJ. SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS Genet. 2007;3:e131. doi: 10.1371/journal.pgen.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y-K, Chong HK, Infante AM, Xie X, Osborne TF. Genome-wide analysis of SREBP-1 binding in mouse liver chromatin reveals a preference for promoter proximal binding to a new motif. Proc Natl Acad Sci. 2009;106:13765–13769. doi: 10.1073/pnas.0904246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever N, Yang T, Brown MS, Goldstein JL, DeBose-Boyd RA. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol Cell. 2003;11:25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Shimano H, Horton JD, Shimomura L, Hammer RE, Brown MS, Goldstein JL. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and cultured cells. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Hammer RE, Richardson JA, Ikemoto S, Bashmakov Y, Goldstein JL, Brown MS. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: Model for congenital generalized lipodystrophy. Genes & Dev. 1998;12:3182–3194. doi: 10.1101/gad.12.20.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Bashmakov Y, Ikemoto S, Horton JD, Brown MS, Goldstein JL. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci. 1999a;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J Biol Chem. 1999b;274:30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- Shin DJ, Osborne TF. Thyroid hormone regulation and cholesterol metabolism are connected through sterol regulatory element-binding protein-2 (SREBP-2) J Biol Chem. 2003;278:34114–34118. doi: 10.1074/jbc.M305417200. [DOI] [PubMed] [Google Scholar]
- Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. Origin and diversification of the basic helix–loop–helix gene family in metazoans: Insights from comparative genomics. BMC Evol Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JR, Osborne TF, Brown MS, Goldstein JL, Gil G. Multiple sterol regulatory elements in promoter for hamster 3-hydroxy-3-methylglutaryl coenzyme A synthase. J Biol Chem. 1988;263:18480–18487. [PubMed] [Google Scholar]
- Sun LP, Seemann J, Goldstein JL, Brown MS. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc Natl Acad Sci. 2007;104:6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist A, Bengoechea-Alonso MT, Ye X, Lukiyanchuk V, Jin J, Harper JW, Ericsson J. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell Metab. 2005;1:379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Taghibiglou C, Martin HG, Rose JK, Ivanova N, Lin CH, Lau HL, Rai S, Wang YT, Rankin CH. Essential role of SBP-1 activation in oxygen deprivation induced lipid accumulation and increase in body width/length ratio in Caenorhabditis elegans. FEBS Lett. 2009;583:831–834. doi: 10.1016/j.febslet.2009.01.045. [DOI] [PubMed] [Google Scholar]
- Taniguchi CM, Kondo T, Sajan M, Luo J, Bronson R, Asano T, Farese R, Cantley LC, Kahn CR. Divergent regulation of hepatic glucose and lipid metabolism by phosphoinositide 3-kinase via Akt and PKCλ/ζ. Cell Metab. 2006;3:343–353. doi: 10.1016/j.cmet.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes & Dev. 2006;20:1137–1149. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theopold U, Ekengren S, Hultmark D. HLH106, a Drosophila transcription factor with similarity to the vertebrate sterol responsive element binding protein. Proc Natl Acad Sci. 1996;93:1195–1199. doi: 10.1073/pnas.93.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd BL, Stewart EV, Burg JS, Hughes AL, Espenshade PJ. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol Cell Biol. 2006;26:2817–2831. doi: 10.1128/MCB.26.7.2817-2831.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Kim JB, Graves RA, Spiegelman BM. ADD1: A novel helix–loop–helix transcription factor associated with adipocyte determination and differentiation. Mol Cell Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth JI, Datta S, Athanikar JN, Freedman LP, Osborne TF. Selective co-activator interactions in gene activation by SREBP-1a and -1c. Mol Cell Biol. 2004;24:8288–8300. doi: 10.1128/MCB.24.18.8288-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Briggs MR, Hua X, Yokoyama C, Goldstein JL, Brown MS. Nuclear protein that binds sterol regulatory element of low density receptor promoter II: Purification and characterization. J Biol Chem. 1993;268:14497–14504. [PubMed] [Google Scholar]
- Wang X, Zelenski NG, Yang J, Sakai J, Brown MS, Goldstein JL. Cleavage of sterol regulatory element binding proteins (SREBPs) by CPP32 during apoptosis. EMBO J. 1996;15:1012–1020. [PMC free article] [PubMed] [Google Scholar]
- Werstuck GH, Lentz SR, Dayal S, Hossain GS, Sood SK, Shi YY, Zhou J, Maeda N, Krisans SK, Malinow MR, et al. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J Clin Invest. 2001;107:1263–1273. doi: 10.1172/JCI11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA., Jr A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008;4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc Natl Acad Sci. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS. Liver-specific mRNA for Insig-2 down-regulated by insulin: Implications for fatty acid synthesis. Proc Natl Acad Sci. 2003;100:3155–3160. doi: 10.1073/pnas.0130116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS. Crucial step in cholesterol homeostasis: Sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S, et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Yellaturu CR, Deng X, Cagen LM, Wilcox HG, Mansbach CM, II, Siddiqi SA, Park EA, Raghow R, Elam MB. Insulin enhances post-translational processing of nascent SREBP-1c by promoting its phosphorylation and association with COPII vesicles. J Biol Chem. 2009;284:7518–7532. doi: 10.1074/jbc.M805746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, Goldstein JL, Brown MS. SREBP-1, a basic-helix–loop–helix-zipper protein that controls transcription of the low density receptor gene. Cell. 1993;75:185–197. [PubMed] [Google Scholar]
- Zhang CZ, Shin D-J, Osborne TF. A simple promoter containing two Sp1 sites controls expression of sterol regulatory element binding protein-1a (SREBP-1a) Biochem J. 2005;386:1–8. doi: 10.1042/BJ20041514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Qi C, Korenberg JR, Chen XN, Noya D, Rao MS, Reddy JK. Structural organization of mouse peroxisome proliferator-activated receptor γ (mPPAR γ) gene: Alternative promoter use and different splicing yield two mPPAR γ isoforms. Proc Natl Acad Sci. 1995;92:7921–7925. doi: 10.1073/pnas.92.17.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]