Abstract
Previous studies of epithelial cell growth and differentiation in the prostate gland have identified the homeodomain protein Nkx3.1 as a central regulator of prostate development and carcinogenesis. To understand the molecular mechanisms of Nkx3.1 function, we have used yeast two-hybrid analysis to identify Nkx3.1 interacting proteins, and have isolated Fem1b, a mammalian homolog of the C. elegans sex-determining gene Fem-1. In mice, the Fem1b and Nkx3.1 genes encode proteins that interact in glutathione-S-transferase (GST) pull-down and co-immunoprecipitation assays, and are co-expressed in the prostate and testis of neonatal mice. Null mutants for Fem1b generated by gene targeting display defects in prostate ductal morphogenesis and secretory protein expression, similar to phenotypes found in Nkx3.1 mutants. We propose that Fem1b may have a conserved role in the generation of sexual dimorphism through its interaction with Nkx3.1 in the developing prostate gland.
Keywords: prostate gland, testis, homeodomain, sexual dimorphism, gene targeting, yeast two-hybrid assay
In mammals, the generation of sexual dimorphism in secondary sexual tissues is governed by the activities of androgenic and estrogenic hormones during fetal development. In particular, the expression of the sex-determining genes Sry and Sox9 in the XY germline leads to male sexual differentiation through the activation of MIS and subsequent Mullerian duct regression, together with the production of androgens by Leydig cells of the testis (Kim and Capel, 2006; Wilhelm and Koopman, 2006). However, the molecular mechanisms involved in specification of sexually dimorphic cell types downstream of androgen signaling remain poorly understood.
The prostate gland is a male-specific tissue that arises from the primitive urogenital sinus during fetal development. The earliest known marker of prostate epithelial cells is the homeobox gene Nkx3.1, which is expressed by the nascent prostate buds, as well as by the urogenital sinus epithelium prior to bud formation (Sciavolino et al., 1997; Bhatia-Gaur et al., 1999). Nkx3.1 is a transcription factor that can bind specific DNA sequences and displays both transcriptional activator and repressor activity in vitro and in vivo (reviewed in (Shen and Abate-Shen, 2003)). Notably, the expression of Nkx3.1 in the urogenital sinus is male-specific, both before and during prostate bud formation, yet expression of Nkx3.1 in other embryonic tissues is not sexually-dimorphic (Sciavolino et al., 1997; Tanaka et al., 1999). Null mutations of Nkx3.1 in mice result in defects in prostate ductal morphogenesis and secretory protein production, together with an epithelial hyperplasia that displays progressive severity with aging (Bhatia-Gaur et al., 1999; Schneider et al., 2000; Tanaka et al., 2000; Kim et al., 2002a).
In contrast to mammalian sex determination, C. elegans sex is governed by the ratio of X chromosomes to autosomes, and utilizes a complex cascade of sex determining genes that have been well-characterized in genetic analyses (reviewed in (Hansen and Pilgrim, 1999; Kuwabara, 1999)). In particular, the fem genes play central roles in sex determination of C.elegans by promoting male development in the soma of XO animals as well as spermatogenesis in the germ lines of both males and hermaphrodites. Although the three C. elegans fem genes play similar roles in specification of male development, with null mutants displaying feminization of XX and XO animals (Kimble et al., 1984; Hodgkin, 1986), they encode unrelated intracellular proteins. FEM-2 is related in sequence to protein serine/threonine phosphatase of Type 2C (PP2C), and exhibits magnesium-dependent casein phosphatase activity in vitro (Pilgrim et al., 1995; Chin-Sang and Spence, 1996). FEM-3 interacts directly with FEM-2, but its sequence is currently unrelated to any other known proteins (Ahringer and Kimble, 1991; Chin-Sang and Spence, 1996). Finally, FEM-1 is a soluble, intracellular protein whose N-terminus contains 6 ankyrin repeats, a 33-amino acid motif that mediates specific protein–protein interactions (Spence et al., 1990). Notably, recent studies have shown that the three C. elegans FEM proteins together with Cullin-2 (Cul2), Elongin B, and Elongin C form a multi-subunit complex that has E3 ubiquitin ligase activity, and negatively regulates TRA-1 through ubiquitylation and subsequent proteasomal degradation (Starostina et al., 2007).
Several studies have shown that FEM-1 is highly conserved throughout evolution. Three distinct mammalian Fem1 genes have been identified that encode proteins that have greater than 30% amino acid identity with C. elegans FEM-1 and greater than 40% amino acid identity with each other (Ventura-Holman et al., 1998; Chan et al., 2000; Ventura-Holman and Maher, 2000; Ventura-Holman et al., 2003). Although the sequence similarities between Fem1 family members mainly occur in the ankyrin repeats at their N-terminus, the individual Fem1 orthologs share 99% amino acid identity between human and mouse (Ventura-Holman et al., 1998; Ventura-Holman et al., 2003).
Below, we identify human FEM1B as a protein that interacts with NKX3.1 in prostate epithelium. We show that mouse Fem1b and Nkx3.1 are co-expressed in the prostate epithelium as well as testicular germ cells during organogenesis. Furthermore, we show that null mutation for Fem1b results in defects in prostate ductal morphogenesis and secretory protein expression resembling those observed in Nkx3.1 null mutants. Our results suggest that Fem1b may have conserved functions in the generation of mammalian sexual dimorphism, in part through its interactions with Nkx3.1.
Results
Interaction of Fem1b and Nkx3.1 proteins
To identify proteins that interact with human NKX3.1, we employed a yeast two-hybrid assay using the E. coli LexA interaction trap system (Golemis et al., 2008). For this purpose, we constructed a bait plasmid expressing a full-length human NKX3.1–LexA fusion protein to screen an adult human prostate library. In three rounds of screening a total of 2 × 105 independent clones, we identified 24 positive clones that were galactose-independent and expressed LacZ (Table 1). Importantly, among these clones was Prostate derived Ets factor (PDEF), which has been previously shown to interact with Nkx3.1 (Chen et al., 2002).
Table 1.
Clones isolated in yeast two-hybrid assay.
| Symbol | Gene | Entrez ID | Mating test* | # isolates |
|---|---|---|---|---|
| Heterogenous nuclear ribonucleoprotein A0 | 15080450 | − | 21 | |
| Menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 12654032 | − | 7 | |
| ISG20L1 | Interferon stimulated exonuclease gene 20KDa-like 1 | 21362093 | − | 6 |
| LIM and cysteine-rich domains 1 | 33875679 | − | 5 | |
| TEX264 | testis expressed sequence 264 | 34147580 | − | 3 |
| CCT7 | Chaperonin containing TCP1, subunit 7 | 58331183 | − | 2 |
| PDEF | Ets transcription factor PDEF | 4007417 | − | 2 |
| Ribosomal Protein L37a | 347385 | + | 2 | |
| FEM1B | Fem-1 (C. elegans) homolog b | 52851431 | +++ | 1 |
| CHPF | Chondroitin polymerizing factor | 168229166 | − | 1 |
| CKAP4 | Cytoskeleton-associated protein 4 | 19920316 | − | 1 |
| C9orf86 | Chromosome 9 open reading frame 86 | 186700622 | − | 1 |
| Mannosidase, alpha, class 2B, member 1 | 37588991 | − | 1 | |
| NADH dehydrogenase subunit 4 | 21717330 | − | 1 | |
| Pp8857 mRNA, unknown | 18027823 | ++ | 1 | |
| PBP | Phosphatidyl-ethanolamine binding protein homologue | 435637 | − | 1 |
| PTMS | Parathymosin | 46276862 | − | 1 |
| STRA13 | bHLH transcription factor DEC1 | 13926075 | +++ | 1 |
| RARG | Retinoic acid receptor, gamma | 112181192 | − | 1 |
| Ribosomal protein SA | 33872545 | − | 1 | |
| STMN3 | Stathmin like-3 | 14670374 | − | 1 |
| HSP90B1 | Heat shock protein 90kDa beta (Grp94), member 1 | 4507676 | ++ | 1 |
| TPT1 | Tumor protein, translationally controlled 1 | 4507668 | − | 1 |
Relative intensity of blue staining on (gal)-his-ura-trp+X-gal plates is indicated.
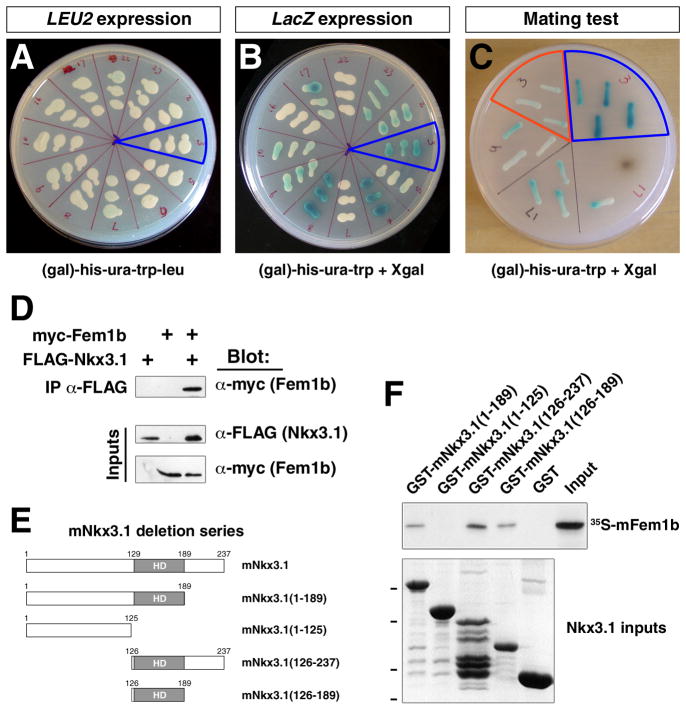
Among the 24 positive clones, one was of particular interest since it corresponded to a 1.8 kb clone of human FEM1B that contained 5′ UTR sequence and most of the open reading frame. We validated the interaction of FEM1B with NKX3.1 by showing that the yeast strain EGY48 containing the prey plasmid pJG4-5-hFEM1B, the bait plasmid pEG202-hNKX3.1, and the LexA-operator-lacZ reporter pSH18-34 was galactose-dependent for growth and could activate lacZ expression (Fig. 1A, B). In addition, the pJG4-5-hFEM1B clone passed a further mating test that distinguishes more stringent interactions since it reduces transcriptional activation of reporters (Golemis et al., 2008). Thus, we observed that mating of the RFY206 strain containing the prey plasmid pJG4-5-hFEM1b with the EGY48 strain containing the bait plasmid pEG202-hNKX3.1 and the LexA-operator-lacZ reporter pSH18-34 resulted in galactose-dependent growth and lacZ expression (Fig. 1C). In contrast, mating of RFY206 containing pJG4-5-hFEM1b with EGY48 containing pSH18-34 together with a negative-control plasmid that produces an inactive fusion protein were unable to express lacZ after 48 hours of growth (Fig. 1C).
Figure 1.
Interaction of Nkx3.1 and Fem1b proteins. A, B: Yeast two-hybrid assay. The yeast strain EGY48 containing the prey plasmid pJG4-5-hFEM1B, the bait plasmid pEG202-hNKX3.1, and the LexA-operator-LacZ reporter pSH18-34 is able to grow on (gal)-his-ura-trp-leu plates (blue outline in A) and can activate β-galactosidase expression as detected by X-gal staining (blue outline in B). C: Mating test for interaction of FEM1B and NKX3.1 using single-copy plasmids. Mating of the RFY206 strain containing the prey plasmid pJG4-5-hFEM1B with the EGY48 strain containing the bait plasmid pEG202-hNKX3.1 and reporter plasmid pSH18-34 results in growth on (gal)-his-ura-trp-leu plates and expresses high levels of β-galactosidase after 48 hr of growth (blue outline). The negative control (red outline) corresponds to RFY206 containing pJG4-5-hFEM1b mated with EGY48 containing pSH18-34 and the plasmid pRFHM1 containing the ADH promoter expressing an inactive LexA-bicoid homeodomain fusion protein. D: Co-immunoprecipitation of mFem1b and mNkx3.1. The indicated expression constructs were co-transfected into 293T cells, followed by immunoprecipitation of epitope-tagged Nkx3.1 from cell lysates with anti-FLAG antiserum. Western blots of immunoprecipitated and input proteins are shown, detected using α-FLAG (Nkx3.1, 27 kDa) and α-myc (Fem1b, 70 kDa) antibodies. E, F: Deletion mapping of the region of mouse Nkx3.1 that interacts with mFem1b in GST (glutathione-S-transferase) interaction assays. E: Schematic representation of mouse Nkx3.1 deletion constructs and location of the homeodomain (HD). F: Top panel shows interaction of in vitro translated 35S-labeled mFem1b following incubation with the indicated GST-mNkx3.1 fusion proteins or GST alone as a negative control. Bottom panel shows SDS-PAGE analysis of purified GST-Nkx3.1 fusion proteins; dashes indicate positions of molecular size standards at 49, 37, 26, and 20 kDa.
Following verification of the interaction between human NKX3.1 and FEM1B in yeast, our subsequent studies employed mouse Fem1b and Nkx3.1 proteins since we were interested in investigating the expression and function of Fem1b in mice. To examine the potential interaction of mouse Fem1b and Nkx3.1 proteins in mammalian cells, we performed co-immunoprecipitation experiments in transfected HEK 293T cells. When Myc-tagged mouse Fem1b was co-expressed with FLAG-tagged mNkx3.1 in HEK 293T cells, we could immunoprecipitate myc-Fem1b fusion protein from whole cell lysates and detect FLAG-Nkx3.1 by Western blotting (Fig. 1D).
To map the domain of Nkx3.1 responsible for its interaction with Fem1b, we performed in vitro GST (glutathione-S-transferase) interaction assays. In these assays, we produced GST fusions of purified human or mouse Nkx3.1 proteins with internal deletions, and assessed their binding to in vitro translated 35S-labeled mouse Fem1b protein. We found that mouse Fem1b protein could bind to N- and C-terminal deletions of mouse Nkx3.1 containing the homeodomain (mNkx3.1(1-189)) and mNkx3.1(126-237)) or to the homeodomain alone (mNkx3.1(126-189)), but not to the N-terminal region alone (mNkx3.1(1-125)) (Fig. 1E); similar results were observed using deletion mapping of human NKX3.1 (data not shown). Taken together, these results indicate that Nkx3.1 can directly bind to Fem1b, most likely through an interaction mediated by its homeodomain.
Expression of Fem1b in urogenital tissues
Previous studies using Northern blot analysis of adult mouse tissues have suggested widespread expression of Fem1b (Ventura-Holman et al., 1998); in contrast, Nkx3.1 is expressed in limited domains during embryogenesis and adulthood (Sciavolino et al., 1997; Tanaka et al., 1999). In our initial experiments, we performed quantitative real-time RT-PCR using RNA extracted from seminal vesicles and all three prostate lobes from 8-week-old male mice, and found that the level of Fem1b expression is highest in the ventral lobe within the prostate (Fig. 2A). Next, to determine whether Fem1b and Nkx3.1 display co-localized expression, we examined their expression during prostate development using in situ hybridization on adjacent sections. Although Nkx3.1 is expressed in the urogenital sinus epithelium prior to initial prostate budding at 17.5 dpc (Bhatia-Gaur et al., 1999), Fem1b expression was not detected at these stages. However, we found that Fem1b and Nkx3.1 expression were co-localized during organogenesis in the epithelial cells of the anterior and ventral prostate ducts at post-natal day 2 and 9 (Fig. 2B-D; data not shown).
Figure 2.
Expression of Fem1b in developing prostate and testis. A: Quantitative real-time RT-PCR analysis of Fem1b expression in prostate lobes and seminal vesicles from 8-week old Swiss-Webster mice. Expression levels are normalized to β-actin expression at 1 × 106 arbitrary units; error bars correspond to one standard deviation. B: Expression of Fem1b in the urogenital sinus at post-natal day 2. Abbreviations: ap, anterior prostate; bl, bladder; dd, ductus deferens. C-G: Expression of Nkx3.1 and Fem1b in adjacent sections using in situ hybridization. C, D: Expression of Nkx3.1 and Fem1b in epithelial cells of anterior prostate ducts at post-natal day 9. E-H: Expression of Nkx3.1 and Fem1b in seminiferous tubules of the testis at post-natal day 0, as shown at low-power (E, F) and high-power (G, H). Note that expression is limited to the germ cells (arrowheads in G, H) and is not detected in the surrounding Sertoli cells (arrows). Scale bars correspond to 100 microns.
We also observed that Fem1b is specifically expressed in the germ cells within seminiferous tubules of the testis, with no detectable expression in Sertoli cells (Fig. 2D). Interestingly, we found that Fem1b and Nkx3.1 are co-localized in germ cells in the testis at post-natal day 0 (Fig. 2D-G). In adult testis, Fem1b is highly expressed in all types of spermatogonia (Fig. 3E), consistent with previously published data for the adult rat testis (Oyhenart et al., 2005), while Nkx3.1 expression is only weakly detected in germ cells (data not shown).
Figure 3.
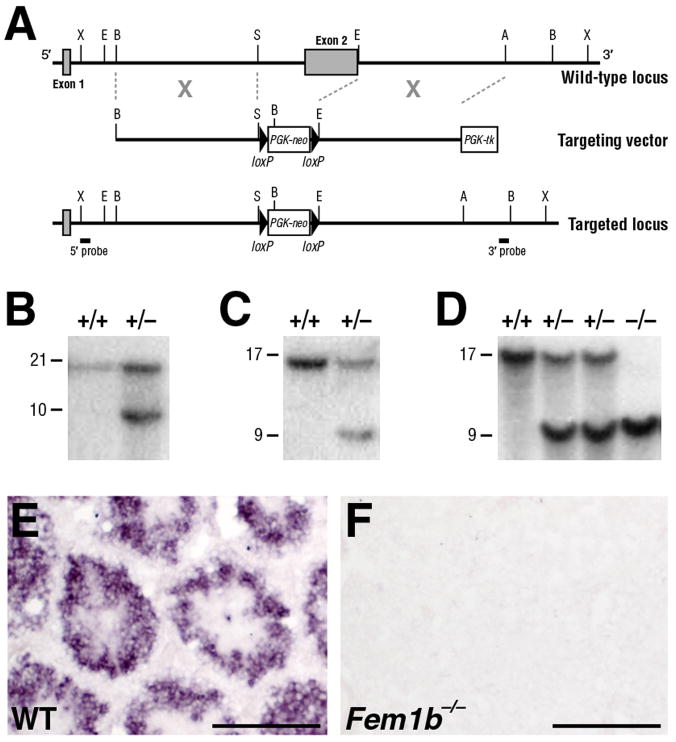
Targeting of the Fem1b locus. A: Schematic depiction of the targeting strategy. The mouse Fem1b genomic locus comprises two exons (shaded boxes), with most of the coding region (aa 83 – 628) in the second exon. Homologous recombination with the targeting vector deletes the second exon, which includes most of the ankyrin repeats. The positions of the 5′- and 3′- flanking probes used for Southern blot analysis are shown. B: Southern blot analysis of genomic DNA from ES cell clones using the 5′-flanking probe, which detects a 21 kb XbaI wild-type fragment and a 10 kb fragment from the targeted allele. C: Southern blot analysis of ES cell clones using the 3′-flanking probe, which detects a 17 kb BamHI wild-type fragment and a 9 kb fragment from the targeted allele. D: Southern blot analysis of genomic DNA from wild-type, heterozygous and homozygous adult mice using the 3′-flanking probe. F, G: Section in situ hybridization analysis shows that Fem1b expression is detected at 14 weeks of age in wild-type testis (F), but not in the Fem1b homozygous testis (G). Scale bars in F, G correspond to 100 microns.
Generation of Fem1b deficient mice
To investigate the in vivo function of Fem1b, we performed gene targeting using a vector that deletes the second exon, which encodes 87% of the mature protein, which should thereby result in a null mutation (Fig. 3A). Intercrosses between heterozygous mutant mice resulted in the generation of viable and fully fertile mice of both sexes that were homozygous for the targeted Fem1b allele. Southern blot analyses demonstrated the Fem1b allelic status in wild type, heterozygous, and homozygous mutant ES cells and mice (Fig. 3B-D). As a control, we confirmed that Fem1b mRNA expression is absent in the testis of Fem1b homozygotes (Fig. 3E, F).
To determine whether Fem1b might affect sex determination in mice, we examined the sex distribution of progeny from intercrosses of Fem1b heterozygous mice. We found that the Fem1b allele segregated in a normal Mendelian ratio (27% wild type, 50% heterozygous and 22% homozygous mutant mice, n = 318). Although the number of female Fem1b homozygous mutants was slightly greater than expected (10% Fem1b homozygous males vs. 12.6% female), this result was not statistically significant (P = 0.75 [χ2 test]); genotyping of these Fem1b mice confirmed that these were XX animals.
Lack of testis phenotype in Fem1b mutant mice
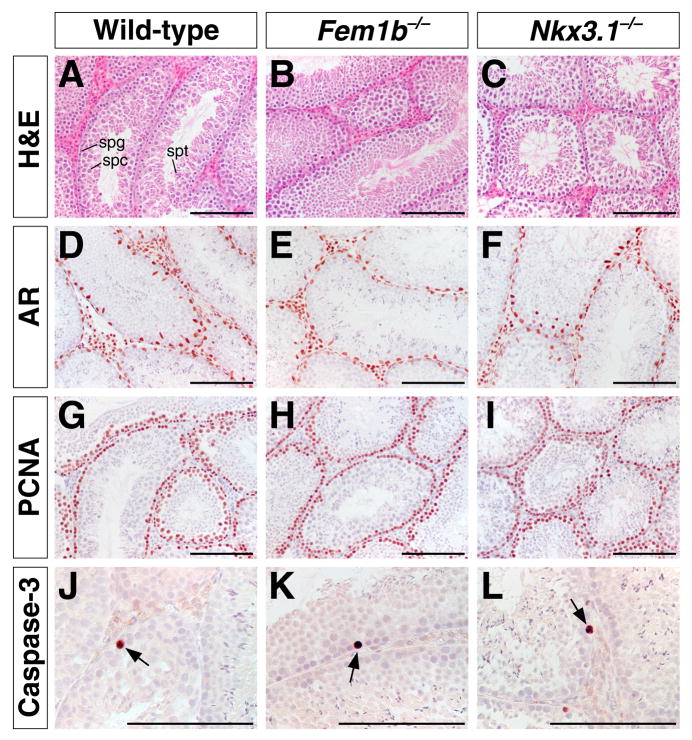
To evaluate whether Fem1b inactivation affects the development of male germ cells, we investigated testis histology by hematoxylin-eosin staining and marker analysis; in parallel, we performed a similar analysis with Nkx3.1 mutant males. Overall, we did not detect any histological abnormalities in the testes of either Fem1b or Nkx3.1 mutant mice. In particular, there were no observable differences in the number or localization of Sertoli cells, as determined by androgen receptor (AR) immunostaining (Fig. 4D-F). We also did not find any defects in cellular proliferation during spermatogenesis by immunohistochemical staining with proliferating cell nuclear antigen (PCNA) (Fig. 4G-I).
Figure 4.
Histological and immunohistochemical analysis of Fem1b and Nkx3.1 mutant testis. A-C: Hematoxylin-eosin staining of sections from wild type (A), Fem1b homozygous mutant (B), and Nkx3.1 homozygous mutant testis (C), with positions of spermatogonial cells (spg), spermatocytes (spc), and spermatids (spt) indicated in A. D-F: Immunohistochemical staining for androgen receptor (AR) in 8-week-old testis, showing normal localization of Sertoli cells (arrows). G-I: Immunohistochemical detection of PCNA (proliferating cell nuclear antigen) in 11-week-old testis, showing normal localization of germ cells. J-L: Immunohistochemical detection of activated caspase-3 in 2-week-old testis, showing apoptotic cells (arrows). Scale bars correspond to 100 microns.
Since FEM1B has been reported to interact with members of a death receptor family that mediates apoptosis, and overexpression of Fem1b can induce apoptosis in mammalian cells in culture (Chan et al., 2000), we performed immunohistochemical staining of 2-week old and adult testes with antibodies against activated caspase-3. No difference in apoptosis in the testis was observed between age-matched Fem1b mutant and wild type mice (Fig. 4J-L). Moreover, we did not find significant differences in the size and wet weight of the testes, or in the number and motility of mature sperm obtained from the cauda epididymis (data not shown).
Prostate phenotype of Fem1b mutant mice
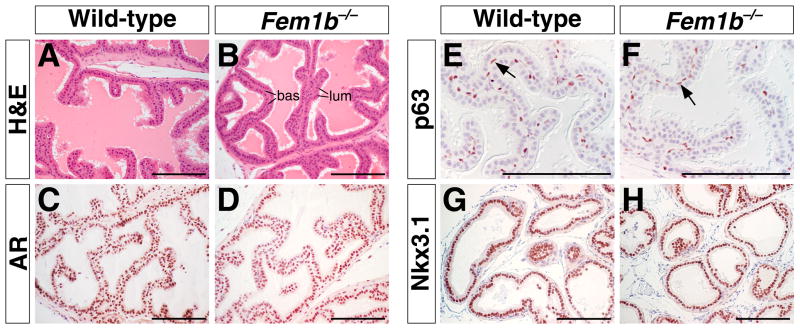
In the prostate, we observed normal histology of the three prostatic lobes of Fem1b mutants compared to wild-type littermates (Fig. 5A, B). We also found that AR, which is predominantly expressed in luminal cells, displayed normal patterns of expression in homozygous mutant males, as was also the case for the basal cell marker p63 (Fig. 5C-F). Importantly, wild-type expression patterns of Nkx3.1 were found in all three prostatic lobes of Fem1b mutants (Fig. 5G, H), indicating that loss of Fem1b has no effect on Nkx3.1 expression.
Figure 5.
Histological and immunohistochemical analysis of Fem1b mutant prostate at 8 weeks of age. A, B: Hematoxylin-eosin staining of sections from wild type (A) and Fem1b homozygous mutant (B) anterior prostate, with positions of basal (bas) and luminal (lum) epithelial cells indicated in B. C, D: Immunohistochemical staining for androgen receptor (AR) in anterior prostate, with positive immunoreactivity predominantly in luminal cells. E, F: Immunohistochemical detection of p63 in basal cells. G, H: Expression of Nkx3.1 in luminal cells of wild-type and Fem1b mutant anterior prostate. Scale bars correspond to 100 microns.
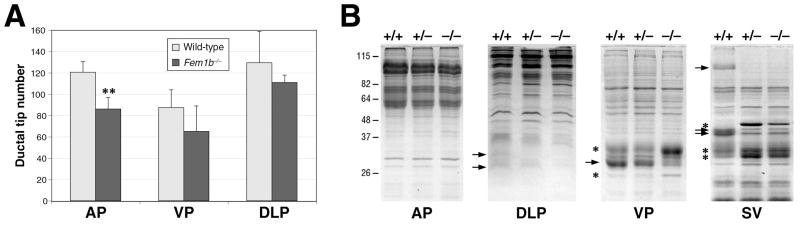
Since our in situ hybridization analysis revealed that Fem1b is expressed in the prostate ducts during organogenesis, we examined whether loss of Fem1b might lead to defects in ductal branching. We performed a quantitative analysis of ductal tip number using all three prostate lobes from Fem1b homozygotes and wild-type littermates at 6 weeks of each (n=6 each). We found a significant reduction of ductal tip numbers in the anterior prostate lobe (p = 0.0009), but not in the dorsolateral or ventral lobes (Fig. 6A).
Figure 6.
Decreased ductal tip branching and altered prostatic secretory protein expression in Fem1b mutants. A: Quantitation of ductal tip number in the prostatic lobes of Fem1b mutants and wild-type littermate controls. Error bars correspond to one standard deviation; the difference in ductal tip number between mutant and wild-type anterior prostate is significant (**, p = 0.0009). B: Altered expression of secretory proteins in Fem1b heterozygous and homozygous anterior prostate (AP), dorsolateral prostate (DLP), ventral prostate (VP), and seminal vesicle (SV). Protein secretions were collected and resolved by SDS-PAGE, with 15 μg protein loaded in each lane. Arrows indicate proteins whose levels decrease in Fem1b heterozygotes and/or homozygotes; asterisks indicate proteins whose levels increase.
Finally, we investigated whether there were possible defects in secretory protein production in the prostate, bulbourethral glands (BUG), and seminal vesicles (SV) of Fem1b mutants. Analysis of total secreted proteins from wild type, heterozygous and homozygous mutant mice (n=3 each) by SDS-PAGE revealed the altered expression of several secretory proteins in the dorsolateral and ventral prostate as well as seminal vesicles (Fig. 6B). Notably, these prostate ductal branching and secretory protein defects observed in Fem1b mutants resemble the phenotypes of Nkx3.1 null mutant mice (Bhatia-Gaur et al., 1999).
Discussion
We have identified a mammalian homolog of C. elegans Fem-1 as a protein expressed in prostate epithelium that interacts with the Nkx3.1 homeodomain protein. We have confirmed this putative interaction using GST pull-down and co-immunoprecipitation analyses, and have shown that Fem1b and Nkx3.1 are co-expressed in prostate epithelial cells as well as in germ cells of the neonatal testis. Moreover, we have shown that Fem1b and Nkx3.1 mutant mice display similar defects in ductal branching morphogenesis and prostate secretory protein expression. These data suggest that Nkx3.1 and Fem1b interact in the regulation of prostate epithelial growth and differentiation, possibly through activities in the same or related pathways.
Previous studies have provided disparate views of the function of Fem1 proteins in mammalian cells and embryos. For example, a putative homeodomain transcription factor 1 (Phtf1) has been shown to associate with Fem1b in male germ cells, and can recruit Fem1b to the endoplasmic reticulum membrane (Oyhenart et al., 2005). Other studies have reported that C. elegans Fem-1 and human FEM1B induce apoptosis when overexpressed in breast carcinoma cells, consistent with an interaction of mouse Fem1b protein with Fas and tumor necrosis factor receptor 1, two members of the “death receptor family” (Chan et al., 1999; Chan et al., 2000). However, analyses of mouse Fem1c in retinal ganglion cell death have not supported a role for Fem1c in apoptosis (Schlamp et al., 2004); a similar lack of apoptotic function was shown in the male testis (Oyhenart et al., 2005); moreover, our immunohistochemical analysis also fails to demonstrate altered apoptosis in the testes of Fem1b mutant mice. Finally, independent studies of Fem1b null mutant mice have indicated its expression in pancreatic islets and its function in insulin secretion and plasma glucose levels, suggesting potential links between sex-determining genes and insulin pathway regulation (Lu et al., 2005). Perhaps relevant for these findings, we have noted a slight decrease in body weight in Fem1b mutants (X. W. and M. M. S., unpublished observations), but have not characterized this defect further. Overall, the relatively weak phenotypes observed in Fem1b mice might be due to partial functional redundancy with Fem1c, which has a similar expression pattern as Fem1b in adult tissues (Ventura-Holman et al., 1998; Ventura-Holman et al., 2003), and has minimal null mutant phenotypes (Schlamp et al., 2004).
Although Fem1b null mutants are homozygous viable and overtly healthy, they display specific defects in prostate organogenesis and secretory function. These prostate defects observed in Fem1b mutant mice are reminiscent of the defects found in young Nkx3.1 mutant mice, consistent with a functional protein interaction between Fem1b and Nkx3.1. In particular, Fem1b mutants display a 30% reduction in ductal tip number in the anterior prostate, with less significant differences in the ventral and dorsolateral prostate, which is comparable to the 25-35% reduction in ductal tip number observed in all three prostate lobes in Nkx3.1 mutants (Bhatia-Gaur et al., 1999). In addition, at least one secretory protein that is altered in the Fem1b ventral prostate (Fig. 6B, arrow in VP panel) is also affected in Nkx3.1 mutants (Bhatia-Gaur et al., 1999; Economides and Capecchi, 2003); based on its molecular weight and abundance, this protein appears to correspond to the prostatic spermine-binding protein, which is a major component of ventral prostate secretions (Mills et al., 1987). Notably, the secretory protein defects suggest that the prostate phenotype of Fem1b mice is the consequence of incomplete/mis-specified epithelial differentiation during organogenesis. However, in contrast with Nkx3.1 mutants, we have not observed epithelial hyperplasia in Fem1b mutants up to 1 year of age (X. W. and M. M. S., unpublished observations).
Based on our findings, we can envision two possible roles for an interaction between Nkx3.1 and Fem1b. One possible model is that Nkx3.1 may modify the function of Fem1b in a multi-subunit E3 ubiquitin ligase complex (Starostina et al., 2007). Notably, although there is no mammalian ortholog of Fem3, human Fem1b has been isolated as part of a Cul2-containing E3 ubiquitin ligase complex (Kamura et al., 2004), which may potentially also contain the human ortholog of Fem2 (Tan et al., 2001). Another possibility is that Nkx3.1 activity may itself be altered through ubiquitination mediated by a Fem1b-containing Cul2 complex. Although such ubiquitination might lead to proteasomal degradation of Nkx3.1, it could alternatively result in modification of protein localization or function, leading to increased Nkx3.1 activity (reviewed in (Mukhopadhyay and Riezman, 2007)). In this regard, it is notable that Nkx3.1 can indeed be ubiquitylated on residues within its homeodomain by the TOPORS E3 ubiquitin ligase (Guan et al., 2008).
Our results suggest that regulatory genes such as Nkx3.1 that play critical roles in the specification of secondary sexual organs can also be considered as downstream effectors in the sex determination pathway. Although mechanisms of sex determination appear to be highly diverse throughout the animal kingdom, there is evidence for evolutionary conservation of regulatory genes in the generation of sexual dimorphism. In particular, the Drosophila doublesex and C. elegans mab-3 genes encode proteins with a non-classical “zinc finger” DNA binding motif (DM domain) that regulate sex-specific differentiation (Baker and Ridge, 1980; Shen and Hodgkin, 1988; Raymond et al., 1998). In vertebrates, several members of the family of DM domain-containing (Dmrt) proteins are involved in regulation of gonadal development and germline differentiation (Raymond et al., 2000; Balciuniene et al., 2006; Kawamata and Nishimori, 2006; Hong et al., 2007; Kim et al., 2007). These similarities suggest that there may be greater evolutionary conservation of sex-determining genes than hitherto recognized.
Experimental Procedures
Yeast two-hybrid screening
Yeast two-hybrid screening was performed using the LexA interaction trap methodology (Golemis et al., 2008). To generate a LexA-fused bait protein, full-length human NKX3.1 cDNA was subcloned into the pEG202 vector using the primers: 5′-CGG AAT TCC TCA GGG TTC CGG AGC C -3′ and 5′-CCG CTC GAG TTA CCA AAA AGC TGG GCT -3′. The resulting pEG202-NKX3.1 was transformed into the yeast strain EGY48, and the absence of auto-activation of lacZ and LEU2 reporter genes was confirmed. For library screening, EGY48 cells containing pEG202-NKX3.1 and the reporter plasmid pSH18-34 were transformed with a human adult prostate cDNA library constructed in the GAL1-AD fusion vector pJG4-5 (DupLEX-A, OriGene Technologies). Transformants were selected for LEU2 expression on YNB (gal)-his-ura-trp-leu plates and tested for galactose-dependence and lacZ expression on YNB (glu)-his-ura-trp-leu, YNB (gal)-his-ura-trp-leu, YNB (glu)-his-ura-trp+X-gal and YNB (gal)-his-ura-trp+X-gal plates. The specificity of candidate interactions was further assessed by a mating test, in which the recovered library plasmid DNA was transformed into RFY206 and pEG202-NKX3.1 together with the reporter plasmid pSH18-34 into EGY48, followed by mating of the two haploid strains to examine galactose-dependence and lacZ expression.
Protein interaction analysis
35S-labeled partial Fem1b protein was obtained by in vitro transcription/translation using TNT Coupled Reticulocyte Lysate System (Promega). GST (glutathione-S-transferase) fusion proteins expressed from the pGEX-2T vector (Amersham Pharmacia) were generated and purified using standard methods. GST-fusion protein pull-down assays were performed using 20 μl glutathione-agarose beads, 2 μg GST or GST-fusion proteins on glutathione-agarose beads, in vitro translated and 35S-labeled Fem1b, and 200 μl binding buffer (20 mM Tris-HCl, pH 7.9 at 4°C; 0.2 mM EDTA, 0.1 M NaCl, 0.2% NP-40). The mixture was incubated at 4°C for 1 hour, and washed with 20 mM Tris-HCl, pH 7.9, 0.2 mM EDTA, 0.5 M NaCl, 0.4% NP-40, followed by SDS-PAGE and autoradiography.
A full-length clone of mouse Fem1b was isolated from cDNA obtained from adult testis of Swiss Webster mice. Full-length Fem1b cDNA was isolated from total RNA of the testes from 2-month-old Swiss Webster mice, and cloned with 3′ myc epitope tags in pcDNA3.1 using primers: 5′-CGG AAT TCG CCG CCA TGG AGG GCC TGC CTG GCT ATG-3′ and 5′-CCG CTC GAG TTA CAG ATC TTC TTC AGA AAT AAG TTT TTG TTC ATG AAA TCC AAC AAA CTC TTC-3′. Human embryonic kidney (HEK) 293T cells were transfected using Lipofectamine and Plus reagents (Invitrogen) and lysed with RIPA buffer (0.1% SDS, 1% sodium deoxycholate, 1% Triton X-100, 150 mM NaCl, 10 mM Tris-HCl, pH 7.5, and 1 mM EDTA) 48 hours after transfection. Supernatants from cell lysates were incubated with anti-FLAG (Sigma) or anti-myc antibody coupled to agarose beads (Sigma) at 4°C overnight. Protein detection was performed with SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Quantitative real-time RT-PCR analysis
Total RNA was isolated from mouse tissues using Trizol (Invitrogen); first-strand cDNA synthesis was performed using standard protocols. For real-time RT-PCR analysis, LUX (Invitrogen) primers labeled with FAM or JOE were added with 25 μl of PCR Supermix (Invitrogen), 2 μl of cDNA, 1 mM MgCl2 and ROX reference dye. The following primers were used: for Fem1b, unlabeled 5′-GCC TTC ACA GCT TCA TTG AGG T-3′ and FAM-labeled 5′-GAC TTG AAT CTG CTT CGG TTT GCT CAA G-3′; for β-actin, unlabeled 5′-GGT TGG CCT TAG GGT TCA GG-3′ and JOE-labeled 5′-CAC GCC ACC TTC TAC AAT GAG CTG CGT G-3′. Results for Fem1b levels were normalized to β-actin levels. The reaction was carried out on an Mx4000 thermal cycler and data were analyzed using Mx4000 software (Stratagene).
Gene targeting
A 32P-labeled 1 kb murine Fem1b cDNA subcloned from an IMAGE clone (ID #4937245) was used to screen a 129Sv mouse BAC DNA library (BACPAC Resources), resulting in isolation of 4 genomic clones that contain the coding region. A 5.1 kb NotI-XhoI fragment and a 4.5 kb BamHI-KpnI fragment were subcloned into pPNTloxp (Shalaby et al., 1995). ES cell culture, gene targeting, and blastocyst injection was performed as previously described (Ding et al., 1998). 1 out of 96 clones was identified as targeted. Thirteen chimeric males were obtained, and used to obtain germline transmission of the targeted allele. The Fem1b null mutation has been maintained in a C57BL6-129SvEvTac mixed background and on an inbred 129SvEvTac background, with no differences in phenotype observed.
Genotyping was performed by Southern blotting or by PCR using genomic DNA from tails. Primers for PCR genotyping were following: for wild type allele: 5′-GGC CAT GGG ATG ACA CCA TTG AAG GTG-3′ and 5′-CAT TGA GGT GTA TCA TCT GAG AGA AG-3′; for mutant allele: 5′-CTG CCG CGC TGT TCT CCT CTT CCT -3′ and 5′-ACA CCC AGC CGG CCA CAG TCG-3′.
Histology, in situ hybridization, and immunohistochemistry
Hematoxylin-eosin staining was performed using standard protocols on 6 μm paraffin sections. For in situ hybridization, digoxigenin-labeled RNA antisense probes were made by reverse transcription of cDNA sequences encoding mouse Nkx3.1 (Bhatia-Gaur et al., 1999), or mouse Fem1b, corresponding to nt 543-1519 within the second exon. Tissues were fixed using 4% paraformaldehyde at 4°C overnight, and embedded in OCT compound. Section in situ hybridization was performed as previously described (Bhatia-Gaur et al., 1999).
For immunohistochemical staining, tissues were fixed in 10% formalin overnight and embedded in paraffin. The following primary antibodies were used: androgen receptor rabbit polyclonal antiserum (Affinity Bioreagents; 1:250 dilution), Nkx3.1 rabbit polyclonal antiserum (Kim et al., 2002b) (1:6000 dilution), PCNA mouse monoclonal antibody (Santa Cruz, clone PC10; 1:500 dilution), p63 mouse monoclonal antibody (Santa Cruz, clone 4A4; 1:600 dilution) and caspase-3 rabbit monoclonal antibody (BD Biosciences, clone C92-605; 1:100 dilution). In brief, 6 μm paraffin sections were dewaxed in xylene, blocked in 10% goat serum or with blocking reagents provided in the M.O.M. kit (Vector Labs) for mouse primary antibodies, then incubated together with primary antibodies overnight at 4°C. Secondary antibodies were obtained from Vectastain ABC kits (Vector Labs) and diluted 1:250 or 1:500. Signal was enhanced using the Vectastain ABC system and visualized with NovaRed Substrate Kit (Vector Labs). Stained sections were imaged using a Nikon Eclipse E800 microscope equipped with a Nikon DXM1200 digital camera.
Prostate phenotype analysis
Ductal tip numbers were quantitated as previously described (Bhatia-Gaur et al., 1999). Secretory proteins were isolated by a published method, with minor modifications (Economides and Capecchi, 2003). In brief, tissues were washed in PBS and minced in PBS containing protease inhibitor cocktail (Invitrogen). The minced tissue was pelleted by centrifugation at 14,000 rpm for 5 minutes at 4°C, and the supernatants analyzed by SDS-PAGE and Commassie blue staining.
Acknowledgments
We thank Leif Austenberg for generating Nkx3.1 deletion constructs, Janet Rossant for providing plasmids, and Marianna de Julio for comments on the manuscript. This work was supported by NIH grants CA67501 (C. A.-S.) and CA115985 (M. M. S.).
References
- Ahringer J, Kimble J. Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3′ untranslated region. Nature. 1991;349:346–348. doi: 10.1038/349346a0. [DOI] [PubMed] [Google Scholar]
- Baker BS, Ridge KA. Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics. 1980;94:383–423. doi: 10.1093/genetics/94.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciuniene J, Bardwell VJ, Zarkower D. Mice mutant in the DM domain gene Dmrt4 are viable and fertile but have polyovular follicles. Mol Cell Biol. 2006;26:8984–8991. doi: 10.1128/MCB.00959-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Young P, Norton CR, Gridley T, Cardiff RD, Cunha GR, Abate-Shen C, Shen MM. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Tan KO, Zhang L, Yee KS, Ronca F, Chan MY, Yu VC. F1Aalpha, a death receptor-binding protein homologous to the Caenorhabditis elegans sex-determining protein, FEM-1, is a caspase substrate that mediates apoptosis. J Biol Chem. 1999;274:32461–32468. doi: 10.1074/jbc.274.45.32461. [DOI] [PubMed] [Google Scholar]
- Chan SL, Yee KS, Tan KM, Yu VC. The Caenorhabditis elegans sex determination protein FEM-1 is a CED-3 substrate that associates with CED-4 and mediates apoptosis in mammalian cells. J Biol Chem. 2000;275:17925–17928. doi: 10.1074/jbc.C000146200. [DOI] [PubMed] [Google Scholar]
- Chen H, Nandi AK, Li X, Bieberich CJ. NKX-3.1 interacts with prostate-derived Ets factor and regulates the activity of the PSA promoter. Cancer Res. 2002;62:338–340. [PubMed] [Google Scholar]
- Chin-Sang ID, Spence AM. Caenorhabditis elegans sex-determining protein FEM-2 is a protein phosphatase that promotes male development and interacts directly with FEM-3. Genes Dev. 1996;10:2314–2325. doi: 10.1101/gad.10.18.2314. [DOI] [PubMed] [Google Scholar]
- Ding J, Yang L, Yan YT, Chen A, Desai N, Wynshaw-Boris A, Shen MM. Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature. 1998;395:702–707. doi: 10.1038/27215. [DOI] [PubMed] [Google Scholar]
- Economides KD, Capecchi MR. Hoxb13 is required for normal differentiation and secretory function of the ventral prostate. Development. 2003;130:2061–2069. doi: 10.1242/dev.00432. [DOI] [PubMed] [Google Scholar]
- Golemis EA, Serebriiskii I, Finley RL, Jr, Kolonin MG, Gyuris J, Brent R. Interaction trap/two-hybrid system to identify interacting proteins. Curr Protoc Mol Biol Chapter 20. 2008 doi: 10.1002/0471142727.mb2001s46. Unit 20 21. [DOI] [PubMed] [Google Scholar]
- Guan B, Pungaliya P, Li X, Uquillas C, Mutton LN, Rubin EH, Bieberich CJ. Ubiquitination by TOPORS regulates the prostate tumor suppressor NKX3.1. J Biol Chem. 2008;283:4834–4840. doi: 10.1074/jbc.M708630200. [DOI] [PubMed] [Google Scholar]
- Hansen D, Pilgrim D. Sex and the single worm: sex determination in the nematode C. elegans. Mech Dev. 1999;83:3–15. doi: 10.1016/s0925-4773(99)00024-6. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics. 1986;114:15–52. doi: 10.1093/genetics/114.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CS, Park BY, Saint-Jeannet JP. The function of Dmrt genes in vertebrate development: it is not just about sex. Dev Biol. 2007;310:1–9. doi: 10.1016/j.ydbio.2007.07.035. [DOI] [PubMed] [Google Scholar]
- Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata M, Nishimori K. Mice deficient in Dmrt7 show infertility with spermatogenic arrest at pachytene stage. FEBS Lett. 2006;580:6442–6446. doi: 10.1016/j.febslet.2006.10.066. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Bhatia-Gaur R, Banach-Petrosky WA, Desai N, Wang Y, Hayward SW, Cunha GR, Cardiff RD, Shen MM, Abate-Shen C. Nkx3.1 mutant mice recapitulate early stages of prostate carcinogenesis. Cancer Res. 2002a;62:2999–3004. [PubMed] [Google Scholar]
- Kim MJ, Cardiff RD, Desai N, Banach-Petrosky WA, Parsons R, Shen MM, Abate-Shen C. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci USA. 2002b;99:2884–2889. doi: 10.1073/pnas.042688999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Namekawa SH, Niswander LM, Ward JO, Lee JT, Bardwell VJ, Zarkower D. A mammal-specific Doublesex homolog associates with male sex chromatin and is required for male meiosis. PLoS Genet. 2007;3:e62. doi: 10.1371/journal.pgen.0030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Capel B. Balancing the bipotential gonad between alternative organ fates: a new perspective on an old problem. Dev Dyn. 2006;235:2292–2300. doi: 10.1002/dvdy.20894. [DOI] [PubMed] [Google Scholar]
- Kimble J, Edgar L, Hirsh D. Specification of male development in Caenorhabditis elegans: the fem genes. Dev Biol. 1984;105:234–239. doi: 10.1016/0012-1606(84)90279-3. [DOI] [PubMed] [Google Scholar]
- Kuwabara PE. Developmental genetics of Caenorhabditis elegans sex determination. Curr Top Dev Biol. 1999;41:99–132. doi: 10.1016/s0070-2153(08)60271-9. [DOI] [PubMed] [Google Scholar]
- Lu D, Ventura-Holman T, Li J, McMurray RW, Subauste JS, Maher JF. Abnormal glucose homeostasis and pancreatic islet function in mice with inactivation of the Fem1b gene. Mol Cell Biol. 2005;25:6570–6577. doi: 10.1128/MCB.25.15.6570-6577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JS, Needham M, Parker MG. Androgen regulated expression of a spermine binding protein gene in mouse ventral prostate. Nucleic Acids Res. 1987;15:7709–7724. doi: 10.1093/nar/15.19.7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Oyhenart J, Benichou S, Raich N. Putative homeodomain transcription factor 1 interacts with the feminization factor homolog fem1b in male germ cells. Biol Reprod. 2005;72:780–787. doi: 10.1095/biolreprod.104.035964. [DOI] [PubMed] [Google Scholar]
- Pilgrim D, McGregor A, Jackle P, Johnson T, Hansen D. The C. elegans sex-determining gene fem-2 encodes a putative protein phosphatase. Mol Biol Cell. 1995;6:1159–1171. doi: 10.1091/mbc.6.9.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CS, Murphy MW, O'Sullivan MG, Bardwell VJ, Zarkower D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000;14:2587–2595. doi: 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–695. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- Schlamp CL, Thliveris AT, Li Y, Kohl LP, Knop C, Dietz JA, Larsen IV, Imesch P, Pinto LH, Nickells RW. Insertion of the beta Geo promoter trap into the Fem1c gene of ROSA3 mice. Mol Cell Biol. 2004;24:3794–3803. doi: 10.1128/MCB.24.9.3794-3803.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Brand T, Zweigerdt R, Arnold H. Targeted disruption of the Nkx3.1 gene in mice results in morphogenetic defects of minor salivary glands: parallels to glandular duct morphogenesis in prostate. Mech Dev. 2000;95:163–174. doi: 10.1016/s0925-4773(00)00355-5. [DOI] [PubMed] [Google Scholar]
- Sciavolino PJ, Abrams EW, Yang L, Austenberg LP, Shen MM, Abate-Shen C. Tissue-specific expression of murine Nkx3.1 in the male urogenital system. Dev Dyn. 1997;209:127–138. doi: 10.1002/(SICI)1097-0177(199705)209:1<127::AID-AJA12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, Breitman ML, Schuh AC. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- Shen MM, Abate-Shen C. Roles of the Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Dev Dyn. 2003;228:767–778. doi: 10.1002/dvdy.10397. [DOI] [PubMed] [Google Scholar]
- Shen MM, Hodgkin J. mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell. 1988;54:1019–1031. doi: 10.1016/0092-8674(88)90117-1. [DOI] [PubMed] [Google Scholar]
- Spence AM, Coulson A, Hodgkin J. The product of fem-1, a nematode sex-determining gene, contains a motif found in cell cycle control proteins and receptors for cell-cell interactions. Cell. 1990;60:981–990. doi: 10.1016/0092-8674(90)90346-g. [DOI] [PubMed] [Google Scholar]
- Starostina NG, Lim JM, Schvarzstein M, Wells L, Spence AM, Kipreos ET. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev Cell. 2007;13:127–139. doi: 10.1016/j.devcel.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan KM, Chan SL, Tan KO, Yu VC. The Caenorhabditis elegans sex-determining protein FEM-2 and its human homologue, hFEM-2, are Ca2+/calmodulin-dependent protein kinase phosphatases that promote apoptosis. J Biol Chem. 2001;276:44193–44202. doi: 10.1074/jbc.M105880200. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Komuro I, Inagaki H, Jenkins NA, Copeland NG, Izumo S. Nkx3.1, a murine homolog of Drosophila bagpipe, regulates epithelial ductal branching and proliferation of the prostate and palatine glands. Dev Dyn. 2000;219:248–260. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1054>3.3.co;2-5. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Lyons GE, Izumo S. Expression of the Nkx3.1 homobox gene during pre and postnatal development. Mech Dev. 1999;85:179–182. doi: 10.1016/s0925-4773(99)00084-2. [DOI] [PubMed] [Google Scholar]
- Ventura-Holman T, Lu D, Si X, Izevbigie EB, Maher JF. The Fem1c genes: conserved members of the Fem1 gene family in vertebrates. Gene. 2003;314:133–139. doi: 10.1016/s0378-1119(03)00712-1. [DOI] [PubMed] [Google Scholar]
- Ventura-Holman T, Maher JF. Sequence, organization, and expression of the human FEM1B gene. Biochem Biophys Res Commun. 2000;267:317–320. doi: 10.1006/bbrc.1999.1942. [DOI] [PubMed] [Google Scholar]
- Ventura-Holman T, Seldin MF, Li W, Maher JF. The murine fem1 gene family: homologs of the Caenorhabditis elegans sex-determination protein FEM-1. Genomics. 1998;54:221–230. doi: 10.1006/geno.1998.5569. [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Koopman P. The makings of maleness: towards an integrated view of male sexual development. Nat Rev Genet. 2006;7:620–631. doi: 10.1038/nrg1903. [DOI] [PubMed] [Google Scholar]