Abstract
It remains unclear whether a microRNA (miRNA) affects a given phenotype via concomitant down-regulation of its entire repertoire of targets or instead by suppression of only a modest subset of effectors. We demonstrate that inhibition of breast cancer metastasis by miR-31—a miRNA predicted to modulate >200 mRNAs—can be entirely explained by miR-31's pleiotropic regulation of three targets. Thus, concurrent re-expression of integrin-α5, radixin, and RhoA abrogates miR-31-imposed metastasis suppression. These effectors influence distinct steps of the metastatic process. Our findings have implications concerning the importance of pleiotropy for the biological actions of miRNAs and provide mechanistic insights into metastasis.
Keywords: MicroRNA, metastasis, miR-31, breast cancer, colonization, pleiotropy
MicroRNAs (miRNAs) are an evolutionarily conserved family of regulatory RNAs that inhibit their mRNA targets post-transcriptionally, leading to modulation of diverse biological processes, including the development and progression of cancer (Ambros 2004; Bartel 2009; Ventura and Jacks 2009). An individual miRNA is capable of regulating dozens of distinct mRNAs (Baek et al. 2008; Selbach et al. 2008), and it is thought that pleiotropic suppression of multiple downstream effectors may underlie the phenotypic changes observed upon perturbing the levels of certain miRNAs (Rodriguez et al. 2007; Thai et al. 2007; van Rooij et al. 2007; Zhao et al. 2007; Johnnidis et al. 2008; Ventura et al. 2008). It remains unclear, however, whether these consequences depend on simultaneous deregulation of the entire repertoire of targets of a given miRNA or instead on the altered activity of only a small subset of effectors.
Metastases, which are responsible for 90% of human cancer deaths, arise via a complex series of events, collectively termed the invasion–metastasis cascade (Fidler 2003; Gupta and Massagué 2006). In order to metastasize, cells in a primary tumor must become motile, degrade surrounding extracellular matrix (local invasion), intravasate into the vasculature, retain viability during transit through the circulation, extravasate into the parenchyma of a distant tissue, survive in this foreign microenvironment to form micrometastases, and, finally, thrive in their new milieu and establish macroscopic secondary tumors (colonization) (Fidler 2003). Colonization is the rate-limiting step of the invasion–metastasis cascade, yet the molecular underpinnings of this process are poorly understood (Gupta and Massagué 2006).
We determined recently that expression of the miRNA miR-31 was both necessary and sufficient to inhibit the metastasis of human breast cancer xenografts, and that miR-31 levels correlated inversely with metastatic relapse in breast carcinoma patients (Valastyan et al. 2009). We attributed these effects to miR-31's ability to pleiotropically suppress a cohort of prometastatic targets; however, we did not identify a minimal set of downstream effectors whose concomitant re-expression is sufficient to fully override miR-31's influences on metastasis. For this reason, we sought to determine whether the impact of miR-31 on metastasis could be explained by its ability to pleiotropically modulate a defined subset of its >200 predicted targets.
Results and Discussion
We demonstrated previously that miR-31 regulates six mRNAs that encode proteins with roles in cell motility and tumor progression: frizzled3 (Fzd3), integrin-α5 (ITGA5), matrix metallopeptidase 16 (MMP16), myosin phosphatase-Rho-interacting protein (M-RIP), radixin (RDX), and RhoA (Valastyan et al. 2009). To begin to address whether miR-31-imposed inhibition of one or more of these effectors might be responsible for mediating miR-31's anti-metastatic influences, we stably suppressed these six mRNAs individually in otherwise metastatic MDA-MB-231 human breast cancer cells (“231 cells”) using shRNAs. 231 cells are largely devoid of endogenous miR-31 and robustly express these six effectors; moreover, ectopic miR-31 impairs metastasis by these cells (Valastyan et al. 2009).
For each gene, we derived multiple cell lines that stably expressed a distinct shRNA targeting unique sequences in the encoded mRNA in order to minimize confounding influences from shRNA off-target effects (Supplemental Figs. 1A, 2A). At least one shRNA against each of the six effectors reduced its target's level by a factor comparable with that elicited by miR-31 expression (Valastyan et al. 2009). This allowed us to reasonably approximate the consequences of miR-31's actions on each individual downstream effector.
These shRNA-expressing 231 cells were subjected to in vitro assays that model traits important for metastasis. We observed that individual suppression of ITGA5, RDX, or RhoA reduced invasion, motility, and resistance to anoikis-mediated cell death in vitro; in contrast, the Fzd3, MMP16, or M-RIP shRNAs failed to substantially affect these behaviors (Supplemental Figs. 1B–D, 2B–D). For shRNAs that conferred measurable responses, the magnitude of these responses was directly correlated with the extent of knockdown achieved, suggesting that these effects arose as a specific consequence of reduced levels of the targeted protein. Inhibition of Fzd3, ITGA5, MMP16, M-RIP, RDX, or RhoA failed to affect in vitro proliferation (Supplemental Figs. 1E, 2E). Also, the responses evoked by the ITGA5, RDX, and RhoA shRNAs could not be ascribed to saturation of the miRNA biogenesis machinery, as mature levels of eight control miRNAs were unaffected in these cells (Supplemental Fig. 3).
We determined whether suppression of these six mRNAs altered metastatic capacity in vivo by intravenously injecting the shRNA-expressing 231 cells into mice. One month later, cells bearing shRNAs targeting ITGA5, RDX, or RhoA had generated 80%, 85%, and 55% fewer lung metastases than controls, respectively; however, down-regulation of Fzd3, MMP16, or M-RIP did not affect the number of metastases spawned (Supplemental Fig. 4). Thus, inhibition of ITGA5, RDX, or RhoA—but not Fzd3, MMP16, or M-RIP—affects in vitro surrogates of metastatic capacity as well as in vivo metastasis.
To extend these analyses, we stably re-expressed miRNA-insensitive versions of the mRNAs encoding Fzd3, ITGA5, MMP16, M-RIP, RDX, or RhoA individually in 231 cells that already expressed either miR-31 or control vector (Supplemental Fig. 5A). This allowed us to gauge the ability of each of these effectors—when re-expressed—to reverse miR-31's impact on in vivo metastasis. When introduced into the venous circulation of mice, miR-31-expressing cells formed 85% fewer lung metastases than controls 1 mo post-injection (Supplemental Fig. 5B), consistent with our prior findings (Valastyan et al. 2009). Individual re-expression of ITGA5, RDX, or RhoA restored the number of lung metastases in miR-31-expressing cells to 55%, 50%, and 65% of control levels, respectively; in contrast, Fzd3, MMP16, or M-RIP failed to increase lesion number (Supplemental Fig. 5B). Overexpression of ITGA5, RDX, or RhoA did not further enhance metastasis in control 231 cells (Supplemental Fig. 5B), suggesting that signaling from these pathways was already saturated in 231 cells, as has been established previously for RhoA-controlled networks (Pillé et al. 2005). Together, these findings implied that, although miR-31 is capable of suppressing numerous mRNA species, its ability to regulate only a subset of these effectors appears to be crucial for its capacity to impair metastasis.
In support of this notion, when stably re-expressed in 231 cells, Fzd3, MMP16, or M-RIP failed to reverse miR-31-imposed attenuation of invasion, motility, and anoikis resistance in vitro (Supplemental Fig. 6); in contrast, our prior work revealed that restored levels of ITGA5, RDX, or RhoA rescued, at least partially, miR-31-evoked defects in these phenotypes (Valastyan et al. 2009). Based on these in vitro and in vivo re-expression data, as well as the above-described in vitro and in vivo loss-of-function findings, we focused our subsequent analyses on the ability of inhibition of ITGA5, RDX, and RhoA to account for miR-31's anti-metastatic activities.
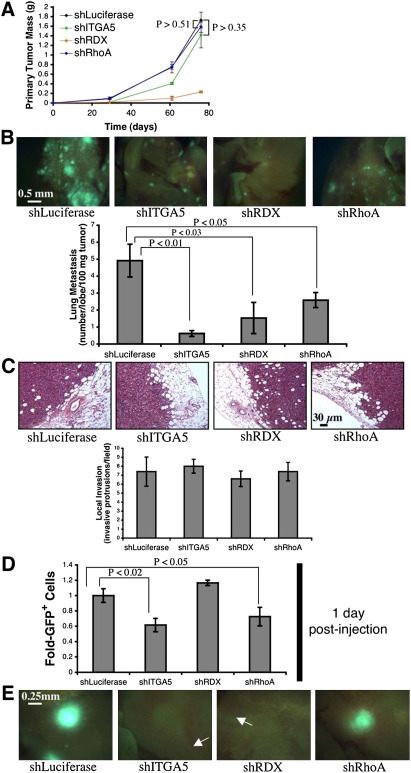
To this end, we investigated the consequences of suppressing ITGA5, RDX, or RhoA individually in an orthotopic injection assay. Accordingly, we implanted 231 cells expressing shRNAs targeting either ITGA5, RDX, or RhoA into the mammary fat pads of mice. Suppression of ITGA5 or RhoA did not affect primary tumor growth; conversely, inhibition of RDX reduced the size of resulting mammary tumors (Fig. 1A). After normalizing for differences in primary tumor growth, cells expressing shRNAs against ITGA5, RDX, or RhoA formed 85%, 70%, and 50% fewer lung metastases than controls 2.5 mo after injection, respectively (Fig. 1B). Thus, inhibition of ITGA5, RDX, or RhoA each impedes metastasis; however, this assay did not reveal the particular step(s) of the invasion–metastasis cascade that were impaired due to suppression of ITGA5, RDX, or RhoA.
Figure 1.
Individual suppression of ITGA5, RDX, or RhoA impairs metastasis in vivo. (A) Primary tumor growth upon orthotopic injection of the indicated GFP-labeled 231 cells into NOD/SCID mice. The assay was terminated after 11 wk due to primary tumor burden. n = 5 per time point. (B, top panels) Fluorescent images of murine lungs to visualize 231 cells 76 d after orthotopic implantation. (Bottom panel) Quantification of metastatic burden. n = 5. (C, top panels) H&E stain of 231 cell primary mammary tumors 57 d after injection. (Bottom panel) Quantification of local invasion. n = 5. All P-values are >0.67 relative to shLuciferase. (D) Prevalence of GFP-labeled 231 cells in the lungs 1 d after intravenous introduction into NOD/SCID mice. n = 4. (E) Fluorescent images of murine lungs to visualize 231 cells 89 d after intravenous injection. (Arrows) Micrometastases. shRNAs used in these assays were shITGA5 #4, shRDX #3, and shRhoA #5. All error bars represent mean ± SEM.
In our previous work, we observed that miR-31 impinges on three steps of the invasion–metastasis cascade in vivo: local invasion, early post-intravasation events (intraluminal viability, extravasation, and/or initial survival in distant tissues), and colonization (Valastyan et al. 2009). Consequently, we evaluated whether the individual suppression of ITGA5, RDX, or RhoA was sufficient to recapitulate one or more of miR-31's multiple effects on the metastatic process. We found that 231 cells containing shRNAs against either ITGA5, RDX, or RhoA formed primary tumors that appeared histologically invasive and were indistinguishable from controls (Fig. 1C). Thus, inhibition of ITGA5, RDX, or RhoA alone does not abolish local invasion in vivo.
Putative effects on early post-intravasation events were examined by quantifying shRNA-expressing 231 cells in the lungs 1 d after intravenous injection. Cells with either suppressed ITGA5 or RhoA were 40% and 30% less prevalent than controls, respectively; however, RDX knockdown did not reduce persistence in the lungs (Fig. 1D). These effects were not attributable to a differential ability of the cells to become lodged initially in the lung microvasculature, as equal numbers of cells were detected in the lungs 10 min after intravenous injection (Supplemental Fig. 7). These data indicated that inhibition of either ITGA5 or RhoA impairs early post-intravasation events in vivo.
To investigate potential effects on colonization (i.e., the capacity of disseminated single cells to yield large, multicellular metastases), the sizes of lung metastases in intravenously injected animals were analyzed 3 mo after implantation. 231 cells expressing either ITGA5 or RDX shRNAs formed only small micrometastases, while RhoA shRNA-containing cells generated macroscopic metastases comparable with those spawned by control cells (Fig. 1E). Hence, suppression of either ITGA5 or RDX alone prevents colonization in vivo.
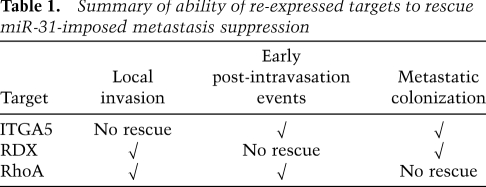
Together, these observations revealed that, while individual suppression of ITGA5, RDX, or RhoA impairs one or more steps of the invasion–metastasis cascade, inhibition of any one of these proteins alone is unable to phenocopy the full spectrum of miR-31's impact on metastasis. This suggested that miR-31 may achieve its influences on multiple distinct stages of the metastatic process via concomitant suppression of several downstream effectors. Provocatively, our loss-of-function analyses indicated that ITGA5, RDX, and RhoA act during at least partially distinct steps of the invasion–metastasis cascade (e.g., RhoA affected early post-intravasation events but not colonization, while RDX had no impact on early post-intravasation events but altered colonization); hence, their concurrent regulation provides a plausible mechanism by which miR-31 might elicit its multiple anti-metastatic effects.
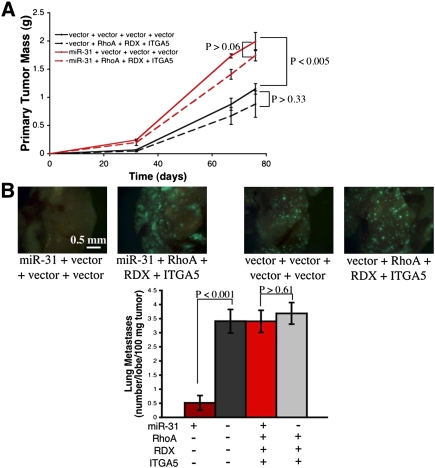
To test this hypothesis, we stably re-expressed miRNA-insensitive mRNAs encoding ITGA5, RDX, and RhoA together in combination—along with either miR-31 or control vector—in 231 cells. When these cells were orthotopically injected into mice, miR-31 enhanced primary tumor growth, recapitulating our prior findings (Valastyan et al. 2009); simultaneous re-expression of ITGA5, RDX, and RhoA failed to alter the size of miR-31-containing or control primary tumors (Fig. 2A). Despite their ability to generate larger primary tumors, miR-31-expressing 231 cells were impaired by >80% in their ability to spawn lung metastases (Fig. 2B). ITGA5, RDX, and RhoA did not enhance metastasis in control 231 cells; however, concomitant re-expression of ITGA5, RDX, and RhoA in 231 cells containing miR-31 completely abrogated miR-31-imposed metastasis suppression (Fig. 2B). These data implied that the impact of miR-31 on in vivo metastasis can be explained by miR-31's capacity to inhibit a cohort of three downstream effectors. This was quite surprising, as computational algorithms predict that miR-31 regulates >200 mRNAs, many of which encode proteins that function in metastasis-relevant processes (Krek et al. 2005; Grimson et al. 2007).
Figure 2.
Simultaneous re-expression of ITGA5, RDX, and RhoA abrogates miR-31-imposed metastasis suppression in vivo. (A) Primary tumor growth upon orthotopic injection of the indicated GFP-labeled 231 cells into NOD/SCID mice. The assay was terminated after 11 wk due to primary tumor burden. n = 5 per time point. (B, top panels) Fluorescent images of murine lungs to visualize 231 cells 67 d after orthotopic implantation. (Bottom panel) Quantification of metastatic burden. n = 5. All error bars represent mean ± SEM.
Since the combined re-expression of ITGA5, RDX, and RhoA entirely abolished miR-31-evoked metastasis suppression, we also determined whether these three effectors were able to reverse a subset of miR-31's influences on metastasis when re-expressed either individually or in different combinations. Thus, we created 231 cells stably expressing miR-31 or control vector plus all possible permutations of zero, one, two, or three of these miR-31 targets (all rendered miRNA-resistant) (Supplemental Fig. 8). miR-31, ITGA5, RDX, and RhoA failed to affect cell proliferation in vitro (Supplemental Fig. 9A). However, individual re-expression of ITGA5, RDX, or RhoA rescued, at least partially, in vitro defects in invasion, motility, and anoikis resistance conferred by ectopic miR-31; the extent of reversal was more pronounced when multiple effectors were re-expressed in combination (Supplemental Fig. 9B–D).Thus, ITGA5, RDX, and RhoA control in vitro behaviors important for metastasis downstream from miR-31.
To assay the respective abilities of all possible combinations of re-expressed ITGA5, RDX, and/or RhoA to reverse miR-31's influences on in vivo metastasis, 231 cells expressing miR-31, ITGA5, RDX, and/or RhoA were orthotopically implanted into mice. miR-31 generally promoted primary tumor growth, while restored levels of ITGA5, RDX, and RhoA failed to consistently affect the growth of primary tumors (Fig. 3A; Supplemental Table 1). miR-31 reduced the incidence of metastatic lesions in the lungs by >90% (Fig. 3B). When individually re-expressed in miR-31-containing cells, ITGA5, RDX, or RhoA increased metastasis to 40%, 45%, and 65% of control levels, respectively; re-expression of any two of these targets in miR-31-positive cells yielded 85% as many metastases as controls (Fig. 3B). As before, concomitant re-expression of ITGA5, RDX, and RhoA in cells containing miR-31 restored the number of lung metastases to 100% of that observed in controls (Fig. 3B). Hence, these three effectors make distinct contributions to in vivo metastasis that can collaborate to explain miR-31's influence on this process; however, these observations failed to delineate the specific step(s) of the invasion–metastasis cascade affected by various combinations of re-expressed ITGA5, RDX, and/or RhoA.
Figure 3.
Re-expression of ITGA5, RDX, and/or RhoA affords both unique and partially overlapping reversal of miR-31-evoked inhibition of spontaneous metastasis in vivo. (A) Primary tumor growth upon orthotopic implantation of the indicated GFP-labeled 231 cells into nude mice. The assay was terminated after 13 wk due to primary tumor burden. n = 5. (B,top panels) Fluorescent images of murine lungs to visualize 231 cells 88 d after orthotopic injection. (Bottom panel) Quantification of metastatic burden. n = 5. (C) H&E stain of 231 cell primary mammary tumors 54 d after injection. (Bottom panel) Quantification of local invasion. n = 5. All error bars represent mean ± SEM.
miR-31 affects three steps of the invasion–metastasis cascade in vivo: local invasion, early post-intravasation events, and colonization (Valastyan et al. 2009). To investigate whether ITGA5, RDX, and RhoA—when overexpressed—could synergize to reverse miR-31's effects on local invasion, we examined the histological appearance of primary tumors that developed in orthotopically injected mice. Whereas control 231 cell tumors displayed clear evidence of invasion, miR-31-expressing tumors were well-confined (Fig. 3C), as we documented previously (Valastyan et al. 2009). While ITGA5, RDX, and RhoA did not alter invasion in control 231 cell tumors, combined re-expression of these three targets abolished the previously well-encapsulated phenotype of miR-31-expressing tumors (Fig. 3C). miR-31-containing cells with restored levels of either RDX or RhoA alone formed primary tumors that appeared invasive, although reversal of miR-31-imposed invasion defects was incomplete; ITGA5 did not affect encapsulation (Fig. 3C). These observations revealed that miR-31-dependent attenuation of local invasion can be attributed to miR-31's ability to regulate RDX and RhoA. Ostensibly, in light of our shRNA studies (Fig. 1C), RDX and RhoA function redundantly—with either one another or additional, still-unidentified miR-31 targets—to promote invasion in vivo.
We also examined whether re-expression of these three targets could reverse the impact of miR-31 on early post-intravasation events. To do so, we introduced 231 cells into the venous circulation of mice and assayed the number of cells in the lungs 1 d after injection. Consistent with our previous findings (Valastyan et al. 2009), miR-31-expressing cells were fivefold impaired in their ability to persist in the lungs (Fig. 4A), indicating that miR-31 impeded one or more early post-intravasation events. ITGA5, RDX, and RhoA failed to affect early post-intravasation events in control 231 cells (Fig. 4A). In contrast, individual re-expression of either ITGA5 or RhoA restored the number of miR-31-expressing cells in the lungs to 50% of control levels; RDX did not augment the ability of cells containing miR-31 to persist in the lungs at this time point (Fig. 4A). Simultaneous reintroduction of ITGA5 and RhoA in miR-31-expressing cells sufficed to completely override miR-31-imposed obstruction of early post-intravasation events (Fig. 4A). These effects were not a consequence of an altered ability of ITGA5-, RDX-, RhoA-, and/or miR-31-expressing cells to become lodged initially in the lung microvasculature, as equal numbers of cells were detected in the lungs 10 min after intravenous injection (Supplemental Fig. 10). These data provided evidence that miR-31-evoked suppression of early post-intravasation events can be ascribed to miR-31's ability to modulate ITGA5 and RhoA.
Figure 4.
Re-expression of ITGA5, RDX, and/or RhoA affords both unique and partially overlapping reversal of miR-31-mediated inhibition of experimental metastasis in vivo. (A) Prevalence of the indicated GFP-labeled 231 cells in the lungs 1 d after intravenous introduction into NOD/SCID mice. n = 4. (B) Fluorescent images of murine lungs to visualize 231 cells 84 d after tail vein injection. (C) Lung metastatic burden 84 d subsequent to intravenous injection. n = 5. All error bars represent mean ± SEM.
Three months after intravenous injection, control 231 cells generated large macroscopic metastases while miR-31-expressing cells yielded only small micrometastases (Fig. 4B). Hence, miR-31 prevented disseminated tumor cells from reinitiating their proliferative program at the site of metastasis, in consonance with miR-31's reported influence on colonization (Valastyan et al. 2009). Concomitant re-expression of ITGA5, RDX, and RhoA in miR-31-containing cells abrogated miR-31-imposed suppression of colonization, yet overexpression of these three targets in control 231 cells failed to increase lesion size (Fig. 4B). Individually restored levels of either ITGA5 or RDX in miR-31-expressing cells reversed miR-31's effects on colonization; RhoA did not affect this parameter (Fig. 4B). Thus, the ability of miR-31 to inhibit colonization can derive from its capacity to suppress ITGA5 and RDX.
In this same assay, miR-31-expressing 231 cells formed 20-fold fewer lung metastases than controls (Fig. 4C). When individually re-expressed in miR-31-containing cells, ITGA5, RDX, or RhoA increased the number of metastases formed to 60%, 60%, and 50% of control levels, respectively (Fig. 4C). Restored levels of pairwise combinations of these three targets in miR-31-expressing cells enhanced lesion number to >70% of controls; importantly, simultaneous re-expression of ITGA5, RDX, and RhoA in miR-31-containing cells completely abolished miR-31-mediated metastasis suppression (Fig. 4C). Taken together, the preceding experiments indicated that the impact of miR-31 on metastasis can be entirely explained by miR-31's capacity to regulate ITGA5, RDX, and RhoA; these three targets act at partially overlapping steps of the invasion–metastasis cascade downstream from miR-31 in vivo (Table 1).
Table 1.
Summary of ability of re-expressed targets to rescue miR-31-imposed metastasis suppression
It remained possible that the ability of ITGA5, RDX, and RhoA to override miR-31's actions arose due to some peculiarity of the 231 cell system. To address this, we extended our analyses to SUM-159 human breast cancer cells. Like 231 cells, SUM-159 cells lack endogenous miR-31, are highly aggressive in vitro, and display impaired invasion, motility, and anoikis resistance upon ectopic miR-31 (Valastyan et al. 2009). We created SUM-159 cells stably expressing all 16 potential combinations of either miR-31 or control vector plus miRNA-resistant mRNAs encoding ITGA5, RDX, and/or RhoA; all lines displayed comparable in vitro proliferative kinetics (Supplemental Fig. 11A,B). Consistent with our observations in 231 cells, individual re-expression of ITGA5, RDX, or RhoA in miR-31-containing SUM-159 cells rescued, at least partially, in vitro defects in invasion, motility, and anoikis resistance attributable to ectopic miR-31; as before, the extent of rescue was more pronounced when multiple effectors were concomitantly re-expressed (Supplemental Fig, 11C–E). Hence, the ability of ITGA5, RDX, and RhoA re-expression to override the actions of miR-31 is not confined to 231 cells.
Whereas individual re-expression of ITGA5, RDX, or RhoA largely reversed certain miR-31-imposed metastasis-relevant defects in vitro (Supplemental Figs. 9, 11), individual restoration of ITGA5, RDX, or RhoA levels only partially rescued miR-31's effects on metastasis in vivo (Figs. 3, 4). This underscores the fact that available in vitro assays inadequately model the full complexity of in vivo metastasis; caution must therefore be exercised when deploying these techniques, particularly in the absence of parallel in vivo analyses.
Collectively, the findings of the present study suggest that a miRNA's effects on a given phenotype can be explained by its ability to suppress a relatively modest number of downstream targets. In the present case, the relevant effectors comprise only a small percentage of the total roster of mRNAs targeted by the miRNA under investigation. Our observations are confined to a single miRNA and a single biological endpoint; accordingly, the extent to which this phenomenon is generalizable awaits future investigation. Nevertheless, several recent studies describe strong, but partial, effects on miRNA-mediated phenotypes by modulating individual targets of miRNAs of interest (Ma et al. 2007; Xiao et al. 2007; Yu et al. 2007; Kumar et al. 2008). Such reports suggest the existence of other similarly organized miRNA response networks, in which a miRNA's impact on a biological process can be attributed to that miRNA's ability to inhibit only a small subfraction of its targets.
While our data indicate that ITGA5, RDX, and RhoA represent a minimal cohort of effectors whose regulation is sufficient to account for miR-31's impact on metastasis, these observations do not preclude the existence of additional miR-31 targets that impinge on metastasis-relevant pathways in a manner that ostensibly is functionally redundant with the actions of ITGA5, RDX, and/or RhoA. Also, it is possible that one or more bona fide targets of miR-31 that have metastatic relevance fail to be significantly down-regulated by this miRNA in 231 cells. Overall, due to the fact that metastases are responsible for the overwhelming majority of patient mortality from carcinomas, this study highlights the idea that modulation of miR-31 and its effectors may prove clinically useful.
Materials and methods
Cell culture
Green fluorescent protein (GFP)-labeled 231 cells have been described (Valastyan et al. 2009). SUM-159 cells were provided by S. Ethier (Ma et al. 2007). Stable expression was achieved via retroviral (expression constructs) or lentiviral (shRNAs) transduction, followed by selection with puromycin, neomycin, hygromycin, and/or zeocin (Elenbaas et al. 2001).
Animal studies
All research involving animals complied with protocols approved by the Massachusetts Institute of Technology (MIT) Committee on Animal Care. Age-matched NOD/SCID (propagated on site) or nude (Taconic) mice were used in the xenograft studies, as indicated. For spontaneous metastasis assays, the indicated female mice were bilaterally injected into the mammary fat pads with 1.0 × 106 tumor cells resuspended in 1:2 Matrigel (BD Biosciences) plus normal growth media. In spontaneous metastasis assays employing nude mice, primary tumor diameter was measured every 7 d using precision calipers; tumor volume was calculated according to the formula V = (4/3)∏r3. For experimental metastasis assays, the indicated mice were injected intravenously with 5.0 × 105 tumor cells (in PBS) via the tail vein. Lung metastasis was quantified using a fluorescent dissecting microscope within 3 h of specimen isolation. Tumor histology was assessed by staining paraffin-embedded tissue sections with hematoxylin and eosin (H&E).
Statistical analysis
Data are presented as mean ± SEM; Student's two-tailed t-test was used for comparisons, with P < 0.05 considered significant.
Acknowledgments
We thank Julie Valastyan and Sandra McAllister for critical reading of this manuscript; M. Saelzler, L. Waldman, and other members of the Weinberg laboratory for discussions; G. Bokoch, S. Crouch, P. Klein, S. Kuwada, H. Surks, and S. Weiss for reagents; and M. Brown and the Koch Institute Histology Facility for tissue sectioning. This research was supported by the NIH (RO1 CA078461), MIT Ludwig Center for Molecular Oncology, U.S. Department of Defense, Breast Cancer Research Foundation, and DoD BCRP Idea Award. S.V. and R.A.W. are inventors on a patent application in part based on findings detailed in this manuscript. S.V. is a U.S. Department of Defense Breast Cancer Research Program Predoctoral Fellow. R.A.W. is an American Cancer Society Research Professor and a Daniel K. Ludwig Foundation Cancer Research Professor.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1832709.
Supplemental material is available at http://www.genesdev.org.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC, Weinberg RA. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes & Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler IJ. The pathogenesis of cancer metastasis: The ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GP, Massagué J. Cancer metastasis: Building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Pillé JY, Denoyelle C, Varet J, Bertrand JR, Soria J, Opolon P, Lu H, Pritchard LL, Vannier JP, Malvy C, et al. Anti-RhoA and anti-RhoC siRNAs inhibit the proliferation and invasiveness of MDA-MB-231 breast cancer cells in vitro and in vivo. Mol Ther. 2005;11:267–274. doi: 10.1016/j.ymthe.2004.08.029. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Ventura A, Jacks T. MicroRNAs and cancer: Short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]