Abstract
Sustained extracellular signal-regulated kinase (ERK)-signaling plays a critical role in T-cell-mediated IL-2 production. Although many downstream targets are known for ERK, details remain unknown about which molecules play functional roles in IL-2 production. Here, we addressed this question using proteomic analysis of nuclear proteins from TCR-activated T cells and identified hnRNP-K as one of the ERK targets essential for IL-2 production. hnRNP-K was previously shown by others to be a direct substrate of ERK and form complexes with multiple signaling proteins as well as DNA and RNA. Our data showed a clear ERK-dependent increase in one form of hnRNP-K after TCR stimulation. Small interfering RNA-mediated gene knockdown of hnRNP-K expression abrogated IL-2 production by T cells. Moreover, reduction of hnRNP-K expression caused a notable increase in proteolysis of Vav1, a binding target of hnRNP-K. Since Vav1 is an essential molecule for T-cell activation, the data suggest that ERK signaling is required for T-cell activation partly by inhibiting activation-induced proteolysis of Vav1.
Keywords: ERK, hnRNP-K, IL-2, signal transduction, T cell, TCR, Vav1
Introduction
Antigen receptor stimulation in T cells causes multiple signaling events that lead to the activation and differentiation of mature T cells. One of the most significant events induced by TCR activation is the production of IL-2, a lymphokine critical for the expansion and survival of antigen-activated effector and regulatory T cells (1). The lack of IL-2 production leads to loss of regulatory T cells and causes severe autoimmune disease (2). Accumulating evidence shows that sustained TCR engagement is essential for IL-2 production by primary T cells (3–6). These studies showed that the disruption of antigen receptor engagement as late as 10 h after the initiation of T-cell stimulation abrogates IL-2 production.
We previously demonstrated that the Shc- and extracellular signal-regulated kinase (ERK)-mediated signaling plays critical roles in the regulation of IL-2 expression. A kinetic study on T-cell activation revealed that IL-2 production requires ERK signal 4–6 h after the start of stimulation (termed herein ‘late phase’), whereas inhibition of ERK during the first 2 h (termed ‘early phase’) showed no effect on IL-2 production. On the other hand, ERK signal was not required for expression of CD25, the IL-2 receptor a chain. The data demonstrated that there is a qualitative difference between signaling events that take place during the early and late hours of T-cell activation.
The need for signal accumulation is one explanation detailing why sustained activation is required for full T-cell activation (4). Sustained signaling will lead to the accumulation of transcription factors necessary for IL-2 gene expression. As a result, transcription begins once the amount of accumulated molecules exceeds the point of threshold. Another explanation is that sustained signaling leads to the activation of signaling molecules that are not present in resting T cells but are essential for IL-2 gene transcription (7). mRNA production of IL-2 requires de novo synthesis of proteins after TCR stimulation, whereas CD25 mRNA synthesis does not (5). Further, when ERK signal was inhibited during the late phase, IL-2, but not CD25 expression, was blocked. Based on these data, we proposed a ‘two tier model’ that hypothesized ERK transduces a signal during the late phase of T-cell activation that is essential for IL-2 production (8).
To test this hypothesis, we searched for molecules that mediate signals during the late phase of T-cell activation in co-operation with ERK. ERK is known to translocate into the nucleus after stimulation. Thus, we isolated nuclear proteins from activated T cells with or without ERK signal inhibition and compared the levels of proteins separated by 2-dimensional gel electrophoresis. We identified five proteins that display differences in expression when ERK signal is inhibited and found that one of the proteins, hnRNP-K, plays a critical role in IL-2 production. hnRNP-K binds to RNA, DNA and protein partners and is strongly suggested to act as a docking platform to facilitate communication among molecules involved in signal transduction and gene expression (9). Our data indicate that hnRNP-K may be required for IL-2 production, in part, for its ability to protect Vav from activation-induced proteolysis.
Methods
Antibodies and reagents
Anti-TCR antibody (C305) was a generous gift from A. Weiss (University of California, San Francisco, CA, USA). Anti-ERK1/2 antibody, anti-c-Rel antibody and anti-phosphotyrosine antibody (4G10) were purchased from Upstate (Charlottesville, VA, USA). Phospho-p44/42 mitogen-activated protein kinase (MAPK) (Thr202/Tyr204) mAb (E10), phospho-ERK kinase (MEK1/2) (Ser217/221) antibody, anti-mouse IgG, HRP-linked antibody, anti-rabbit IgG, HRP-linked antibody and PD98059 (a MEK1 inhibitor) were purchased from Cell Signaling Technology, Inc. (Beverly, MA, USA). BCA Assay Kit and Coomassie Plus protein assay reagent were purchased from Pierce. Iodoacetamide and dimethyl pimelimidate were purchased from Sigma-Aldrich (Saint Louis, MO, USA). Anti-Lck antibody (MOL171) was kindly provided by Y. Koga (Tokai University, Kanagawa, Japan). Anti-hnRNP-K antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and ImmuQuest Ltd (Cleveland, UK). Anti-MEK1 antibody was also from Santa Cruz Biotechnology. Anti-Vav1, anti-IL-2, anti-mouse CD3 (2C11) and anti-mouse CD8 antibodies, PE-labeled anti-human CD25 and anti-human CD69, allophycocyanin-labeled anti-human CD154 antibodies, Z-DEVD-FMK, Z-VAD-FMK and Streptavidin–HRP were from BD Biosciences (San Diego, CA, USA). HRP-labeled anti-goat antibody and goat anti-mouse IgG antibody were purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA, USA). Phorbol 12-myristate 13-acetate (PMA) and ionomycin were from Calbiochem. Protein G Sepharose® 4 Fast Flow, CyDye DIGE Cy2, Cy3, Cy5, Immobiline DryStrip, Urea, CHAPS, IPG buffer and dithiothreitol were purchased from Amersham Biosciences (Piscataway, NJ, USA). SYPRO Ruby protein gel stain was purchased from Bio-Rad (Hercules, CA, USA). Sequencing grade-modified trypsin and Dual-Luciferase® Reporter Assay System were purchased from Promega (Madison, WI, USA). Mouse mAb anti-hemagglutinin (HA) (clone 12CA5) was from Roche Diagnostics Corporation (Indianapolis, IN, USA). Opti-MEM I Reduced Serum Medium and Geneticin® were purchased from Invitrogen Corporation (Carlsbad, CA, USA).
Mice and cells
Eight-week-old BALB/cJ mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). Jurkat cells (a gift from A. Weiss, UCSF) were maintained in RPMI 1640 medium supplemented with 5% FCS (Hyclone, Logan, UT, USA), 100 IU ml−1 penicillin, 100 μg ml−1 streptomycin and 2 mM L-glutamine. Jurkat cells expressing SV40 large T antigen (10) (Jurkat Tag cells, a gift from G. Crabtree, Stanford) were maintained in RPMI 1640 medium as for Jurkat cells J.V cells and J.HA-K cells were obtained by selecting Geneticin®-resistant stable clones from pCDNA3 or HA-hnRNP-K-transfected Jurkat cells. A 2 mg ml−1 Geneticin® was used for initial selection, and 0.1 mg ml−1 Geneticin® was used for maintenance of stable transfectants.
DNA
IL-2 (Sb)-luciferase was constructed by inserting luciferase complementary DNA into the first exon of the IL-2 gene at the translation initiation site. Nuclear factor kappa-light-chain enhancer of activated B cells (NF-kappa B)-Luciferase was from J. Fujisawa (Kansai Medical University, Osaka, Japan). Nuclear factor of activated T cells-luciferase was obtained from G. Crabtree (Stanford University, Stanford, CA, USA). Activator protein (AP)-1-Luciferase was described previously (11). Heat shock protein (Hsp) 90–luciferase construct was a kind gift from N. Mivechi (Medical College of Georgia, Augusta, GA, USA). Small RNA interference construct pBS/U6-hnRNP-Ki (siK) was designed by inserting short sense and antisense oligonucleotides specifically targeted to human hnRNP-K (GGGATTTCCCATGCGGGGAAGA) with a spacer between them into pBS/U6 vector (12) (a gift from Y. Shi, Harvard Medical School, Boston, MA, USA). A fragment containing the EGFP gene driven by the CMV promoter was inserted into siK to construct pBS/U6-hnRNP-Ki-EGFP (siK-EGFP). For mouse samples, siK-EGFP was constructed by replacing human siK-EGFP oligonucleotides fragment with GGTGAATTTGGTAAACGCCCTG, which are specific to mouse hnRNP-K. HA-hnRNP-K is a gift from Z. Ronai (Mount Sinai School of Medicine, New York, NY, USA). pEFVav1 is a gift from A. Weiss.
Stimulation of T cells
Jurkat cell stimulation was performed as described (13). Mouse CD4 + T cells were stimulated with antigen presenting cells (APC) alone or APC plus anti-CD3 antibody 2C11 (1 μg ml−1). The ratio of APC (syngenic splenocytes irradiated 2000 rad) to CD4 + T cells was 1.5:1. For experiments with the MEK inhibitor, cells were pre-incubated with 25 μM PD98059 or dimethyl sulfoxide (DMSO) (solvent control) at 37°C for 30 min before stimulation and the media were replaced with medium not containing inhibitors at the indicated time points.
Protein preparation
Cells were harvested at the indicated time point and lysed with Nonidet P40 lysis buffer [10 mM Tris (pH 7.8), 150 mM NaCl, 1% Nonidet P40] with protease and phosphatase inhibitors (1 mM phenylmethylsulfonyl fluoride, 50 μg ml−1 leupeptin, 10 μg ml−1 aprotinin, 1 μg ml−1 pepstatin, 0.4 mM ethylenediaminetetraacetic acid, 0.4 mM sodium orthovanadate and 10 mM NaF), unless otherwise specified. For preparation of cytoplasmic and nuclear extracts, cells were first swelled in Buffer A [20 mM HEPES (pH 7.9), 300 mM sucrose, 10 mM KCl, 1.5 mM MgCl2] with protease and phosphatase inhibitors. Cells were then lysed with Buffer A containing 0.5% Nonidet P40. Supernatants were collected as cytoplasmic fractions. The resulting pellets were washed once with buffer A containing 0.5% Nonidet P40 before lysing with twice the pellet volume of buffer C [20 mM HEPES (pH 7.9), 300 mM sucrose, 420 mM NaCl, 1.5 mM MgCl2 and 0.5% Nonidet P40] containing protease and phosphatase inhibitors as described above. Lysates were centrifuged for 10 min and supernatants containing nuclear extracts were collected. For two-dimensional differential in-gel electrophoresis (2D-DIGE) analyses, pelleted nuclei were lysed with urea lysis buffer (30 mM Tris, 2 M thiourea, 7 M urea, 4% CHAPS and adjusted to pH 8.5) plus protease and phosphatase inhibitors as described above. Lysates were sonicated, centrifuged and supernatants were collected as nuclear extracts.
Immunoblot analysis
Protein assays were performed using the BCA Assay Kit and an equal amount from each sample was applied for gel separation. Separated proteins were transferred to Immobilon-P membrane (Millipore Corporate, Billerica, MA, USA) and incubated with primary and subsequently secondary antibodies as indicated for each experiment. Quantitative analysis of each band was performed using NIH Image 1.62.
Two-dimensional differential in-gel electrophoresis
Samples containing thiourea were first diluted 10-fold prior to protein determination using Coomassie Plus protein assay reagent. CyDye labeling and 2D-DIGE were performed according to the Ettan DIGE user manual (Amersham Biosciences). Each sample (50 μg) was labeled with 400 pmol CyDye DIGE fluors: Cy2, Cy3 or Cy5. Labeling was performed on ice for 30 min in the dark and the reaction was terminated by the addition of lysine to a final concentration of 1 mM. The samples were kept on ice for another 10 min. After labeling, an equal volume of 2× sample buffer (2 M thiourea, 7 M urea, 2% IPG buffer, 2% dithiothreitol (DTT) and 4% CHAPS) was added to each sample and incubated on ice for 15 min. Labeled samples (Cy2, Cy3 or Cy5 labeled) were pooled (a total of 150 μg of protein) and combined with 150 μg of unlabeled proteins of each sample. A total of 600 mg of proteins was applied to an Immobiline DryStrip (24 cm, pH 3-10NL) and focused using an Ettan IPGphor isoelectric focusing (IEF) system (Amersham Biosciences). After 12–14 h of rehydration, the IEF parameters used were as follows: 100 V for 100 Vhr; 500 V for 500 Vhr; 1000 V for 1000 Vhr and 8000 V for 64 000 Vhr. After IEF, strips were equilibrated with 0.5% (w/v) DTT in equilibration solution [50 mM Tris–Cl (pH 8.8), 6 M urea, 30% (v/v) glycerol, 2% (w/v) SDS] at room temperature for 15 min. Strips were further equilibrated with 4.5% iodoacetamide in equilibration solution for 15 min at room temperature. Samples were separated by SDS–PAGE using an Ettan DALT system (for 24 cm strips; Amersham Biosciences) or Hoefer SE 600 (for 13 cm strips; Amersham Biosciences). Fluorescent images were obtained by scanning 2D gels with a Typhoon variable mode imager (Amersham Biosciences). Images acquired were then normalized and analyzed using DeCyder DIA software (Amersham Biosciences). After scanning, 2D gels were immediately fixed with 30% methanol/7.5% acetic acid with gentle swirling for 2 h at room temperature. Gels were then incubated overnight with Sypro Ruby at room temperature to stain total proteins. Gels were destained with 10% methanol/6% acetic acid at room temperature for 2 h and scanned with a Typhoon variable mode imager. Spots that showed differential expression in all three samples (or in two samples in anti-Shc IP/2D-DIGE experiment) were selected based on Sypro Ruby staining using an Ettan Spot Picker (Amersham Biosciences).
Trypsin digestion
Gel plugs were first washed with 20 mM ammonium bicarbonate in 50% methanol three times for 20 min each at room temperature. Plugs were washed with water twice and dehydrated with acetonitrile for 5 min twice. Plugs were further dried in a speed vac until plugs appeared ‘flake like’. Trypsin (reconstituted in 20 mM ammonium bicarbonate) was used to digest gel plugs (0.5 mg for one plug). Digestion was performed at 37°C for 2.5 h and stopped by 2% trifluoroacetic acid (TFA). Peptides were extracted by adding 0.1% TFA in 75% methanol and dried.
Mass spectrometry and protein identification
Peptides were re-suspended in 1 μl of 0.1% TFA. Samples were mixed with an equal amount of freshly prepared matrix [10 mg ml−1 α-cyano-hydroxy-cinnamic acid in acetonitrile: 0.1% TFA (1:1; v: v)]. Mixtures were applied to the target and dried at room temperature prior to mass spectrometry analysis (Voyager, MALDI-TOF or ABI 4700 Proteomics Analyser, Applied Biosystems, Foster City, CA, USA). Protein identifications were obtained by a probability-based database search by Medical College of Georgia Proteomics Core Facility (14).
Cell transfection, promoter assay, immunoblot analysis and flow cytometry
Jurkat cells and Jurkat Tag cells were transfected using the Gene Pulser II electroporation system (Bio-Rad) as described (13). Primary T-cell transfection was performed as described previously (15). Luciferase activity was determined using the Dual-Luciferase® Reporter Assay System following the manufacturer’s directions. Transfection efficiency was normalized using the activity of the co-transfected Renilla Luciferase (pRL-CMV) in unstimulated samples. Flow cytometry and immunoblot were performed as described (13).
Results
Identification of downstream targets of ERK by 2D-DIGE/mass spectrometry
To identify the molecule that mediates ERK signaling for IL-2 production, Jurkat cells were stimulated for 6 h with an anti-TCR antibody in the presence or absence of a MEK inhibitor, PD98059. Nuclear extracts were prepared from these samples for 2D-DIGE analysis. Unstimulated samples were labeled with Cy2. Stimulated samples were labeled with Cy3 and stimulated samples in the presence of PD98059 were labeled with Cy5. Labeled samples were pooled into one and separated by IEF, followed by SDS–PAGE. Fluorescent images were obtained from gels and spots that showed statistically significant differences among the three samples were picked for trypsin digestion. Digested peptides were analyzed by MALDI mass spectrometry (MS)-MS analysis for sequence determination.
The summary of results is listed in Table 1. In total, five proteins were identified from spots that showed statistically significant differences between PD98059-treated and untreated samples. Spots corresponding to the heterogeneous nuclear ribonucleoprotein A2/B1 [hnRNP-A2/B1 (isoforms A2)] and acidic ribosomal phosphoprotein P2 were significantly increased after TCR stimulation and the increase was blocked by PD98059. On the contrary, spots containing heterogeneous nuclear ribonucleoprotein K (hnRNP-K), beta actin and aldolase A showed decreases after TCR stimulation and the decrease was blocked by MEK inhibitor treatment.
Table 1.
List of proteins identified by nuclear proteome analysis
| Protein ID | Fold change by stimulation | Fold change by PD treatment | pI | Molecular weight | Peptides sequence matched | Coverage (%) |
| hnRNP-A2/B1 | 1.96** | −1.3** | 8.7 | 36 | 16 | 55 |
| Acidic ribosomal phosphoprotein P2 | 1.33 | −1.36** | 4.4 | 11.7 | 5 | 69 |
| hnRNP-K | −2.25** | 1.42** | 5.2 | 51.3 | 13 | 34 |
| β-Actin | −1.65** | 1.35** | 5.3 | 41.7 | 9 | 27 |
| Aldolase A | −3.36** | 1.72** | 8.39 | 39.7 | 14 | 40 |
**P < 0.05. pI, isoelectric point.
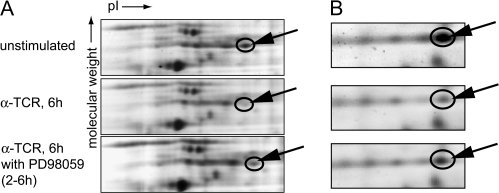
Figure 1 shows gel images for two proteins identified through this approach. The spot identified as hnRNP-K (arrow) showed a 2.25-fold decrease (P < 0.05) when anti-TCR-stimulated samples were compared with the resting cell sample (Fig. 1A, upper and middle panels). PD98059 treatment of TCR-stimulated samples induced a 1.42-fold (P < 0.05) increase over PD98059-untreated sample (Fig. 1A, middle and lower panels). Similar changes were observed for this spot when total cell lysates were analyzed in the same manner (data not shown).
Fig. 1.
Identification of nuclear target of ERK signal in T cells. Jurkat cells were stimulated with plate-bound anti-TCR antibody. PD98059 (25 μM) or DMSO (carrier for PD98059) was added to the culture at 2–6 h during stimulation. Nuclear extracts were prepared 6 h after stimulation. Samples were labeled with Cy2 (unstimulated), Cy3 (TCR stimulated) and Cy5 (TCR stimulated with PD98059), mixed and loaded on one gel and separated by 2D-PAGE. Images acquired for each sample were normalized using the standard and compared for the level of proteins. Spots that showed differences in expression among three samples were isolated and analyzed by Mass spectrometry. Protein identifications were obtained by probability-based database search. Arrows point to proteins identified as (A) hnRNP-K (B) aldolase. pI, isoelectric point.
Another spot that showed a notable change was identified as aldolase A (Fig. 1B). Aldolase A (shown by arrow) showed a 3.4-fold decrease (P < 0.05) when resting and anti-TCR-stimulated samples were compared (Fig. 1B, upper and middle panels). This spot showed a 1.72-fold (P < 0.05) increase when PD98059-treated and untreated samples were compared (Fig. 1B, middle and lower panels).
hnRNP-K is required for TCR-induced IL-2 production
We chose to further analyze the role of hnRNP-K in T-cell activation. hnRNP-K was identified as a downstream target of ERK upon PMA stimulation of K562 cells (16). hnRNP-K associates with multiple signaling proteins, including Itk (17), Vav (18), Src (19), Fyn, Lyn (20) and PKCδ (21). hnRNP-K contains both a nuclear localization signal and a nuclear shuttling domain (KNS) (22). Moreover, hnRNP-K was shown to bind to the NF-kappaB enhancer element (23) and has the transactivating function (24, 25). In vivo and in vitro studies have shown that hnRNP-K forms a complex with Vav1, a critical signaling molecule for T-cell activation and differentiation (18, 26).
To test the validity of changes identified by 2D-DIGE in hnRNP-K protein, we examined the level of hnRNP-K protein in the nucleus and found no significant change in the amount of hnRNP-K in the nucleus before and after TCR stimulation (Fig. 2A). However, western blot analysis of nuclear proteins separated by 2D gels showed substantial changes of several spots that correspond to the effect of TCR stimulation and PB 98059 treatment (Fig. 2B). At least two spots of hnRNP-K showed decrease and two spots showed increase after TCR stimulation in an ERK-dependent manner.
Fig. 2.
Changes of nuclear hnRNP-K protein by TCR-induced ERK signal. (A) Jurkat cells were stimulated for 6 h with anti-TCR antibody in the medium alone (αTCR) or with PD98059 between 2 and 6 h after start of stimulation (PD). Cells were lysed and separated into soluble fractions (cytoplasm) and insoluble fractions, which were used to prepare nuclear extracts (nuclear). Same amount of proteins from each fraction was used for analysis by western blot with antibody against hnRNP-K (upper panel) and phosphorylated ERK (lower panel). (B) Nuclear fractions from sample prepared as in (A) were separated by 2D-gel electrophoresis and analyzed by western blot with anti-hnRNP-K antibody. Dotted lines show the standard isoelectric point (pI) point to align three gels from unstimulated (top), stimulated (middle) and stimulated with PD98059 from 2 to 6 h (bottom). Circles and arrows show spots that show differences among these three samples. Closed arrowheads: spots increased after stimulation, open arrowheads: spots decreased after TCR stimulation.
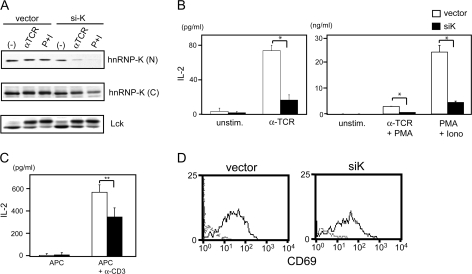
Next, we tested the function of hnRNP-K in T-cell signaling using an small interfering RNA (siRNA) expression construct against hnRNP-K (siK). First, the effectiveness and specificity of siK was tested. Jurkat cells were transfected with siK or vector control. After 1 day, cells were stimulated for 24 h. Cytoplasmic proteins and nuclear extracts were prepared and analyzed by western blot for the level of hnRNP-K protein. The reduction of hnRNP-K expression was clearly detectable in cells transfected with siK (Fig. 3A). In nuclear extracts of unstimulated cells, hnRNP-K protein level was decreased to 62% of the control transfected in siK-transfected cells. Reduction of hnRNP-K expression was more prominent in anti-TCR-stimulated samples (84% reduction) and in phorbol 12-myristate 13-acetate plus ionomycin (PMA + iono)-stimulated samples (94% reduction). A less significant reduction in hnRNP-K protein was observed in cytoplasmic fractions (Fig. 3A, lower panel). In both cases, the reduction of hnRNP-K protein was greater in TCR and PMA + iono-stimulated samples than unstimulated ones, suggesting that hnRNP-K was more labile after stimulation. The same set of cell lysates was also immunoblotted for Lck to show that protein expression levels and loading of these samples were comparable in the vector control and siK-transfected samples.
Fig. 3.
Decrease of hnRNP-K expression impairs IL-2 production in Jurkat cells and primary T cells. (A) Reduction of hnRNP-K protein by siRNA. Jurkat cells were transfected with siRNA expression vector for hnRNP-K (siK) or control vector (vector). Cells were left unstimulated (−), stimulated with anti-TCR antibody (αTCR) or stimulated with PMA plus ionomycin (Iono) (P + I) 24 h after transfection. Cytoplasmic proteins or nuclear extracts were prepared 24 h after stimulation. Nuclear (N) and cytoplasmic (C) fractions were immunoblotted with antibodies with anti-hnRNP-K antibody. Cytoplasmic fractions were also blotted with anti-Lck antibody (lower panel) to confirm the loading levels of proteins. (B) IL-2 production by siK-transfected Jurkat cells. Jurkat cells were transfected with siRNA expression vector for hnRNP-K (siK, closed bar) or control vector (vector, open bar). Left panel show the results from cells unstimulated (unstim.) or stimulated by anti-TCR antibody (αTCR). Right panel shows the results from cells stimulated by anti-TCR and PMA (αTCR + PMA) or PMA + ionomycin (P + I). Data are shown separately because of the differences in the level of IL-2; *P < 0.005. (C) IL-2 production by CD4+ T cells transfected with control vector (open bar) or siK (closed bar). Transfected cells were stimulated with gamma-irradiated splenocytes (APC) alone or APC plus 1 μg ml−1 anti-CD3 antibody. Supernatants were harvested 24 h after stimulation and the amount of IL-2 was determined by ELISA; **P < 0.05. (D) CD69 expression by Jurkat cells transfected with siK expression vector that also expresses EGFP (siK-GFP) or with control vector (vector) and stimulated with anti-TCR antibody. Twenty-four hours after stimulation, cells were harvested and stained with ant-CD69 antibody. Data are shown for cells that expressed EGFP.
To examine the functional relevance of hnRNP-K in T-cell activation, we tested the effect of siK on IL-2 production and surface antigen expression. When cells were transfected with siK, anti-TCR, anti-TCR + PMA and PMA + ionoinduced IL-2 production was severely impaired (Fig. 3B). The data suggest that siK blocked the signaling process that is distal to T-cell activation since stimulation with PMA + iono did not restore the level of IL-2 production. A reduction in IL-2 production was also observed when mouse primary CD4 T cells were used for analysis (Fig. 3C). The level of IL-2 production by siK (mouse)-transfected CD4 T cells was significantly reduced compared with the control vector transfectants (P < 0.05). The effect of siK was less prominent compared with Jurkat cells likely due to lower transfection efficiency of primary T cells (15–20% of cells were constantly transfected compared with ∼50% of transfection efficiency of Jurkat cells).
In contrast to the outcomes from IL-2 production, expression levels of CD25 and CD69 were similar between vector-transfected cells and siK-transfected Jurkat cells (Fig. 3D). Together, these results show that, as observed in ERK signaling, hnRNP-K plays a selective and functional role in IL-2 gene expression.
Transcription regulation by hnRNP-K
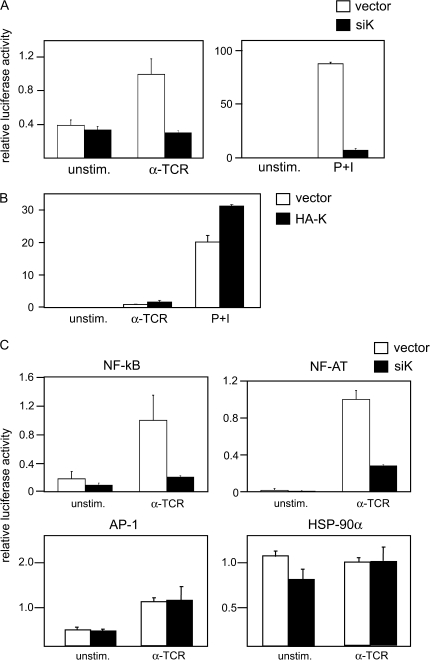
To further understand the role of hnRNP-K in the regulation of IL-2 gene expression, we measured the effect of siK on the promoter activity of IL-2 gene using an IL-2–luciferase reporter construct. When hnRNP-K expression was reduced by siK, IL-2 promoter activity was greatly impaired both in anti-TCR antibody- and PMA + iono-stimulated cells (Fig. 4A). These data suggest that hnRNP-K regulates IL-2 production at the transcriptional level. We also confirmed this result by over-expression of hnRNP-K. HA-tagged full-length hnRNP-K (pCDNA3-HA-K) or control vector (pCDNA3) were transfected into Jurkat cells along with the IL-2-Luciferase reporter vector. As presented in Fig. (4B), over-expression of hnRNP-K significantly enhanced IL-2 promoter activity upon stimulation (P < 0.005). Together, these data strongly support the hypothesis that hnRNP-K plays a critical role in IL-2 gene activation in response to T-cell stimulation at the transcriptional level.
Fig. 4.
Requirement of hnRNP-K for activation of the IL-2 promoter. (A) Jurkat cells were transfected with IL-2–Luciferase reporter construct and siRNA expression vector against hnRNP-K (siK, open bar) or control vector (vector, closed bar). Twenty-four hours later, cells were stimulated with anti-TCR antibody (αTCR) or PMA plus ionomycin (P + I). Cell were harvested 18 h later and used for luciferase assay. Error bars show SD n = 3; *P < 0.005. (B) Jurkat Tag cells were co-transfected with expression vector for HA-tagged hnRNP-K (HA-K, closed bar) or control vector (vector, open bar). Twenty-four hours after transfection, cells were stimulated and analyzed as described in (A); *P < 0.005. (C) Jurkat cells were transfected with NF-κB-luciferase, NF-AT-luciferase, AP-1 luciferase or HSP-90α luciferase reporter constructs with siRNA expression construct for hnRNP-K (siK, closed bar) or control vector (vector, open bar). Transfected cells were stimulated (α-TCR) or left unstimulated (unstim.) and analyzed for luciferase activity; *P < 0.005, **P < 0.05.
To further investigate the mechanism of hnRNP-K-mediated activation of IL-2 gene transcription, we tested the effects of siK on transcription factors that play critical roles in IL-2 gene expression, namely NF-kappaB, NF-AT and AP-1. Reporter constructs for each factor was co-transfected with siK or control vector and transfectants were stimulated with an anti-TCR antibody (Fig. 4C) and PMA + iono (data not shown). When hnRNP-K expression was decreased by siK transfection, NF-kappaB and NF-AT activities were severely reduced (Fig. 4A and B). In contrast, siK had no effect on the AP-1 activity when T cells were stimulated with anti-TCR and only a minor reduction was observed when cells were stimulated PMA + iono (Fig. 4C). siK showed no effect on a constitutively active promoter of Hsp90. Together, these data suggested that hnRNP-K selectively plays roles in activation of two critical transcription factors, NF-kappaB and NF-AT, in T-cell activation.
TCR-induced MEK/ERK activation was impaired by inhibition of hnRNP-K expression
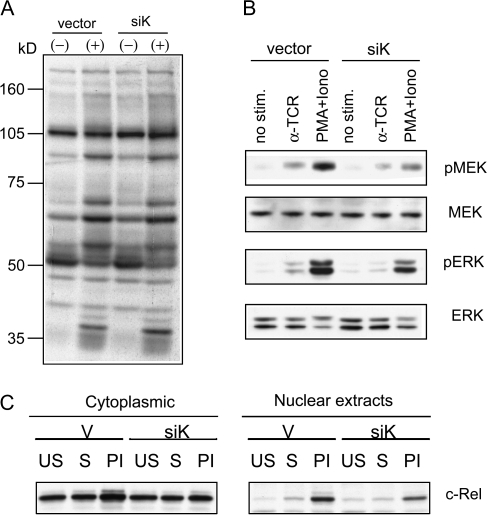
To elucidate the mechanism of how hnRNP-K contributes to the transcriptional activity of TCR-induced NF-AT and NF-kappaB, we examined whether siK impaired early TCR activation-induced events. Twenty-four hours after transfection with vector control or siK, transfected Jurkat cells were stimulated with soluble anti-TCR antibody for 5 min or left unstimulated. Cell lysates from these samples were analyzed by western blot with an anti-phosphotyrosine antibody (Fig. 5A). Anti-TCR-induced induction of tyrosine phosphorylated proteins were clearly observed in both vector-transfected and siK-transfected cells. The patterns of protein tyrosine phosphorylation were indistinguishable between these two samples suggesting that the proximal events of TCR signaling were not affected by knockdown of hnRNP-K.
Fig. 5.
Impaired ERK activation by TCR in siK-treated Jurkat cells. Jurkat cells were transfected with siRNA expression construct for hnRNP-K (siK) or control vector (vector). (A) Transfected cells were stimulated (indicated with + above lanes) with 1 μg ml−1 soluble anti-TCR antibody or left unstimulated (−) for 5 min. Cell lysates were prepared and immunoblotted with anti-phospho-tyrosine antibody. (B) Transfected cells were stimulated with anti-TCR antibody (αTCR), PMA + iono or left unstimulated (no stim.) for 5 min. Same amount of cell lysates from each sample were analyzed by western blot with antibodies shown on the side of each panel. (C) siK- or vector-transfected cells were left unstimulated (US), stimulated with anti-TCR antibody (S) or with PMA plus ionomycin (PI). After 24 h, cells were lysed and cytoplasm and nuclear extracts were prepared for western blot analysis with anti-c-Rel antibody.
In contrast, activation of MEK was greatly reduced as detected by the level of phospho-MEK1/2 (Fig. 5B). MEK1 protein level was comparable between these two samples. The activation of ERK was also reduced in siK-transfected cells. These data suggest that although the ERK signaling regulates hnRNP-K, its presence is also important for ERK activation.
We previously identified an NF-kappaB family member c-Rel as a downstream target of ERK signaling (13). c-Rel is a critical factor for IL-2 production and requires ERK signal for nuclear accumulation. Activation-induced upregulation of c-Rel protein in the nucleus was significantly reduced in the siK-transfected cells (Fig. 5C), showing that nuclear translocation and/or accumulation of c-Rel requires the presence of hnRNP-K.
Regulation of Vav1 proteolysis by hnRNP-K
Both in vivo and in vitro studies have shown the association of hnRNP-K and Vav1 (18, 26). Vav1 has been shown to play critical roles in T-cell activation and is also required for the activation of ERK signaling (27) and transcription factors such as NF-kappaB and NF-AT (28). These functional effects observed by mutagenesis of Vav1 overlaps significantly with what we have observed for gene knockdown of hnRNP-K. Thus, we examined whether Vav1 mediates the effect of hnRNP-K reduction on T-cell activation. In siK-transfected cells, we observed notable changes of Vav1 protein both in the cytoplasm and the nucleus fractions. Full-length Vav1 protein (95 KDa) levels were reduced considerably in siK-transfected cells relative to the control. The reduction was more prominent in stimulated samples. Like hnRNP-K itself, reduction of full-length Vav1 was more substantial in nuclear extracts than in cytoplasmic fractions (Fig. 6B). As full-length Vav1 disappears in these samples, another band of lower molecular weight (∼76 KDa) Vav1 was detected. In contrast to the 95 KDa band, the intensity of the 76 KDa band was greatly increased in siK-transfected cells. The ratio of the intensity of the lower band to the upper band in each lane was determined and shown below each lane. In PMA + iono-stimulated nuclear extracts, the ratio was 16-fold higher in siK transfected relative to vector control-transfected cells. It was previously shown that 95 KDa Vav1 is cleaved by caspase 3 into two cleavage products, Vav1p19 and Vav1p76 (29). Thus, the data suggest that Vav1 undergoes accelerated proteolytic cleavage in cells transfected with siK and hnRNP-K plays a significant role in the maintenance of Vav1 protein integrity.
Fig. 6.
Increase of Vav1 proteolysis by reduction of hnRNP-K expression. (A) Jurkat cells were transfected with siRNA expression vector for hnRNP-K (siK) or control vector (vector). Twenty-four hours later, cells were stimulated with anti-TCR antibody (αTCR) or PMA plus ionomycin (P + I), or left unstimulated (N.S.). Cytoplasmic proteins (upper panel, C) or nuclear extracts (lower panel, N) were prepared 24 h after stimulation and blotted with anti-Vav1 antibody. The relative amount of protein in each sample was determined by densitometry. The ratio of lower band to upper band in each lane was calculated, and the ratio of vector-transfected unstimulated sample was arbitrary determined as 1. Arrows point to corresponding full-length Vav1 (p95) and cleaved form of Vav1 (p76), respectively. (B) Jurkat cells were stimulated with plate-bound anti-TCR antibody in the medium alone (αTCR) or in the presence of PD98059 for the first 2 h (PD0–2) or 2–24 h (PD2–24) during stimulation. Nuclear extracts were prepared 24 h after stimulation and blotted with anti-Vav1 antibody. The relative amount of p76 in each sample was determined by densitometry and is shown below each lane. The density of unstimulated sample was arbitrary determined as 1. Arrow marks cleaved form of Vav1. (C) Jurkat cells were transfected with siRNA expression vector for hnRNP-K (siK, closed bar) or with control vector (vector, open bar) with control the expression vector for Vav1 (Vav1) or control vector (vector). IL-2–luciferase reporter construct was included in each transfection. Error bars show SD n = 3; *P < 0.005. (D) Caspase inhibitors partially restored NF-AT activity in hnRNP-K knockdown cells. Control vector (pBS/U6) or pBS/U6-hKi was co-transfected with NF-AT–Luciferase reporter construct into Jurkat cells. Transfected cells were pre-treated with DMSO, 20 μM Z-DEVD-FMK or 20 μM Z-VAD-FMK for 30 min at 37°C before stimulation. Twenty-four hours after transfection, cells were stimulated with anti-TCR antibody or left unstimulated, with the presence of DMSO or inhibitors. Cell lysates were harvested after 14 h of stimulation for Luciferase assay. Promoter activities were normalized by Renilla luciferase activity from co-transfected vector (pRL-CMV). Luciferase activity from cells transfected with control vector and stimulated in the presence of DMSO was assigned as one, and relative luciferase activity of each sample was calculated. n = 3; *P < 0.005.
Since we identified hnRNP-K as a downstream target of ERK signaling, we tested whether inhibition of ERK signaling causes increases in Vav1 proteolysis. To test this, Jurkat cells were stimulated with anti-TCR antibody in the presence or absence of MEK inhibitor, PD98059. Nuclear and cytoplasmic proteins were prepared at 24 h after stimulation and analyzed by immunoblotting with an anti-Vav1 antibody. Cleavage of full-length Vav1 was observed in anti-TCR-stimulated cells to a small degree (Fig. 6B). Inhibition of ERK activation significantly enhanced this cleavage of Vav1, specifically when PD98059 was added during the late phase of activation. Interestingly, addition of PD98059 during the early hours of stimulation reduced this cleavage, indicating that the signal induces Vav1 cleavage is also dependent on ERK.
To further examine the role of hnRNP-K in Vav1 proteolysis and IL-2 gene expression, we tested the effect of siK on Vav1-mediated enhancement of IL-2 promoter activity. Over-expression of Vav1 increased both basal- and TCR-induced IL-2 promoter activity (Fig. 6C). However, when Vav1 and hKi were co-transfected, Vav1-mediated IL-2 promoter activity was severely reduced. Reduction of the promoter activity was most significant in TCR-stimulated samples.
Taken together, these data show that hnRNP-K plays a critical role in the function of Vav1 in T-cell activation, possibly by protecting Vav1 from activation-induced proteolysis. Thus, we tested if inhibition of Vav1 proteolysis would rescue the impairment caused by hKi. We co-transfected hKi or control vector (pBS/U6) with NF-AT-Luciferase. Transfected cells were pre-treated with 20 μM caspase-3 inhibitor Z-DEVD-FMK, general caspase inhibitor Z-VAD-FMK or DMSO as a control, at 37°C for 30 min before stimulation. Cells were then stimulated with plate-bound anti-TCR antibody in the presence of inhibitors or DMSO. Z-VAD-FMK effectively restored the inhibitory effect of hKi on NF-AT activity and Z-DEVD-FMK partially restored the inhibitory effect of hKi. The effectiveness of caspase inhibitors was also tested by immunoblotting against Vav1. The treatment of caspase inhibitors minimized the proteolysis of Vav1 (data not shown). Together, the data confirmed that caspase-mediated protein cleavage is required for inhibitory functions of siK.
Discussion
In this study, we performed proteomic analysis using nuclear extracts from activated T cells and analyzed the ERK-signaling pathway during the late phase of T-cell activation. In total, five different proteins were identified as downstream targets of ERK upon TCR stimulation by our 2D-DIGE/MS-based proteomics approach. To validate these, we performed functional analyses of hnRNP-K. We selected hnRNP-K not only because this protein showed substantial changes in response to TCR simulation and MEK inhibitor treatment but also because previous reports support the notion that hnRNP-K may play an important role in T-cell activation. The data show that hnRNP-K is a downstream target of ERK and hnRNP-K regulates IL-2 production at the transcriptional level and that Vav is one of the downstream target molecules for hnRNP-K. The data suggest that hnRNP-K is exported from the nucleus in response to ERK signaling and protects full-length Vav from caspase-dependent proteolytic cleavage and allows sustained presence of Vav during T-cell activation.
hnRNP-K as a target of ERK-signaling pathway
Previous studies have shown hnRNP-K is a downstream target of ERK upon PMA stimulation in K562 cells (16). ERK phosphorylates hnRNP-K both in vitro and in vivo at serine 284 and 353. Serine phosphorylation of hnRNP-K by ERK causes the cytoplasmic accumulation of hnRNP-K and promotes the subsequent inhibition of translation by degradation of mRNA that binds hnRNP-K (30). These results suggest that the MAPK signal transduction pathway can directly control the cellular distribution of hnRNP-K and consequently influence the ability of hnRNP-K to regulate translation.
Our data showed that treatment of activated Jurkat cells with MEK inhibition altered the state of nuclear hnRNP-K. Two spots that disappear after PD98059 treatment correspond to hnRNP-K. A potential explanation for this change is that some forms of hnRNP-K translocates to the cytoplasm after TCR-induced ERK stimulation and this translocation is blocked by MEK inhibition. Phosphorylation by ERK may cause altered interactions of hnRNP-K with the target proteins due to the changes in subcellular localization of hnRNP-K, resulting in impaired IL-2 gene expression. A majority of hnRNP-K is present in the nucleus in the resting cells and moves out of nucleus after TCR stimulation. ERK signaling could promote hnRNP-K-Vav interactions by increasing the cytoplasmic hnRNP-K. Previous studies showed that the VAV–hnRNP-K association is mediated by proline-rich sequence of hnRNP-K with the SH3 domain of Vav (18). Thus, the interactions between these proteins may be reduced by inhibition of ERK signaling due to the lowered level of hnRNP-K in the cytoplasm but not due to the effect of molecular interactions between these proteins.
Protection of Vav1 by hnRNP-K during T-cell activation
Full-length Vav1 protein levels were significantly reduced in Jurkat cells when hnRNP-K expression was reduced. The data suggest that hnRNP-K plays a critical role in protecting Vav1 protein after TCR stimulation. A previous report showed that cleavage site of Vav1 localizes within the acidic domain of Vav1 (29). This cleaves removes a majority of the acidic region of Vav1 protein and rendered Vav1 protein incapable of activating IL-2 promoter, NF-kappaB or NF-AT. Vav1 is an integral component of the transcriptionally active NF-AT and NF-kappaB-like complexes (28). Together, these data suggest that enhanced cleavage of Vav1 in the absence of hnRNP-K may impair the activity of IL-2 promoter.
The carboxyl terminal SH3 domain of Vav1 mediates its cytoplasmic retention and deletion of the carboxyl SH3 domain of Vav1 results in a constitutive nuclear localization of Vav1 (28). Interestingly, this mutant of Vav1 is not capable of potentiating IL-2 promoter. The same SH3 domain is the site of interaction with hnRNP-K and another heterogeneous nuclear ribonucleoprotein, hnRNP-C, both in vivo and in vitro (18, 26, 31). Thus, another potential mechanism of how hnRNP-K helps IL-2 promoter activity is to assist export of Vav1 from the nucleus for its function in the cytoplasm. Indeed, our data showed that reduction of hnRNP-K profoundly increased cleavage of Vav1 in the cytoplasm and impaired MEK activation. Vav1 has been shown to catalyze GDP-GTP exchange on Rho family GTPases and plays a critical role in the activation pathway for Ras/MAPK (32). Despite the normal activation of Lck and ZAP-70, Vav1−/− T cells have defects in TCR-induced intracellular calcium fluxes and activation of ERK and NF-kappaB (33). Vav1/2/3-null mice showed that the loss of Vav1 proteins leads to a disruption of Ras/MAPK signaling in both immature and mature T cells (34). More recently, it was shown that Vav1 transduces TCR signals to activate ERK1 and ERK2 in double-positive thymocytes via LAT, Sos and RasGRP1 (35). Therefore, reduction of hnRNP-K may affect activation of ERK via impaired Vav1 activity and blocks positive feedback loop to maintain ERK activation.
Although the effect was relatively mild, we observed decrease of ERK activation in siK-treated Jurkat cells (Fig. 5), while activation of AP-1 was not impaired. This is not unprecedented. For example, we have previously shown that loss of Shc caused reduced ERK activation in T cells but does not affect induction of c-Fos expression (13). ERK is known to activate multiple downstream targets and the data suggest that hnRNP-K maybe involved in the ERK pathway that is distinctive from AP-1 activation. Since hnRNP-K is also in a downstream target of ERK, the data presented here suggest that hnRNP-K plays a role in a positive feedback loop for ERK activation and this feedback may function in a manner specific to a certain subset of ERK in the cells. ERK signaling is activated by small GTPase Ras. There are three isoforms of Ras expressed in T cells, namely, hRas, kRas and nRas (36). It has been well documented that these isoforms compartmentalize differently in T cell and thymocytes and carry distinctive functions (37–39). Thus, hnRNP-K might play a role in the activation process of a specific Ras isoform and a subset of ERK-signaling pathway, which is not involved in the transcriptional regulation of AP-1. Alternatively, cytoplasmic hnRNP-K may be involved in an ERK activation pathway that is involved in the process unrelated to AP-1 activation. Since hnRNP-K interacts with various molecules, detailed mechanism underlies this observation requires further studies.
In summary, upon TCR stimulation, hnRNP-K is modified by late ERK activation. This modification may enable hnRNP-K to protect Vav1 from proteolysis by caspases and potentiate the ability of Vav1 both in the cytoplasm and the nucleus to activate early- and late T-cell activation events. Though the data show that Vav is a critical target for hnRNP-K, existence of other factors is expected and possible because hnRNP-K has been shown to interact with various proteins, DNA and RNA, suggesting its versatile functions in cells.
Funding
US National Institute of Health (AI49398, AI47266) to M.I.
Acknowledgments
Authors thank Weiss, Crabtree, Koga, Fujisawa, Mivechi and Ronai for reagents. Markowitz for carefully editing the manuscript.
Glossary
Abbreviations
- AP-1
activator protein
- APC
antigen presenting cells
- ERK
extracellular signal-regulated kinase
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- Hsp
heat shock protein
- HA
hemagglutinin
- IEF
isoelectric focusing
- MAPK
mitogen-activated protein kinase
- MEK
ERK kinase
- MS
mass spectrometry
- NF-AT
nuclear factor of activated T cells
- NF-kappa B
nuclear factor kappa-light-chain enhancer of activated B cells
- PMA
phorbol 12-myristate 13-acetate
- PMA plus iono
phorbol 12-myristate 13-acetate plus ionomycin
- siRNA
small interfering RNA
- TFA
trifluoroacetic acid
References
- 1.Waldmann TA, Dubois S, Tagaya Y. Contrasting roles of IL-2 and IL-15 in the life and death of lymphocytes: implications for immunotherapy. Immunity. 2001;14:105. [PubMed] [Google Scholar]
- 2.Schorle H, Holtschke T, Hunig T, Schimpl A, Horak I. Development and function of T cells in mice rendered interleukin-2 deficient by gene targeting. Nature. 1991;352:621. doi: 10.1038/352621a0. [DOI] [PubMed] [Google Scholar]
- 3.Lanzavecchia A, Lezzi G, Viola A. From TCR engagement to T cell activation: a kinetic view of T cell behavior. Cell. 1999;96:1. doi: 10.1016/s0092-8674(00)80952-6. [DOI] [PubMed] [Google Scholar]
- 4.Rachmilewitz J, Lanzavecchia A. A temporal and spatial summation model for T-cell activation: signal integration and antigen decoding. Trends Immunol. 2002;23:592. doi: 10.1016/s1471-4906(02)02342-6. [DOI] [PubMed] [Google Scholar]
- 5.Weiss A, Shields R, Newton M, Manger B, Imboden J. Ligand-receptor interactions required for commitment to the activation of the interleukin 2 gene. J. Immunol. 1987;138:2169. [PubMed] [Google Scholar]
- 6.Huppa JB, Gleimer M, Sumen C, Davis MM. Continuous T cell receptor signaling required for synapse maintenance and full effector potential. Nat. Immunol. 2003;4:749. doi: 10.1038/ni951. [DOI] [PubMed] [Google Scholar]
- 7.Iwashima M. Kinetic perspectives of T cell antigen receptor signaling. Immunol. Rev. 2003;192 doi: 10.1034/j.1600-065x.2003.00024.x. [DOI] [PubMed] [Google Scholar]
- 8.Leonard WJ, Kronke M, Peffer NJ, Depper JM, Greene WC. Interleukin 2 receptor gene expression in normal human T lymphocytes. Proc. Natl Acad. Sci. USA. 1985;82:6281. doi: 10.1073/pnas.82.18.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bomsztyk K, Denisenko O, Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26:629. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 10.Northrop JP, Ullman KS, Crabtree GR. Characterization of the nuclear and cytoplasmic components of the lymphoid-specific nuclear factor of activated T cells (NF-AT) complex. J. Biol. Chem. 1993;268:2917. [PubMed] [Google Scholar]
- 11.Iwashima M, Takamatsu M, Yamagishi H, et al. Genetic evidence for Shc requirement in TCR-induced c-Rel nuclear translocation and IL-2 expression. Proc. Natl Acad. Sci. USA. 2002;99:4544. doi: 10.1073/pnas.082647499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl Acad. Sci. USA. 2002;99:5515. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koike T, Yamagishi H, Hatanaka Y, et al. A novel ERK-dependent signaling process that regulates interleukin-2 expression in a late phase of T cell activation. J. Biol. Chem. 2003;278:15685. doi: 10.1074/jbc.M210829200. [DOI] [PubMed] [Google Scholar]
- 14.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 15.Lai W, Chang CH, Farber DL. Gene transfection and expression in resting and activated murine CD4 T cell subsets. J. Immunol. Methods. 2003;282:93. doi: 10.1016/j.jim.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 16.Lewis TS, Hunt JB, Aveline LD, et al. Identification of novel MAP kinase pathway signaling targets by functional proteomics and mass spectrometry. Mol. Cell. 2000;6:1343. doi: 10.1016/s1097-2765(00)00132-5. [DOI] [PubMed] [Google Scholar]
- 17.Bunnell SC, Henry PA, Kolluri R, Kirchhausen T, Rickles RJ, Berg LJ. Identification of Itk/Tsk Src homology 3 domain ligands. J. Biol. Chem. 1996;271:25646. doi: 10.1074/jbc.271.41.25646. [DOI] [PubMed] [Google Scholar]
- 18.Hobert O, Jallal B, Schlessinger J, Ullrich A. Novel signaling pathway suggested by SH3 domain-mediated p95vav/heterogeneous ribonucleoprotein K interaction. J. Biol. Chem. 1994;269:20225. [PubMed] [Google Scholar]
- 19.Taylor SJ, Shalloway D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- 20.Weng Z, Thomas SM, Rickles RJ, et al. Identification of Src, Fyn, and Lyn SH3-binding proteins: implications for a function of SH3 domains. Mol. Cell. Biol. 1994;14:4509. doi: 10.1128/mcb.14.7.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schullery DS, Ostrowski J, Denisenko ON, et al. Regulated interaction of protein kinase Cdelta with the heterogeneous nuclear ribonucleoprotein K protein. J. Biol. Chem. 1999;274:15101. doi: 10.1074/jbc.274.21.15101. [DOI] [PubMed] [Google Scholar]
- 22.Siomi H, Matunis MJ, Michael WM, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostrowski J, Van Seuningen I, Seger R, et al. Purification, cloning, and expression of a murine phosphoprotein that binds the kappa B motif in vitro identifies it as the homolog of the human heterogeneous nuclear ribonucleoprotein K protein. Description of a novel DNA-dependent phosphorylation process. J. Biol. Chem. 1994;269:17626. [PubMed] [Google Scholar]
- 24.Ladomery M. Multifunctional proteins suggest connections between transcriptional and post-transcriptional processes. Bioessays. 1997;19:903. doi: 10.1002/bies.950191010. [DOI] [PubMed] [Google Scholar]
- 25.Lee MH, Mori S, Raychaudhuri P. trans-Activation by the hnRNP K protein involves an increase in RNA synthesis from the reporter genes. J. Biol. Chem. 1996;271:3420. doi: 10.1074/jbc.271.7.3420. [DOI] [PubMed] [Google Scholar]
- 26.Bustelo XR, Suen KL, Michael WM, Dreyfuss G, Barbacid M. Association of the vav proto-oncogene product with poly(rC)-specific RNA-binding proteins. Mol. Cell. Biol. 1995;15:1324. doi: 10.1128/mcb.15.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tybulewicz VL. Vav-family proteins in T-cell signalling. Curr. Opin. Immunol. 2005;17:267. doi: 10.1016/j.coi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Houlard M, Arudchandran R, Regnier-Ricard F, et al. Vav1 is a component of transcriptionally active complexes. J. Exp. Med. 2002;195:1115. doi: 10.1084/jem.20011701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofmann TG, Hehner SP, Droge W, Schmitz ML. Caspase-dependent cleavage and inactivation of the Vav1 proto-oncogene product during apoptosis prevents IL-2 transcription. Oncogene. 2000;19:1153. doi: 10.1038/sj.onc.1203406. [DOI] [PubMed] [Google Scholar]
- 30.Habelhah H, Shah K, Huang L, et al. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell Biol. 2001;3:325. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- 31.Romero F, Germani A, Puvion E, et al. Vav binding to heterogeneous nuclear ribonucleoprotein (hnRNP) C. Evidence for Vav-hnRNP interactions in an RNA-dependent manner. J. Biol. Chem. 1998;273:5923. doi: 10.1074/jbc.273.10.5923. [DOI] [PubMed] [Google Scholar]
- 32.Crespo P, Schuebel KE, Ostrom AA, Gutkind JS, Bustelo XR. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature. 1997;385:169. doi: 10.1038/385169a0. [DOI] [PubMed] [Google Scholar]
- 33.Costello PS, Walters AE, Mee PJ, et al. The Rho-family GTP exchange factor Vav is a critical transducer of T cell receptor signals to the calcium, ERK, and NF-kappaB pathways. Proc. Natl Acad. Sci. USA. 1999;96:3035. doi: 10.1073/pnas.96.6.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujikawa K, Miletic AV, Alt FW, et al. Vav1/2/3-null mice define an essential role for Vav family proteins in lymphocyte development and activation but a differential requirement in MAPK signaling in T and B cells. J. Exp. Med. 2003;198:1595. doi: 10.1084/jem.20030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds LF, de Bettignies C, Norton T, Beeser A, Chernoff J, Tybulewicz VL. Vav1 transduces T cell receptor signals to the activation of the Ras/ERK pathway via LAT, Sos, and RasGRP1. J. Biol. Chem. 2004;279:18239. doi: 10.1074/jbc.M400257200. [DOI] [PubMed] [Google Scholar]
- 36.Mor A, Philips MR. Compartmentalized Ras/MAPK signaling. Annu. Rev. Immunol. 2006;24:771. doi: 10.1146/annurev.immunol.24.021605.090723. [DOI] [PubMed] [Google Scholar]
- 37.Perez de Castro I, Bivona TG, Philips MR, Pellicer A. Ras activation in Jurkat T cells following low-grade stimulation of the T-cell receptor is specific to N-Ras and occurs only on the Golgi apparatus. Mol. Cell. Biol. 2004;24:3485. doi: 10.1128/MCB.24.8.3485-3496.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rebollo A, Perez-Sala D, Martinez AC. Bcl-2 differentially targets K-, N-, and H-Ras to mitochondria in IL-2 supplemented or deprived cells: implications in prevention of apoptosis. Oncogene. 1999;18:4930. doi: 10.1038/sj.onc.1202875. [DOI] [PubMed] [Google Scholar]
- 39.Daniels MA, Teixeiro E, Gill J, et al. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]