Mouse and human fibroblasts were the first cell types successfully reprogrammed by ectopic expression of OCT4, SOX2, KLF4 and c-MYC (OSKM)(Lowry et al., 2008; Maherali et al., 2007; Park et al., 2008; Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Yu et al., 2007). Further studies have shown that the age, origin and cell type used have a deep impact on the reprogramming efficiency, eventually requiring the expression of fewer factors and/or reducing the timing of the whole process. In general, stem cells are rare, and difficult to access and isolate in large numbers [neural stem cells, for instance (Kim et al., 2009c; Kim et al., 2008)], and therefore represent a complicated target for reprogramming. However, cord blood (CB) represents an alternative and readily-accessible source of stem cells. Here we describe reprogramming of CD133+ CB cells to pluripotency by retroviral transduction of four (OSKM), three (OSK) and as few as two (OS) transcription factors, without the need for additional chemical compounds.
Cord Blood (CB) cells are considered an alternative to bone marrow (BM) as a source of haematopoietic stem cells for transplantation. CB cells can be collected without any risk for the donor, are young cells expected to carry minimal somatic mutations and possess the immunological immaturity of newborn cells (Rocha et al., 2004). These properties allow for less stringent criteria for HLA-donor-recipient selection, which represents a decisive benefit for transplantation and have resulted in more than 400,000 immunologically characterized CB units being currently available worldwide through a network of CB banks (Gluckman and Rocha, 2009).
For these experiments, CB-derived stem cells were isolated using standard CD133 immuno-magnetic selection, obtaining a purity range of 90–94% (Figure S1A). Because the integration and the expression of retroviral constructs requires mitotic division of the target cells, we first culture the quiescent CB cells, for 24 hours in presence of Stem Cell Factor (SCF), Trombopoietin (TPO), Flt ligand 3 (FLT-3) and Interleukin 6 (IL-6)(Akkina et al., 1996). Then, the cells were seeded over retronectin-coated plates previously pre-adsorbed with the viral particles, as previously described (Gammaitoni et al., 2006). Using this approach, we obtained an infection efficiency of approximately 28% as monitored by a constitutive GFP reporter retrovirus. Within the GFP+ population, 61% were CD133+, while 39% were CD133− (Figure S1B).
We first asked if maintaining CB cells under hES cell culture conditions could be sufficient to induce reprogramming, as it has recently been shown to be the case for spermatogonial stem cells (Conrad et al., 2008). After three weeks of culture under these conditions, CB cells formed no colonies. Flow Cytometry analysis revealed that the resulting cells no longer expressed the haematopoietic stem cells markers CD133, CD34, and CD38, remained positive for the haematopoietic marker CD45, but did not acquire the embryonic markers SSEA-3, SSEA-4 or TRA1-60, suggesting that untransduced CB cells differentiate into mature haematopoietic cells when cultured in hES cell conditions (Figure S1C).
We then attempted to reprogram CB cells using OSKM, OSK (either a combination of single factors or a polycistronic constructs, see Figure S2A, 2B), and OS. Three days post transduction, cells were plated onto irradiated Human Foreskin Fibroblasts (HFF-1) feeder cells and cultured in hES medium. As early as 9 days post infection, small colonies started to appear in cells transduced with OSKM, OSK and OS. At 12 to 15 days post infection, some of the colonies exhibited typical hES cell morphology, with sharp borders, and were comprised of a small, tightly packed cell population with large nuclei and clearly visible nucleoli (Figure 1A). On average, 8×104 infected CD133+ cells gave rise to 5 hES-like colonies that we named CBiPS. We successfully repeated the experiment with 6 independent CB units testing all three conditions (OSKM, OSK, OS) and generating 27 CBiPS cell lines, of which 20 lines have been expanded and characterized for endogenous expression of pluripotency markers and pluripotent differentiation ability in vitro. Furthermore, 6 independent CBiPS lines (2 lines from each of the 3 reprogramming condition: CBiPS 4F-3, CBiPS 4F-5, CBiPS 3F-10, CBiPS 3F-12, CBiPS 2F-1, CBiPS 2F-4) have been fully characterized. In parallel, as control, we infected fibroblasts and keratinocytes from a variety of independent donors with OSK and OS as previously described (Aasen et al., 2008). Overall, the reprogramming efficiency, as judged by the number of iPS cell-like colonies per 104 cells, of CB-derived cells using OSK was between that of keratinocytes and fibroblasts (0.45 ± 0.27, n = 5; 1.38 ± 0.51, n = 12; and 0.15 ± 0.14, n= 6, for CB-derived cells, keratinocytes and fibroblasts, respectively). Importantly, however, unlike the case of CB-derived cells, we never succeeded at generating iPS cell-like colonies from keratinocytes or fibroblasts using OS, despite numerous attempts (12 and 6, respectively) performed in parallel.
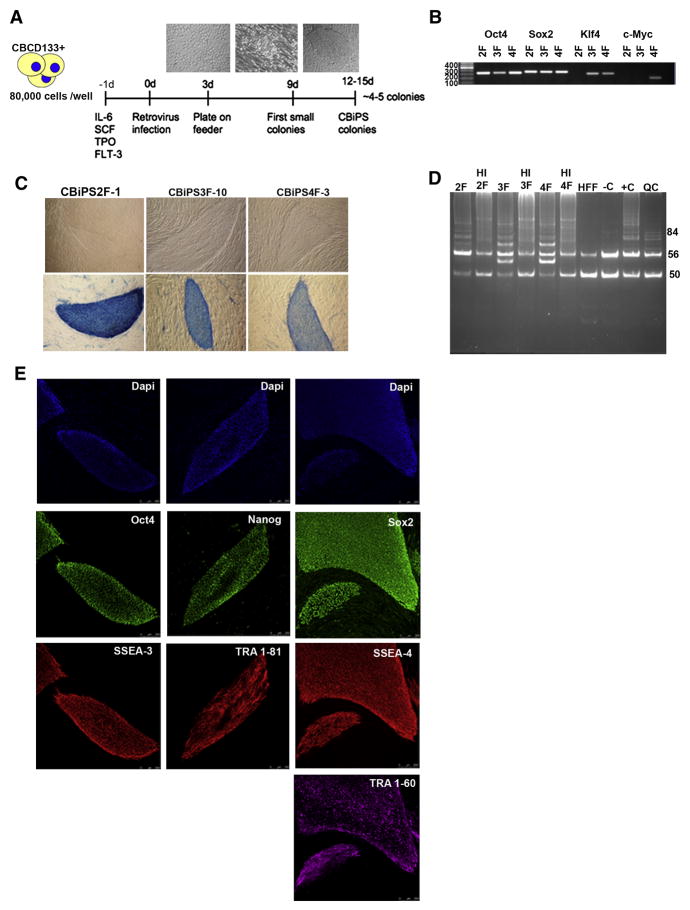
Figure 1. Generation of CBiPS cell lines using only OCT4 and SOX2 factors.
(A) Timeline of cord blood stem cells reprogramming. Three days post infection, CB CD133+ cells are transferred on feeders. Small adherent colonies are observed around day 9. Typical hES-like colonies are clearly visible after 12 days
(B) Genomic DNA PCR confirming the insertion of 4, 3, and only 2 transgenes
(C) Representative phase contrast images and Alkaline Phosphatase (AP) staining of CBiPS2F-1, 3F-10 and 4F-3 cell lines
(D) Representative Telomerase activity in CBiPS2F, 3F and 4F cell lines (HI: Heat Inactivation, HFF: Human Foreskin Fibroblast, −C: lysis buffer as negative control, +C: positive control and QC: Quantitative Control)
(E) Immunofluorescence analysis of CBiPS2F-1 cell line for pluripotency markers. The colonies express the embryonic markers SSEA-4, SSEA-3, TRA-1–60, TRA-1–81 and the transcription factors OCT4, SOX2 and NANOG. Underlying fibroblasts provide a negative control. Scale bars, 250 μm
In addition, since banked CB units are stored in a cryopreserved status, in 2 independent experiments we have generated 5 CBiPS Frozen (CBiPSFr) cell lines from thawed CB units that had been stored frozen for more than 5 years. The CBiPSFr cell lines were characterized for expression of pluripotency associated transcription factors and surface markers, and pluripotent differentiation ability in vitro. These data showed that the standard cryopreservation protocol does not affect the reprogramming ability of these cells.
The presence of each retroviral transgene was confirmed by PCR genotyping, demonstrating the insertion of the expected 4, 3 or 2 transcription factors in CBiPS 4F, CBiPS 3F, CBiPS 2F, respectively (Figure 1B).
All the CBiPS lines tested showed strong alkaline phosphatase (Figure 1C) and had reactivated the enzyme telomerase (Figure 1D) activity. Immunofluorescence of 6 CBiPS cell lines revealed expression of pluripotency markers such as OCT4, SOX2, TRA-1-81, TRA-1-60, SSEA3, SSEA4, and NANOG (Figure 1E and Figure S3A, 3B). With CBiPS cells lines derived from frozen CB units, we obtained similar results (Figure S3C). In addition, flow cytometry analysis revealed that CBiPS cells were negative for the haematopoietic stem cell markers CD45, CD34, but still positive for CD133, a common marker of haematopoietic and hES cells (Figure S4).
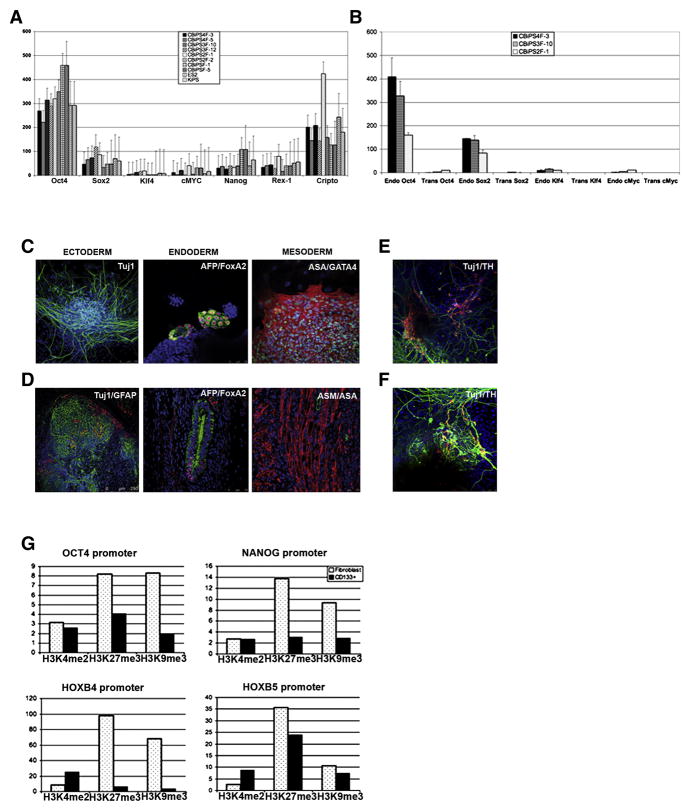
Quantitative RT- PCR showed that all CBiPS lines tested, expressed a set of pluripotency genes including OCT4, SOX2, NANOG, CRIPTO and REX1, uncovering a gene-expression profile comparable to other iPS (Aasen et al., 2008) and hES [2] cell lines (Raya et al., 2008) (Figure 2A). Accordingly, the genome wide transcriptional profile of CBiPS and hES lines was similar, as shown by microarray analysis (Figure S5A, 5B). Quantitative RT-PCR also revealed that the expression of the retroviral transgenes was reduced to low or undetectable levels and that they correctly up-regulated the endogeneous expression of OCT4, SOX2, KLF4 and c-MYC (Figure 2B). Silencing of the retroviral transgenes was further confirmed, in the CBiPS2F-1 line by immunofluorescence staining using antibodies specific for FLAG-tagged transgenic factors (Figure S6). Consistent with these observations, bisulfite sequencing revealed an extensive demethylation of CpG dinucleotides of the OCT4 promoter, reflecting the transcriptional reactivation of this key pluripotency gene, and the epigenetic reprogramming of CBiPS cells (Figure S7).
Figure 2. Characterization of CBiPS cell lines.
(A) Quantitative RT-PCR analysis for pluripotency markers OCT4, SOX2, NANOG, REX1, CRIPTO, KLF4 and c-MYC. ES[2] and Keratinocyte-iPS (KiPS) cell lines were analysed together with the different CBiPS cell lines derived from fresh and frozen samples. Error bars indicate the s.d. generated from triplicates.
(B) Quantitative RT-PCR showing the repression of the OCT4, SOX2, KLF4 and c-MYC transgenes in the CBiPS cell lines.
(C) In vitro differentiation of CBiPS 2F-1 into the three primary germ cell layers (Ectoderm-Tuj1, Endoderm-AFP and FOXA2, and Mesoderm-ASA and GATA4).
(D) Immunofluorescence analysis of teratoma sections 60 days after intra-testicular injection of CBiPS2F-1 showing Tuj1/GFAP positive ectoderm, AFP/FoxA2 positive endoderm and ASM/ASA positive mesoderm. Scale bar 75–250 μm.
(E) Specific in vitro differentiation of CBiPS2F-1 and (F) CBiPS3F-12 into dopaminergic neurons (Tuj1/TH tyrosine hydroxilase), which are immunophenotypically mature.
(G) Chromatin immuno-precipitation assays comparing the levels of histone H3 methylation at K4 (H3K4me2), K27 (H3K27me3) and K9 (H3K9me3) in the promoters of OCT4, NANOG, HOXB4 and HOXB5 in human fibroblasts and CD133+ cells.
We confirmed the clonal origin of CBiPS2F by subcloning one of this line and finger printing the resulting sub-clones by Southern blot using probes recognizing OCT4, SOX2, KLF4 and c-MYC. As expected, the KLF4 and c-MYC probes only recognized the endogenous genomic sequences, whereas OCT4 and SOX2 probes revealed in the original clone and two randomly picked sub-clones (CBiPS2F-1a and CBiPS2F-1b) identical additional bands: one for SOX2 and one for OCT4 (Figure S8).
Cytogenetic analysis showed that the CBiPS cell lines maintained a normal 46XY or 46 XX karyotype after more than 10 passages and could be maintained in culture for, at least, 20 passages. In addition, the male chromosomal content in the CBiPS4F and 2F cell lines excludes the possibility that the reprogrammed cells arise from a small fraction of contaminating mother cells known to be present in the initial cord blood sample (Figure S9A, 9B, 9C).
Next, we evaluated the differentiation potential of CBiPS lines. CBiPS cells were able to form embryoid bodies (EBs) with high efficiency (Figure S10A), which could be differentiated into derivatives of the three embryonic germ layers, including Tuj1 positive ectoderm, α-fetoprotein (AFP) and FoxA2 positive endoderm, α-sarcomeric actin (ASA) and GATA4 positive mesoderm, (Figure 2C and Figure S10B, 10C, 10D). Upon injection into immuno-compromised SCID beige mice, CBiPS cells generated complex intra-testicular teratomas, comprising structures and tissues derived from the three embryonic germ layers, as evidenced by the expression of Tuj1 and GFAP for ectoderm, AFP and FoxA2 for endoderm, and α–smooth muscle actin (ASM) and ASA for mesoderm (Figure 2D and Figure S10E, 10F). Following specific in vitro differentiation protocols, CBiPS cells gave rise also to specialized mesoderm-derived cell types such as rhythmically beating cardiomyocytes (Supplementary movie 1) and ectodermal cells such as dopaminergic neurons (Figure 2E, 2F). Our results confirm that CBiPS cells are transcriptionally reprogrammed to a state similar to hiPS and hES cells, are karyotypically stable, and show a differentiation potential consistent with pluripotency.
Since CB cells can be reprogrammed with just 2 factors, and are thus more amenable to reprogramming than fibroblasts (Takahashi et al., 2007) or keratinocytes (Aasen et al., 2008), we tested whether their global transcriptional profile was closer to that of pluripotent stem cells. We performed a global comparison of the transcriptomes of CD133+ cells, fibroblasts, keratinocytes, hESC, KiPS, and CBiPS cells. Interestingly, the overall transcriptional profile of CD133+ cells was not closer to that of pluripotent stem cells than those of fibroblasts or keratinocytes (Figure S11A). Although we cannot formally exclude the possibility that a rare cell subpopulation with a transcriptome similar to ES cells exists within CD133+ cells, several lines of evidence indicate that the increased reprogramming susceptibility of CD133+ cells is a characteristic of the majority of cells, rather than of a rare cell population. First, CD133+ cells express a set of pluripotency associated genes (Kucia et al., 2007; Nikolova et al., 2007; Zhao et al., 2006) such as OCT4, NANOG, SOX2, REX1, CRIPTO, SALL2, DPPA4, ZNF589 and DNMT3A/B (Figure S11B, 11C), albeit at much lower levels than ES cells (data not shown). Second, we could not detect a subpopulation of CB cells expressing high levels of OCT4 or NANOG by flow cytometry, but rather a normal distribution of cells expressing low levels of either factor (data not shown). Finally, the overall levels of histone repressive marks (methylation at H3K27 and H3K9) at the OCT4 and NANOG promoters were much lower in CB derived stem cells than in fibroblasts (Figure 2G). These results indicate that the increased reprogramming susceptibility of CB cells may be the result of transcriptional differences in a small subset of genes (which we tried to uncover by comparing the genome-wide transcriptional profiles of all these cell types, see Supplementary text 1 for a more exhaustive description of these analyses and their results) and a more permissive chromatin organization. On the other hand, the combination of high levels of KLF4 and c-MYC in CD133+ cells compared to fibroblasts and keratinocytes (Figure S11D) further underlies our previous hypothesis that endogenous expression of these factors may allow for enhanced reprogramming of those cells (Aasen et al., 2008). In support of this notion, neuronal stem cells, which express endogenously high levels of SOX2, can be reprogrammed to pluripotency with only OCT4 (Kim et al., 2009b; Kim et al., 2009c).
It has recently been shown that mobilized peripheral blood (mPB) cells can be reprogrammed to pluripotency (Loh et al., 2009). However, compared to newborn CB stem cells, adult mPB cells will have the potential disadvantages that they may have accumulated genomic alterations as a result of ageing or disease, and that the pharmacological treatment used to mobilize the adult haematopoietic stem cell compartment represents a health risk for the donor (Anderlini, 2009). In turn, CB derived cells are readily available (not requiring mobilization or biopsy and establishment of primary cultures), young cells (minimizing the risk of having accumulated genetic mutations), and already banked along with immunological information. These characteristics offer evident logistic advantages over the use of adult somatic cell types or adult stem cells for the purpose of creating iPS cell banks (Rocha et al., 2004). To date, more than 400,000 CB units are available worldwide in a comprehensive network of CB banks, facilitating a rapid and effective search for compatible donors for CBiPS generation (Gluckman and Rocha, 2009). Even though the generation of patient specific iPS lines has been pursued in the context of devising autologous cell therapy strategies (Raya et al., 2009), this approach may be unfeasible in many instance. Specifically, treatment of acute conditions or situations in which the patient’s somatic cells are altered as a consequence of the disease or ageing would benefit from off-the-shelf allogeneic approaches. Large-scale production and banking of CBiPS lines representing a wide panel of HLA haplotypes, organized in a publicly available network could therefore represent an alternative for future clinical applications. Moreover, selection of donors homozygous for common HLA haplotypes could be easily accomplished using banked CB units and would significantly reduce the number of CBiPS lines needed to provide a perfect HLA match for a large percentage of the population (Taylor et al., 2005). Together with the recent developments on reprogramming strategies using non-integrative or excisable approaches (Kaji et al., 2009; Vandendriessche et al., 2009; Woltjen et al., 2009; Yu et al., 2009) or direct protein transduction (Kim et al., 2009a) the studies presented here should facilitate the clinical translation of iPSC-based therapies.
Supplementary Material
Acknowledgments
We are grateful to Tiscornia Gustavo, Menendez Sergio, Garreta Elena and Eguizabal Cristina for the assistance during the reprogramming process, José Miguel Andrés Vaquero for assistance with flow cytometry, Meritxell Carrió and Vanesa Tobajas Fernandez for expert assistance with cell culture techniques, Esther Melo, Lola Mulero Pérez, and Mercé Gaudes Martí for bioimaging assistance, and Yvonne Richaud and Yolanda Muñoz Santos for excellent technical assistance. MJB was partially supported by the Ramón y Cajal program. This work was partially supported by grants from Ministerio de Educación y Ciencia grants BFU2006-12251, the Fondo de Investigaciones Sanitarias (RETIC-RD06/0010/0016), TERCEL, the G. Harold and Leila Y. Mathers Charitable Foundation, and Fundación Cellex.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilic J, Pekarik V, Tiscornia G, et al. Nature biotechnology. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Akkina RK, Walton RM, Chen ML, Li QX, Planelles V, Chen IS. Journal of virology. 1996;70:2581–2585. doi: 10.1128/jvi.70.4.2581-2585.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderlini P. Current opinion in hematology. 2009;16:35–40. doi: 10.1097/MOH.0b013e328319913c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad S, Renninger M, Hennenlotter J, Wiesner T, Just L, Bonin M, Aicher W, Buhring HJ, Mattheus U, Mack A, et al. Nature. 2008;456:344–349. doi: 10.1038/nature07404. [DOI] [PubMed] [Google Scholar]
- Eminli S, Utikal J, Arnold K, Jaenisch R, Hochedlinger K. Stem cells (Dayton, Ohio) 2008;26:2467–2474. doi: 10.1634/stemcells.2008-0317. [DOI] [PubMed] [Google Scholar]
- Gammaitoni L, Lucchi S, Bruno S, Tesio M, Gunetti M, Pignochino Y, Migliardi G, Lazzari L, Aglietta M, Rebulla P, et al. Stem cells (Dayton, Ohio) 2006;24:1201–1212. doi: 10.1634/stemcells.2005-0408. [DOI] [PubMed] [Google Scholar]
- Gluckman E, Rocha V. Haematologica. 2009;94:451–454. doi: 10.3324/haematol.2009.005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Biostatistics (Oxford, England) 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, et al. Cell stem cell. 2009a;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Greber B, Arauzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Scholer HR. Nature. 2009b doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, et al. Cell. 2009c;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M, et al. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Kucia M, Halasa M, Wysoczynski M, Baskiewicz-Masiuk M, Moldenhawer S, Zuba-Surma E, Czajka R, Wojakowski W, Machalinski B, Ratajczak MZ. Leukemia. 2007;21:297–303. doi: 10.1038/sj.leu.2404470. [DOI] [PubMed] [Google Scholar]
- Loh YH, Agarwal S, Park IH, Urbach A, Huo H, Heffner GC, Kim K, Miller JD, Ng K, Daley GQ. Blood. 2009 doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Nikolova T, Wu M, Brumbarov K, Alt R, Opitz H, Boheler KR, Cross M, Wobus AM. Differentiation; research in biological diversity. 2007;75:100–111. doi: 10.1111/j.1432-0436.2006.00119.x. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Raya A, Rodriguez-Piza I, Aran B, Consiglio A, Barri PN, Veiga A, Izpisua Belmonte JC. Cold Spring Harbor symposia on quantitative biology. 2008 doi: 10.1101/sqb.2008.73.038. [DOI] [PubMed] [Google Scholar]
- Raya A, Rodriguez-Piza I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castella M, Rio P, Sleep E, et al. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha V, Labopin M, Sanz G, Arcese W, Schwerdtfeger R, Bosi A, Jacobsen N, Ruutu T, de Lima M, Finke J, et al. The New England journal of medicine. 2004;351:2276–2285. doi: 10.1056/NEJMoa041469. [DOI] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. PLoS biology. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Taylor CJ, Bolton EM, Pocock S, Sharples LD, Pedersen RA, Bradley JA. Lancet. 2005;366:2019–2025. doi: 10.1016/S0140-6736(05)67813-0. [DOI] [PubMed] [Google Scholar]
- Vandendriessche T, Ivics Z, Izsvak Z, Chuah MK. Blood. 2009 doi: 10.1182/blood-2009-04-210427. [DOI] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, et al. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Science (New York, NY) 2009 doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Science (New York, NY) 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Wang H, Mazzone T. Experimental cell research. 2006;312:2454–2464. doi: 10.1016/j.yexcr.2006.04.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.