Abstract
Background
Integrase inhibitors are currently being incorporated into highly active antiretroviral therapy (HAART). Due to high HIV variability, integrase inhibitor efficacy must be evaluated against a range of integrase enzymes from different subtypes.
Methods
This study compares the enzymatic activities of HIV-1 integrase from subtypes B and C as well as susceptibility to various integrase inhibitors in vitro. The catalytic activities of both enzymes were analyzed in regard to each of 3' processing and strand transfer activities both in the presence and absence of the integrase inhibitors raltegravir (RAL), elvitegravir (EVG), and MK-2048.
Results
Our results show that integrase function is similar with enzymes of either subtype and that the various integrase strand transfer inhibitors (INSTIs) that were employed possessed similar inhibitory activity against both enzymes.
Conclusion
This suggests that the use of integrase inhibitors against HIV-1 subtype C will result in comparable outcomes to those obtained against subtype B infections.
Background
Integration of viral cDNA into the host genome is one of the definitive features of retroviral replication. Integration is mediated by the HIV pol-encoded integrase enzyme. Recently, integrase inhibitors have been added to the arsenal of antiviral drugs used in therapy. RAL (Merck) was the first integrase inhibitor to be approved by the US Food and Drug Administration (FDA) after clinical trials demonstrated that this drug promoted a rapid and sustained antiretroviral effect [1]. EVG (GS-9137, Gilead), another integrase inhibitor, is currently in phase III clinical trials [2]. Other integrase inhibitors, such as MK-2048 (Merck), are still in pre-clinical development.
Integrase inhibitors are active against both B- and non-B subtypes in therapy [3,4]. Subtype C variants are responsible for approximately 50% of global infections, mostly in Sub-Saharan Africa and India [5]. It is therefore important to determine whether the integrase enzymes of different HIV-1 subtypes behave in a parallel manner to one another and whether they respond similarly to the use of integrase inhibitors of HIV-1 replication.
After viral entry and reverse transcription, reverse-transcribed double-stranded blunt-ended DNA is incorporated into the host cell genome through two catalytic activities mediated by integrase: 3' end processing and strand transfer [6,7]. During 3' end processing, a dinucleotide adjacent to the conserved 3' terminal CA is excised from the 3' end of the recently reverse transcribed HIV-1 DNA genome, generating 3' hydroxyl ends. During the strand transfer reaction, both newly generated 3' ends are covalently linked to target DNA in a concerted fashion via a one-step transesterification reaction [8]. In vitro, integrase can also catalyze two additional reactions: disintegration and specific internal endonucleolytic cleavage [9,10].
Variability between different HIV-1 integrases at an amino acid level is low, ≈ 8-12%. However, sites of amino acid differences between subtypes are often close to resistance-related amino acids. We were therefore interested in analyzing whether such minor differences might be important in differential acquisition of INSTI resistance mutations in a subtype-specific manner [11]. Furthermore, natural polymorphisms in non-B integrase proteins might alter INSTI binding or activity [12,13]. An in silico comparison of subtype B and CRF A/G integrase predicted that polymorphisms within subtypes might affect structure and substrate binding characteristics of IN enzymes [13]. In this study, we compared the enzymatic activities of subtype B and C recombinant integrases in the context of inhibition by RAL, EVG, and the novel INSTI MK-2048.
Results
Purification of active subtype C integrase
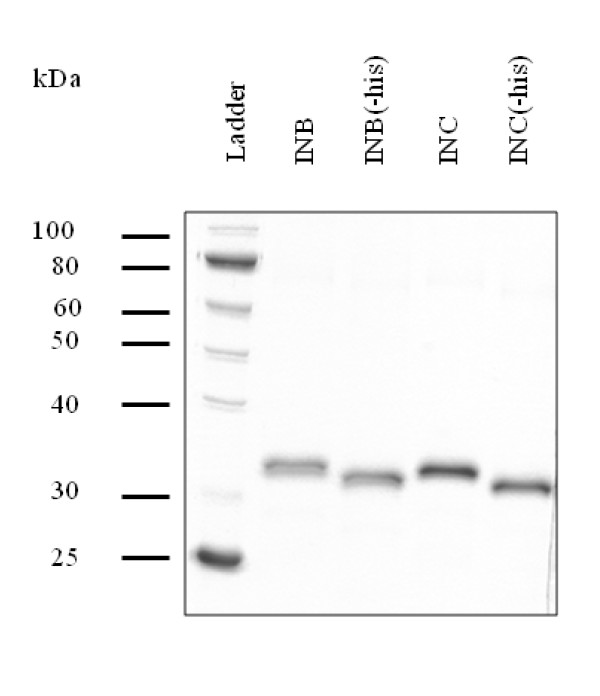
Subtype C integrase was PCR amplified from the pINDIE-C1 molecular clone and introduced into the expression vector pET-15B, replacing the ORF of subtype B integrase previously cloned by Bushman et al. [14]. To increase the solubility of subtype C recombinant proteins, two amino acid changes were introduced: a phenylalanine at codon 185 was changed to a histidine, and a cysteine at codon 280 was changed to a serine. These changes mimic those previously introduced into subtype B integrase to increase solubility and are known to not affect catalytic activity [15,16]. Expression and purification of the subtype B and C integrase enzymes were performed simultaneously as previously described for subtype B integrase [15] with minor modifications. Subtype B and C integrases were successfully purified to > 95% homogeneity (Figure 1). The N-terminal His tag was removed from recombinant integrase enzymes by thrombin cleavage (Figure 1). When the enzymatic activities of both subtype B and C purified recombinant proteins in the presence or absence of the N-terminal His tag were compared, no difference was detected (data not shown). Therefore, all further experiments were orchestrated using recombinant integrase that did not undergo His tag removal.
Figure 1.
Purification of recombinant subtype B and C integrase enzymes. N-terminal His tags of the enzymes were removed from purified subtype B and C recombinant proteins by thrombin cleavage. Lane 1, protein ladder (10-250 kDa) (New England Biolabs); INB, subtype B integrase; INC, subtype C integrase.
Biochemical properties of subtype C integrase
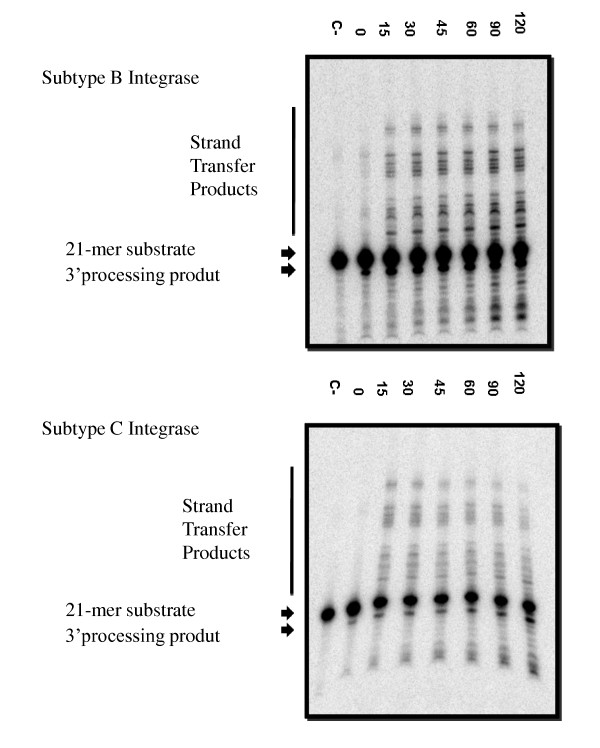
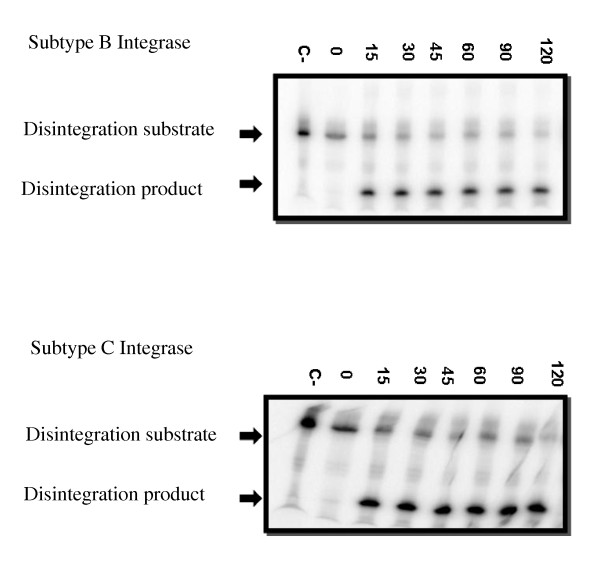
Integrase mediates the insertion of viral cDNA into host chromatin through two unique enzymatic activities: 3' processing and strand transfer [6,17]. Oligonucleotides that mimic the viral LTR ends can be utilized to analyze these two catalytic activities in vitro. First, subtype B and C integrases were tested for their ability to perform 3' processing (Figure 2) and strand transfer (Figure 3). Time course experiments show similar results for both enzymes. Disintegration was also analyzed and subtype C recombinant protein catalyzed this activity to a similar extent as did subtype B recombinant protein (Figure 4). These experiments confirm the activity of our subtype C purified recombinant protein.
Figure 2.
3' Processing assay. One representative reaction (out of five reactions) is illustrated. Recombinant enzyme was incubated at 37°C with templates (radiolabeled double stranded oligonucleotide INT1/2) for the indicated times up to 120 minutes. The 21-mer substrate and 19-mer 3' processing products are indicated.
Figure 3.
Strand transfer assay. One representative reaction (out of five reactions) is depicted. Recombinant integrase enzyme was incubated at 37°C for 3 minutes for the initial 3' processing reaction. T35/SK70, double stranded oligonucleotide substrate, was added and reaction tubes were incubated at 37°C for the indicated times up to 120 minutes. The 21-mer substrate, 19-mer 3' processing, and strand transfer products are indicated.
Figure 4.
Disintegration assay. One representative reaction (out of five reactions) is portrayed. Recombinant enzyme was incubated at 37°C for the indicated times up to 120 minutes with disintegration template (radiolabeled oligonucleotide D). Disintegration template and product are indicated. (C-), Negative control lane without integrase enzyme. Top panel, subtype B integrase; bottom panel, subtype C integrase.
Subtype B and C enzymes are inhibited to a similar extent by RAL, MK-2048 and EVG
RAL and EVG are INSTIs with high specific activity against strand transfer [18,19]. MK-2048 is a prototype second-generation INSTI with a resistance profile that is distinct from RAL and EVG [20,21]. These three drugs have been reported to be approximately 100-fold less specific for the inhibition of 3' processing activity compared to strand transfer [18,22,23].
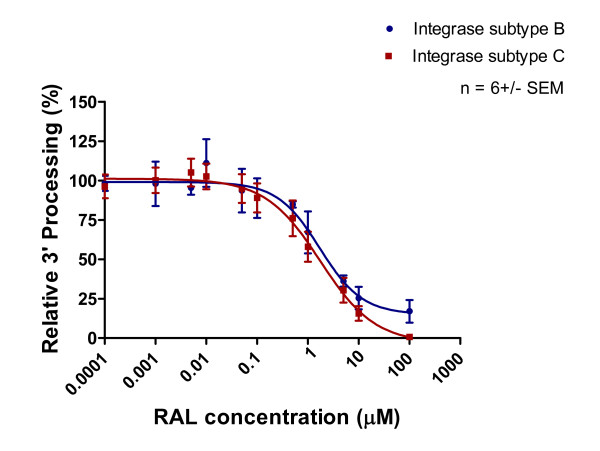
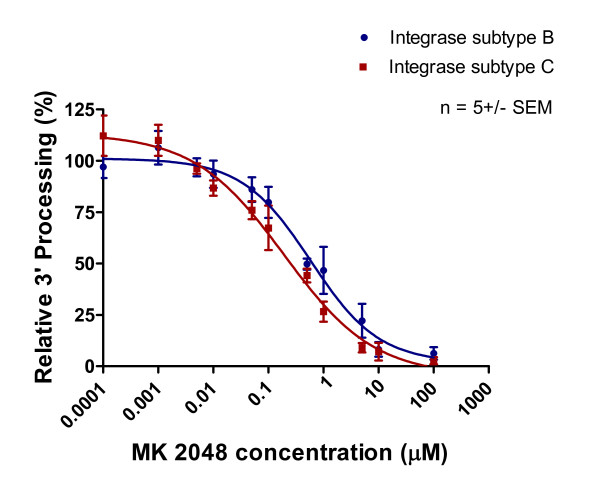
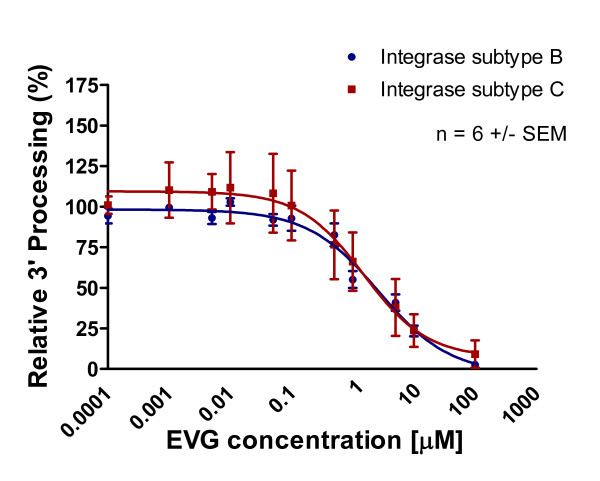
Purified recombinant subtype B and C integrase enzymes were incubated with increasing concentrations of integrase inhibitors and corresponding templates. The results of Table 1 and Figures 5, 6 and 7 show that 3' processing mediated by recombinant enzymes of both subtypes was inhibited to a similar extent (p > 0.05) by all three drugs in the presence of MnCl2. The inhibition of 3' processing required much higher concentrations of integrase inhibitors than those needed to block strand transfer for both subtype enzymes (Table 1), consistent with previously reported data for subtype B integrase [18].
Table 1.
IC50 values for RAL, EVG and MK-2048 for subtype B and subtype C integrase in Mn2+-based enzymatic assays.
| 3' Processing IC50a | Strand Transfer IC50a | |||
| Subtype B | Subtype C | Subtype B | Subtype C | |
| RAL(μM) | 1.71(0.7-4.5) | 1.75(0.7-3.9) | 0.37(0.2-0.8) | 0.15(0.09-0.3) |
| MK-2048(μM) | 0.58(0.28-1.20) | 0.19(0.09-0.39) | 0.075(0.04-0.14) | 0.08(0.03-0.2) |
| EVG(μM) | 2.66(1.44-4.91) | 1.5(0.29-7.74) | 0.014(0.003-0.07) | 0.018(0.006-0.05) |
aAll differences between subtypes were not statistically significant. 95% Confidence Intervals are indicated.
Figure 5.
Inhibition of 3' processing as a function of increasing RAL concentration. Subtype B and C 3' processing activity (presented as relative percentage) in relation to increasing RAL concentration. This graph was prepared with GraphPad Prism 4.0, the combined result of quantification and analyses of at least 3 independent experiments.
Figure 6.
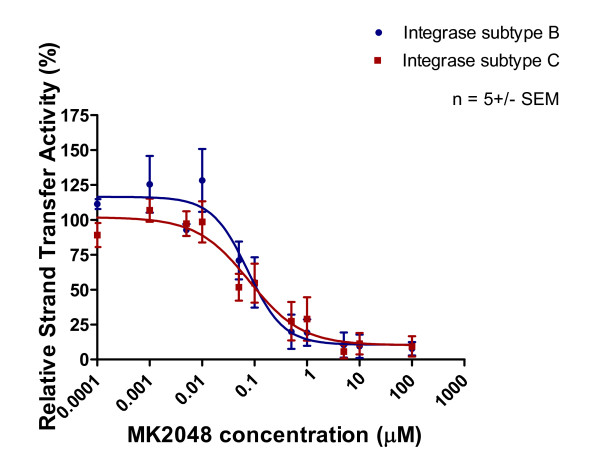
Inhibition of 3' processing as a function of increasing MK-2048 concentration. Subtype B and C 3' processing activity (presented as relative percentage) in relation to increasing MK-2048 concentration. This graph was prepared with GraphPad Prism 4.0, the combined result of quantification and analyses of at least 3 independent experiments.
Figure 7.
Inhibition of 3' processing as a function of increasing EVG concentration. Subtype B and C 3' processing activity (presented as relative percentage) in relation to increasing EVG concentration. This graph was prepared with GraphPad Prism 4.0, the combined result of quantification and analyses of at least 3 independent experiments.
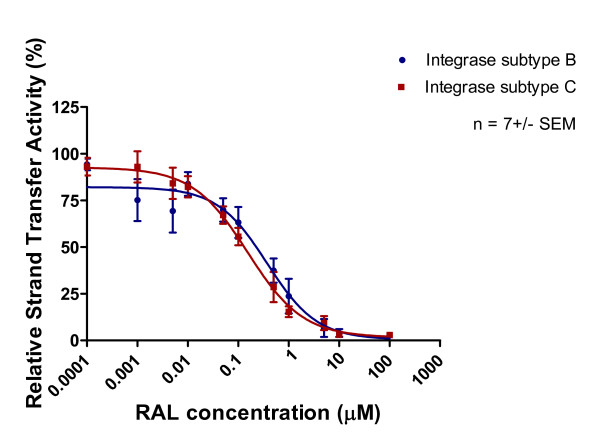
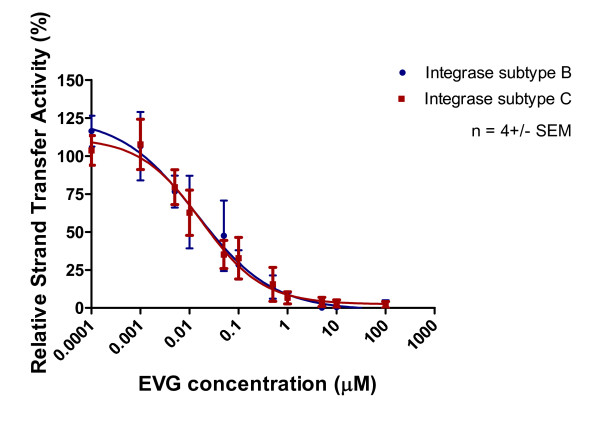
The strand transfer activity of subtype B and C recombinant proteins was inhibited by all three inhibitors. The IC50 values of RAL for subtype B and C integrase strand transfer were 0.37 μM and 0.15 μM, respectively, in assays that employed Mn2+ as the cation (Figure 8, Table 1). The IC50 values for EVG inhibition of strand transfer in Mn2+-based assays were 0.014 μM and 0.018 μM for the subtype B and C enzymes, respectively (Figure 9, Table 1). The IC50 values for MK-2048 against subtype B and C enzymes were 0.075 μM and 0.08 μM, respectively (Figure 10, Table 1).
Figure 8.
Inhibition of strand transfer as a function of increasing RAL concentration. Subtype B and C strand transfer activity (presented as relative percentage) in relation to increasing RAL concentration. This graph was prepared with GraphPad Prism 4.0, the combined result of quantification and analyses of at least 3 independent experiments.
Figure 9.
Inhibition of strand transfer as a function of increasing EVG concentration. Subtype B and C strand transfer activity (presented as relative percentage) in relation to increasing EVG concentration. This graph was prepared with GraphPad Prism 4.0, the combined result of quantification and analyses of at least 3 independent experiments.
Figure 10.
Inhibition of strand transfer as a function of increasing MK-2048 concentration. Subtype B and C strand transfer activity (presented as relative percentage) in relation to increasing MK-2048 concentration. This graph was prepared with GraphPad Prism 4.0, the combined result of quantification and analyses of at least 3 independent experiments.
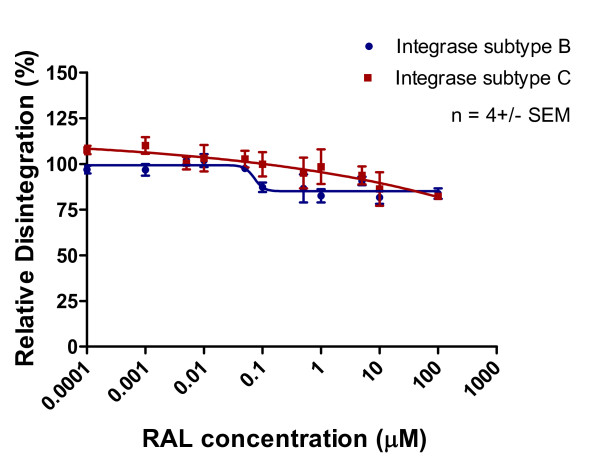
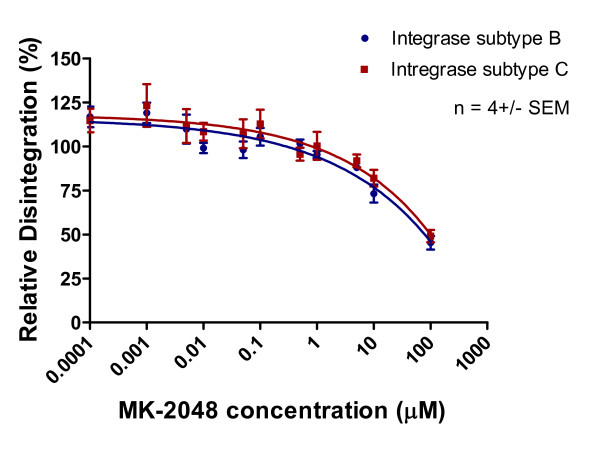
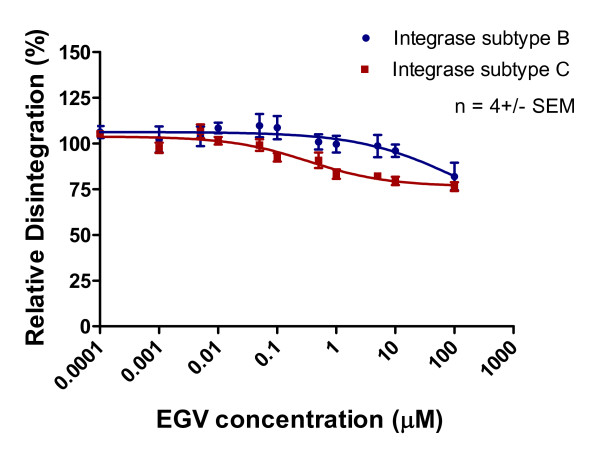
Disintegration was inhibited by high concentrations of MK-2048 to a comparable extent with both subtype B and C enzymes (Figures 11, 12, 13). In contrast, neither RAL nor EVG had much effect on this process, which is a discovery that is consistent with work by others [22]. We also evaluated strand transfer in the presence of MgCl2 rather than MnCl2 and obtained similar IC50 values (p > 0.05) for each of subtype B versus C enzymes with each of RAL, EVG and MK-2048 in a microtiter plate system [24] (Table 2). Consistent with previous observations, IC50 values were lower when these reactions were performed with MgCl2 than with MnCl2 [2,24].
Figure 11.
Inhibition of disintegration as a function of increasing RAL concentration. Subtype B and C disintegration activity (presented as relative percentage) in reaction to increasing RAL concentration. This graph was prepared with GraphPad Prism 4.0, the combined result of quantification and analyses of 3 independent experiments.
Figure 12.
Inhibition of disintegration as a function of increasing MK-2048 concentration. Subtype B and C disintegration activity (presented as relative percentage) in reaction to increasing MK-2048 concentration. This graph was prepared with GraphPad Prism 4.0, the combined result of quantification and analyses of 3 independent experiments.
Figure 13.
Inhibition of disintegration as a function of increasing EVG concentration. Subtype B and C disintegration activity (presented as relative percentage) in reaction to increasing EVG concentration. This graph was prepared with GraphPad Prism 4.0, the combined result of quantification and analyses of 3 independent experiments.
Table 2.
IC50 values for RAL, EVG and MK-2048 for subtype B and subtype C integrase in Mg2+-based enzymatic assays.
| Strand Transfer IC50 | ||
| Subtype Ba | Subtype Ca | |
| RAL (μM) | 0.0047 (0.0013-0.0064) | 0.0037 (0.0011-0.0086) |
| MK-2048 (μM) | 0.0047 (0.0021-0.010) | 0.0023 (0.001-0.007) |
| EVG (μM) | 0.0017 (0.0009-0.0051) | 0.0011 (0.0002-0.021) |
aAll differences between subtypes were not statistically significant. 95% Confidence Intervals are indicated.
Inhibition of replication by integrase inhibitors was also evaluated in cell culture based assays using cord blood mononuclear cells (Table 3). Subtype B and C clinical isolates were inhibited to a similar extent by each of RAL, EVG and MK-2048.
Table 3.
EC50 values for RAL, MK-2048 and EVG for subtype B and C viruses cultured in cord blood mononuclear cells.
| Subtype B | EC50 Values (μM) | ||
|---|---|---|---|
| Viruses | RAL (μM) | MK-2048 (μM) | EVG (μM) |
| 5326 | 0.0243 ± 0.0025 | 0.0072 ± 0.0021 | NDa |
| 5331 | 0.0056 ± 0.0022 | 0.0010 ± 0.0002 | 0.0001 ± 0.00007 |
| BK 132 | 0.0301 ± 0.0054 | 0.0148 ± 0.0032 | NDa |
| pNL4-3 | 0.0082 ± 0.0013 | 0.0027 ± 0.0009 | 0.0009 ± 0.0003 |
| IIIb | 0.0012 ± 0.0005 | 0.0003 ± 0.0001 | 0.0018 ± 0.0006 |
| Subtype C | EC50 Values (μM) | ||
| Viruses | RAL (μM) | MK-2048 (μM) | EVG (μM) |
| Mole 03 | 0.0046 ± 0.0002 | 0.0011 ± 0.0001 | NDa |
| 96USNG31 | 0.0087 ± 0.0007 | 0.0015 ± 0.0001 | 0.0008 ± 0.0006 |
| 4742 | 0.0206 ± 0.0090 | 0.0033 ± 0.0020 | 0.0022 ± 0.0002 |
| BG-05 | 0.0067 ± 0.0009 | 0.0022 ± 0.0007 | 0.0008 ± 0.00004 |
| HB-1 | 0.0015 ± 0.0004 | 0.0007 ± 0.0005 | 0.0001 ± 0.00001 |
aND = not done
Discussion
Most HIV-1 patients are infected with non-B subtypes, most commonly subtype C [5], and subtype-specific differences in the development of drug resistance have been reported [25]. Therefore, it is important to understand the activity of enzymes of different subtypes. In our study, subtype B and C integrase enzymes were evaluated; and the activity of integrase inhibitors against them were compared, since a role for polymorphisms and structure-function differences between subtypes in drug resistance has been demonstrated [11,12,23].
Strand transfer inhibitors have been shown as efficient inhibitors of integration amongst a wide range of retroviruses [26]. In silico observations suggest that subtype-specific differences in regard to key amino acids in integrase, including those close to the catalytic site, may pose an effect on the binding of RAL [13,27,28]. Therefore, subtype-specific variations in DNA-binding domains could also affect the affinity of RAL for integrase. In vitro, subtype B and C recombinant proteins retain similar enzymatic capacities in the absence of drug (Figures 2, 3, 4), with comparable strand transfer, 3' processing and disintegration activities, as measured by time course experiments. We also show that RAL and EVG had similar effects against both subtype B and C integrase enzymes, regardless of whether Mg2+ or Mn2+ was used as a cation (Tables 1 and 2). In addition to the foregoing, we have evaluated the IC50 values of RAL, EVG and MK-2048 in cell-based assays using clinical isolates of viruses of either subtype B or subtype C origin (Table 3). No significant differences were observed between subtypes in regard to drug susceptibility. These findings are consistent with recent results on similarities vis-à-vis biochemical activity and susceptibility to antiretroviral drugs of reverse transcriptase enzymes derived from HIV-1 subtypes B and C [29].
Conclusion
Our results provide biochemical and tissue culture evidence that integrase enzymes from HIV-1 subtypes B and C are inhibited by each of RAL, EVG and MK-2048 to a similar extent. These findings are supportive of the use of these inhibitors in patients infected with subtype C virus.
Materials and methods
Oligonucleotides and reagents
Oligonucleotides were purchased from Invitrogen and then PAGE purified. A 21-mer duplex was formed by annealing INT1 (5'-TGTGGAAAATCTCTAGCAGT-3') and INT2 (5'-ACTGCTAGAGATTTTCCACA-3'). The 35-mer duplex was produced by annealing T35 (5'-ACTATACCAGACAATAATTGTCTGGCCTGTACCGT-3') and SK70 (5'-ACGGTACAGGCCAGACAATTATTGTCTGGTATAGT-3'). The disintegration primer (5'-TGCTAGTTCTAGCAGGCCCTTGGGCCGGCGGCGCTTGCGCC-3') was heated to 95°C and slowly cooled to achieve its secondary structure [30].
RAL and MK-2048 were obtained from Merck Pharmaceuticals, Inc. EVG was obtained from Gilead Biosciences.
Cloning and Mutagenesis
Subtype C integrase was PCR amplified from the molecular clone pINDIE-C1 (accession number: AB023804) and subcloned into the bacterial expression vector pET15B, replacing the subtype B integrase ORF kindly obtained from Dr. Robert Craigie, NIH.
The QuickChange II (Stratagene) site-directed mutagenesis kit was utilized for the introduction of the solubility mutations F185H and C280S into subtype C integrase. The primers utilized for this mutagenesis were INC-FF185H (5'-GCAGTATTCATTCACAATCATAAAAGAAAAGGGGGG-3'), INC-RF185H (5'-CCCCCCTTTTCTTTTATGATTGTGAATGAATACTGC-3'), INC-FC280S (5'-GCAGGTGCTGATTCTGTGGCAGGTAGACAG-3') and INC-RC280S (5'-CTGTCTACCTGCCACAGAATCAGCACCTGC-3').
Protein purification
Wild type and mutant His-tagged integrase proteins were expressed in Escherichia coli BL21(DE3) and purified under non-denaturing conditions. Bacterial cultures were grown at 37°C. When cultures of BL21 achieved an optical density of 0.5 at 600 nm, protein expression was induced by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. The cultures were incubated for 3 hours at 37°C, centrifuged (5000 rpm for 12 min), and frozen at -80°C. The pellets were resuspended in lysis buffer (20 mM Hepes pH 7.5, 100 mM NaCl, 2 mM β-ME, protease inhibitors) and lysed by sonication. The lysates were centrifuged (12 500 rpm for 30 min), the supernatants discarded, and the pellets resuspended in binding buffer (1 M NaCl, 20 mM imidazole, 20 mM Hepes pH 7.5, 2 mM β-ME, 100 μM ZnCl2, protease inhibitors). Following centrifugation at 12500 rpm for 30 min, the supernatants were incubated with nickel-nitrioltriacetic acid (Ni-NTA) agarose beads (Qiagen) for 1 hour at 4°C with mild agitation. Proteins were purified utilizing propylene columns (Qiagen). His-tagged integrase protein was then eluted, utilizing a gradient of increasing imidazole concentration (0-2 M) in elution buffer (1 M NaCl, 20 mM Hepes pH7.5, 10% glycerol, 2 mM β-ME, 100 μM ZnCl2). The eluates were analyzed on 10% SDS-polyacrylamide gels with Coomassie staining (Sigma-Aldrich). Proteins were dialyzed overnight at 4°C against 1 M NaCl, 200 mM Hepes pH 7.5, 100 μM ZnCl2, 10% glycerol and 2 mM DTT in dialysis cassettes (10,000 MWCO, ThermoScientific). The samples were aliquoted and fast frozen at -80°C. Protein concentration was measured by Bradford assay utilizing the Bradford Protein Assay kit (Bio-Rad Laboratories).
Thrombin His tag exclusion
The His tags were removed from purified recombinant subtype B and C integrases using the Thrombin CleanCleave kit (Sigma) according to manufacturers' instructions. Proteins were dialyzed overnight at 4°C against 1 M NaCl, 200 mM Hepes pH 7.5, 100 μM ZnCl2, 10% glycerol and 2 mM DTT in dialysis cassettes (10,000 MWCO, ThermoScientific).
3' processing, strand transfer and disintegration assays evaluated in urea gels
Oligonucleotide-based assays were performed to measure integrase enzymatic activities. All oligonucleotide probes were gel purified and phenol-chloroform extracted. INT1 oligonucleotide was radiolabeled using the T4 Polynucleotide Kinase kit (Ambion) with [γ-32P] ATP (Perkin Elmer). Unincorporated nucleotides were discarded utilizing 'NucAway columns' (Ambion). A double stranded oligonucleotide substrate was obtained by mixing equal concentrations of INT1 and INT2, heating to 95°C and stepwise cooling to 37°C in 100 mM NaCl. The oligonucleotides T35 and SK70 were annealed to form a second double stranded oligonucleotide. INT1/2 mimicked the HIV-1 U5 long terminal repeat (LTR) and acted as a substrate for 3' processing. T35/SK70 acted as a site for integration of the processed INT1/2 fragment and mimicked host DNA.
Concentrations of enzymes used in the following reactions were optimized in a series of preliminary experiments. Integrase reactions were performed in a buffer containing 20 mM Hepes (pH 7.5), 30 mM NaCl, 1 mM dithiothreitol, 125 μM ZnCl2 and 0.125 pmol dsDNA substrate (INT1/2) in a final volume of 10 μl. Recombinant integrase (3.1 μM), and varying concentrations of integrase inhibitors or water/DMSO as control, were mixed and preincubated at 37°C for 15 min. 3' processing reactions were initiated by addition of 7.5 mM of MnCl2 and incubated at 37°C for 3 min unless otherwise indicated. For strand transfer reactions, 1.25 pmol of dsDNA template (T35/SK70) were added and the reaction mixture was further incubated at 37°C for 1 hour unless otherwise indicated. Reactions were stopped by adding 5 times the volume of gel loading dye (formamide containing 1% SDS, 0.25% bromophenol blue, and 0.25% xylene cyanol) and heating to 95°C. Reaction products were separated on 6% acrylamide, 7 M urea sequencing gels. Gels were dried and exposed utilizing phosphorimager screens (GE Healthcare) and scanned in a Molecular Dynamics Typhoon Phosphorimager (GE Healthcare). Product analysis and quantification were conducted using ImageQuant and GraphPad Prism 4.0 software. Quantification of standard error of the mean (SEM) was performed with GraphPad Prism 4.0 software.
The use of microtiter plates for strand transfer analysis
A microtiter plate assay was utilized to evaluate MgCl2-mediated strand transfer as previously described [24,31]. Briefly, biotinylated oligonucleotides mimicking LTR donor DNA (5'-biotin-ACCCTTTTAGTCAGTGTGGAAAATCTCTAGCAGT and 5'-ACTGCTAGAGATTTTCCACACTGACTAAAAG) were immobilized onto black-colour Reacti-Bind Streptavidin-coated plates (ThermoFisher). 313 nM recombinant enzyme was bound to donor DNA on the plates in the presence of 25 mM MnCl2 and the plates were then washed to remove excess unbound enzyme. 3'-FITC labelled dsDNA (5'-TGACCAAGGGCTAATTCACT-FITC-3' and 3'-FITC-ACTGGTTCCCGATTAAGTGA-5'), used as a reaction target, was added to the wells in 25 mM Hepes (pH 7.8), 25 mM NaCl, 2.5 mM MgCl2, and 50 μg/mL BSA [24,32] and the plate was incubated at 37°C for 1 hour. Covalently linked target DNA was detected through use of an anti-FITC antibody conjugated to alkaline phosphatase (Roche) and a chemiluminescence substrate (CSPD Sapphire II, Applied Biosystems). Integrase inhibitors were added at increasing concentrations shortly before the addition of target DNA. Strand transfer was evaluated by chemiluminescence.
Statistical Analysis
Unpaired two-tailed t-tests were used to examine statistical significance in subtype B versus subtype C integrase enzymatic assays using GraphPad Prism 4.0 software.
Determination of activity of integrase inhibitors in cell culture
Recombinant viruses (subtype B) (pNL4-3 and IIIb) and viruses obtained from either our primary HIV infection cohort or from long-term infected patients (subtype B and C) were amplified as previously described [33,34]. Drug susceptibility was measured in cell culture-based phenotypic assays using cord blood mononuclear cells to determine the extent of in vivo HIV replication blockage by integrase inhibitors. 50% drug effective concentrations (EC50s) were determined for each of RAL, EVG and MK-2048 by monitoring the production of p24 antigen, as previously elaborated [33].
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
TB designed all of the biochemistry and enzyme experiments performed in this assay. She wrote the first draft of the manuscript. RDS, DAD and BDK contributed to data management and interpretation. These individuals also contributed to the writing of the manuscript. VHF performed some of the biochemical analyses. HX was involved in preparation and purification of integrase enzymes. MO was responsible for tissue culture analyses. MAW supervised the project, secured funding toward its implementation and contributed to the writing of the manuscript.
Contributor Information
Tamara Bar-Magen, Email: tamara.bar-magen@mail.mcgill.ca.
Richard D Sloan, Email: richard.sloan@mail.mcgill.ca.
Verena H Faltenbacher, Email: v.faltenbacher@web.de.
Daniel A Donahue, Email: daniel.donahue@mail.mcgill.ca.
Björn D Kuhl, Email: Bjorn.kuhl@mail.mcgill.ca.
Maureen Oliveira, Email: moliveira@ldi.jgh.mcgill.ca.
Hongtao Xu, Email: hongtaoxu_00@yahoo.com.
Mark A Wainberg, Email: mark.wainberg@mcgill.ca.
Acknowledgements
We thank Dr. Robert Craigie for the pET15B IN subtype B expression vector, Dr. Yudong Quan and Dr. Amnon Hizi for helpful discussions, Ms. Estrella Moyal and Ms. Bonnie Spira for assistance with digital artwork and Ms. Emily I. McDonough for technical support. This work was supported by a grant from the Canadian Institutes of Health Research (CIHR). RDS is the recipient of a post-doctoral fellowship from the CIHR Canadian HIV Trials Network and the Canadian Foundation for AIDS Research (CANFAR). DAD is the recipient of a pre-doctoral fellowship from the Fonds de la recherche en santé du Québec (FRSQ). We also thank Dr. Daria Hazuda of Merck Inc. for incisive comments as well as for providing drugs and Dr. Lei Zhang of Merck-Frosst for supplying drugs and for research support.
References
- Grinsztejn B, Nguyen BY, Katlama C, Gatell JM, Lazzarin A, Vittecoq D, Gonzalez CJ, Chen J, Harvey CM, Isaacs RD. Safety and efficacy of the HIV-1 integrase inhibitor raltegravir (MK-0518) in treatment-experienced patients with multidrug-resistant virus: a phase II randomised controlled trial. Lancet. 2007;369:1261–1269. doi: 10.1016/S0140-6736(07)60597-2. [DOI] [PubMed] [Google Scholar]
- Shimura K, Kodama E, Sakagami Y, Matsuzaki Y, Watanabe W, Yamataka K, Watanabe Y, Ohata Y, Doi S, Sato M, Kano M, Ikeda S, Matsuoka M. Broad Antiretroviral Activity and Resistance Profile of the Novel Human Immunodeficiency Virus Integrase Inhibitor Elvitegravir (JTK-303/GS-9137) J Virol. 2008;82:764–774. doi: 10.1128/JVI.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DA, Steigbigel RT, Gatell JM, Rockstroh JK, Katlama C, Yeni P, Lazzarin A, Clotet B, Kumar PN, Eron JE, Schechter M, Markowitz M, Loutfy MR, Lennox JL, Zhao J, Chen J, Ryan DM, Rhodes RR, Killar JA, Gilde LR, Strohmaier KM, Meibohm AR, Miller MD, Hazuda DJ, Nessly ML, DiNubile MJ, Isaacs RD, Teppler H, Nguyen BY. BENCHMRK Study Teams. Subgroup and Resistance Analyses of Raltegravir for Resistant HIV-1 Infection. N Engl J Med. 2008;359:355–365. doi: 10.1056/NEJMoa0708978. [DOI] [PubMed] [Google Scholar]
- Briz V, Garrido C, Poveda E, Morello J, Barreiro P, de Mendoza C, Soriano V. Raltegravir and etravirine are active against HIV type 1 group O. AIDS Res Hum Retroviruses. 2009;25:225–227. doi: 10.1089/aid.2008.0222. [DOI] [PubMed] [Google Scholar]
- Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- Sherman PA, Fyfe JA. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-C. [DOI] [PubMed] [Google Scholar]
- Delelis O, Carayon K, Saib A, Deprez E, Mouscadet JF. Integrase and integration: biochemical activities of HIV-1 integrase. Retrovirology. 2008;5:114. doi: 10.1186/1742-4690-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow SA, Vincent KA, Ellison V, Brown PO. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- Myers RE, Pillay D. Analysis of Natural Sequence Variation and Covariation in Human Immunodeficiency Virus Type 1 Integrase. J Virol. 2008;82:9228–9235. doi: 10.1128/JVI.01535-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Danovich R, Witmer M, Nguyen B-Y, Teppler H, Zhao J, Barnard R, Hazuda D, for the HIV-1 Integrase Inhibitor Devt Teams, Protocol 004 Team, and BENCHMRK-1 and -2 Teams. Emerging Patterns of Resistance to Integrase Inhibitors. 16th Conference on Retroviruses and Opportunistic Infections. Montreal. 2009.
- Malet I, Soulie C, Tchertanov L, Derache A, Amellal B, Traore O, Simon A, Katlama C, Mouscadet JF, Calvez V, Marcelin AG. Structural effects of amino acid variations between B and CRF02-AG HIV-1 integrases. J Med Virol. 2008;80:754–761. doi: 10.1002/jmv.21169. [DOI] [PubMed] [Google Scholar]
- Bushman FD, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TM, Engelman A, Ghirlando R, Craigie R. A Soluble Active Mutant of HIV-1 Integrase. J Biol Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- Bischerour J, Leh H, Deprez E, Brochon J-C, Mouscadet J-F. Disulfide-Linked Integrase Oligomers Involving C280 Residues Are Formed In Vitro and In Vivo but Are Not Essential for Human Immunodeficiency Virus Replication. J Virol. 2003;77:135–141. doi: 10.1128/JVI.77.1.135-141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. Inhibitors of Strand Transfer That Prevent Integration and Inhibit HIV-1 Replication in Cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Johnson AA, Marchand C. Integrase inhibitors to treat HIV/Aids. Nat Rev Drug Discov. 2005;4:236–248. doi: 10.1038/nrd1660. [DOI] [PubMed] [Google Scholar]
- Vacca JWJ, Fisher T, Embrey M, Hazuda D, Miller M, Felock P, Witmer M, Gabryelski L, Lyle T. Discovery of MK-2048 - subtle changes confer unique resistance properties to a series of tricyclic hydroxypyrrole integrase strand transfer inhibitors. 4th IAS Conference. Sydney, Australia. 2007.
- Wai JTF, Embrey M, Egbertson M, Vacca J, Hazuda D, Miller M, Witmer M, Gabryelski L, Lyle T. Next generation of inhibitors of HIV-1 integrase strand transfer inhibitor: structural diversity and resistance profiles. Conf Retrovir Opportunistic Infect 2007. Los Angeles, California. 2007.
- Marinello J, Marchand C, Mott BT, Bain A, Thomas CJ, Pommier Y. Comparison of raltegravir and elvitegravir on HIV-1 integrase catalytic reactions and on a series of drug-resistant integrase mutants. Biochemistry. 2008;47:9345–9354. doi: 10.1021/bi800791q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malet I, Delelis O, Valantin M-A, Montes B, Soulie C, Wirden M, Tchertanov L, Peytavin G, Reynes J, Mouscadet J-F, Katlama C, Calvez V, Marcelin AG. Mutations Associated with Failure of Raltegravir Treatment Affect Integrase Sensitivity to the Inhibitor In Vitro. Antimicrob Agents Chemother. 2008;52:1351–1358. doi: 10.1128/AAC.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobler JA, Stillmock K, Hu B, Witmer M, Felock P, Espeseth AS, Wolfe A, Egbertson M, Bourgeois M, Melamed J, Wai JS, Young S, Vacca J, Hazuda DJ. Diketo acid inhibitor mechanism and HIV-1 integrase: Implications for metal binding in the active site of phosphotransferase enzymes. PNAS. 2002;99:6661–6666. doi: 10.1073/pnas.092056199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Cajas JL, Pai NP, Klein MB, Wainberg MA. Differences in resistance mutations among HIV-1 non-subtype B infections: a systematic review of evidence (1996-2008) J Int AIDS Soc. 2009;12:11. doi: 10.1186/1758-2652-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazuda DJ, Anthony NJ, Gomez RP, Jolly SM, Wai JS, Zhuang L, Fisher TE, Embrey M, Guare JP Jr, Egbertson MS, Vacca JP, Huff JR, Felock PJ, Witmer MV, Stillmock KA, Danovich R, Grobler J, Miller MD, Espeseth AS, Jin L, Chen IW, Lin JH, Kassahun K, Ellis JD, Wong BK, Xu W, Pearson PG, Schleif WA, Cortese R, Emini E, Summa V, Holloway MK, Young SD. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. PNAS. 2004;101:11233–11238. doi: 10.1073/pnas.0402357101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarino A. In-Silico docking of HIV-1 integrase inhibitors reveals a novel drug type acting on an enzyme/DNA reaction intermediate. Retrovirology. 2007;4:21. doi: 10.1186/1742-4690-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loizidou EZ, Kousiappa I, Zeinalipour-Yazdi CD, Vijver DAMC Van de, Kostrikis LG. Implications of HIV-1 M Group Polymorphisms on Integrase Inhibitor Efficacy and Resistance: Genetic and Structural in Silico Analyses. Biochemistry. 2009;48:4–6. doi: 10.1021/bi8019349. [DOI] [PubMed] [Google Scholar]
- Xu HT, Martinez-Cajas JL, Ntemgwa ML, Coutsinos D, Frankel FA, Brenner BG, Wainberg MA. Effects of the K65R and K65R/M184V reverse transcriptase mutations in subtype C HIV on enzyme function and drug resistance. Retrovirology. 2009;6:14. doi: 10.1186/1742-4690-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debyser Z, Cherepanov P, Pluymers W, Clercq E. Assays for the Evaluation of HIV-1 Integrase Inhibitors. 2001. pp. 139–155. [DOI] [PubMed]
- Hazuda DJ, Hastings JC, Wolfe AL, Emini EA. A novel assay for the DNA strand-transfer reaction of HIV-1 integrase. Nucleic Acids Res. 1994;22:1121–1122. doi: 10.1093/nar/22.6.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debyser Z, Cherepanov P, Pluymers W, De Clercq E. Assays for the evaluation of HIV-1 integrase inhibitors. Methods Mol Biol. 2001;160:139–155. doi: 10.1385/1-59259-233-3:139. [DOI] [PubMed] [Google Scholar]
- Brenner BG, Oliveira M, Doualla-Bell F, Moisi DD, Ntemgwa M, Frankel F, Essex M, Wainberg MA. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. AIDS. 2006;20:F9–13. doi: 10.1097/01.aids.0000232228.88511.0b. [DOI] [PubMed] [Google Scholar]
- Cotte L, Trabaud MA, Tardy JC, Brochier C, Gilibert RP, Miailhes P, Trepo C, Andre P. Prediction of the virological response to etravirine in clinical practice: Comparison of three genotype algorithms. J Med Virol. 2009;81:672–677. doi: 10.1002/jmv.21461. [DOI] [PubMed] [Google Scholar]