SUMMARY
Golgi membranes, from yeast to humans, are uniquely enriched in phosphatidylinositol-4-phosphate (PtdIns(4)P), although the role of this lipid remains poorly understood. Using a proteomic lipid binding screen, we identify the Golgi protein GOLPH3 (also called GPP34, GMx33, MIDAS, or yeast Vps74p) as a PtdIns(4)P-binding protein that depends upon PtdIns(4)P for its Golgi localization. We further show that GOLPH3 binds the unconventional myosin MYO18A, thus connecting the Golgi to F-actin. We demonstrate that this linkage is necessary for normal Golgi trafficking and morphology. The evidence suggests that GOLPH3 binds to PtdIns(4)P-rich trans-Golgi membranes and MYO18A conveying a tensile force required for efficient tubule and vesicle formation. Consequently, this tensile force stretches the Golgi into the extended ribbon observed by fluorescence microscopy and the familiar flattened form observed by electron microscopy.
INTRODUCTION
Nearly all secretory traffic and the luminal components of most organelles pass through the Golgi. In some secretory cells, the rate of trafficking through the Golgi replaces the entire surface area of the plasma membrane (PM) every 10–20 minutes (Kristen and Lockhausen, 1983; Barr and Warren, 1996). This remarkable rate of trafficking requires that the Golgi employ a highly efficient mechanism to generate vesicles to carry traffic to its destination. Many Golgi components have been shown to be required for trafficking, including small GTPases, coat proteins, and lipids (reviewed in De Matteis and Luini, 2008), but a coherent picture of the general process of tubulation and budding to form vesicles has yet to emerge.
A perplexing feature of the Golgi is its characteristic flattened morphology. The physical properties of lipids, their asymmetric distribution across the bilayer, curvature-inducing lipid binding proteins, and Golgi matrix proteins have all been proposed to contribute to the unique shape of the Golgi (Peter et al., 2004; Puthenveedu and Linstedt, 2005; Short et al., 2005). Although these may be factors in Golgi structure, their contribution, if any, to producing flat cisternae remains uncertain. Beyond the question of the mechanism that maintains Golgi morphology, there remains a larger question of the relationship between this unique morphology and Golgi secretory function.
PtdIns(4)P is required for Golgi function from yeast to humans. Genetic experiments in S. cerevisiae demonstrate a requirement for the Pik1p PI-4-kinase (Walch-Solimena and Novick, 1999; Audhya et al., 2000) and PtdIns(4)P (Hama et al., 1999) in Golgi secretion. In mammalian cells PI-4-kinase-IIIβ (Wong et al., 1997) and PI-4-kinase-IIα (Nakagawa et al., 1996) localize to the Golgi, and reporters for the subcellular localization of PtdIns(4)P (composed of a PtdIns(4)P binding domain fused to GFP, reviewed in Varnai and Balla, 2007) indicate that PtdIns(4)P is found predominantly at the Golgi (Godi et al., 2004). In HeLa cells, a dominant-negative PI-4-kinase-IIIβ interferes with reformation of the Golgi complex after washout of Brefeldin A (BFA) (Godi et al., 1999). HEK 293 cells overexpressing PI-4-kinase-IIIβ show enhanced trafficking from the Golgi to the PM (Hausser et al., 2005) and knockdown of PI-4-kinase-IIα impairs trafficking from the Golgi to the PM (Wang et al., 2003). Taken together, the evidence for a role for PtdIns(4)P in Golgi function is compelling, however the crucial targets of PtdIns(4)P important for Golgi function remain unclear.
A few direct PtdIns(4)P binding proteins are known, including OSBP (OxySterol Binding Protein), FAPP (phosphatidylinositol-Four-P AdaPtor Protein), CERT (CERamide Transporter), and their homologs. All contain PH domains that bind specifically to PtdIns(4)P, mediating the Golgi localization of these proteins (Dowler et al., 2000; Levine and Munro, 2002). Recent evidence demonstrates a role for these PtdIns(4)P binding proteins in non-vesicular trafficking of lipids. OSBP transports cholesterol (Im et al., 2005; Raychaudhuri and Prinz, 2006), FAPP2 transports glucosylceramide (Halter et al., 2007; D’Angelo et al., 2007), and CERT transports ceramide (Hanada et al., 2003). However, the FAPP proteins and CERT are not conserved in S. cerevisiae, and simultaneous mutation of all PH domain-containing OSBPs in yeast has little effect, certainly not copying lethality from deletion of the PIK1 PI-4-kinase (Beh et al., 2001).
We hypothesized that there may exist undiscovered PtdIns(4)P binding proteins mediating the effects of PtdIns(4)P at the Golgi and that these may provide new insight into the biology of the Golgi. Here we use proteomic screening to identify GOLPH3 as a PtdIns(4)P binding protein. GOLPH3 is an abundant protein conserved from yeast to humans but contains no previously known phosphoinositide binding domains. We show that GOLPH3 and the yeast homolog Vps74p localize to the Golgi by binding PtdIns(4)P. We further demonstrate that GOLPH3 interacts with the unconventional myosin, MYO18A, linking Golgi membranes to the actin cytoskeleton. Our data indicate that this interaction provides a tensile force that is required for normal Golgi vesicle trafficking and architecture, demonstrating an unexpected role for PtdIns(4)P at the Golgi.
RESULTS
An in vitro Lipid Binding Screen Identifies GOLPH3 as a PtdIns(4)P Binding Protein
To identify phosphoinositide binding proteins we devised a high throughput proteomic screen based on the lipid blot assay of Dowler et al., 2000. This assay involves spotting phosphoinositides on a membrane, blotting with a protein of interest, washing, and detecting bound protein. We optimized the assay for use with proteins produced by in vitro transcription and translation (IVT) with 35S-methionine to allow detection.
Critical to the reliability of the assay is the quality of the phosphoinositides. We screened individual lots of lipids from commercial suppliers by thin layer chromatography (TLC). We also validated each binding assay with a panel of positive control lipid binding proteins.
We screened the D. melanogaster proteome, which is compact but contains examples of the known phosphoinositide-modifying enzymes and phosphoinositide-dependent signaling pathways found in higher organisms. The Drosophila Gene Collection provides an arrayed set of 15,466 cDNAs of known sequence cloned behind T7 promoters allowing IVT (Stapleton et al., 2002). To date, we have screened ~4000 unique cDNAs from this collection scoring positive hits for many previously identified PH, PX, FYVE, Tub, and Proppin family proteins, validating the method (Figure 1A). The screen has also identified unique phosphoinositide binding proteins. Here we describe one of these, a PtdIns(4)P binding protein (clone ID LD23816, FBgn0010704) which lacks homology to known phosphoinositide binding proteins. The mammalian homolog has been named GOLPH3 (Genbank), GMx33 (Wu et al., 2000), GPP34 (Bell et al., 2001), or MIDAS (Nakashima-Kamimura et al., 2005), and the yeast homolog is Vps74p (Bonangelino et al., 2002).
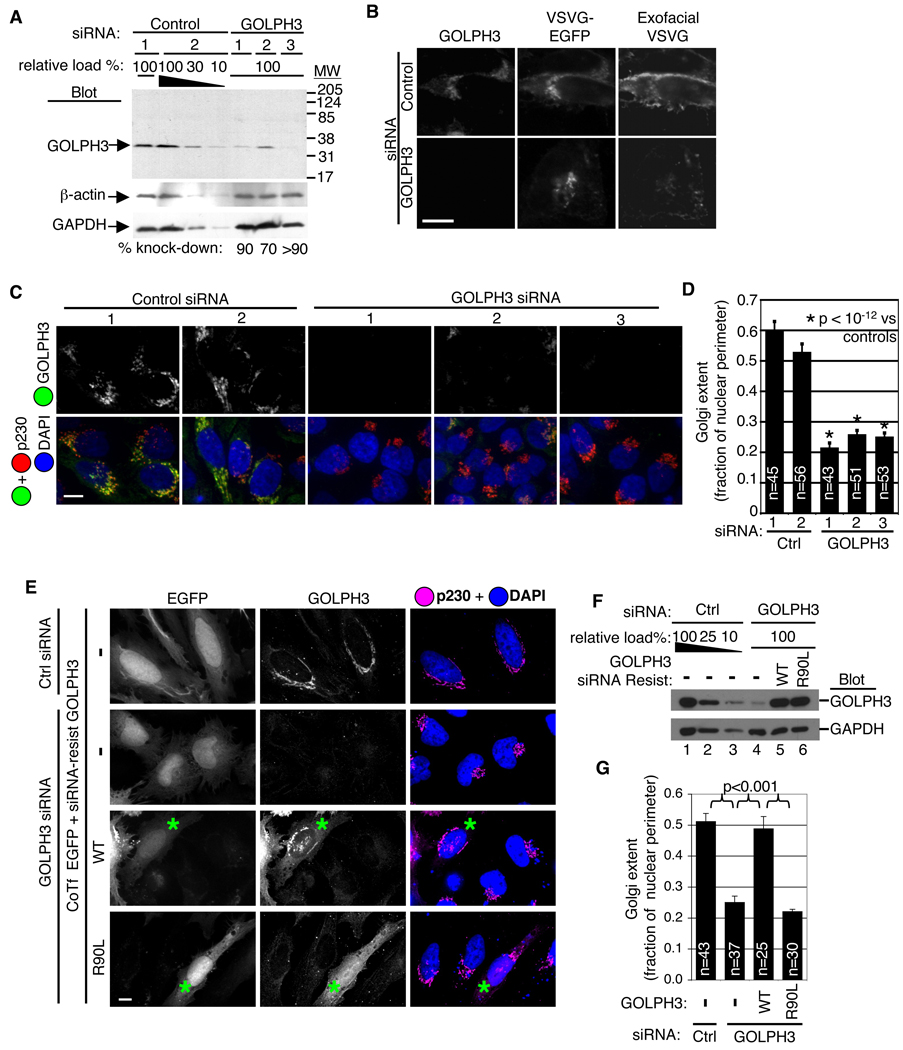
Figure 1. Proteomic screening identifies GOLPH3 as a PtdIns(4)P binding protein that requires PtdIns(4)P for Golgi localization.
(A) Screening of Drosophila Gene Collection for lipid binding. Example hits correspond to previously well-validated lipid binding proteins with known binding domains: AKT (PtdIns(3,4)P2 and PtdIns(3,4,5)P3, clone SD10374); CERT (PtdIns(4)P, clone GH07688); TAPP1 (PtdIns(3,4)P2, clone SD10969); SNX29 (PtdIns(3)P and PA, clone LD35592); SARA (PtdIns(3)P, clone LD33044); Tubby (PtdIns(4,5)P2, clone GH04653); ATG18 (PtdIns(3,5)P2, PtdIns(3)P, clone LD32381). Screen also identified unknown lipid binding proteins. Shown is PtdIns(4)P binding of GOLPH3 (clone LD23816). Yeast (Vps74p) and human orthologs also bind PtdIns(4)P. (B) Depletion of PtdIns(4)P results in dissociation of GOLPH3 from the Golgi. HeLa cells expressing EGFP-tagged phosphatase Sac1-K2A (constitutively at Golgi, Rohde et al., 2003) or EGFP (control) stained for endogenous GOLPH3 (red), TGN marker p230 (yellow), and DAPI (blue). In controls and untransfected cells (*), GOLPH3 and p230 co-localize at the Golgi, but EGFP-Sac1-K2A renders GOLPH3 staining diffuse and distinct from p230. (C) Overexpression of EYFP-FAPP1-PH displaces endogenous GOLPH3 from the Golgi. White arrowheads indicate cells expressing EYPF-FAPP1-PH (green) in which anti-GOLPH3 staining (red) is diffuse vs adjacent untransfected cells. (D) S. cerevisiae expressing temperature sensitive allele pik1-83ts, lose PtdIns(4)P upon shift to 37°C. In wild type, GFP-Vps74p remains colocalized with late Golgi marker Sec7p-dsRed at both temperatures. In pik1-83ts, GFP-Vps74p dissociates from the Golgi upon shift to 37°C (also see Movie S2). (Figure 1–Figure 5 and Figure 7, scale bars=10 µm)
GOLPH3 is conserved from yeast to humans. It was originally identified in a proteomic characterization of rat Golgi and later suggested to be a Golgi matrix protein (Wu et al., 2000; Bell et al., 2001; Snyder et al., 2006), although its mechanism of interaction with the Golgi and function were not determined. Deletion of VPS74 in S. cerevisiae results in defective trafficking of vacuolar proteases (Bonangelino et al., 2002). More recently Vps74p was shown to be Golgi-localized and necessary for localization of glycosyltransferases, glycosylation, and secretory function of the Golgi in yeast (Schmitz et al., 2008; Tu et al., 2008).
To validate our screen, we sought to confirm that GOLPH3 binds PtdIns(4)P and found that not only the D. melanogaster, but also the H. sapiens and S. cerevisiae orthologs expressed in rabbit reticulocyte lysates all bind tightly and specifically to PtdIns(4)P in our lipid blot assay (Figure 1A). The binding to PtdIns(4)P is direct, as shown by using E. coli expressed and purified GOLPH3 (Figure S1). Binding of GOLPH3 to small unilamellar vesicles further demonstrates specific binding to PtdIns(4)P (Figure 2D).
Figure 2. Identification of PtdIns(4)P binding domain and pocket required for Golgi localization of GOLPH3.
(A, B, C) Truncations of Drosophila GOLPH3 show PtdIns(4)P binding in vitro co-maps with Golgi localization. (A) Segments indicated expressed in vitro for lipid blot or (C) as EGFP-fusions in HEK 293 cells for fluorescence microscopy. (B) Expression confirmed by Western blot for EGFP. (D) Lipid vesicle binding shows specificity of wild type GOLPH3 for PtdIns(4)P. R90L and R171A/R174L mutants do not bind. p40phox-PX control binds PtdIns(3)P. (V=vesicle, S=soluble) (E) In HeLa, R90L and R171A/R174L mutations of GOLPH3 (magenta) mislocalize to cytosol, but coexpressed wild type EGFP-GOLPH3 (green) remains at the Golgi as marked by TGN46 (red). DAPI in blue. (F) Western blot showing coexpression of 3xHA-tagged GOLPH3 mutants and EGFP-GOLPH3.
PtdIns(4)P is Required for GOLPH3 Localization to the Golgi
If GOLPH3 binding to PtdIns(4)P is functionally important in vivo, then it should localize to the Golgi, as other groups have previously reported (Wu et al., 2000; Bell et al., 2001; Snyder et al., 2006; Schmitz et al., 2008). Specifically, GOLPH3 was shown by immunofluorescence microscopy (IF) to colocalize with trans Golgi markers and found by immuno-electron microscopy at the rim of the trans Golgi (Wu et al., 2000) and enriched in budding vesicles and tubules (Snyder et al., 2006). With our antibodies to GOLPH3 we too see that endogenous GOLPH3 colocalizes with the endogenous trans Golgi network (TGN) markers TGN46 and p230 (Figure S2A), but somewhat less with the cis Golgi marker GM130, in several mammalian cell lines (data not shown). In yeast, EGFP-Vps74p colocalizes with the late Golgi marker Sec7p-dsRed (Figure S2B).
If GOLPH3 binds PtdIns(4)P in vivo, then the subcellular localization of the two should coincide. By coexpressing the PtdIns(4)P reporter EYFP-FAPP1-PH (Godi et al., 2004) with mCherry-GOLPH3 in HeLa cells, time-lapse confocal fluorescence microscopy shows that the two proteins fully colocalize over space and time (Movie S1). This colocalization is tighter than any other pair of Golgi markers we examined (e.g., Figure S7), consistent with GOLPH3 binding to PtdIns(4)P in vivo. Both EYFP-FAPP1-PH and mCherry-GOLPH3 were enriched on tubules and vesicles leaving the Golgi, similar to previous results localizing PtdIns(4)P (Godi et al., 2004) and GOLPH3 (Snyder et al., 2006).
If GOLPH3 binds PtdIns(4)P in vivo, then reduction of PtdIns(4)P at the Golgi should result in GOLPH3 dissociation from the Golgi. We depleted PtdIns(4)P by expressing a mutant of the lipid phosphatase Sac1 (Sac1-K2A) that constitutively localizes to the Golgi, dephosphorylating PtdIns(4)P without affecting other phosphoinositide pools (Rohde et al., 2003 and Figure S3). In HeLa cells, expression of EGFP-Sac1-K2A resulted in loss of GOLPH3 from the Golgi (Figure 1B), although the Golgi remained intact and other markers remained at the Golgi including p230, TGN46, β-1,4-galactosyltransferase-EYFP, and α-mannosidase II-EGFP (Figure 1B and data not shown).
In S. cerevisiae the PI-4-kinase Pik1p produces PtdIns(4)P at the Golgi. Using a temperature sensitive mutant allele of PIK1, pik1-83ts (Audhya et al., 2000), we show that EGFP-Vps74p dissociates from the Golgi within five minutes after shift to the non-permissive temperature (Figure 1D and Movie S2).
Finally, highly overexpressed EYFP-FAPP1-PH displaces endogenous GOLPH3 from the Golgi (Figure 1C). We conclude that PtdIns(4)P must be present and available for interaction for GOLPH3 to localize to the Golgi.
GOLPH3 binds PtdIns(4)P via a positively charged binding pocket on the hydrophobic face of the protein
Because GOLPH3 contains no domains found in previously characterized phosphoinositide binding proteins, we mapped the domain required for PtdIns(4)P binding. A series of truncations of Drosophila GOLPH3 tested for binding to PtdIns(4)P by lipid blot assay showed that binding to PtdIns(4)P requires amino acids 30–293 (Figure 2A). This corresponds to the most evolutionarily conserved region, and has been termed the GPP34 domain by PFAM. This series of truncations, fused to EGFP and expressed in HEK 293 cells (Figure 2B), shows that the GPP34 domain is also required for localization to the Golgi (Figure 2C).
Using the crystal structure of yeast Vps74p (Schmitz et al., 2008), we identified a positively charged pocket on the hydrophobic face of the GPP34 domain (Figure S4). To test the role of this pocket we made mutations in GOLPH3 designed to neutralize its charge yet retain proper folding. Two independent mutations of the pocket (R90L and R171A/R174L) each impaired binding to PtdIns(4)P in the lipid blot assay (data not shown) and the vesicle binding assay (Figure 2D). IF of 3xHA-tagged wild type and mutant GOLPH3 expressed in HeLa cells shows binding pocket mutants lose Golgi localization (Figure 2E). This further demonstrates the requirement for PtdIns(4)P binding for Golgi localization.
Our data show that GOLPH3 binds tightly, specifically, and directly to PtdIns(4)P in vitro in two assays, binding is evolutionarily conserved from yeast to humans, depletion or blocking of PtdIns(4)P place GOLPH3 downstream of PtdIns(4)P in vivo, and mutational analysis reveals that GOLPH3 requires the ability to bind PtdIns(4)P to localize to the Golgi. Taken together we conclude that GOLPH3 and Vps74p localize to the Golgi by binding PtdIns(4)P.
GOLPH3 is an Abundant Protein
To determine a role for GOLPH3 in Golgi function, we began by quantifying its abundance using quantitative Western blotting of HeLa whole cell lysates. We compared signal intensity from lysates of known numbers of HeLa cells to known quantities of purified bacterial expressed GOLPH3. We estimate that each HeLa cell expresses 25+/−5 fg (mean+/−SEM, n=4) or 500,000+/−100,000 molecules of GOLPH3, making it a highly abundant protein (Figure S5). This abundance suggests that GOLPH3 is a major target of PtdIns(4)P, likely to be of general importance in Golgi function.
GOLPH3 is Required for Trafficking from Golgi to PM
To determine the requirement for GOLPH3 in Golgi function, we examined the effect of reducing its levels by siRNA knockdown. By Western blot, three different GOLPH3-specific siRNA oligos reduced GOLPH3 levels by at least 70–90% in HeLa or HEK 293 cells (Figure 3A and data not shown).
Figure 3. GOLPH3 is required for trafficking and extended ribbon morphology of the Golgi.
(A) Western blot of HeLa lysates shows GOLPH3 knocked down 70–90% by each specific siRNA. GOLPH3 antiserum recognizes a single band at 34 kDa in control lysates. Decreasing amounts of control lysate loaded to allow quantification. Blots for β-actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) verify equal loading. (B) Trafficking of ts045-VSVG-EGFP, through Golgi to PM impaired by knockdown of GOLPH3. HeLa cells serially transfected with GOLPH3 or control siRNA, then ts045-VSVG-EGFP expression vector, incubated at 40°C overnight, then shifted to 32°C for 2 hours. Arrival at PM determined with antibody to extracellular domain applied to unpermeabilized cells (exofacial VSVG). (C) Knockdown of GOLPH3 causes condensation of Golgi ribbon. Control siRNA transfected cells show normal extended Golgi ribbon detected by GOLPH3 (green) and p230 (red). DAPI in blue. GOLPH3 siRNA reduces GOLPH3 and causes condensation of Golgi (p230). Imaging parameters identical in all. (D) Quantification of Golgi extent (Golgi length relative to nuclear perimeter, mean/SEM graphed) demonstrates significant change (p<10−12, t-test). (E) Expression of siRNA-resistant GOLPH3 (expressed near endogenous level, cotransfected with EGFP) rescues Golgi ribbon, validating GOLPH3 siRNA specificity. Expression of siRNA-resistant GOLPH3 PtdIns(4)P binding pocket mutant does not rescue Golgi ribbon phenotype. Asterisks indicate transfected cells. (F) Western blot shows knockdown of GOLPH3 (lane 4) and restoration by siRNA-resistant wild type (lane 5) and R90L (lane6) GOLPH3. Blot for GAPDH shows equal loading. (G) Quantification of Golgi extent (mean/SEM) demonstrates significant rescue by wild type, but not R90L GOLPH3 (p<0.001 for indicated comparisons, t-test).
Using the anterograde cargo ts045-VSVG-EGFP (Hirschberg et al., 2000), we examined trafficking in cells depleted of GOLPH3. Transport of ts045-VSVG-EGFP from ER to Golgi was unaltered by knockdown of GOLPH3 (data not shown). IF of unpermeabilized cells using an antibody specific to the extracellular domain of VSVG, allowing unambiguous detection at the PM (Bossard et al., 2007), showed that knockdown of GOLPH3 impaired trafficking from the Golgi to PM (Figure 3B). Delivery to the PM was quantified by summing exofacial fluorescence over the volume of each cell and normalizing to total ts045-VSVG-EGFP fluorescence. We observed a highly significant (p<10−7, t-test) defect in delivery of ts045-VSVG-EGFP to the PM in GOLPH3 knockdown cells, [control=6.5+/−0.3 (n=13), GOLPH3 knockdown=3.5+/−0.2 (n=12), arbitrary normalized fluorescence units, mean+/−SEM]. Thus, GOLPH3 is required for anterograde trafficking from the Golgi to the PM.
GOLPH3 is Required for the Normal Extended Golgi Ribbon
We next examined Golgi morphology upon GOLPH3 knockdown. Depletion of GOLPH3 altered the Golgi ribbon, changing its normal appearance of extending partially around the nucleus, to condensing at one end of the nucleus (Figure 3C). Quantification of Golgi extent, measured as the fraction of nuclear perimeter encompassed by the Golgi, demonstrated a highly significant condensation of the Golgi upon knockdown of GOLPH3 by each of three specific siRNA oligos (p<10−12 for each versus control, t-test, Figure 3D). We examined several peripheral membrane and transmembrane Golgi markers, and all revealed condensation of cis, medial, and trans Golgi as well as the TGN upon knockdown of GOLPH3 in HeLa, HEK 293, and NIH 3T3 cells (Figures S6, S7, S8, S9, S10, and S11). We conclude that the entire Golgi condenses upon knockdown of GOLPH3.
Recently published data indicate that Vps74p is responsible for the stable cis and medial Golgi localization of glycosyltransferases involved in yeast cell wall biosynthesis (Schmitz et al., 2007; Tu et al., 2007). We examined the localization of three mammalian glycosyltransferases in GOLPH3 knockdown cells and found that their localization indicated condensation of the Golgi, but they still colocalized with other Golgi markers, indicating that GOLPH3 is not necessary for localization of these glycosyltransferases to the Golgi in mammalian cells (Figures S8, S9, S10, and S11).
Rescue of GOLPH3 Knockdown Requires the Ability to Bind PtdIns(4)P
To further confirm specificity of the siRNA knockdown phenotype, we made silent mutations in GOLPH3 allowing expression resistant to our strongest siRNA oligo. Expression of siRNA-resistant wild type GOLPH3 completely rescued the normal extended Golgi ribbon morphology (Figures 3E, 3F, and 3G). We also produced an siRNA-resistant construct with the R90L PtdIns(4)P binding mutation. This mutant, as expected did not localize to the Golgi, and was incapable of rescuing Golgi morphology. These results validate that the effects we see on the Golgi upon knockdown of GOLPH3 are specific to the function of GOLPH3 and further demonstrate the necessity of PtdIns(4)P binding in GOLPH3 function.
GOLPH3 Links the Golgi to the Actin Cytoskeleton
The condensation of the Golgi observed upon knockdown of GOLPH3 raised the suggestion that GOLPH3 may serve to mediate the application of a tensile force to the Golgi, pulling the ribbon around the nucleus. We figured a cytoskeletal motor would likely be needed to apply such a force. We considered the dyneins and kinesins acting on microtubules or the myosins acting on F-actin. The effect of using nocodazole to depolymerize microtubules results in dispersal of the Golgi (Rogalski and Singer, 1984), quite different from the effect of GOLPH3 knockdown.
The role of actin at the Golgi is poorly understood. To examine the effect of actin depolymerization on Golgi morphology, HEK 293 cells expressing a Golgi marker were treated with Latrunculin B (LatB) while performing live cell imaging. Treatment with LatB led to rapid condensation of the Golgi (Movie S3), recapitulating a published report (Lázaro-Diéguez et al., 2006) and the effect of knockdown of GOLPH3.
We compared the effects on the Golgi of LatB treatment or GOLPH3 knockdown. Knockdown of GOLPH3 results in compaction of the Golgi without altering F-actin (Figure 4A). Depolymerization of actin by LatB leads to compaction of the Golgi without affecting the Golgi association of GOLPH3. Simultaneous knockdown of GOLPH3 and treatment with LatB produced a similar compaction of the Golgi, but without an additive effect (Figures 4A and 4B). These results raised the possibility that GOLPH3 may in some way link the Golgi to the actin cytoskeleton, exerting a tensile force on the Golgi.
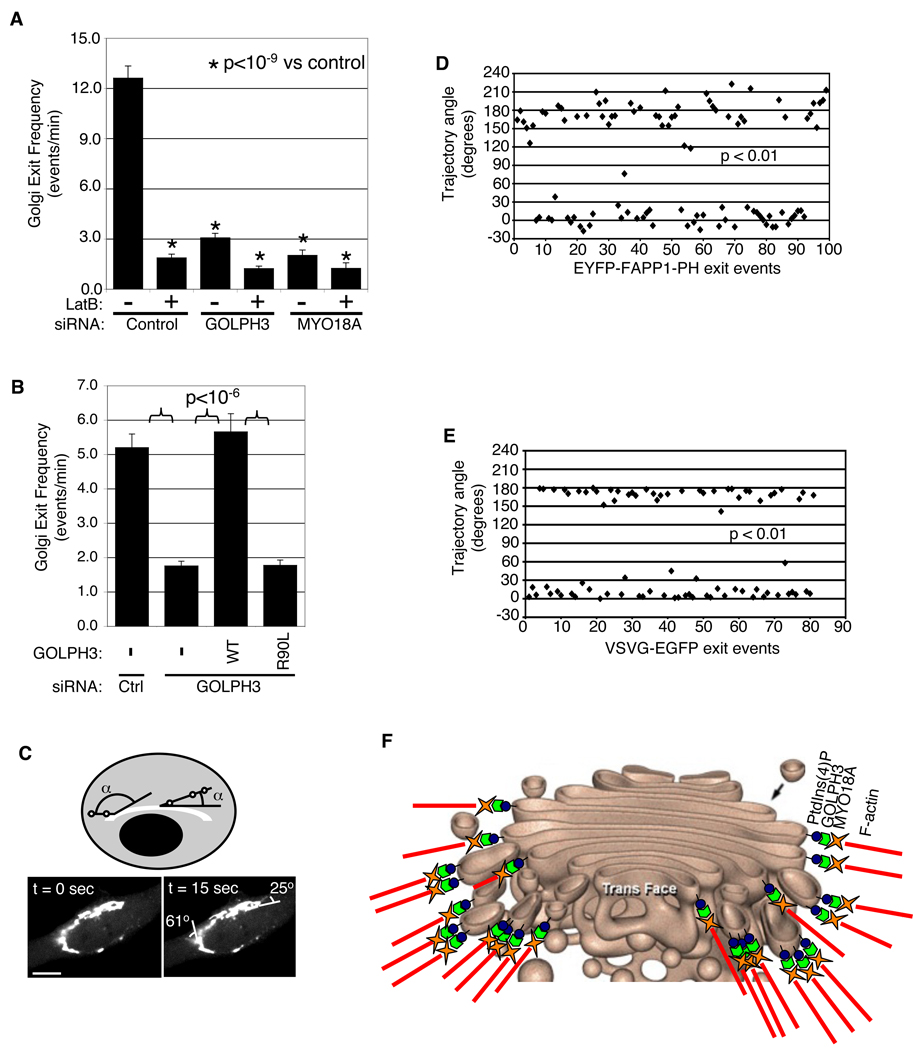
Figure 4. GOLPH3 links the Golgi to F-actin via MYO18A.
(A) HeLa cells treated with control siRNA and DMSO show normal extended Golgi morphology (p230, green and GOLPH3, yellow) and normal F-actin (Texas red phalloidin, magenta). Control siRNA + LatB causes loss of F-actin (stress fibers and peripheral actin) and Golgi condensation, but GOLPH3 remains at Golgi. GOLPH3 siRNA + DMSO causes loss of GOLPH3, Golgi condensation, but F-actin remains normal. Combined GOLPH3 siRNA + LatB causes loss of F-actin and GOLPH3, with Golgi condensation. (B) Measurement of Golgi extent (from A) indicates no additional effects on Golgi seen in combining GOLPH3 siRNA and LatB. (C) Western blot shows MYO18A coIPs specifically with GOLPH3 even when actin depolymerized by LatB (Figure S13). Golgi-localized myosins, MYO2B and MYO6, and MRCKβ did not coIP with GOLPH3. (D) Proposed model: GOLPH3 binds PtdIns(4)P and MYO18A, linking Golgi to cytoskeleton. (E) Purified, bacterial expressed GOLPH3 (but not GST) binds His-SUMO-tagged N-terminal and Middle fragments of MYO18A but not C-terminal fragment or His-SUMO alone bound to Ni beads. (F) MYO18A (green) colocalizes with GOLPH3 (red) and p230 (magenta) in HeLa cells. DAPI in blue. Knockdown of MYO18A shows specificity of MYO18A antibody for IF. Knockdown of GOLPH3 causes loss of MYO18A from Golgi.
GOLPH3 Binds MYO18A
To determine how GOLPH3 might link the Golgi to actin and affect Golgi structure, we took an open-ended approach performing large-scale immunoprecipitation (IP) of GOLPH3 from HeLa whole-cell lysates. Analysis by mass spectrometry of prominent bands near 250 kDa identified the unconventional myosin, MYO18A as a major GOLPH3 interacting protein (Figure S12).
MYO18A (type XVIIIA, also known as MysPDZ) has been observed to localize to the Golgi (Furusawa et al., 2000; Mori et al., 2003; Mori et al., 2005) and to bind actin filaments (Isogawa et al., 2005). Recently, MYO18A was shown to form a tripartite complex with MRCK and LRAP35a regulating actomyosin retrograde flow in cell protrusion and migration (Tan et al., 2008).
The interaction between GOLPH3 and a Golgi-localized myosin suggested a model to explain a role for GOLPH3 in linking the Golgi to the actin cytoskeleton (Figure 4D). Our model builds upon GOLPH3 binding to PtdIns(4)P at the Golgi and suggests that GOLPH3 further interacts with MYO18A, bringing it to the Golgi and thus linking to the actin cytoskeleton.
To confirm interaction between GOLPH3 and MYO18A, we IP’d GOLPH3 from whole cell lysates and Western blotted with a MYO18A-specific antibody. MYO18A coIP’d specifically with GOLPH3 (Figure 4C). The interaction is not mediated by actin, as it was not disrupted by LatB (Figure 4C and Figure S13). The Golgi-localized myosins MYO2B or MYO6 did not coIP with GOLPH3 (Figure 4C), indicating that GOLPH3 interacts uniquely with MYO18A. The GOLPH3/MYO18A complex is likely distinct from the MYO18A/LRAP35a/MRCK complex (Tan et al., 2008), as we did not detect MRCKβ in GOLPH3 IP’s and only a fraction of MYO18A coIP’d with GOLPH3 (data not shown).
We next tested if GOLPH3 and MYO18A interact directly. We tagged N-terminal, Middle, and C-terminal segments of MYO18A with 6xHis-SUMO, expressed them in bacteria, and purified on Ni beads. Incubation of each MYO18A fragment with purified bacterial expressed GOLPH3 and GST (negative control) showed that GOLPH3 was specifically pulled down by the N-terminal and Middle fragments of MYO18A, but not by the C-terminal fragment or 6xHis-SUMO alone (Figure 4E). This pull-down of GOLPH3 with two fragments of MYO18A indicates a bipartite interaction, suggesting the interaction with intact MYO18A may be particularly strong.
We next examined subcellular localization of MYO18A by IF. Endogenous MYO18A was observed at the Golgi, colocalizing with endogenous GOLPH3 and p230 in cells treated with control siRNA (Figure 4F). Localization of GOLPH3 at the Golgi is independent of MYO18A, as shown by knockdown of MYO18A. However, MYO18A no longer localized to the Golgi upon knockdown of GOLPH3, showing that MYO18A depends on GOLPH3 for Golgi localization, providing genetic confirmation of the biochemical interaction.
Depletion of PtdIns(4)P Phenocopies Knockdown of GOLPH3
Our model predicts that depletion of PtdIns(4)P should also result in a condensed Golgi ribbon. In Figure 1B, depletion of PtdIns(4)P by expression of EGFP-Sac1-K2A caused dissociation of GOLPH3 from the Golgi. p230 IF revealed that depletion of PtdIns(4)P also caused the Golgi ribbon to condense into a compact ball (Figure 1B and data not shown). Quantification of Golgi extent around the nuclear perimeter confirmed that the Golgi compaction was highly significant (p<10−8, t-test, Figure S14). Similar results were observed with the Golgi reporter α-mannosidase II-EGFP (data not shown) and indicate that depletion of PtdIns(4)P phenocopies knockdown of GOLPH3 and both are essential for an extended Golgi ribbon.
Knockdown of MYO18A Phenocopies Knockdown of GOLPH3
To determine if knockdown of MYO18A causes the same condensed Golgi phenotype, we used siRNA to knock down MYO18A expression in HeLa cells (Figure 5A). IF to endogenous GOLPH3 and p230 showed that each of three MYO18A-specific siRNAs produced the condensed Golgi phenotype (Figure 5B). Quantification of Golgi extent confirmed that the Golgi compaction in MYO18A knockdown cells was highly significant and similar to GOLPH3 knockdown (p<10−10, t-test, Figure 5C). Similar results were observed in HEK 293 cells (data not shown).
Figure 5. MYO18A knockdown phenocopies GOLPH3 compact Golgi phenotype.
(A) Western blot of HeLa lysates shows MYO18A knocked down >90% by each specific siRNA. GAPDH blot verifies equal loading. (B) Knockdown of MYO18A with each specific siRNA results in condensed Golgi shown by IF to GOLPH3 (green) and p230 (red). DAPI in blue. Similar results seen in HeLa (shown) and HEK 293 cells. (C) Quantification of Golgi extent (length relative to nuclear perimeter, mean/SEM graphed) demonstrates significant compaction of Golgi (p<10−10, t-test). (D) GFP-tagged wild type mouse MYO18A, but not ATP-binding mutant nor EGFP alone, rescues Golgi phenotype of MYO18A knockdown in HeLa cells, shown by IF to GOLPH3 and p230 (magenta). (E) Quantification of Golgi extent (mean/SEM graphed) demonstrates significant rescue of Golgi morphology by wild type MYO18A, but not ATP-binding mutant (p<0.02 for indicated comparisons, t-test).
Rescue of MYO18A Knockdown Requires an Intact MYO18A ATPase Pocket
To confirm the specificity of the MYO18A siRNA phenotype, we expressed GFP-tagged wild type mouse MYO18A, predicted to be resistant to our human siRNA oligos, to rescue the compact Golgi phenotype. We found that overexpressed mouse MYO18A-EGFP was diffuse in the cell, but fully rescued Golgi morphology resulting from knockdown of human MYO18A (Figures 5D and 5E).
Mutation of the MYO18A ATPase pocket was recently demonstrated to result in a dominant negative effect on actomyosin retrograde flow (Tan et al., 2008). We used our ability to rescue human MYO18A knockdown with mouse MYO18A-EGFP to test the function of a mutation in conserved residues in the ATPase pocket. The G520S/K521A ATPase mutant failed to rescue the condensed Golgi phenotype (Figures 5D and 5E). MYO18A’s ability to generate force on actin has not been formally demonstrated, however since MYO18A is homologous to other myosins, especially in important functional regions, and fails to function when the ATPase pocket is mutated (Figures 5D and 5E and Tan et al., 2008), we favor that MYO18A functions, like other myosins, to generate force on F-actin.
trans Golgi Cisternae are Dilated in Cells Depleted of GOLPH3 or MYO18A
Our data demonstrate that GOLPH3 functions to bind specifically to Golgi membranes by binding to PtdIns(4)P and also to MYO18A, which we propose generates a pulling force, one consequence of which is to stretch the Golgi ribbon around the nucleus. We considered that interfering with this apparatus may also produce ultrastructural changes. Depolymerization of actin has been shown to cause rounding of the normally flat Golgi cisternae (Lázaro-Diéguez et al., 2006). We examined Golgi ultrastructure in control, GOLPH3, or MYO18A knockdown cells and found that knockdown of GOLPH3 or MYO18A produced dilated Golgi cisternae, frequently dramatically so (Figure 6A). Cells appeared similar to those observed upon depolymerization of actin (Lázaro-Diéguez et al., 2006). The dilation persists despite cycloheximide treatment, suggesting it is not due to filling of cisternae with protein (data not shown). We also observed that cisternal dilation was asymmetrically localized to one side of the Golgi (Figure 6A). Since PtdIns(4)P (Godi et al., 2004) and GOLPH3 (Wu et al., 2000) localize to the trans Golgi, we infer that these dilated cisternae represent the trans Golgi. The difference in cisternal thickness at the trans Golgi was highly statistically significant (p<10−4, unpaired t-test, Figure 6B), so we conclude that GOLPH3, MYO18A, and F-actin are required to maintain the familiar flattened appearance of the trans Golgi.
Figure 6. Knockdown of GOLPH3 or MYO18A produces dilated Golgi cisternae.
(A) Electron micrographs of negative control, GOLPH3, and MYO18A siRNA-treated HeLa cells (two different oligos for each). Control cells show flat Golgi cisternae arranged in neat stacks. Knockdown of GOLPH3 or MYO18A (verified by Western blot of parallel samples) results in dilated, frequently completely aberrant, Golgi cisternae, especially on trans face (see text). (scale bar=200 nm) (B) Cisternae in each stack divided into the cis-medial and medial-trans halves and thickness of each cisterna measured. Differences between control and knockdown highly significant for medial-trans (p<10−4, t-test, mean/SEM graphed).
PtdIns(4)P/GOLPH3/MYO18A/F-actin are Required for Efficient Vesicle Budding
Our data argue that PtdIns(4)P/GOLPH3/MYO18A/F-actin form a molecular apparatus to pull on trans Golgi membranes. We considered the purpose of this apparatus, and noted that it may contribute to the efficient extraction of tubules or vesicles from the Golgi, and thus may be required for normal Golgi vesicle trafficking. If true, we predicted that interfering with any component of the apparatus depicted in Figure 4D would impair trafficking. Indeed, knockdown of GOLPH3 significantly impaired trafficking of ts045-VSVG-EGFP from the Golgi to the PM (Figure 3B).
We next examined the effect of actin depolymerization, GOLPH3 knockdown, or MYO18A knockdown on the rate of tubule or vesicle exit from the Golgi by measuring the effect on vesicles bearing PtdIns(4)P. We performed fluorescence microscopy of cells expressing low levels of EYFP-FAPP1-PH. Live time-lapse images were obtained and tubules or vesicles emanating from the Golgi identified (Movie S4). The number of vesicles or tubules leaving the Golgi was counted from the same cells before and after treatment with LatB. Depolymerization of actin caused a dramatic reduction in the frequency of Golgi exit events (Figure 7A), consistent with previously reported results (Lázaro-Diéguez et al., 2007). Furthermore, knockdown of GOLPH3 or MYO18A each caused a similar reduction in the frequency of tubule and vesicle exit from the Golgi. We further validated these results by measuring the rate of exit of tubules and vesicles marked with the cargo ts045-VSVG-EGFP (Figure S15). Thus, GOLPH3, MYO18A, and F-actin are required for efficient exit from the Golgi.
Figure 7. GOLPH3/MYO18A/F-actin are necessary for Golgi vesiculation.
(A) Knockdown of GOLPH3, MYO18A, or depolymerization of actin impair Golgi vesicle exit. HeLa cells expressing low levels of EYFP-FAPP1-PH to mark PtdIns(4)P-rich membranes imaged live before and after LatB treatment. Vesicles or tubules exiting Golgi counted (10–15 cells per siRNA, counting 100–1500 exit events per siRNA, pooling two experiments). Differences from control highly significant (p<10−9, t-test). (B) siRNA-resistant wild-type GOLPH3 rescues Golgi trafficking defect, but not the R90L mutant defective in binding PtdIns(4)P. Differences with p<10−6 by t-test (20–30 cells each, counting 900–2400 exit events, pooling two experiments). (C) Measurement of exit angle of tubules/vesicles from Golgi. Live imaging of EYFP-FAPP1-PH shows two PtdIns(4)P-rich tubules leaving Golgi (see also Movie S4) and measurement of initial trajectory angles relative to direction of Golgi ribbon. (D) Initial trajectories of PtdIns(4)P-rich tubules or vesicles plotted for 99 exit events collected from independent imaging of 8 HeLa cells. Clustering around 0° and 180° highly significant (p<0.01, Kolmogorov-Smirnov test). (D) Initial trajectories of vesicles and tubules carrying ts045-VSVG-GFP cluster near 0° and 180°, parallel to Golgi ribbon (p<0.01, Kolmogorov-Smirnov test). (E) Diagram of Golgi (used with permission (Davidson, 2004)) superimposed with Model: GOLPH3 binds PtdIns(4)P and links to MYO18A, transducing a tensile force necessary for Golgi vesiculation, contributing to its flattened form (see also Figure 4D).
We tested the requirement for GOLPH3 binding to PtdIns(4)P for efficient Golgi trafficking. We attempted rescue of siRNA knockdown of GOLPH3 with siRNA-resistant wild type or R90L mutant GOLPH3 and observed trafficking of EYFP-FAPP1-PH. Wild type GOLPH3 rescued efficient trafficking, but the R90L PtdIns(4)P binding mutant did not (Figure 7B). We thus conclude that efficient exit from the Golgi depends on the ability of GOLPH3 to bind PtdIns(4)P.
Our model argues that the extension of the Golgi ribbon around the nucleus is a consequence of a tensile force applied via GOLPH3, and that this force is necessary for extracting tubules and vesicles from the Golgi. If true, the model predicts that tubules and vesicles exit the Golgi on a trajectory parallel to the path of the Golgi ribbon around the nucleus. As above, we expressed low levels of EYFP-FAPP1-PH to mark PtdIns(4)P-bearing vesicles and tubules and measured the angle between the trajectory of exit from the Golgi and a tangent to the Golgi ribbon at the point of exit (Figure 7C and Movie S4). The trajectory angles were graphed for 99 exit events observed in eight cells (Figure 7D). These angles clustered highly significantly near 0° and 180°, or parallel to the Golgi (p<0.01, Kolmogorov-Smirnov). We also observed trafficking of ts045-VSVG-EGFP, and the angles again clustered around 0° and 180°, as predicted (p<0.01, Kolmogorov-Smirnov, Figure 7E).
Taken together, our data indicate that exit of traffic from the Golgi depends on interaction of PtdIns(4)P, GOLPH3, MYO18A, and F-actin. The initial trajectory of vesicle exit follows the orientation of the tensile force that extends the Golgi ribbon and flattens Golgi cisternae. We argue that this force is produced by MYO18A and F-actin and transmitted to the Golgi by GOLPH3 and PtdIns(4)P. We propose that the molecular apparatus diagrammed in Figure 4C and Figure 7F functions to attach the Golgi to the actin cytoskeleton and is necessary to pull tubules and vesicles from it.
DISCUSSION
The GOLPH3 Family of Phosphoinositide Binding Proteins
By proteomic screening we have identified GOLPH3 as a PtdIns(4)P binding protein. The specificity of binding is conserved among yeast, flies, and humans. We have shown that GOLPH3 Golgi localization is a consequence of its interaction with PtdIns(4)P. The highly conserved GOLPH3 proteins represent a family of phosphoinositide binding proteins with a unique structure (Schmitz et al., 2008) unlike other known phosphoinositide binding proteins. Their ability to bind PtdIns(4)P could not have been predicted before this screen. The confirmation of GOLPH3 as a PtdIns(4)P binding protein and characterization of its critical role at the Golgi provides strong validation of the screen and the hypothesis that many undiscovered phosphoinositide binding proteins exist and knowing their identity and properties will illuminate fundamental cell functions.
Function of GOLPH3 at the Golgi
We have shown that PtdIns(4)P, GOLPH3, MYO18A, and F-actin are all necessary for extension of the Golgi around the nucleus. The compact Golgi phenotype is unusual and not described in many other manipulations of the Golgi, including in response to nocodazole, BFA, knockdown of Golgi proteins such as Arl1 (Lu et al., 2004), GCC185 (Derby et al., 2007), p230, (Yoshino et al., 2005), GM130 or GRASP65 (Puthenveedu et al., 2006), COG1, COG2, or COG5 (Oka et al., 2005), or knockdown of other PtdIns(4)P binding proteins such as FAPP1, FAPP2 (Godi et al., 2004), or CERT (Giussani et al., 2008). Golgi compaction is a distinct result of depletion of PtdIns(4)P, knockdown of GOLPH3, knockdown of MYO18A, or depolymerization of F-actin, consistent with the interactions depicted in our model (Figure 4D).
Our data indicate that GOLPH3 functions at the Golgi by binding directly and specifically to Golgi membranes through PtdIns(4)P, although it remains formally possible that an additional Golgi factor may help recruit GOLPH3. Golgi GOLPH3 also binds to MYO18A, linking the Golgi to actin filaments. Based on homology and the requirement for an intact ATPase pocket, we argue that MYO18A is likely functioning as an active motor, although it remains possible that it functions only to link to F-actin, and the generation of tension involves a more complicated mechanism.
Actin and Myosin at the Golgi
A role for F-actin at the Golgi has been suggested previously. In S. cerevisiae, vesicles trafficking from the Golgi in the polarized secretory pathway travel along actin cables in a process dependent on the Pik1p PI-4-kinase (Finger and Novick, 1998; Walch-Solimena and Novick, 1999; Hsu et al., 2004; Pruyne et al., 2004).
In mammalian cells, F-actin and actin-associated proteins have been found at the Golgi (for reviews see Egea et al., 2006 and De Matteis and Luini, 2008). Egea and colleagues have published a series of studies showing that depolymerization of actin alters Golgi morphology and trafficking, although a compelling mechanism was not identified (Egea et al., 2006; Lázaro-Diéguez et al., 2007). Our data and model provide an explanation for their results.
Previous studies have identified other myosins at the Golgi. In mammalian cells, MYO1, MYO2, and MYO6 have been implicated in Golgi secretory function (for review see Allan et al., 2002). Our mass spectrometry and coIP data argue that GOLPH3 couples specifically to MYO18A in mammalian cells.
In mammalian cells, the demonstration that vesicles leaving the Golgi follow a path that colocalizes with microtubules (Cooper et al., 1990) has led to a popular model whereby vesicles are carried by microtubules. A growing number of proteins serve to bridge microtubules and F-actin (reviewed in Rodriguez et al., 2003), providing potential synthesis between the F-actin model favored in yeast and by our data, and the microtubule model that has been favored in the mammalian system.
Conservation to S. cerevisiae
Our data demonstrate that Vps74p binds PtdIns(4)P in vitro and is downstream of PIK1 in vivo. Furthermore, human GOLPH3 can partially rescue a vps74 mutant (Tu et al., 2008), and so we believe they function similarly. In mammals, we observe that GOLPH3 functions through its interaction with MYO18A, but the S. cerevisiae genome does not encode an ortholog of MYO18A, having only type I, II, and V myosins (Berg et al., 2001). We note that there are many examples in S. cerevisiae where a small number of primordial myosins perform the range of functions that in higher organisms are partitioned between a larger number of more specialized myosins (for review see Berg et al., 2001), and thus we expect that Vps74p will link to one of these. In fact, Pik1p-dependent polarized secretion is proposed to be dependent on the type V myosin, Myo2p (Walch-Solimena and Novick, 1999; Pruyne et al., 2004), making it a good candidate.
Two groups recently reported that in yeast Vps74p is involved in maintaining the steady-state localization of cis and medial Golgi-localized glycosyltransferases that function in cell wall biosynthesis (Schmitz et al., 2007, Tu et al., 2007). In mammalian cells, GOLPH3 localizes to the trans Golgi (Wu et al., 2000, Snyder et al., 2006 and current study). The specific glycosyltransferases studied in yeast are not conserved beyond fungi. We have observed several mammalian glycosyltransferases and seen neither interaction with GOLPH3 (data not shown) nor mislocalization upon knockdown of GOLPH3. It is possible that this represents a species difference.
Role of PtdIns(4)P at the Golgi
Based on high abundance of GOLPH3 (~500,000 molecules per HeLa cell) and high conservation from S. cerevisiae to humans, it is a major target of PtdIns(4)P. Other well-documented targets of PtdIns(4)P include OSBP, FAPP, and CERT which function in the non-vesicular transport of cholesterol, glucosylceramide, and ceramide, respectively (Im et al., 2005; Raychaudhuri and Prinz, 2006; Halter et al., 2007; D’Angelo et al., 2007; Hanada et al., 2003). Notably, these proteins all function to regulate the composition of Golgi membranes and therefore are expected to regulate the mechanical properties of the membrane. Thus, PtdIns(4)P has targets that simultaneously regulate the tensile force on the membrane and its mechanical properties, including presumably its elasticity and elastic limit, thereby determining behavior of the membrane when subjected to tension. We speculate that the coordinate regulation of a tensile force and the mechanical properties of the membrane provides an ideal arrangement to control the ability to pull vesicles from the Golgi and consequently, to generate the morphology uniquely characteristic of the Golgi. We believe this model provides a parsimonious explanation for all available data and an appealing link between the process of vesicle budding and Golgi morphology.
During preparation of this manuscript, a study was published identifying GOLPH3 as an oncogene amplified in many human cancers (Scott et al., 2009). It will be interesting to determine how the molecular function of GOLPH3 that we have identified is linked to the regulation of cell proliferation.
EXPERIMENTAL PROCEDURES
Lipid Blots
Lipids (Cell Signals, Columbus, OH and Echelon Biosciences, Salt Lake City, UT) were dissolved in DMSO/20% CHCl3/50 mM HCl, validated by TLC, and 100 nl spotted on PVDF. Blots blocked overnight in 3% fatty acid-free BSA in TBST (150 mM NaCl, 50 mM tris pH 7.5, 0.03% Tween-20) were probed in refreshed block with 35S-labeled protein from a 20 µl IVT mix (Promega TNT Gold) with 33 µCi translabel, using 1 µl for SDS-PAGE. After 2 hours, blots washed five times were exposed to phosphorimager (Molecular Dynamics), including 4–6 positive controls in every experiment.
Lipid Vesicle Pull-down
Small unilamellar lipid vesicles of 55 nmol PE or 55 nmol PE + 2.5 nmol PtdIns(3)P or PtdIns(4)P in 30 µl buffer (100 mM NaCl, 20 mM Hepes pH 7.2, 2 mM EGTA, and 100 ng/mL BSA). Vesicles prepared by sonicating 1 hour and ten cycles of freezing in liquid nitrogen and thawing at 60°C. IVT proteins were pre-cleared, incubated with lipid vesicles 20 min at 30°C, and pelleted at 100K × g for 25 min at 4°C, washed, and boiled in SDS sample buffer.
Immunoprecipitation
HeLa cells lysed on ice in 150 mM NaCl, 10 mM NaHPO4, 2 mM EDTA, 10 mM CHAPS, 5 mM DTT, and protease inhibitors were cleared and incubated with anti-GOLPH3 or pre-immune serum, precipitated with Protein A Sepharose (GE Healthcare), washed extensively, and boiled in SDS sample buffer.
Mass Spectrometry
Bands excised from polyacrylamide stained with SafeStain SimplyBlue (Invitrogen), subjected to in-gel digestion, and analyzed as in Zhou et al., 2004.
Pull-down of GOLPH3 with 6xHis-tagged MYO18A
6xHis-SUMO-tagged proteins bound to Ni-NTA-agarose beads (Qiagen) by rotating 2 hrs at 4°C in 50mM NaH2PO4, 300mM NaCl, 20mM imidazole pH8.0, followed by washing. Equimolar amounts of purified GST and GOLPH3 added to each set of beads and rotated 2 hrs at 4°C in 10 mM NaHPO4, 150 mM NaCl, 20 mM imidazole, 10 mM CHAPS pH 7.4, followed by washing and boiling in SDS sample buffer.
Fluorescence Microscopy
Fluorescence microscopy performed on an Olympus IX81-ZDC spinning disk confocal microscope and analyzed with Slidebook and ImageJ software.
Electron Microscopy
Samples processed as in Takeda et al. (2001), viewed using a JEOL 1200EX II transmission electron microscope, and photographed using a Gatan Orius 600 digital camera. Pixel size calibrated by Pelco grating replica #607 (Ted Pella, Inc.), with measurements performed in ImageJ.
Supplementary Material
ACKNOWLEDGEMENTS
We thank L. Cantley, S. Emr, G. Rosenfeld, and C. Min for helpful advice, reagents, and support. We thank R. Tsien, P. Mayinger, J. Lippincott-Schwartz, V. Malhotra, O. Ohara, and Z. Chroneos for generous gifts of reagents. We thank K. Messer of the UCSD-Moores Cancer Center for help with statistical analyses. We acknowledge the following support: MMN by NIDDK Postdoctoral Training Grant and American Cancer Society Fellowship; SEF-K by NIDDK Pre-Doctoral Training Grant; and SJF by the Burroughs Wellcome Fund, the V Foundation, NIDDK, and NIH New Innovator Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allan VJ, Thompson HM, McNiven MA. Motoring around the Golgi. Nat. Cell Biol. 2002;4:E236–E242. doi: 10.1038/ncb1002-e236. [DOI] [PubMed] [Google Scholar]
- Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FA, Warren G. Disassembly and reassembly of the Golgi apparatus. Semin. Cell Dev. Biol. 1996;7:505–510. [Google Scholar]
- Beh CT, Cool L, Phillips J, Rine J. Overlapping functions of the yeast oxysterol-binding protein homologs. Genetics. 2001;157:1117–1140. doi: 10.1093/genetics/157.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AW, Ward MA, Blackstock WP, Freeman HNM, Choudhary JS, Lewis AP, Chotai D, Fazel A, Gushue JN, Paiement J, Palcy S, Chevet E, Lafrenière-Roula M, Solari R, Thomas DY, Rowley A, Bergeron JJM. Proteomics characterization of abundant Golgi membrane proteins. J. Biol. Chem. 2001;276:5152–5165. doi: 10.1074/jbc.M006143200. [DOI] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol. Biol. Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossard C, Bresson D, Polishchuk RS, Malhotra V. Dimeric PKD regulates membrane fission to form transport carriers at the TGN. J. Cell Biol. 2007;179:1123–1131. doi: 10.1083/jcb.200703166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MS, Cornell-Bell AH, Chernjavsky A, Dani JW, Smith SJ. Tubulovesicular processes emerge from trans-Golgi cisternae, extend along microtubules, and interlink adjacent trans-Golgi elements into a reticulum. Cell. 1990;61:135–145. doi: 10.1016/0092-8674(90)90221-y. [DOI] [PubMed] [Google Scholar]
- D’Angelo G, Polishchuk E, Di Tullio G, Santoro M, Di Campli A, Godi A, West G, Bielawski J, Chuang C-C, van der Spoel AC, Platt FM, Hannun YA, Polishchuk R, Mattjus P, De Matteis MA. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 2007;449:62–67. doi: 10.1038/nature06097. [DOI] [PubMed] [Google Scholar]
- Davidson MW. Molecular expressions cell biology: The Golgi apparatus. 2004 Website: http://micro.magnet.fsu.edu/cells/golgi/golgiapparatus.html.
- De Matteis MA, Luini A. Exiting the Golgi complex. Nat. Rev. Mol. Cell Biol. 2008;9:273–284. doi: 10.1038/nrm2378. [DOI] [PubMed] [Google Scholar]
- Derby MC, Lieu ZZ, Brown D, Stow JL, Goud B, Gleeson PA. The trans-Golgi network golgin, GCC185, is required for endosome-to-Golgi transport and maintenance of Golgi structure. Traffic. 2007;8:758–773. doi: 10.1111/j.1600-0854.2007.00563.x. [DOI] [PubMed] [Google Scholar]
- Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea G, Lázaro-Diéguez F, Vilella M. Actin dynamics at the Golgi complex in mammalian cells. Curr. Opin. Cell Biol. 2006;18:168–178. doi: 10.1016/j.ceb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Finger FP, Novick P. Spatial regulation of exocytosis: lessons from yeast. J. Cell Biol. 1998;142:609–612. doi: 10.1083/jcb.142.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa T, Ikawa S, Yanai N, Obinata M. Isolation of a novel PDZ-containing myosin from hematopoietic supportive bone marrow stromal cell lines. Biochem. Biophys. Res. Commun. 2000;270:67–75. doi: 10.1006/bbrc.2000.2377. [DOI] [PubMed] [Google Scholar]
- Giussani P, Colleoni T, Brioschi L, Bassi R, Hanada K, Tettamanti G, Riboni L, Viani P. Ceramide traffic in C6 glioma cells: evidence for CERT-dependent and independent transport from ER to the Golgi apparatus. Biochim. Biophys. Acta. 2008;1781:40–51. doi: 10.1016/j.bbalip.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA. ARF mediates recruitment of PtdIns-4-OH kinase-β and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Halter D, Neumann S, van Dijk SM, Wolthoorn J, de Mazière AM, Vieira OV, Mattjus P, Klumperman J, van Meer G, Sprong H. Pre- and post-Golgi translocation of glucosylceramide in glysosphingolipid synthesis. J. Cell Biol. 2007;179:101–115. doi: 10.1083/jcb.200704091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- Hausser A, Storz P, Märtens S, Link G, Toker A, Pfizenmaier K. Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIβ at the Golgi complex. Nat. Cell Biol. 2005;7:880–886. doi: 10.1038/ncb1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg K, Phair RD, Lippincott-Schwartz J. Kinetic analysis of intracellular trafficking in single living cells with vesicular stomatitis virus protein G-green fluorescent protein hybrids. Methods Enzymol. 2000;327:69–89. doi: 10.1016/s0076-6879(00)27268-6. [DOI] [PubMed] [Google Scholar]
- Hsu S-C, TerBush D, Abraham M, Guo W. The exocyst complex in polarized exocytosis. Int. Rev. Cytol. 2004;233:243–265. doi: 10.1016/S0074-7696(04)33006-8. [DOI] [PubMed] [Google Scholar]
- Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogawa Y, Kon T, Inoue T, Ohkura R, Yamakawa H, Ohara O, Sutoh K. The N-terminal domain of MYO18A has an ATP-insensitive actin-binding site. Biochemistry. 2005;44:6190–6196. doi: 10.1021/bi0475931. [DOI] [PubMed] [Google Scholar]
- Kristen U, Lockhausen J. Estimation of Golgi membrane flow rates in ovary glands of Aptenia cordifolia using cytochalasin B. Eur. J. Cell Biol. 1983;29:262–267. [PubMed] [Google Scholar]
- Lázaro-Diéguez F, Jiménez N, Barth H, Koster AJ, Renau-Piqueras J, Llopis JL, Burger KNJ, Egea G. Actin filaments are involved in the maintenance of Golgi cisternae morphology and intra-Golgi pH. Cell Motil. Cytoskeleton. 2006;63:778–791. doi: 10.1002/cm.20161. [DOI] [PubMed] [Google Scholar]
- Lázaro-Diéguez F, Colonna C, Cortegano M, Calvo M, Martínez SE, Egea G. Variable actin dynamics requirement for the exit of different cargo from the trans-Golgi network. FEBS Lett. 2007;581:3875–3881. doi: 10.1016/j.febslet.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and –independent components. Curr. Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- Lu L, Guihua T, Hong W. Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the trans-Golgi network. Mol. Biol. Cell. 2004;15:4426–4443. doi: 10.1091/mbc.E03-12-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Furusawa T, Okubo T, Inoue T, Ikawa S, Yanai N, Mori KJ, Obinata M. Genome structure and differential expression of two isoforms of a novel PDZ-containing myosin (MysPDZ) (Myo18A) J. Biochem. 2003;133:405–413. doi: 10.1093/jb/mvg053. [DOI] [PubMed] [Google Scholar]
- Mori K, Matsuda K, Furusawa T, Kawata M, Inoue T, Obinata M. Subcellular localization and dynamics of MysPDZ (Myo18A) in live mammalian cells. Biochem. Biophys. Res. Comm. 2005;326:491–498. doi: 10.1016/j.bbrc.2004.11.058. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Goto K, Kondo H. Cloning, expression, and localization of 230-kDa phosphatidylinositol 4-kinase. J. Biol. Chem. 1996;271:12088–12094. doi: 10.1074/jbc.271.20.12088. [DOI] [PubMed] [Google Scholar]
- Nakashima-Kamimura N, Asoh S, Ishibashi Y, Mukai Y, Shidara Y, Oda H, Munakata K, Goto Y, Ohta S. MIDAS/GPP34, a nuclear gene product, regulates total mitochondrial mass in response to mitochondrial dysfunction. J. Cell Sci. 2005;118:5357–5367. doi: 10.1242/jcs.02645. [DOI] [PubMed] [Google Scholar]
- Oka T, Vasile E, Penman M, Novina CD, Dykxhoorn DM, Ungar D, Hughson FM, Krieger M. Genetic analysis of the subunit organization and function of the conserved oligomeric Golgi (COG) complex. J. Biol., Chem. 2005;280:32736–32745. doi: 10.1074/jbc.M505558200. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- Puthenveedu MA, Linstedt AD. Subcompartmentalizing the Golgi apparatus. Curr. Opin. Cell Biol. 2005;17:369–375. doi: 10.1016/j.ceb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat. Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Prinz WA. Uptake and trafficking of exogenous sterols in Saccharomyces cerevisiae. Biochem. Soc. Trans. 2006;34:359–362. doi: 10.1042/BST0340359. [DOI] [PubMed] [Google Scholar]
- Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 2003;5:559–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- Rogalski AA, Singer SJ. Association of elements of the Golgi apparatus with microtubules. J. Cell Biol. 1984;99:1092–1100. doi: 10.1083/jcb.99.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde HM, Cheong FY, Konrad G, Paiha K, Mayinger P, Boehmelt G. The human phosphatidylinositol phosphatase SAC1 interacts with the Coatomer I complex. J. Biol. Chem. 2003;278:52689–52699. doi: 10.1074/jbc.M307983200. [DOI] [PubMed] [Google Scholar]
- Schmitz KR, Liu J, Li S, Setty TG, Wood CS, Burd CG, Ferguson KM. Golgi localization of glycosyltransferases requires a Vps74p oligomer. Dev. Cell. 2008;14:523–534. doi: 10.1016/j.devcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KL, Kabbarah O, Liang M-C, Ivanova E, Anagnostou V, Wu J, Dhakal S, Wu M, Chen S, Feinberg T, Huang J, Saci A, Widlund HR, Fisher DE, Xiao Y, Rimm DL, Protopopov A, Wong K-K, Chin L. GOLPH3 modulates mTOR signaling and rapamycin sensitivity in cancer. Nat. 2009;459:1085–1090. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short B, Haas A, Barr FA. Golgin and GTPases, giving identity and structure to the Golgi apparatus. Biochim. Biophys. Acta. 2005;1744:383–395. doi: 10.1016/j.bbamcr.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Snyder CM, Mardones GA, Ladinsky MS, Howell KE. GMx33 associates with the trans-Golgi matrix in a dynamic manner and sorts within tubules exiting the Golgi. Mol. Biol. Cell. 2006;17:511–524. doi: 10.1091/mbc.E05-07-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton M, Liao G, Brokstein P, Hong L, Carninci P, Shirake T, Hayashizaki Y, Champe M, Pacleb J, Wan K, Yu C, Carlson J, George R, Celniker S, Rubin GM. The Drosophila Gene Collection: Identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 2002;12:1294–1300. doi: 10.1101/gr.269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, McQuistan T, Orlando RA, Farquhar MG. Loss of glomerular foot processes is associated with uncoupling of podocalyxin from the actin cytoskeleton. J. Clin. Invest. 2001;108:289–301. doi: 10.1172/JCI12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan I, Yong J, Dong JM, Lim L, Leung T. A tripartite complex containing MRCK modulates lamellar actomyosin retrograde flow. Cell. 2008;135:123–136. doi: 10.1016/j.cell.2008.09.018. [DOI] [PubMed] [Google Scholar]
- Tu L, Tai WCS, Chen L, Banfield DK. Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science. 2008;321:404–407. doi: 10.1126/science.1159411. [DOI] [PubMed] [Google Scholar]
- Varnai P, Balla T. Visualization and manipulation of phosphoinositide dynamics in live cells using engineered protein domains. Pflugers Arch. 2007;455:69–82. doi: 10.1007/s00424-007-0270-y. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase Pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Wang J, Sun HQ, Martinez M, Sun YX, Macia E, Kirchhausen T, Albanesi JP, Roth MG, Yin HL. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- Wong K, Meyers R, Cantley LC. Subcellular locations of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 1997;272:13236–13241. doi: 10.1074/jbc.272.20.13236. [DOI] [PubMed] [Google Scholar]
- Wu CC, Taylor RS, Lane DR, Ladinsky MS, Weisz JA, Howell KE. GMx33: a novel family of trans-Golgi proteins identified by proteomics. Traffic. 2000;1:963–975. [PubMed] [Google Scholar]
- Yoshino A, Rao Gangi Setty S, Poynton C, Whiteman EL, Saint-Pol A, Burd CG, Johannes L, Holzbaur EL, Koval M, McCaffery JM, Marks MS. tGolgin-1 (p230, golgin-245) modulates Shiga-toxin transport to the Golgi and Golgi motility towards the microtubule-organizing centre. J. Cell Sci. 2005;118:2279–2293. doi: 10.1242/jcs.02358. [DOI] [PubMed] [Google Scholar]
- Zhou W, Ryan JJ, Zhou H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:32262–32268. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.