Abstract
Chronic inflammation predisposes to a variety of human cancers. Affected tissues slowly accumulate mutations, some of which affect growth regulation and drive successive waves of clonal evolution, whereas a far greater number are functionally neutral and serve only to passively mark expanding clones. Ulcerative colitis (UC) is an inflammatory bowel disease, in which up to 10% of patients eventually develop colon cancer. Here we have mapped mutations in hypermutable intergenic and intronic polyguanine tracts in patients with UC to delineate the extent of clonal expansions associated with carcinogenesis. We genotyped colon biopsies for length altering mutations at 28 different polyguanine markers. In eight patients without neoplasia, we detected only two mutations in a single individual from among 37 total biopsies. In contrast, for 11 UC patients with neoplasia elsewhere in the colon, we identified 63 mutations in 51 nondysplastic biopsies, and every patient possessed at least one mutant clone. A subset of clones were large and extended over many square centimeters of colon. Of these, some occurred as isolated populations in nondysplastic tissue, considerably distant from neoplastic lesions. Other large clones included regions of cancer, suggesting that the tumor arose within a preexisting clonal field. Our results demonstrate that neutral mutations in polyguanine tracts serve as a unique tool for identifying fields of clonal expansions, which may prove clinically useful for distinguishing a subset of UC patients who are at risk for developing cancer.
Keywords: field effect, lineage mapping, neoplastic evolution
Cancer is a disease of somatic cellular evolution characterized by successive waves of mutation, selection, and clonal expansion (1–3). In many malignancies, most of this process is thought to occur within a relatively confined location, such as in the well studied adenoma-to-carcinoma sequence of sporadic colorectal cancer (4). However, cancers arising within the context of certain predisposing and preneoplastic conditions, including oral leukoplakia (5), Barrett's esophagus (6), and inflammatory bowel disease (7), among others, appear to evolve more diffusely. The concept of “field effect,” first articulated by Slaughter (8) more than half a century ago, describes the observation that cells within an area surrounding some tumor types display abnormal, yet not fully cancerous properties. More recently, it has been appreciated that clonally derived cell populations bearing a subset of the genetic and epigenetic abnormalities found in the tumor itself frequently form the basis for such fields (9). Recognition that cancer-causing mutations may first emerge as widespread clones within non-neoplastic tissue has motivated efforts to identify the unique genetic changes that precede cancer for use in predicting its future development.
Ulcerative colitis (UC) is a chronic inflammatory disease of the colon that predisposes to colorectal cancer and affects approximately half a million individuals in the United States alone (10). After 8 years of disease, a patient's risk of cancer increases 0.5–1% per year, reaching nearly one in five after 30 years (11). Longstanding UC presents a formidable clinical challenge; although cancer risk is markedly increased relative to an age-matched population, the absolute risk is not sufficiently high to justify the morbidity, cost, and quality of life issues associated with prophylactic colectomy if management of symptoms is otherwise satisfactory. Because UC-derived dysplasias can be flat and hard to visualize endoscopically, current surveillance measures entail performing colonoscopy every 1 to 2 years to procure 30–60 biopsies for histological assessment in the hope that if cancer or advanced dysplasia exists, it will be found by random sampling (12). This practice is expensive, insufficiently sensitive, and only detects a neoplastic process once it has progressed to a morphologically recognizable stage.
We have previously demonstrated that genetic abnormalities common to UC-associated adenocarcinoma, including TP53 mutations (13), ploidy abnormalities (14, 15), and chromosomal losses and gains (14, 16) can be found as large clonal fields in normal-appearing UC tissue outside of cancer sites. Some of these clonal lesions predict risk of future histological progression in individuals currently without dysplasia (15). A subset of individuals, however, progress in the absence of any of these markers. Recent cancer genome sequencing studies suggest that the genetic alterations responsible for driving tumorigenesis are highly diverse and unique to every tumor (17–19). Although some genes are commonly mutated in specific cancers, others are mutated infrequently. Widespread clonal evolution could occur in nondysplastic colon before all UC-associated cancers, yet sometimes be undetectable by standard markers when clonal expansions are driven by mutation of unsuspected genes or regulators elsewhere within the (epi)genome.
We hypothesize that the general phenotype of clonal expansion, rather than expansion of specific drivers, might serve as a more sensitive biomarker of prehistological neoplastic processes in UC. During normal mitosis, mutations occur at low frequency throughout the genome of all cells (20), bestowing each cell with a unique fingerprint. While some mutations produce phenotypic changes, the vast majority occur outside of genes and regulatory regions and are likely to be functionally silent “passengers.” Irrespective of the specific mutation driving a clonal expansion, in theory, the progeny of any such event will be distinguishable from nearby cells by virtue of sharing the neutral mutational signature of the founding cell. The challenge to such a detection approach lies in the difficulty of locating these rare passenger mutations within a 6-Gb diploid genome.
Short, repetitive sequences are replicated with significantly lower fidelity than other portions of the genome. Polyguanine tracts, in particular, undergo insertion and deletion mutation with rates on the order of ≈10−4 per cell generation (21). These mutational hotspots serve as likely candidates for bearing lineage-identifying somatic variants. We have recently developed a high-throughput genomic approach to screen for mitotically acquired mutations at polyguanine sites (22, 23) and have used this to produce cell fate maps of mouse development (22, 24). In the present study, we adapt this technique to identify clonal expansions in UC colon. Our results indicate that the method is highly effective at detecting discrete clones and that the presence of these clones in nondysplastic tissue distinguishes patients who have progressed to advanced histological disease from those who have not. We demonstrate that the cell lineage information encoded in the genome by neutral mutant markers provides a useful tool for studying histologically invisible neoplastic processes and a potentially powerful method of identifying patients at greatest risk for developing cancer.
Results
Polyguanine Tract Genotyping.
Microsatellite genotyping by capillary electrophoresis produces an analog signal that reflects the predominant allele lengths within a DNA sample. We have previously identified somatic mutations in polyguanine alleles that are unique to single mouse cells by clonally expanding their genome in vitro before analysis (22, 24). Such mutations cannot be detected in bulk-tissue, because rare alleles are obscured by more prevalent ones. Initial experiments in the present study sought to establish the feasibility of detecting polyguanine genotypes in the human colon where clonal expansion has occurred in vivo.
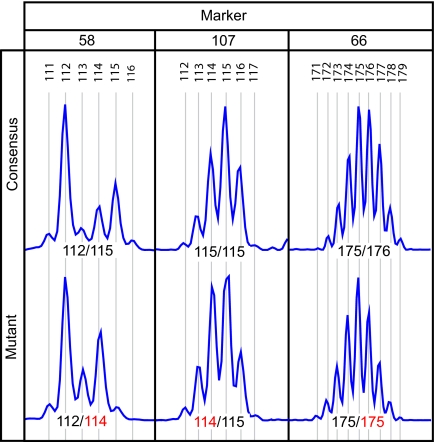
Fluorescently labeled PCR primers were designed to flank 35 non-coding polyguanine tracts, arbitrarily selected from throughout the human genome (Fig. S1). The amplified product of each marker displayed either one or two maximal peaks depending on whether the individual was heterozygous or homozygous for repeat length at the locus (22) (Fig. 1). Adjacent submaximal peaks (“stutter”) are an artifact of PCR amplification, resulting from strand slippage leading to insertions and deletions in a subset of amplicon molecules (25). To confirm reproducibility of genotyping based on maximal peak position, a single DNA sample was used to initiate 15 replicate PCRs for each primer set. Genotype assignments made by a blinded observer were 100% concordant among replicates.
Fig. 1.
Example electropherograms showing polyguanine tract genotype variation between a spatially separated pair of UC colon biopsies in three individuals. For each polyguanine marker, the “consensus” genotype is that most commonly observed among biopsies from a single patient. Mutant genotypes are those that differ from the consensus with respect to the length of at least one allele. X-axis indicates product length (bp), y-axis represents signal intensity. Allele lengths are indicated, with mutant alleles in red. Non-indicated peaks are an artifact of PCR amplification (“stutter”).
To determine the threshold for detection of mutants within a heterogeneous population of mutants and adjacent nonmutant cells, we carried out mixing experiments. For a subset of markers, DNA from two individuals whose germline genotypes differed by a single base pair in one allele were combined in ratios of 20% increments. Electropherograms from these mixtures were then compared to those from the original, unmixed DNAs. The fractional abundance required to reliably identify a simulated mutant clone varied by marker, but consistently fell between 40–60%. Thus, identification of a mutant allele by this technique indicates that a minimum of 40% of harvested cells must share the mutation.
Clonal Expansions in Nondysplastic UC.
We next investigated whether polyguanine tract mutations could be used to identify clonally expanded cell populations within UC tissue. Between 44 and 144 biopsies were harvested in an evenly spaced grid along the colon of 19 UC patients (15 after colectomy, four during colonoscopy). Eight individuals with no histological evidence of high-grade dysplasia (HGD) or cancer anywhere in the colon were classified as “Non-Progressors.” Eleven individuals with at least one biopsy with HGD or cancer were classified as “Progressors.” An average of 4.6 non-dysplastic biopsies were arbitrarily selected (average spacing of 20.1 cm) along each colon for genotyping. Of exception were three Non-Progressor and one Progressor cases where genotyped biopsies were limited to the rectum. DNA separately purified from epithelial and stromal cell fractions of an ≈9-mm2 portion of each biopsy was genotyped at 28 polyguanine sites and four “Bethesda Panel” markers used to diagnose microsatellite instability (MSI) in DNA mismatch repair (MMR)-deficient states (26). Personnel performing the genotyping were blinded to all clinical information.
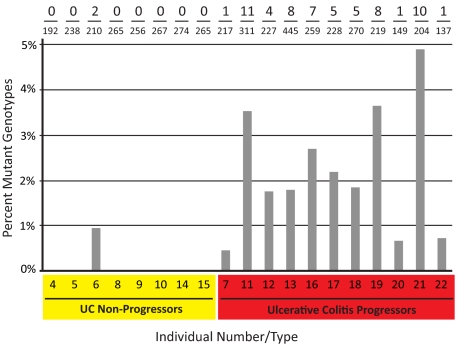
Within each patient, the majority of genotypes for a given marker were identical across all samples. Occasionally, however, the allele pattern of a biopsy tissue fraction differed from the predominant, “consensus genotype” identified elsewhere in the colon, indicating that a rare, clonally expanded mutation had been sampled (Fig. 1). A complete listing of genotype calls is provided in Fig. S2. The presence of detectable clones was strongly correlated with progression status (Fig. 2). Of the 63 mutations identified, 97% occurred among Progressors. Whereas 100% of Progressors had at least one identifiable mutation, only one out of eight (12.5%) Non-Progressors showed any mutations (P < 0.001, one-tailed Fisher's exact test). Although the overall prevalence of detectable mutations was low (≈1.4% of all successful genotypings), within the Progressor group, 63% of biopsies carried at least one mutant marker. Thus, at least two-thirds of the nondysplastic biopsies obtained from Progressor colon arose from postzygotic clonal expansions. As a screening tool for detecting HGD or cancer elsewhere in the colon, identification of one or more mutant markers in nondysplastic tissue was 100% sensitive and 88% specific.
Fig. 2.
Frequency of mutant polyguanine genotypes by disease status. An average of 4.6 histologically nondysplastic biopsies were obtained from eight individuals with UC and no histological evidence of cancer or HGD anywhere in the colon (UC Non-Progressors) and 11 with UC and at least one site with adenocarcinoma and/or HGD (UC Progressors). Biopsies were divided into epithelial and stromal fractions, and both fractions were genotyped at 28 polyguanine markers. For each individual, the number of mutant genotypes out of the total number of successful genotypes (Top), and the percentage of mutant genotypes (bars) are reported. Genotyping was performed under fully blinded conditions.
Of the clonal mutations identified, 78% (49 of 63) occurred in biopsy epithelium, while 22% (14 of 63) were found in biopsy stroma. In the majority of cases (94%), a matching mutation from the epithelium was not found in the corresponding stroma. Besides affirming the robustness of tissue separation, this finding suggests that clonal expansions infrequently cross compartment boundaries. Interestingly, of the two instances of a dual compartment mutation, one pair represented the only mutations found in the Non-Progressor group (patient 7) and might represent an embryonically derived clonal lineage.
Of biopsies with a detectable clone, the majority (69%) were identified by a mutation in a single marker. In one-third of mutant biopsies, however, from two to six markers were simultaneously altered. Based on an evolutionary model of cancer, clones having accumulated multiple independent mutations might have undergone more rounds of selection and expansion than clones with single mutations and potentially be more “cancer-like.” We plotted the number of mutations identified among all progressors as a cumulative function of distance to the nearest site of cancer or HGD (Fig. S3). The presence of detectable clones was independent of proximity to neoplasia. No significant associations (by Wilcox Rank Sum test) were found between the frequency of clonal mutations and age (P = 0.73, dichotomized by median), gender (P = 0.31), presence of primary sclerosing cholangitis (PSC; a positive modifier of cancer risk in UC) (P = 0.20), duration (P = 0.14, dichotomized by median) or clinical severity of UC (P = 0.72), or whether progression was to HGD or cancer (P = 0.72) (Fig. S4). The limited number of Progressors in this pilot study, however, is insufficient (estimated power of 7% at 0.05 alpha level) to conclude that a difference between observed mean mutation frequencies existed between different subgroups of Progressors.
Mutations were found in some marker sites more frequently than in others (Fig. S5). No alterations were identified in half of the polyguanine sites. Significantly, no mutations were detected for any of the four Bethesda panel MSI markers tested indicating that polyguanine marker mutations are independent of DNA MMR deficiency. In contrast, several other polyguanine loci were mutated in about 9% of Progressor samples. This non-normal distribution (P < 0.001, Shapiro-Wilk normality test) likely reflects differences in slippage frequency inherent to particular loci (24, 27).
Different types of genotype alterations were observed among mutant clones (Fig. 1 and Fig. S2). Deletions [51] were about four times more frequent than additions [12] among allele changes. Among mutations, 24 homozygote-to-heterozygote and 11 heterozygote-to-heterozygote alterations resulted from slippage of one allele. Twenty-three apparent heterozygote-to-homozygote mutations could have resulted either from allele slippage in the polyguanine tract or a chromosomal loss-of-heterozygosity (LOH) event, yielding an identical-appearing hemizygote. However, the latter explanation appears unlikely. The strong bias toward deletions was present in heterozygote-to-homozygotes (21 deletions, two additions) to the same degree as in the other two categories. If LOH predominated as a mechanism, the relative size of the allele lost would be random, yet this was not the case (P < 0.001 against the null hypothesis of equal loss probability). Thus, most, if not all mutations observed resulted from slippage in a polyguanine tract with retention of both alleles before clonal expansion.
Clonal Patch Size.
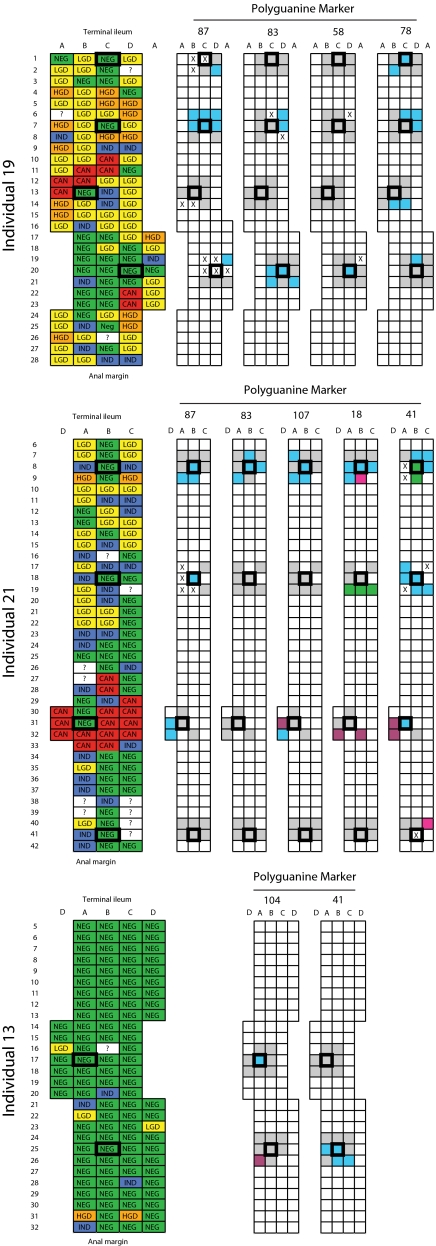
On six occasions, identical mutations were detected in two or more well separated biopsies from the same patient raising the question of whether a clonal patch extends over more than one biopsy site. To further characterize the spatial extent of clonal expansions, we selected 10 biopsy sites from three patients, as regions for further analysis. Epithelial DNA was isolated from portions of four to eight additional biopsies immediately surrounding each of the 10 sites. Surrounding biopsies were of all histological subtypes and ranged from 1–5 cm from the central biopsy. As a control, epithelial DNA was also re-extracted from the 10 original samples. All samples were then genotyped at a subset of markers previously identified as mutant in one or more of the central biopsies for each individual. Fig. 3 graphically summarizes the spatial relationship between mutations identified. Complete genotype information is provided in Fig. S6.
Fig. 3.
Clonal patches identified by polyguanine mapping. Longitudinally opened colectomy specimens from three individuals are diagrammed with small boxes representing individual biopsies (≈9 mm2), taken at evenly spaced intervals within an alphanumeric grid. The histological diagnosis of each biopsy in the colon is indicated at left: NEG, negative for dysplasia; IND, indefinite for dysplasia; LGD, low grade dysplasia; HGD, high grade dysplasia; CAN, cancer; ?, no data. The genotypes of biopsies for specific polyguanine markers are indicated at right. Outlined boxes represent biopsies used in the initial study. Empty boxes represent biopsies not genotyped, “X” indicates unsuccessful genotyping. Gray fields indicate biopsies with the consensus genotype for the marker, and different colors represent distinct mutant genotypes within an individual for each marker. Clustering of identical mutant genotypes in adjacent biopsies suggests large, clonally derived patches.
Blinded regenotyping of central samples was 100% concordant with initial results. A large number of mutations were identified in surrounding biopsies. Many of these were identical to those found in the central biopsies, strongly suggesting derivation from a common clonal “patch.” In total, 30 clones were identified among these three patients, of which 18 encompassed two or more adjacent biopsies. In some instances, patches were small; the mutant genotype of a central sample was completely surrounded by biopsies bearing consensus genotypes at the same marker (i.e., Fig. 3, individual 13, biopsy 17A). In other cases patches were large, extending into multiple biopsies (i.e., individual 19, rows 6–7). Up to four different mutant genotypes were seen in the same marker within a single individual (i.e., individual 21, marker 18). Spatial clustering of discrete variants reinforces the technical precision of genotyping and indicates independent expansions.
Clonal patches were identified among biopsies of all histological diagnoses. Some large patches were found in exclusively nondysplastic areas (i.e., Fig. 3, individual 13, rows 25–26, marker 41), whereas other patches were present in neoplastic mucosa of a variety of grades (i.e., individual 19, rows 6–7, marker 87). Thus, genetically defined cell lineage mapping identifies clones that cannot be discerned from morphologic characteristics. In some instances, adjacent clonal patches defined by different mutant markers could be clearly distinguished from each other (i.e., individual 13, rows 25–26). In other cases, a patch defined by one marker appeared within a larger patch defined by a different marker (i.e., individual 19, rows 6–7). Such serially nested clones provide information regarding the phylogenetic history of expansion. Within individual 19 for example, parsimonious logic dictates that the three-biopsy patch defined by marker 78 most likely arose as a subclone of the six-biopsy patch defined by marker 87.
Complete Colon Map.
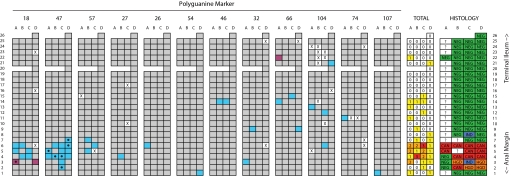
The extent of dysplasia in Progressor colons varied from limited (i.e., individual 13) to widespread (i.e., individuals 19 and 21). To determine the extent of clonal expansions in a best approximation of “early” UC Progressor tissue, we densely sampled an additional colectomy specimen with pancolonic inflammatory disease, where histological changes were limited to the rectosigmoid colon. Epithelial DNA was isolated from a grid of 98 biopsies evenly spaced along the entire colon and blindly genotyped using the complete panel of polyguanine markers (Fig. 4 and complete genotype data in Fig. S7).
Fig. 4.
Polyguanine mapping of a complete UC Progressor colon. A longitudinally opened colectomy specimen is diagramed with boxes representing evenly spaced biopsies measuring ≈9 mm2 within an alphanumeric grid. The histological diagnosis of each biopsy is indicated at far right: NEG, negative for dysplasia; IND, indefinite dysplasia; HGD, high grade dysplasia; CAN, cancer; ?, no data. Biopsy genotypes for various polyguanine marker are indicated in separate grids. “X” indicates unsuccessful genotyping. Gray fields indicate biopsies with the consensus genotype for the marker, and different colors represent distinct mutant genotypes for each marker. Dots indicate biopsies where a mixture of consensus and mutant genotypes were observed, suggesting a mixed population of cells with different genotypes. The total number of mutant genotypes identified across all markers is reported for each biopsy (heat map). Large, clonally derived patches identified by three markers were observed near the cancer site. Numerous smaller patches were detected throughout the nondysplastic portions of the colon.
Mutations were identified in nearly one-third (31 of 98) of biopsies tested. The majority of clones detected in the histologically negative portion of the colon were small in size, with mutant genotypes appearing only in single biopsies. At the cancer focus, however, a large clonal patch extending over at least 13 biopsy sites was found. The abundance of mutant samples in this case made it possible to distinguish samples that contained a mix of both mutant and consensus genotype cells (Fig. S8). These predictably occurred on patch boundaries. In addition to nine cancerous biopsies, a mutation in marker 47 identified two histologically negative and two noncancer dysplasia samples as belonging to the same clone, suggesting that the cancer and dysplasia emerged from a preexisting, nondysplastic clonal field.
Discussion
We have shown that neutral markers of cell lineage can be used to identify clonal expansions in nondysplastic UC colon and effectively distinguish patients who have progressed to histologically advanced disease from those who have not. These clones may occur as fields from which a cancer has arisen or as otherwise invisible populations, more than half a meter from the nearest dysplasia. Our findings reinforce prior evidence that neoplastic evolution in UC is delocalized, multifocal, and involves both epithelium and stroma (7, 13–16, 28, 29). Our results suggest that screening for clonal expansions in UC patients may be able to identify tumorogenic processes before the emergence of histologically recognizable disease.
The presence of large, clonally derived patches in colon represents a divergence from normal cellular homeostasis. The colon is divided into replicative units known as crypts. These invaginated structures contain a small population of stem cells at their base that continually replicate to clonally populate the luminal surfaces with terminally differentiated progeny that are then sloughed off after several days (30). Studies of normal female colons have shown that embryonically originating patches sharing a common inactivated X-chromosome do not exceed 450 crypts in size (31). Spontaneously arising mitochondrial mutations indicate that clonal crypt clusters arise postnatally in normal colon and increase in size with age, yet rarely exceed a dozen units (32). The biopsy portions we genotyped contained ≈2,000–5,000 crypts. Because at least 40% of cells in a sample must share a mutation for it to be detectable, it can be estimated that clones within isolated mutant biopsies comprise at least 800–2,000 crypts. In our study, only one patch was identified in a single Non-Progressor colon. It is possible that this case may, in fact, represent one of the 10% of the general UC population who would have ultimately progressed to cancer.
Using neutral passenger rather than putative driver mutations as lineage markers of clonal expansions has two primary advantages. First, cancer is a stochastic evolutionary process, and not all tumors necessarily arise through mutation of the same set of drivers (18, 19). Neutral mutations offer an unbiased, generalizable way of identifying clones that is independent of molecular causation. Such an approach integrates all possible bases for an abnormal cell/tissue behavior, including mutation of unknown genetic and epigenetic sites as well as nonheritable influences of local environment. Second, screening for known drivers may limit detectability to relatively late-arising clones. Experimental mutation of many of the best characterized oncogenes and tumor suppressors in otherwise untransformed cells induces growth arrest and senescence (33). It is conceivable that mutation in common tumor drivers seen in UC-derived adenocarcinoma, including TP53 and KRAS, may not be tolerated in the earliest arising clones. Similar arguments mitigate against the utility of randomly arising gross chromosomal abnormalities as early clonal markers.
Functionally, neutral mutations serve as useful markers of clonality but are, nevertheless, imperfect. Detection of a clone requires that members share a common mutation, originating in the founding cell, which distinguishes them from the surrounding population (Fig. 5). While every clone is likely to be uniquely marked relative to neighboring cells somewhere in the thousands of polyguanine tracts in its genome, there is no guarantee that a mutation will have occurred in the specific subset of tracts being genotyped, so some clones will go undetected. Although we identified clones in two-thirds of Progressor biopsies, the true proportion is likely to be even greater given that we only examined 28 polyguanine tracts.
Fig. 5.
Model for how mutant genotypes become identifiable as a result of clonal expansion. (A) As cells divide throughout life, they acquire unique somatic mutations at polyguanine tracts. However, because such mutations are rare and independent, for any given locus the majority of cells do not carry a mutation. Consequently, genotyping identifies only the dominant, nonmutated (or “consensus”) genotype in a biopsy, and the individual mutant genotypes carried by single cells or small subclones are not observed. (B) If an individual cell marked by a mutant allele clonally proliferates to populate a relatively large area, a unique genotype can come to dominate the sampled population and mutant alleles become detectable by genotyping.
Although our study relies on microsatellite mutations as lineage markers, we find no evidence for the presence of the extensive MSI that results from deficiencies in MMR (26). No mutations were identified in any of the National Cancer Institute (NCI) Bethesda Panel MSI markers examined, and the low frequency of polyguanine mutations detected (1.4% overall, 2.3% in Progressors) is inconsistent with a vastly increased rate of microsatellite slippage (21). Moreover, whereas MMR-mediated MSI commonly manifests as a “widening” of electropherogram stutter peaks, indicative of a large number of length variants simultaneously present within a sample (34), the rare polyguanine mutations we identified were predominantly the result of single slipped alleles. While there have been reports of “low-level” MSI in UC-derived adenocarcinomas and occasionally in surrounding nondysplastic tissue (29, 35, 36), microsatellite slippage events are expected to be occasionally witnessed in any clonally derived population (22, 37), and the latter may best explain our findings. Nevertheless, it is possible that a moderately increased rate of slippage may occur in UC. Direct measurement of the per-division mutation rate in UC Progressors relative to normal colon would be challenging given that detection of slippage is necessarily coupled to clonal expansion—something that does not occur in normal colon. Although an increased rate of mutagenesis resulting from inflammation-derived reactive oxygen species (38, 39) or defects in replication fidelity acquired during neoplastic transformation (2) would increase the number of somatic variants and facilitate detection of clones, it is not mandatory to invoke such mechanisms to explain our observations.
A unique aspect of our study is that it exclusively relies on neutral markers of cell lineage to identify clinically relevant clonal expansions in a preneoplastic condition. Several groups have previously used neutral markers to study tumor ancestry and tissue dynamics. In combination with suspected driver mutations, Maley and colleagues used slippage in short tandem repeats to show that risk of cancer progression is related to clonal diversity in Barrett's esophagus (40). Although the bulk of clones in the study were identified by likely driver mutations, evolutionary statistics suggested that clonal heterogeneity, in and of itself, is an important cancer predictor. Frumkin et al. used microsatellite slippage events to phylogenetically reconstruct the lineage relationship between single cells in a mouse lymphoma (41). Shibata's group used noncoding microsatellite mutations as a “molecular clock” of somatic division and used this to calculate the number of cell generations between initiation and sampling of human MSI tumors (34). More recently, these investigators have capitalized on the high frequency of epimutations in CpG islands of nontranscribed genes to study the clonal structure of non-MSI tumor development (42). Wright and colleagues have recently used the stainable phenotype of neutral, homoplasmic mutations in a mitochondrially encoded cyclooxygenase gene to study cell lineages in variety of tissues (43). Although the process by which homoplasmy arises is not well understood and takes many years to develop in some tissues (32), the ability to directly visualize the individual cells of a clonal population in an unperturbed tissue context makes this a promising technique with the potential to complement our studies.
The experiments we have undertaken expand on our previous studies of field effects in UC. We have shown that neutral markers of cell lineage can identify large clonally derived patches in normal-appearing colon of patients having histologically recognizable disease but not in those without. Although this ability will be of clinical utility in its own right, a more significant possibility is that such patches may be able to identify individual UC patients at greatest risk for developing colon cancer before the emergence of histological changes (i.e., Future Progressors). Our observation that clones may cover an area with both dysplastic and nondysplastic tissue strongly argues that expansions can predate dysplasia. Prospective studies will be needed to determine efficacy as a predictive biomarker of cancer risk.
From a basic science perspective, we have demonstrated that random mutations can be used to define the boundaries of clonal expansions occurring in vivo and identify subclones within larger clones. Screening a larger number of polyguanine marker sites will enable detailed phylogenetic reconstruction of the relationship between clones and may allow estimation of the number of rounds of selective outgrowths needed for neoplastic transformation. After using these markers to define clone boundaries, it will be possible to screen for candidate driver mutations and determine the fraction of expansions that are driven by factors other than genes already known to play a role in UC-mediated carcinogenesis. Our technique should be adaptable to the study of other preneoplastic conditions or cancers. We believe that the general tactic of tracing cell lineage with spontaneously arising neutral markers of all forms holds promise for better understanding the neoplastic process. While there are thousands of polyguanine tracts in the human genome, there are billions other nucleotides and epigenetic sites for which mutation is likely to be functionally silent. When it becomes technically and economically possible to screen the whole genome with future sequencing technologies, a wealth of developmental history from all forms of normal and abnormal cell proliferation will become available. Not only do random mutations form the fundamental basis of evolutionary biology, they provide a powerful tool for studying its role in human disease.
Materials and Methods
Specimen information as well as sample preparation and polyguanine tract genotyping protocols are detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Eva McMonagle and Allyn Stevens for technical assistance. This work was supported by the National Institutes of Health Grants RO1CA102029 and RO1CA115802 (to L.A.L.), DP1OD003278 and RO1DK078430 (to M.S.H.), RO1CA068124 (to T.A.B., P.S.R., and M.P.B.), F30AG033485 (to J.J.S.), F30AG030316 (to S.J.S.), CA137136 (to R.A.R.), and T32GM007266 (to J.J.S. and S.J.S.); Crohn's and Colitis Foundation of America (T.A.B. and M.P.B.); and ARCS Fellowship (J.J.S and S.J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909428106/DCSupplemental.
References
- 1.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- 3.Merlo LMF, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 5.Brennan JA, et al. Molecular assessment of histopathological staging in squamous-cell carcinoma of the head and neck. N Engl J Med. 1995;332:429–435. doi: 10.1056/NEJM199502163320704. [DOI] [PubMed] [Google Scholar]
- 6.Prevo LJ, Sanchez CA, Galipeau PC, Reid BJ. p53-mutant clones and field effects in Barrett's esophagus. Cancer Res. 1999;59:4784–4787. [PubMed] [Google Scholar]
- 7.Risques RA, Rabinovitch PS, Brentnall TA. Cancer surveillance in inflammatory bowel disease: New molecular approaches. Curr Opin Gastroenterol. 2006;22:382–390. doi: 10.1097/01.mog.0000231812.95525.a7. [DOI] [PubMed] [Google Scholar]
- 8.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 9.Braakhuis BJM, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of Slaughter's concept of field cancerization: Evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]
- 10.Hanauer SB. Inflammatory bowel disease: Epidemiology, pathogenesis, and therapeutic opportunities. Inflamm Bowel Dis. 2006;12(Suppl 1):S3–S9. doi: 10.1097/01.mib.0000195385.19268.68. [DOI] [PubMed] [Google Scholar]
- 11.Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: A meta-analysis. Gut. 2001;48:526–535. doi: 10.1136/gut.48.4.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ullman T, Odze R, Farraye FA. Diagnosis and management of dysplasia in patients with ulcerative colitis and Crohn's disease of the colon. Inflamm Bowel Dis. 2009;15:630–638. doi: 10.1002/ibd.20766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brentnall TA, et al. Mutations in the p53 gene: An early marker of neoplastic progression in ulcerative colitis. Gastroenterology. 1994;107:369–378. doi: 10.1016/0016-5085(94)90161-9. [DOI] [PubMed] [Google Scholar]
- 14.Burmer GC, et al. Neoplastic progression in ulcerative colitis: Histology, DNA content, and loss of a p53 allele. Gastroenterology. 1992;103:1602–1610. doi: 10.1016/0016-5085(92)91184-6. [DOI] [PubMed] [Google Scholar]
- 15.Rubin CE, et al. DNA aneuploidy in colonic biopsies predicts future development of dysplasia in ulcerative colitis. Gastroenterology. 1992;103:1611–1620. doi: 10.1016/0016-5085(92)91185-7. [DOI] [PubMed] [Google Scholar]
- 16.Chen R, Bronner MP, Crispin DA, Rabinovitch PS, Brentnall TA. Characterization of genomic instability in ulcerative colitis neoplasia leads to discovery of putative tumor suppressor regions. Cancer Genet Cytogenet. 2005;162:99–106. doi: 10.1016/j.cancergencyto.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 18.Fox EJ, Salk JJ, Loeb LA. Cancer genome sequencing—an interim analysis. Cancer Res. 2009;69:4948–4950. doi: 10.1158/0008-5472.CAN-09-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salk JJ, Fox EJ, Loeb LA. Mutational heterogeneity in human cancers: Origins and consequences. Ann Rev Pathol: Mech Dis. 2010;5:51–75. doi: 10.1146/annurev-pathol-121808-102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyer JC, et al. Sequence dependent instability of mononucleotide microsatellites in cultured mismatch repair proficient and deficient mammalian cells. Hum Mol Genet. 2002;11:707–713. doi: 10.1093/hmg/11.6.707. [DOI] [PubMed] [Google Scholar]
- 22.Salipante SJ, Horwitz MS. Phylogenetic fate mapping. Proc Natl Acad Sci USA. 2006;103:5448–5453. doi: 10.1073/pnas.0601265103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salipante SJ, Thompson JM, Horwitz MS. Phylogenetic fate mapping: Theoretical and experimental studies applied to the development of mouse fibroblasts. Genetics. 2008;178:967–977. doi: 10.1534/genetics.107.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salipante SJ, Nas A, McMonagle E, Horwitz MS. Phylogenetic analysis of developmental and postnatal mouse cell lineages. Evol Dev. doi: 10.1111/j.1525-142X.2009.00393.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke LA, Rebelo CS, Gonçalves J, Boavida MG, Jordan P. PCR amplification introduces errors into mononucleotide and dinucleotide repeat sequences. Mol Pathol. 2001;54:351–353. doi: 10.1136/mp.54.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boland CR, et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 27.Eckert KA, Hile SE. Every microsatellite is different: Intrinsic DNA features dictate mutagenesis of common microsatellites present in the human genome. Mol Carcinog. 2009;48:379–388. doi: 10.1002/mc.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leedham S, et al. Clonality, founder mutations, and field cancerization in human ulcerative colitis-associated neoplasia. Gastroenterology. 2009;136:542–550. doi: 10.1053/j.gastro.2008.10.086. [DOI] [PubMed] [Google Scholar]
- 29.Yagishita H, Yoshida T, Ishiguro K, Numata Y, Okayasu I. Epithelial and stromal genetic instability linked to tumor suppressor genes in ulcerative colitis-associated tumorigenesis. Scand J Gastroenterol. 2008;43:559–566. doi: 10.1080/00365520701817419. [DOI] [PubMed] [Google Scholar]
- 30.Leedham SJ, Wright NA. Expansion of a mutated clone: From stem cell to tumour. J Clin Pathol. 2008;61:164–171. doi: 10.1136/jcp.2006.044610. [DOI] [PubMed] [Google Scholar]
- 31.Novelli M, et al. X-inactivation patch size in human female tissue confounds the assessment of tumor clonality. Proc Natl Acad Sci USA. 2003;100:3311–3314. doi: 10.1073/pnas.0437825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greaves LC, et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci USA. 2006;103:714–719. doi: 10.1073/pnas.0505903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben-Porath I, Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol. 2005;37:961–976. doi: 10.1016/j.biocel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 34.Tsao JL, et al. Genetic reconstruction of individual colorectal tumor histories. Proc Natl Acad Sci USA. 2000;97:1236–1241. doi: 10.1073/pnas.97.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brentnall TA, et al. Microsatellite instability in nonneoplastic mucosa from patients with chronic ulcerative colitis. Cancer Res. 1996;56:1237–1240. [PubMed] [Google Scholar]
- 36.Ozaki K, et al. Heterogeneous microsatellite instability observed within epithelium of ulcerative colitis. Int J Cancer. 2006;119:2513–2519. doi: 10.1002/ijc.22095. [DOI] [PubMed] [Google Scholar]
- 37.Graham T, Halford S, Page KM, Tomlinson IPM. Most low-level microsatellite instability in colorectal cancers can be explained without an elevated slippage rate. J Pathol. 2008;215:204–210. doi: 10.1002/path.2351. [DOI] [PubMed] [Google Scholar]
- 38.Jackson AL, Chen R, Loeb LA. Induction of microsatellite instability by oxidative DNA damage. Proc Natl Acad Sci USA. 1998;95:12468–12473. doi: 10.1073/pnas.95.21.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofseth LJ, et al. The adaptive imbalance in base excision-repair enzymes generates microsatellite instability in chronic inflammation. J Clin Invest. 2003;112:1887–1894. doi: 10.1172/JCI19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maley CC, et al. The combination of genetic instability and clonal expansion predicts progression to esophageal adenocarcinoma. Cancer Res. 2004;64:7629–7633. doi: 10.1158/0008-5472.CAN-04-1738. [DOI] [PubMed] [Google Scholar]
- 41.Frumkin D, et al. Cell lineage analysis of a mouse tumor. Cancer Res. 2008;68:5924–5931. doi: 10.1158/0008-5472.CAN-07-6216. [DOI] [PubMed] [Google Scholar]
- 42.Siegmund K, Marjoram P, Tavaré S, Shibata D. Many colorectal cancers are “flat” clonal expansions. Cell Cycle. 2009;8:2187–2193. doi: 10.4161/cc.8.14.9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fellous T, et al. A methodological approach to tracing cell lineage in human epithelial tissues. Stem Cells. 2009;27:1410–1420. doi: 10.1002/stem.67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.