Abstract
A variety of DNA sequence motifs including inverted repeats, minisatellites, and the χ recombination hotspot, have been reported in association with gene conversion in human genes causing inherited disease. However, no methodical statistically-based analysis has been performed to formalize these observations. We have performed an in silico analysis of the DNA sequence tracts involved in 27 non-overlapping gene conversion events in 19 different genes reported in the context of inherited disease. We found that gene conversion events tend to occur within (C+G)- and CpG-rich regions and that sequences with the potential to form non-B-DNA structures, and which may be involved in the generation of double-strand breaks that could in turn serve to promote gene conversion, occur disproportionately within maximal converted tracts and/or short flanking regions. Maximal converted tracts were also found to be enriched (p<0.01) in a truncated version of the χ-element (a TGGTGG motif), immunoglobulin heavy chain class switch repeats, translin target sites and several novel motifs including (or overlapping) the classical meiotic recombination hotspot, CCTCCCCT. Finally, gene conversions tend to occur in genomic regions that have the potential to fold into stable hairpin conformations. These findings support the concept that recombination-inducing motifs, in association with alternative DNA conformations, can promote recombination in the human genome.
Keywords: gene conversion, recombination-inducing sequence motifs, non-B DNA
Introduction
Homologous recombination is generally thought to be initiated by double-strand breaks (DSBs), resulting in either gene conversion or crossovers (Chen et al. 2007). Gene conversion occurs frequently between tandemly linked homologous DNA sequences and involves the non-reciprocal transfer of DNA from a ‘donor’ to an ‘acceptor’ sequence. When such transfers inactivate functional human genes, pathological consequences can ensue. To date, a large number of homologous recombination ‘hotspots’ have been described in yeast, mice and humans (Jeffreys et al. 2004; Nishant and Rao 2006). In addition, a variety of DNA sequences, including direct repeats, inverted repeats (sometimes incorrectly termed palindromes), minisatellite repeats, the χ recombination hotspot, and alternating purine-pyrimidine tracts with Z-DNA-forming potential, have been noted in association with gene conversion in human genes (Kilpatrick et al. 1984; Flanagan et al. 1984; Giordano et al. 1997; Chen and Férec 2000; Lopez-Correa et al. 2001; Lee et al. 2002; Rozen et al. 2003; Hallast et al. 2005; Wolf et al. 2009). However, the failure to identify any unique DNA sequence feature associated with ‘hotspot’ activity has fuelled speculation that the determinants of homologous recombination might not only be numerous but also ‘fuzzy’ (Jeffreys et al. 2004; Nishant and Rao 2006). Moreover, the reported gene conversion ‘hotspots’ could be remote from the DSB-initiating sites, since it is the sites of recombinational resolution rather than recombinational initiation that have almost invariably been investigated (Jeffreys et al. 2004).
Recent in vitro studies have revealed that simple repetitive DNA sequences known to be capable of adopting non-B DNA conformations (such as slipped structures, triplexes, tetraplexes, etc) are highly mutagenic and prone to breakage (Bacolla et al. 2004; Bacolla and Wells 2004; Wang and Vasquez 2006). These findings have been augmented by evidence for a hairpin processing activity possessed by the Artemis/DNA-PKcs complex and Holliday-junction resolvase, both in the context of V(D)J recombination (Ma et al. 2002; Raghavan et al. 2006; Lieber et al. 2006) and in the delivery of recombinant adeno-associated virus used for gene therapy (Inagaki at al. 2007). Thus, it may be that it is the ability of a given DNA sequence to adopt a non-B DNA conformation, rather than the DNA sequence per se in the orthodox right-handed Watson-Crick B-form, that induces chromosomal DSBs. Recently, this postulate has received broad support from the convergence of biochemical, genetic and genomic studies in the context of gross genomic deletions, inversions, duplications and translocations (reviewed in Wells 2007).
We speculated that DNA sequences with the potential to form non-B structures might also play an important role in homologous recombination leading to gene conversion. To test this postulate, we employed as a model system a series of known examples of human pathological mutations resulting from gene conversion events that involve the transfer of a short sequence tract from a donor to an acceptor gene. The advantage of this approach was that the extents of the maximal converted tracts (MaxCTs) and minimal converted tracts (MinCTs) (see Figure 1 for term definitions) associated with such pathological events can usually be accurately determined and annotated. To verify the prevailing view that the homologous recombination sites of DSBs that initiate gene conversion reside within MaxCTs (Chen et al. 2007) and to explore their precise location, a series of regions including or spanning the MinCTs and MaxCTs were screened for the presence of specific DNA sequence motifs and for sequences capable of adopting non-B DNA conformations. The basic assumption made in this analysis was that a marked overrepresentation of such DNA sequences either within the gene conversion tracts themselves, or within their short flanking regions, could imply their involvement in DSB formation.
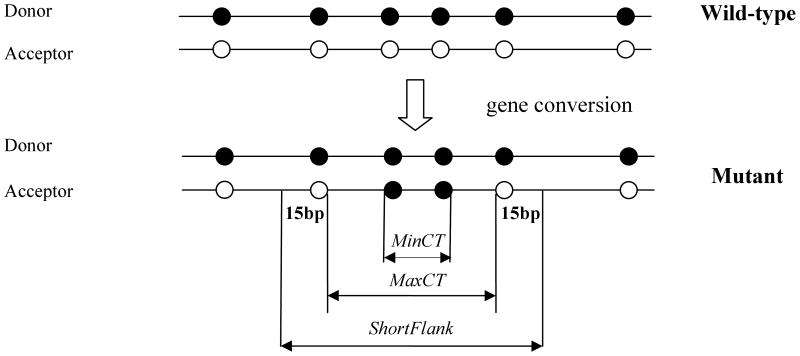
Figure 1.
Schematic representation of an archetypal gene conversion event showing the relative locations of the maximal converted tract (MaxCT), the minimal converted tract (MinCT) and the region spanning MaxCT and including short (15bp) regions flanking MaxCT.
Materials & Methods
Sequence data derived from pathological gene conversion events
Four different sequence datasets were employed comprising minimal converted tracts (MinCTs), maximal converted tracts (MaxCTs) [see Figure 1 for term definitions], and regions spanning MaxCTs but including either ±15 or ±150 bp flanking sequences (henceforth termed ShortFlank and LongFlank datasets respectively). DNA sequence data were derived from 27 non-overlapping interlocus gene conversion events in 19 different genes reported to cause human inherited disease (Table 1, Supp. Figure S1). Whereas the majority of gene conversion events were listed in the collation of Chen et al. (2007), an additional eight cases associated respectively with congenital adrenal hyperplasia (Globerman et al. 1988), increased CYP3A7 gene expression in adult liver and intestine (Kuehl et al. 2001), agammaglobulinemia (Conley et al. 1999) and conversion events in the GYPA (Huang et al. 2000), HBA1 (Law et al. 2006), Sec1 (Soejima et al. 2008), CD46 (Fremeaux-Bacchi et al. 2006) and KRT17 (Hashiguchi et al. 2002) genes were included.
TABLE 1. Inter-locus gene conversion events causing human genetic disease used in the analysis.
| Disease/phenotype | Donor gene* |
Acceptor gene* |
Chromosomal localization |
Mutation | Converted tract length (bp) |
Genic location of | Reference** | |
|---|---|---|---|---|---|---|---|---|
| MaxCT | MinCT | |||||||
| Atypical hemolytic uremic syndrome | CFHR1 | CFH | 1q32 | c.[3572C>T;3590T>C] | 19-331 | ORF/3′ | ORF | Heinen et al. 2006 |
| Congenital adrenal hyperplasia | CYP21A1P | CYP21A2 | 6p21.3 | [-209T>C;-198C>T;-189/-188insT] | 21-155 | 5′ | 5′ | Lee et al. 2006 |
| Intron 2 conversion | 237-520 | ORF | ORF | Globerman et al. 1988 | ||||
| [1380T>A;1383T>A;1389T>A;IVS6+12_13AC>GT] | 42-210 | ORF | ORF | Higashi et al. 1988 | ||||
| [1688G>T;1767_1768insT] | 80-202 | ORF | ORF | Friaes et al. 2006 | ||||
| Increased CYP3A7 expression in adult liver and intestine | CYP3A4 | CYP3A7 | 7q21-q22.1 | Promoter conversion | 60-120 | 5′ | 5′ | Kuehl et al. 2001 |
| Increased 18-hydroxycortisol production | CYP11B1 | CYP11B2 | 8q21-q22 | Conversion of two nucleotides separated by two bases in exon 8 | 4-56 | ORF | ORF | Nicod et al. 2004 |
| Autosomal dominant cataract | CRYBP1 | CRYBB2 | 22q11.23 | c.[475C>T;483C>T] | 9-104 | ORF | ORF | Vanita et al. 2001 |
| Neural tube defects | FOLR1P | FOLR1 | 11q13.3-q14.1 | 7497_7662 including 13 discriminant nucleotides | 166-215 | ORF/3′ | ORF/3′ | De Marco et al. 2000 |
| Gaucher disease | GBAP | GBA | 1q22 | [L444P;A456P;V460V] | 50-475 | ORF | ORF | Eyal et al. 1990; Hong et al. 1990 |
| Short stature | GH2 | GH1 | 17q22-q24 | Conversion involving 12 discriminant nucleotides in the promoter region | 40-218 | 5′ | 5′ | Millar et al. 2003 |
| Novel St glycophorin | GYPE | GYPA | 4q28-q31 | GPA-E-A hybrid gene | 391-431 | ORF | ORF | Huang et al. 2000 |
| Microcytosis | HBA2 | HBA1 | 16p13.3 | α121 patchwork | 8-237 | ORF | ORF | Law et al. 2006 |
| Agammaglobulinemia | IGLL3 | IGLL1 | 22q11.23 | c.[393T>C;420T>C;425C>T] | 33-152 | ORF | ORF | Minegishi et al. 1998 |
| Conversion of exon 2 | 9-80 | ORF | ORF | Conley et al. 1999 | ||||
| Chronic granulomatous disease | NCF1B or NCF1C | NCF1 | 7q11.23 | [ΔGT;ins20bp] | 247-423 | ORF | ORF | Vazquez et al. 2001 |
| Autosomal dominant polycystic kidney disease | ? | PKD1 | 16p13.3 | [8446T>G;8490T>C;8493G>C;8498C>G;8502T>C] | 57-126 | ORF | ORF | Watnick et al. 1998 |
| [8639G>T;8651G>A;8658T>C;8662C>T] | 24-230 | ORF | ORF | Inoue et al. 2002 | ||||
| Chronic pancreatitis | PRSS2 | PRSS1 | 7q34 | Conversion involving 22 discriminant nucleotides | 289-457 | ORF | ORF | Teich et al. 2005 |
| Shwachman-Bodian-Diamond syndrome | SBDSP | SBDS | 7q11.22 | c.[129-443A>G;129-433G>A] | 11-104 | ORF | ORF | Nicolis et al. 2005 |
| c.[141C>T;183_184TA>CT;201A>G;258+2T>C]d | 120-398 | ORF | ORF/3′ | Nicolis et al. 2005 | ||||
| Sec1-FUT2-Sec1 hybrid allele | FUT2 | Sec1 | 19q13.3 | Conversion involving exonic sequence | 54-275 | ORF | ORF | Soejima et al. 2008 |
| von Willebrand disease | VWFP | VWF | 22q11.22-q11.23 (VWFP)/12p13.3 (VWF) | [IVS27-45C>T;IVS27-36C>T;3686T>G;3692A>C] | 63-131 | ORF | ORF | Eikenboom et al. 1998 |
| c.[3789G>A;3797C>T;3835G>A] | 47-195 | ORF | ORF | Eikenboom et al. 1994 | ||||
| c.[3931C>T;3951C>T;4027A>G;4079T>C;4105T>A] | 175-297 | ORF | ORF | Surdhar et al. 2001 | ||||
| Atypical hemolytic uremic syndrome | CR1L | CD46 | 1q32 | D151N + Y155D | 13-153 | ORF | ORF | Fremeaux-Bacchi et al. 2006 |
| Pachyonychia congenita type 2 | KRT17P3 | KRT17 | 17q21.2 | 452G>A and 457T>C | 6-170 | ORF | ORF | Hashiguchi et al. 2002 |
The majority of entries have been taken from an earlier compilation of characterized gene conversion events viz. Table 1 of Chen et al. (2007). Additional entries are shown in bold type.
DNA sequences spanning donor and acceptor genes were retrieved from the reference assembly and are given in Supp. Figure S1.
References are listed in Supp. References
Clearly, if all reported pathological gene conversion events had simply been collated without regard to their frequency of occurrence, the dataset would have included a number of identical DNA sequences which would have been represented multiple times owing to the existence of gene conversion hotspots. This multiple inclusion of specific DNA sequences would then have introduced considerable bias into any subsequent search for sequence motifs involved in promoting gene conversion. Therefore, to avoid this problem we adopted a highly conservative strategy in which no overlapping DNA sequences were allowed within any of the datasets to be analyzed. Thus, in cases where a number of different gene conversion events in the same gene overlapped (e.g. in the GBA, NCF1 and VWF genes), only non-overlapping events with MaxCT≤520 bp were included. The gene conversion events identified in the HBD and OPN1LW genes were also excluded from the analysis since the MinCTs and MaxCTs could not be accurately and unambiguously ascertained.
The locations of MaxCTs and MinCTs were broadly assigned to the following genomic regions: open reading frame (ORF) if the gene conversion region resided within the genomic sequence of a known gene or ORF; ORF/3′ if the gene conversion region started within the genomic sequence of a known gene or ORF but ended within a 3′UTR; 5′/ORF if the gene conversion region began within a 5′UTR but ended within the genomic sequence of a known gene or ORF; 5′ or 3′ if the gene conversion region lay within a 5′ or 3′ flanking region respectively.
Control dataset from an ‘artificial chromosome’
An ‘artificial chromosome’ of ∼7 Mbp was constructed by concatenating the genomic sequences (sense strand) of 100 randomly selected human genes (Supp. Table S1), taken from the complete set of ∼23,000 annotated genes (UCSC hg18 Human Genome Assembly, March 2006), and including the exons and introns plus 1000 bp 5′ flanking region (before the transcriptional start site) and 1000 bp 3′ flanking region (beyond the transcriptional termination site) in each case. Although most of these 100 randomly selected genes have been validated, several of them still have provisional status. For genes characterized by multiple transcripts, the longest transcripts were almost invariably selected. The structure of the artificial chromosome therefore reflects as closely as possible the gene/ORF composition of an actual chromosome albeit without the intergenic regions. The R/Y composition of the transcribed strand of the artificially created chromosome (49%/51%) was found to correlate well with the corresponding nucleotide composition calculated for the human genome as a whole (50%/50%).
Bioinformatics analyses
The DNA sequences from the MaxCT, MinCT and ShortFlank datasets were screened for the presence of 37 DNA sequence motifs of length ≥5 nucleotides (nt) [plus their complements] known to be associated with site-specific cleavage/recombination, high frequency mutation and gene rearrangement (Abeysinghe et al. 2003) as well as various ‘super-hotspot motifs’ found in the vicinity of micro-deletions, micro-insertions and indels (Ball et al. 2005). These DNA sequence datasets were also screened for the presence of 15 additional recombination-associated motifs (plus their complements) identified by Cullen et al. (2002) and a CCTCCCT motif associated with a classical meiotic recombination hotspot (Myers et al. 2005; Myers et al. 2006; Frazer et al., 2007).
In addition, complexity analysis (Gusev et al. 1999) was used to identify perfect direct repeats, inverted repeats and symmetric elements that would be capable of forming non-B DNA structures which are believed to induce DNA breakage (Wells 2007). To this end, the following sequences were sought: direct repeats of length ≥7 nt that were less than 20 nt apart from each other and which could form slipped structures; tetraplexes formed by four GGG, GGGG or GGGGG repeats (termed G-quartets) and separated from each other by up to 5 nt; cruciforms formed by two inverted repeats with the minimum stem size and maximum loop length being set to 7 nt and 20 nt respectively; triplexes formed by two symmetric elements of length ≥7 nt comprising at least 75% R (or Y) bases and separated by up to 20 nt (overlapping direct repeats, inverted repeats or self-symmetrical elements with combined lengths shorter than 14 nt were excluded from the analysis); left-handed Z-DNA formed by at least 6 consecutive RY motifs; triplet repeat sequences of the form GAA•TTC, CGG•CCG and CTG•CAG (the dot separates the complementary strands), of total length ≥9 bp, which are known to induce genetic instability (Napierala et al. 2005; Wells and Ashizawa 2006; Wells et al. 2005; Mirkin 2007). These assumptions about the lengths of the repeats capable of non-B DNA structure formation and their relative locations were derived from parameters observed in in vitro studies and from empirical biochemical data on non-D DNA conformations (Sinden 1994; Bacolla and Wells 2004; Wells 2007; Bacolla et al. 2008).
For each type of analysis, the statistical significance was assessed by comparison with two distinct types of control dataset, generated as follows. Firstly, 1000 control datasets comprising the same number of random sequences as the original dataset, and matching the original dataset in terms of their length and mononucleotide composition were simulated by reshuffling the original sequences. Secondly, for each of the original (case) sequence datasets, 1000 control datasets comprising the same number of sequences as the original dataset, and matching the original dataset in terms of their length and location, were randomly selected from the artificial chromosome. z-scores (Marino-Ramirez et al. 2004) were then calculated for the collection of sequence motifs (plus their complements) and the above-described non-B DNA-forming sequences as follows: where N (R) is the frequency of a specific non-B DNA-forming sequence or sequence motif either in the case dataset or in its matching control dataset generated as described above, and N̅ (R) and V̄ (R) are the mean frequency and its variance estimated from 1000 control datasets. Any sequence/motif with a z-score, calculated for all DNA sequences comprising the case dataset, that exceeded the 99th (99.9th) or 95th (99.5th) percentile of the maximum z-scores found for the corresponding 1000 control datasets, generated either by reshuffling or instead selected at random from the artificial chromosome, was deemed to be statistically significant at the respectively 1% (0.1%) or 5% (0.5%) level. All results were corrected for multiple testing using the Bonferroni correction.
To further validate the reshuffling procedure and to correct for possible asymmetries in nucleotide composition between the case and control datasets, a randomly selected matching dataset of non-gene conversion prone sequences from the artificial chromosome was treated as a mock ‘case dataset’.
The AlignACE3.0 software based on Gibbs sampling (Hughes et al. 2000) available at http://atlas.med.harvard.edu/cgi-bin/alignace.pl was used to search for novel sequence motifs in the MaxCT and MinCT datasets.
The MFold software (Zuker 2003), available at http://mfold.bioinfo.rpi.edu/cgi-bin/dna-form1.cgi, was used to predict the secondary structure of single stranded DNA sequences from the LongFlank dataset. ANOVA tests were performed to assess whether or not there was a significant difference in the mean free energy required to attain the adopted DNA conformations, between the case and control datasets. For each of the single stranded DNA sequences from the LongFlank dataset, 10 matching sequences were generated from the artificial chromosome and their mean free energy was calculated across these 10 generated sequences.
Results and Discussion
Location of gene conversion events
In the case of 21 of the 27 gene conversion events analyzed (Table 1), the MinCT and MaxCT regions were located within the ORF portions of the acceptor gene sequences. In one case (the CFHR1-CFH gene pair), however, the MinCT was confined to within the ORF whereas the MaxCT extended as far as the ORF/3′ portion of the acceptor CFH gene. In 3 other cases (one CYP21A1P-CYP21A2, the CYP3A4-CYP3A7 and GH2-GH1 gene pairs), both the MinCT and MaxCT were located within the promoter (5′) portion of the genes. Hence, >80% of the pathological gene conversion events occurred within the ORF segments of genes, whereas no cases were found within the 5′/ORF or downstream (3′) of the genes.
Gene conversion events tend to occur in (C+G)-rich regions
The basic premise underlying this analysis has been that a marked overrepresentation of certain DNA motifs located within the gene conversion tracts (or within ShortFlank regions spanning MaxCTs) in comparison to 1000 control datasets compiled from the artificial chromosome, could imply their involvement in DSB formation. Although this assumption would appear to have a sound statistical basis, comparison of MinCT, MaxCT and ShortFlank datasets with 1000 matching datasets generated from the artificial chromosome revealed striking asymmetries in their nucleotide compositions. Thus, the relative frequencies of nucleotides T, A, G and C in the MaxCTs from the case datasets, for example, were found to be 23%, 23%, 27% and 27% respectively, whilst the corresponding relative frequencies in the control dataset generated from the artificial chromosome were 31%, 28%, 21% and 20%. These differences were found to be highly significant by means of the χ2-test, with the corresponding p-values for the MinCT, MaxCT and ShortFlank sequences being 5.1×10-11, 6.44×10-47 and 9.2×10-54 respectively.
To determine whether these differences were simply due to the sense strand orientation of the artificial chromosome sequences in contrast to both the sense and antisense strand orientations of the case sequences, the nucleotide frequencies were re-categorized into two groups: (C+G) and (A+T). This analysis revealed that the (C+G) to (A+T) ratio in the MinCT, MaxCT and ShortFlank regions were >1.09 and <0.72 for the case and control sequences, respectively, with the Pearson χ2-test p-values for MinCT, MaxCT and ShortFlank regions being 1.39×10-12, 1.24×10-48 and 1.12×10-55 respectively. Therefore, because the (G+C) and (A+T) frequencies are independent of strand orientation, we conclude that the pathological gene conversion events have tended to occur disproportionately within (C+G)-rich regions.
Site-specific recombination motifs
In the context of assessing the overrepresentation of motifs/repeats in the vicinity of the gene conversion tracts, the observed asymmetries in nucleotide composition render the use of reshuffled sequence controls [which preserve the (C+G)-richness of the original sequences] preferable to those controls generated from the artificial chromosome. Indeed, the use of the artificial chromosome controls may actually increase the number of false positives; (C+G)- or (A+T)-rich motifs could be found to be respectively over-represented and underrepresented as a result of the paucity of (C+G) and relative abundance of (A+T) in the control dataset generated from the artificial chromosome. On the other hand, since analysis of the nucleotide composition of specific motifs used in this study indicated that there are no overall asymmetries in the occurrence of specific nucleotides that were previously observed between the case dataset and controls generated from the artificial chromosome (all nucleotides were found to be equally probable), both types of control were used to assess the overrepresentation of motifs.
Several motifs known to be associated with site-specific cleavage/recombination, high frequency mutation and gene rearrangement (Abeysinghe et al. 2003) were found to be over-represented (p≤0.05) within the MinCTs, MaxCTs and their flanking regions, ShortFlank (Table 2). Specifically, a truncated version of the χ-element, TGGTGG, previously reported as a mutational ‘super-hotspot’ common to micro-deletions, micro-insertions and indels (Ball et al. 2005; Table 2), was found to be significantly over-represented with respect to both control datasets (p<0.01) within MinCTs (6 occurrences), MaxCTs (11 occurrences) and ShortFlank (12 occurrences) regions. [NB. This TGGTGG motif self-evidently comprises two short direct repeats]. The human minisatellite conserved sequence/χ-like element CCWCCWGC (and/or its complement, GCWGGWGG) were found in all three datasets. However, the complementary motif was found to be over-represented in the MaxCTs only at the 5% level using the reshuffling control and at the 1% level for the ShortFlank dataset using both types of controls. Surprisingly, the full-length χ-element, GCTGGTGG, often noted in the vicinity of gene conversion tracts and reported to promote gene conversion (Giordano et al. 1997; Lopez-Correa et al. 2001; Rozen et al. 2003), was observed only once (and only as its complement CCACCAGC) within all three regions; it was found to be over-represented in the MinCTs for both types of control, but only at the 5% level of significance.
TABLE 2. Known motifs and non-B DNA forming sequences found within MinCTs, MaxCTs and their ShortFlank regions.
| Motifs | Consensus sequencea | Number of motifs (its complement) in minimal converted tracts (MinCT) | Number of motifs (its complement) in maximal converted tracts (MaxCT) | Number of motifs (its complement) in regions spanning maximal converted tracts and ±15 bp of flanking sequences (ShortFlank) |
|---|---|---|---|---|
| Site-specific recombination motif | ||||
| Heptamer recombination signal | CACAGTG | 0 (1) | 1 (1) | 1 (1) |
| Nonamer recombination signal | ACAAAAACC | 0 (0) | 0 (0) | 0 (0) |
| Immunoglobulin heavy chainclass switch repeats | GAGCT | 3 (6b) | 9 (15) | 10 (15) |
| TGGGG | 11(2) | 17(16) | 20† (19†) | |
| TGAGC | 1 (4) | 7 (13†) | 6 (13†) | |
| GGGCT | 3 (3) | 15 (9) | 16 (12‡) | |
| GGGGT | 1 (2) | 5 (8) | 6 (10‡) | |
| Translin target sites | ATGCAG | 4 (0) | 6 (0) | 6 (0) |
| GCCCWSSW | 1 (1) | 5‡ (1) | 5 (2) | |
| χ-element | GCTGGTGG | 0 (1) | 0 (1) | 0 (1) |
| Human Fra(X) breakpoint cluster | CGGCGG | 0 (1) | 1(2) | 1 (3‡) |
| Human minisatellite conserved sequence/χ-like element | CCWCCWGC | 1(0) | 2(3†) | 3† (4) |
| Autonomously replicated sequence 2c | WRTTTATTTAW | 1† (0) | 1† (0) | 1† (0) |
| Chinese hamster scaffold attachment site 1c | AATAAAYAAA | 0 (1†) | 0 (1†) | 0 (1†) |
| Classical meiotic recombination hotspotd | CCTCCCT | 0 (0) | 2 (0) | 2 (0) |
| Recurrent sequence motifs | ||||
| Murine LTR recombination hotspot | TGGAAATCC | 1† (0) | 1(0) | 1‡ (0) |
| Deletion hotspot consensus sequence | TGRRKM | 16† (13†) | 35† (37†) | 45† (39†) |
| Hamster and human APRT deletion hotspot | TTCTTC | 2 (4) | 2(5†) | 2 (5†) |
| Hamster deletion hotspot sequence | TGGAG | 6† (5†) | 22 (24) | 25 (25) |
| DNA polymerase arrest site | WGGAG | 8(10†) | 37 (39) | 46 (42) |
| DNA polymerase α frameshift hotspot | CTGGCG | 0(0) | 2 (2‡) | 2 (3‡) |
| DNA polymerase β frameshift hotspot | ACCCWR | 1(3) | 11† (6) | 12† (6) |
| ‘Super-hotspot’ motifs | CCCAG | 6† (6) | 21 (14†) | 25 (16†) |
| TTCWCCCC | 0(2) | 1(2) | 1 (2) | |
| CCACCA | 1(6) | 5† (11) | 5 (12) | |
| GGGACA | 2(2) | 4 (2) | 5 (2) | |
| GCCCCG | 0(2‡) | 3 (3‡) | 3 (3) | |
| AGCTG | 6† (6†) | 12† (15) | 15† (16†) | |
| CCATCT | 0 (2) | 6† (4) | 6 (4) | |
| GGAGAA | 5 (0) | 6 (4) | 7† (4) | |
| Long polypurine/polypyrimidine tracts | R10 | 15† | 28† | 35† |
| Y10 | 4 | 26† | 26† | |
| Non-B DNA forming sequence | ||||
| Tetraplexes | G3-5N1-5G3-5N1-5G3-5N1-7G3-5 | 0 (0) | 0(0) | 0 (0) |
| CTG-motif – slipped structures | (CTG)≥3 | 0 (0) | 1 (0) | 1 (0) |
| GAA-motif – slipped structures | (GAA)≥3 | 1 (0) | 1† (0) | 1† (0) |
| CGG-motif – slipped structures | (CGG)≥3 | 0 (0) | 1 (0) | 1‡ (0) |
| Left-handed Z-DNA | (RY)≥6 | 0 | 1 | 1 |
| Slipped structures | direct repeats | 6 | 15† | 16† |
| Cruciforms | inverted repeats | 3 | 8 | 9 |
| Triplexes | R•Y symmetric elements | 1 | 2 | 2 |
The parentheses show the number of complementary motifs.
R=A/G, Y=C/T, W=A/T, S=G/C, M=A/C, K=G/T, N= any base.
Motifs found to be overrepresented (p≤0.05) by comparison both through reshuffling and with the artificial chromosome are shown in bold.
Motifs listed in Cullen et al. (2002).
Motif discussed in Myers et al. (2005) and Myers et al. (2006).
Motifs found to be overrepresented (p≤0.05) through reshuffling.
Motifs found to be overrepresented (p≤0.05) by comparison with the artificial chromosome.
Two immunoglobulin heavy chain class switch repeats, TGGGG and GGGCT, were found to be over-represented (p<0.01) as direct copies within the MinCTs & MaxCTs and MaxCTs & ShortFlank regions respectively. Motifs complementary to TGGGG and GAGCT were over-represented within the MaxCTs and MaxCTs & ShortFlank regions respectively (p<0.01) in comparison with both types of control (note that TGGGG and GGGGT, and GAGCT and TGAGC represent the same sequence but with different reading frames). Interestingly, the actual number of occurrences of motifs GAGCTc and TGAGCc [the superscript c denotes the complementary sequence] is unchanged when longer sequences (ShortFlank) are analyzed. Analysis of the spatial distributions of the TGGGG motif and its complement, CCCCA, revealed that whereas the majority of the CCCCA motifs occurred in the regions flanking the MinCTs, the majority of the TGGGG motifs were located within the MinCT region (Supp. Figure S2).
One of the translin target sites, ATGCAG, was found to be over-represented (at the 5% level) in all three regions with respect to both types of control; translin is a DNA-binding protein that specifically recognises consensus sequences at the breakpoint junctions of chromosomal translocations, albeit usually involving immunoglobulin/T-cell receptor genes (Abeysinghe et al. 2003; Gajecka et al. 2006). Overall, statistical significance was independent on strand orientation for motifs occurring more frequently (≥12 occurrences), whereas it was frequently associated with strand bias for the more rarely occurring motifs (<12 occurrences). Hence, such strand bias might simply have resulted from the stochastic orientation of repeats along the chromosomes (Bacolla et al., 2008).
Thus, in summary, three types of DNA sequence motif, 1) a truncated version of the χ-element and its relative, the human minisatellite conserved sequence/χ-like element, 2) two immunoglobulin heavy chain class switch repeats and 3) translin target sites were found to be consistently over-represented in either MinCTs, MaxCTs or ShortFlank datasets analysed in our study. These findings were independent of the type of control dataset used.
Recurrent sequence motifs
Several motifs including the ‘hamster deletion hotspot’, TGGAG, and the DNA polymerase arrest site WGGAG and their complements, were found to be over-represented (p≤0.01) in both MaxCTs and ShortFlank regions (Table 2). Analysis of their relative spatial distributions (Supp. Figure S2) indicated that the majority of CTCCA (complement of TGGAG) and CTCCW (complement of WGGAG) motifs occurred in the regions flanking the MinCTs.
Motifs complementary to the DNA polymerase α frameshift hotspot sequence, CTGGCG, found in 2 and 3 copies in MaxCTs and ShortFlank regions, were also over-represented at the 5% and 1% levels respectively but only in comparison with the artificial chromosome. By contrast, the DNA polymerase β frameshift hotspot, ACCCWR, was found to be over-represented in MaxCTs and ShortFlank regions at the 5% level by comparison with the reshuffled controls. MaxCT and ShortFlank regions were enriched (p<0.01) in long R- or Y-tracts but only by comparison with the reshuffled controls.
Several motifs, such as the alternating purine-pyrimidine tract RYRYR, the human Fra(X) breakpoint cluster CGGCGG, and the murine parvovirus recombination hotspot CTWTTY, were found to be underrepresented in MaxCT and ShortFlank regions either in comparison with reshuffled controls and/or an artificial chromosome. On the other hand, a motif complementary to the human Fra(X) breakpoint cluster was overrepresented in ShortFlank regions but only in a comparison made with the artificial chromosome.
Non-B DNA-forming sequences
Sequences capable of non-B DNA slipped structure formation (Table 2; Supp. Figure S1) were found to be significantly over-represented (p<0.001) within MinCTs, MaxCTs and ShortFlank regions when compared with the reshuffled control datasets whereas sequences capable of cruciform structure formation were found to be over-represented (p<0.001) within MaxCTs and ShortFlank regions irrespective of the type of control used. Inspection of these non-B DNA forming sequences indicated that they were also (C+G)-rich, suggesting that (C+G)-richness of the non-B DNA forming sequences may be an important additional feature in rendering such sequences susceptible to gene conversion.
To further validate the reshuffling procedure and to account for (C+G) content, a mock ‘case dataset’ was randomly selected from the artificial chromosome (see Materials and Methods); none of the non-B DNA forming repeats found in the ‘mock case dataset’ were over-represented by comparison with controls generated by reshuffling of the ‘mock’ dataset. In addition, the sequences (GAA)≥3, (CGG)≥3 and (CTG)≥3, present in one copy in at least one of the datasets analyzed and already known to induce genetic instability (Napierala et al. 2005; Wells and Ashizawa 2006; Wells et al. 2005; Mirkin 2007) were found to be over-represented (p<0.01) in MinCTs (GAA) and MaxCTs (CGG) and in both MaxCTs and ShortFlank regions (CTG). By contrast, no triplex-forming (R•Y symmetric elements), tetraplex-forming (G-tetrads) or Z-DNA-forming [(RY)≥6)] motifs were found to be over-represented, in any of the gene conversion regions studied.
Novel motifs
The AlignACE3.0 software was also used to search for novel DNA sequence motifs recurring within the MinCTs and MaxCTs. Motifs sharing at least 5, 6 or 7 positions, with ≤2 positions in the consensus sequence occupied by N (any nucleotide), were considered. The background C+G content was set to 41%, corresponding to that observed in the human genome at large. Sixteen and 23 novel sequence motifs (length ≥5bp) were identified as being overrepresented within the MinCTs and MaxCTs respectively; all were found to be over-represented (in one or both orientations) by comparison with the control dataset generated by reshuffling and with the control dataset derived from the artificial chromosome (p<0.01) (Supp. Tables S2 and S3).
Of the 39 types of motif identified in MinCTs and MaxCTs, 27 contained fragments of known ‘super-hotspots’ for mutation, 21 contained fragments of the immunoglobulin class switch region (Rabbitts 1994; Ohno 1981), 14 included fragments of translin-binding sites (Aoki et al. 1995), 13 contained fragments of the human hypervariable minisatellite core sequence/recombination hotspot (Wahls et al. 1990a) or fragments of the hamster APRT deletion hotspot (Smith and Adair 1996), 10 corresponded to known DNA polymerase frameshift/arrest sites, 5 contained fragments of the PUR protein binding site (Bergemann and Johnson 1992; Smith et al. 1998) or fragments of the classical recombination hotspot CCTCCCCT (Myers et al. 2005; Myers et al. 2006; Frazer et al., 2007), 4 contained fragments of the mariner transposon-like element (Reiter et al. 1996) or fragments of χ/χ-like elements, 2 consisted of (CGG)2 repeats of the human Fra(X) breakpoint cluster, and finally one sequence motif was part of the XY32 R•Y H-palindrome (Rooney and Moore 1995).
Hence, the search for motifs known to occur recurrently at recombination sites (Table 2) and the search for novel motifs (Supp. Tables S2 and S3) were concordant in that they identified a number of similar types of DNA sequence. In addition, the search for novel motifs revealed aptamer-like sequences used during class switch recombination, as well as translin pseudo-binding sites. It is certainly possible that, in these pathological gene conversion events, RAG-associated and translin proteins could have played key roles during the recombination process. By contrast, χ/χ-like elements are likely only to have played a modest role.
Several novel motifs were found to include either a fragment or the entire sequence of the human minisatellite conserved sequence/χ-like element. Specifically, motif CWGSWG was found to be over-represented in both orientations within the MinCTs whereas motifs GGWGGc, CTGGNSc and KGGWGGc were found to be over-represented within the MaxCTs.
One motif, GGSAG, found to be over-represented within MinCTs, and four motifs, GGWGG, SNSWGG, KGGWGG and SNWGNRRSS, found to be over-represented in MaxCTs include either a fragment or the entire sequence of the classical meiotic recombination hotspot CCTCCCT (Myers et al. 2005; Myers et al. 2006.
The second type of information uncovered by the search for novel motifs was the overrepresentation of CpG dinucleotides within the case dataset as compared to the corresponding control generated from the artificial chromosome (χ2-test; corresponding p-values for the MinCT, MaxCT and ShortFlank sequences are 7.5×10-5, 5.1×10-11 and 6.2×10-15 respectively). An association between CpG dinucleotide richness and recombination has been previously reported in different contexts (Kong et al., 2002; Jensen-Seaman et al., 2004; Han et al., 2008; Tsai et al., 2008) as has an association between CpG dinucleotide richness and gene conversion (Högstrand and Böhme 1999). Because CpG dinucleotides may be methylated, these composite observations raise the intriguing possibility of a relationship between CpG methylation and gene conversion.
Numbers of motifs/repeats and the lengths of the corresponding tracts
The lengths of the MinCTs, MaxCTs and ShortFlank regions vary between 4 bp & 289 bp, 56 bp & 520 bp, and 86 bp & 550 bp, respectively. To determine whether the numbers of the various motifs/non-B DNA forming sequences found to be overrepresented in these regions correlated directly with the lengths of the tracts in which they were found, Pearson's correlation was calculated for each of the three datasets. For the majority of motifs found to be overrepresented within the MinCT regions, a strong correlation (r>0.8, p<0.001) was noted between the number of motifs observed and the length of the corresponding tract. One simple explanation is that >30% of the MinCTs have lengths shorter than the motifs or repeats sought. By contrast, motifs/repeats found to be overrepresented in MaxCT and ShortFlank regions did not exhibit any correlation >0.7 between their numbers and tract lengths. For example, the Pearson's correlation coefficient observed between the number of non-B DNA forming sequences and their corresponding tract lengths did not exceed 0.4 for both MaxCT and ShortFlank regions. This indicates that, at least for the MaxCT and ShortFlank regions, enrichment in a given motif/repeat was not dependent upon sequence length.
Stable cruciform structures in LongFlanks
As described above, inverted repeats, which may form hairpin structures, were found to be both abundant and over-represented in the MaxCT and ShortFlank regions (Table 2). To investigate whether these possible secondary structures might have been part of larger and relatively stable DNA conformations encompassing the regions of pathological gene conversion, a comparison was performed of the free energies (–ΔG) required for folding the most thermodynamically stable hairpin structures, of the sequences from the LongFlank dataset with matching controls from the artificial chromosome (see Materials and Methods). The mean –ΔG value for the most stable folded-back hairpin structures from the 27 LongFlank gene conversion cases was -75.7 kcal/mole, whereas the mean free energy for the 270 controls was -53.6 kcal/mole (ANOVA, p=9.5×10-4). This implies that gene conversion has tended to occur within genomic regions that have the potential to fold into stable hairpin conformations (Supp. Figure S3 shows an example). The increased stability of the gene conversion-associated non-B DNA structures may be due to the extended hairpin-stems and/or a greater number of Watson-Crick C•G pairs within the stems, as would be expected from the (G+C)-rich nature of the relevant genomic regions. In summary, our composite analyses revealed that sites of gene conversion frequently comprise recombination hotspots (Hellmann et al. 2005; Kong et al. 2002) associated with non-B DNA-forming repeats.
Conclusions
Our findings have for the first time placed the DNA sequence analysis of gene conversion tracts on a sound statistical footing. This study has provided firm evidence that motifs associated with recombination activity and sequences with the potential to form non-B DNA structures are both over-represented within MaxCTs and ShortFlank sequences. These results strongly support the postulate that the gene conversion events were initiated by DSBs at sites of non-B DNA structure formation, which then activated the proximal recombination-promoting motifs to serve as substrate for the subsequent mutagenic repair process.
Two types of non-B DNA-forming short repeat were found to be consistently over-represented within the MaxCT and ShortFlank regions; in particular, direct repeats which may form slipped structures, and inverted repeats which may form hairpin/cruciform structures (Table 2; Supp. Figure S1). Both types of non-B DNA conformations may be expected to be acted upon by DNA repair proteins and therefore represent an intrinsic source of induced DSBs (Wang and Vasquez 2006). Since the number of direct and inverted repeats in the analysed regions was quite substantial (respectively 16 and 9 in the ShortFlank regions), these results provide strong support for the notion that occasional non-B DNA conformations formed by these repeats can contribute to the initial events (including DSBs) that trigger recombination leading to gene conversion (Wang and Vasquez 2006; Bacolla et al. 2004). Further, our results also support the view that DNA breakage tends to occur within MaxCTs, or at least within MaxCTs with short flanking sequences included.
Maximal converted tracts were found to be enriched (p<0.01) in a truncated version of the χ-element, TGGTGG, previously noted to be over-represented in the vicinity of microdeletions, microinsertions and indels (Ball et al. 2005). Several novel motifs, including either a fragment or the entire sequence of a classical meiotic recombination hotspot, CCTCCCCT, were also found to be over-represented (p<0.01) within MaxCTs. We therefore propose that a) non-B DNA-mediated DSBs occurring within the narrow regions immediately flanking and including the MaxCTs may serve to promote gene conversion and b) a high local density of recombination-promoting motifs may act in combination with these DNA conformations to potentiate gene conversion. This reinforces the early, isolated and purely anecdotal observations of an association between Z-DNA-forming sequences and gene conversion (Kilpatrick et al. 1984; Wahls et al. 1990b) since it would appear that other types of non-B DNA conformations such as slipped structures and cruciform-forming repeats are also likely to be involved in promoting DSBs.
Although most of the biochemical steps underlying the mutational mechanism(s) responsible for gene conversion still remain to be elucidated, our present findings provide additional support to a recently proposed model (Wang and Vasquez 2006; Bacolla et al. 2006; Kurahashi et al. 2006; Wells 2007; Raghavan et al. 2007; Rooms et al. 2007) in which it is the resolution of non-B DNA conformations that induces chromosomal DSBs which can then give rise to gross genomic rearrangements via processes that involve DNA recombination-repair. Most notably, we show that recombination-associated motifs play an integral part in this non-B DNA structure-induced mutational process. Specifically, we postulate that the high density of recombination-related motifs serves as target binding sites for protein complexes, such as translin and RAG-associated proteins, or arrest sites for DNA polymerases, which may assist, induce or indeed be required for the recombination-repair process.
Supplementary Material
Acknowledgments
This work was partially supported by the INSERM (Institut National de la Santé et de la Recherche Médicale), France (to J.-M.C. and C.F.), BIOBASE GmbH (through financial support to D.N.C.) and by grants from the National Institutes of Health (NS37554 and ES11347) and the Robert A. Welch Foundation to R. D. W.
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
References
- Abeysinghe SS, Chuzhanova N, Krawczak M, Ball EV, Cooper DN. Translocation and gross deletion breakpoints in human inherited disease and cancer I: Nucleotide composition and recombination-associated motifs. Hum Mutat. 2003;22:229–244. doi: 10.1002/humu.10254. [DOI] [PubMed] [Google Scholar]
- Aoki K, Suzuki K, Sugano T, Tasaka T, Nakahara K, Kuge O, Omori A, Kasai M. A novel gene, translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat Genet. 1995;10:167–174. doi: 10.1038/ng0695-167. [DOI] [PubMed] [Google Scholar]
- Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. J Biol Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- Bacolla A, Jaworski A, Larson JE, Jakupciak JP, Chuzhanova N, Abeysinghe SS, O'Connell CD, Cooper DN, Wells RD. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc Natl Acad Sci USA. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacolla A, Wojciechowska M, Kosmider B, Larson JE, Wells RD. The involvement of non-B DNA structures in gross chromosomal rearrangements. DNA Repair. 2006;5:1161–1170. doi: 10.1016/j.dnarep.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Bacolla A, Larson JE, Collins JR, Li J, Milosavljevic A, Stenson PD, Cooper DN, Wells RD. Abundance and length of simple repeats in vertebrate genomes are determined by their structural properties. Genome Res. 2008;18:1545–1553. doi: 10.1101/gr.078303.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball EV, Stenson PD, Krawczak M, Cooper DN, Chuzhanova NA. Micro-deletions and micro-insertions causing human genetic disease: common mechanisms of mutagenesis and the role of local DNA sequence complexity. Hum Mutat. 2005;26:205–213. doi: 10.1002/humu.20212. [DOI] [PubMed] [Google Scholar]
- Bergemann AD, Johnson EM. The HeLa Pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and origins of DNA replication. Mol Cell Biol. 1992;12:1257–1265. doi: 10.1128/mcb.12.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Férec C. Origin and implication of the hereditary pancreatitis-associated N21I mutation in the cationic trypsinogen gene. Hum Genet. 2000;106:125–126. doi: 10.1007/s004390051019. [DOI] [PubMed] [Google Scholar]
- Chen JM, Cooper DN, Chuzhanova N, Férec C, Patrinos GP. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 2007;8:762–775. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- Conley ME, Rapalus L, Boylin EC, Rohrer J, Minegishi Y. Gene conversion events contribute to the polymorphic variation of the surrogate light chain gene lambda 5/14.1. 1: Clin Immunol. 1999;93:162–167. doi: 10.1006/clim.1999.4785. [DOI] [PubMed] [Google Scholar]
- Cullen M, Perfetto SP, Klitz W, Nelson G, Carrington M. High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am J Hum Genet. 2002;71:759–776. doi: 10.1086/342973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco P, Moroni A, Merello E, de Franchis R, Andreussi L, Finnell RH, Barber RC, Cama A, Capra V. Folate pathway gene alterations in patients with neural tube defects. Am J Med Genet. 2000;95:216–223. doi: 10.1002/1096-8628(20001127)95:3<216::aid-ajmg6>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Eikenboom JC, Vink T, Briet E, Sixma JJ, Reitsma PH. Multiple substitutions in the von Willebrand factor gene that mimic the pseudogene sequence. Proc Natl Acad Sci U S A. 1994;91:2221–2224. doi: 10.1073/pnas.91.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eikenboom JC, Castaman G, Vos HL, Bertina RM, Rodeghiero F. Characterization of the genetic defects in recessive type 1 and type 3 von Willebrand disease patients of Italian origin. Thromb Haemost. 1998;79:709–717. [PubMed] [Google Scholar]
- Flanagan JG, Lefranc MP, Rabbitts TH. Mechanisms of divergence and convergence of the human immunoglobulin alpha 1 and alpha 2 constant region gene sequences. Cell. 1984;36:681–688. doi: 10.1016/0092-8674(84)90348-9. [DOI] [PubMed] [Google Scholar]
- Eyal N, Wilder S, Horowitz M. Prevalent and rare mutations among Gaucher patients. Gene. 1990;96:277–283. doi: 10.1016/0378-1119(90)90264-r. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. COPYEDITOR: ALLOW TRUNCATED AUTHOR LIST FOR THIS ARTICLE, OVER 100 AUTHORS. Thanks – Managing Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fremeaux-Bacchi V, Moulton EA, Kavanagh D, Dragon-Durey MA, Blouin J, Caudy A, Arzouk N, Cleper R, Francois M, Guest G, Pourrat J, Seligman R, Fridman WH, Loirat C, Atkinson JP. Genetic and functional analyses of membrane cofactor protein (CD46) mutations in atypical hemolytic uremic syndrome. J Am Soc Nephrol. 2006;17:2017–2025. doi: 10.1681/ASN.2005101051. [DOI] [PubMed] [Google Scholar]
- Friaes A, Rego AT, Aragues JM, Moura LF, Mirante A, Mascarenhas MR, Kay TT, Lopes LA, Rodrigues JC, Guerra S, Dias T, Teles AG, Goncalves J. CYP21A2 mutations in Portuguese patients with congenital adrenal hyperplasia: identification of two novel mutations and characterization of four different partial gene conversions. Mol Genet Metab. 2006;88:58–65. doi: 10.1016/j.ymgme.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Gajecka M, Pavlicek A, Glotzbach CD, Ballif BC, Jarmuz M, Jurka J, Shaffer LG. Identification of sequence motifs at the breakpoint junctions in three t(1;9)(p36.3;q34) and delineation of mechanisms involved in generating balanced translocations. Hum Genet. 2006;120:519–526. doi: 10.1007/s00439-006-0222-1. [DOI] [PubMed] [Google Scholar]
- Giordano M, Marchetti C, Chiorboli E, Bona G, Momigliano Richiardi P. Evidence for gene conversion in the generation of extensive polymorphism in the promoter of the growth hormone gene. Hum Genet. 1997;100:249–255. doi: 10.1007/s004390050500. [DOI] [PubMed] [Google Scholar]
- Globerman H, Amor M, Parker KL, New MI, White PC. Nonsense mutation causing steroid 21-hydroxylase deficiency. J Clin Invest. 1988;82:139–144. doi: 10.1172/JCI113562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev VD, Nemytikova LA, Chuzhanova NA. On the complexity measures of genetic sequences. Bioinformatics. 1999;15:994–999. doi: 10.1093/bioinformatics/15.12.994. [DOI] [PubMed] [Google Scholar]
- Hallast P, Nagirnaja L, Margus T, Laan M. Segmental duplications and gene conversion: Human luteinizing hormone/chorionic gonadotropin beta gene cluster. Genome Res. 2005;15:1535–1546. doi: 10.1101/gr.4270505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Su B, Li WH, Zhao Z. CpG island density and its correlations with genomic features in mammalian genomes. Genome Biol. 2008;9:R79. doi: 10.1186/gb-2008-9-5-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi T, Yotsumoto S, Shimada H, Terasaki K, Setoyama M, Kobayashi K, Saheki T, Kanzaki T. A novel point mutation in the keratin 17 gene in a Japanese case of pachyonychia congenita type 2. J Invest Dermatol. 2002;118:545–547. doi: 10.1046/j.0022-202x.2001.01701.x. [DOI] [PubMed] [Google Scholar]
- Heinen S, Sanchez-Corral P, Jackson MS, Strain L, Goodship JA, Kemp EJ, Skerka C, Jokiranta TS, Meyers K, Wagner E, Robitaille P, Esparza-Gordillo J, Rodriguez de Cordoba S, Zipfel PF, Goodship TH. De novo gene conversion in the RCA gene cluster (1q32) causes mutations in complement factor H associated with atypical hemolytic uremic syndrome. Hum Mutat. 2006;27:292–293. doi: 10.1002/humu.9408. [DOI] [PubMed] [Google Scholar]
- Hellmann I, Prüfer K, Ji H, Zody MC, Pääbo S, Ptak SE. Why do human diversity levels vary at a megabase scale? Genome Res. 2005;15:1222–1231. doi: 10.1101/gr.3461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y, Tanae A, Inoue H, Hiromasa T, Fujii-Kuriyama Y. Aberrant splicing and missense mutations cause steroid 21-hydroxylase [P-450(C21)] deficiency in humans: possible gene conversion products. Proc Natl Acad Sci U S A. 1988;85:7486–7490. doi: 10.1073/pnas.85.20.7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Högstrand K, Böhme J. Gene conversion of major histocompatibility complex genes is associated with CpG-rich regions. Immunogenetics. 1999;49:446–455. doi: 10.1007/s002510050518. [DOI] [PubMed] [Google Scholar]
- Hong CM, Ohashi T, Yu XJ, Weiler S, Barranger JA. Sequence of two alleles responsible for Gaucher disease. DNA Cell Biol. 1990;9:233–241. doi: 10.1089/dna.1990.9.233. [DOI] [PubMed] [Google Scholar]
- Huang CH, Chen Y, Blumenfeld OO. A novel St(a) glycophorin produced via gene conversion of pseudoexon III from glycophorin E to glycophorin A gene. Hum Mutat. 2000;15:533–540. doi: 10.1002/1098-1004(200006)15:6<533::AID-HUMU5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- Inagaki K, Ma C, Storm TA, Kay MA, Nakai H. The role of DNA-PKcs and artemis in opening viral DNA hairpin termini in various tissues in mice. J Virol. 2007;81:11304–11321. doi: 10.1128/JVI.01225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Inoue K, Utsunomiya M, Nozaki J, Yamada Y, Iwasa T, Mori E, Yoshinaga T, Koizumi A. Mutation analysis in PKD1 of Japanese autosomal dominant polycystic kidney disease patients. Hum Mutat. 2002;19:622–628. doi: 10.1002/humu.10080. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Holloway JK, Kauppi L, May CA, Neumann R, Slingsby MT, Webb AJ. Meiotic recombination hot spots and human DNA diversity. Philos Trans R Soc Lond B Biol Sci. 2004;359:141–152. doi: 10.1098/rstb.2003.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Seaman MI, Furey TS, Payseur BA, Lu Y, Roskin KM, Chen CF, Thomas MA, Haussler D, Jacob HJ. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004;14:528–538. doi: 10.1101/gr.1970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick MW, Klysik J, Singleton CK, Zarling DA, Jovin TM, Hanau LH, Erlanger BF, Wells RD. Intervening sequences in human fetal globin genes adopt left-handed Z helices. J Biol Chem. 1984;259:7268–7274. [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Inagaki H, Ohye T, Kogo H, Kato T, Emanuel BS. Palindrome-mediated chromosomal translocations in humans. DNA Repair. 2006;5:1136–1145. doi: 10.1016/j.dnarep.2006.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law HY, Luo HY, Wang W, Ho JF, Najmabadi H, Ng IS, Steinberg MH, Chui DH, Chong SS. Determining the cause of patchwork HBA1 and HBA2 genes: recurrent gene conversion or crossing over fixation events. Haematologica. 2006;91:297–302. [PubMed] [Google Scholar]
- Lee HH, Niu DM, Lin RW, Chan P, Lin CY. Structural analysis of the chimeric CYP21P/CYP21 gene in steroid 21-hydroxylase deficiency. J Hum Genet. 2002;47:517–522. doi: 10.1007/s100380200077. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair. 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Lopez-Correa C, Dorschner M, Brems H, Lazaro C, Clementi M, Upadhyaya M, Dooijes D, Moog U, Kehrer-Sawatzki H, Rutkowski JL, Fryns JP, Marynen P, Stephens K, Legius E. Recombination hotspot in NF1 microdeletion patients. Hum Mol Genet. 2001;10:1387–1392. doi: 10.1093/hmg/10.13.1387. [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- Marino-Ramirez L, Spouge JL, Kanga GC, Landsman D. Statistical analysis of over-represented words in human promoter sequences. Nucleic Acids Res. 2004;32:949–958. doi: 10.1093/nar/gkh246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar DS, Lewis MD, Horan M, Newsway V, Easter TE, Gregory JW, Fryklund L, Norin M, Crowne EC, Davies SJ, Edwards P, Kirk J, Waldron K, Smith PJ, Phillips JA, 3rd, Scanlon MF, Krawczak M, Cooper DN, Procter AM. Novel mutations of the growth hormone 1 (GH1) gene disclosed by modulation of the clinical selection criteria for individuals with short stature. Hum Mutat. 2003;21:424–440. doi: 10.1002/humu.10168. [DOI] [PubMed] [Google Scholar]
- Minegishi Y, Coustan-Smith E, Wang YH, Cooper MD, Campana D, Conley ME. Mutations in the human lambda5/14.1 gene result in B cell deficiency and agammaglobulinemia. J Exp Med. 1998;187:71–77. doi: 10.1084/jem.187.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Myers S, Spencer CCA, Auton A, Bottolo L, Freeman C, Donnelly P, McVean G. The distribution and causes of meiotic recombination in human genome. Biochem Soc Trans. 2006;34:526–530. doi: 10.1042/BST0340526. [DOI] [PubMed] [Google Scholar]
- Napierala M, Bacolla A, Wells RD. Increased negative superhelical density in vivo enhances the genetic instability of triplet repeat sequences. J Biol Chem. 2005;280:37366–37376. doi: 10.1074/jbc.M508065200. [DOI] [PubMed] [Google Scholar]
- Nicod J, Dick B, Frey FJ, Ferrari P. Mutation analysis of CYP11B1 and CYP11B2 in patients with increased 18-hydroxycortisol production. Mol Cell Endocrinol. 2004;214:167–174. doi: 10.1016/j.mce.2003.10.056. [DOI] [PubMed] [Google Scholar]
- Nicolis E, Bonizzato A, Assael BM, Cipolli M. Identification of novel mutations in patients with Shwachman-Diamond syndrome. Hum Mutat. 2005;25:410. doi: 10.1002/humu.9324. [DOI] [PubMed] [Google Scholar]
- Nishant KT, Rao MR. Molecular features of meiotic recombination hot spots. Bioessays. 2006;28:45–56. doi: 10.1002/bies.20349. [DOI] [PubMed] [Google Scholar]
- Ohno S. (AGCTG) (AGCTG) (AGCTG) (GGGTG) as the primordial sequence of intergenic spacers: the role in immunoglobulin class switch. Differentiation. 1981;18:65–74. doi: 10.1111/j.1432-0436.1981.tb01106.x. [DOI] [PubMed] [Google Scholar]
- Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- Raghavan SC, Tong J, Lieber MR. Hybrid joint formation in human V(D)J recombination requires nonhomologous DNA end joining. DNA Repair. 2006;5:278–285. doi: 10.1016/j.dnarep.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Raghavan SC, Gu J, Swanson PC, Lieber MR. The structure-specific nicking of small heteroduplexes by the RAG complex: implications for lymphoid chromosomal translocations. DNA Repair. 2007;6:751–759. doi: 10.1016/j.dnarep.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Pentao L, Muzny DM, Gibbs RA, Lupski JR. A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet. 1996;12:288–297. doi: 10.1038/ng0396-288. [DOI] [PubMed] [Google Scholar]
- Rooms L, Reyniers E, Kooy RF. Diverse chromosome breakage mechanisms underlie subtelomeric rearrangements, a common cause of mental retardation. Hum Mutat. 2007;28:177–182. doi: 10.1002/humu.20421. [DOI] [PubMed] [Google Scholar]
- Rooney SM, Moore PD. Antiparallel, intramolecular triplex DNA stimulates homologous recombination in human cells. Proc Natl Acad Sci USA. 1995;92:2141–2144. doi: 10.1073/pnas.92.6.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- Sinden RR. DNA Structure and Function. Academic Press; 1994. [Google Scholar]
- Smith DG, Adair GM. Characterization of an apparent hotspot for spontaneous mutation in exon 5 of the Chinese hamster APRT gene. Mutat Res. 1996;352:87–96. doi: 10.1016/0027-5107(96)00007-3. [DOI] [PubMed] [Google Scholar]
- Smith RA, Ho PJ, Clegg JB, Kidd JR, Thein SL. Recombination breakpoints in the human β-globin gene cluster. Blood. 1998;92:4415–4421. [PubMed] [Google Scholar]
- Soejima M, Fujihara J, Takeshita H, Koda Y. Sec1-FUT2-Sec1 hybrid allele generated by interlocus gene conversion. Transfusion. 2008;48:488–492. doi: 10.1111/j.1537-2995.2007.01553.x. [DOI] [PubMed] [Google Scholar]
- Surdhar GK, Enayat MS, Lawson S, Williams MD, Hill FG. Homozygous gene conversion in von Willebrand factor gene as a cause of type 3 von Willebrand disease and predisposition to inhibitor development. Blood. 2001;98:248–250. doi: 10.1182/blood.v98.1.248. [DOI] [PubMed] [Google Scholar]
- Teich N, Nemoda Z, Kohler H, Heinritz W, Mossner J, Keim V, Sahin-Toth M. Gene conversion between functional trypsinogen genes PRSS1 and PRSS2 associated with chronic pancreatitis in a six-year-old girl. Hum Mutat. 2005;25:343–347. doi: 10.1002/humu.20148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanita, Sarhadi V, Reis A, Jung M, Singh D, Sperling K, Singh JR, Burger J. A unique form of autosomal dominant cataract explained by gene conversion between beta-crystallin B2 and its pseudogene. J Med Genet. 2001;38:392–396. doi: 10.1136/jmg.38.6.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez N, Lehrnbecher T, Chen R, Christensen BL, Gallin JI, Malech H, Holland S, Zhu S, Chanock SJ. Mutational analysis of patients with p47-phox-deficient chronic granulomatous disease: The significance of recombination events between the p47-phox gene (NCF1) and its highly homologous pseudogenes. Exp Hematol. 2001;29:234–243. doi: 10.1016/s0301-472x(00)00646-9. [DOI] [PubMed] [Google Scholar]
- Wahls WP, Wallace LJ, Moore PD. Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell. 1990a;60:95–103. doi: 10.1016/0092-8674(90)90719-u. [DOI] [PubMed] [Google Scholar]
- Wahls WP, Wallace LJ, Moore PD. The Z-DNA motif d(TG)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human cells in culture. Mol Cell Biol. 1990b;10:785–793. doi: 10.1128/mcb.10.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability. Mutat Res. 2006;598:103–119. doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Watnick TJ, Gandolph MA, Weber H, Neumann HP, Germino GG. Gene conversion is a likely cause of mutation in PKD1. Hum Mol Genet. 1998;7:1239–1243. doi: 10.1093/hmg/7.8.1239. [DOI] [PubMed] [Google Scholar]
- Wells RD, Dere R, Hebert ML, Napierala M, Son LS. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 2005;33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RD, Ashizawa T, editors. Genetic Instabilities and Neurological Diseases. 2nd. San Diego: Elsevier/Academic Press; 2006. pp. 1–784. [Google Scholar]
- Wells RD. Non-B DNA conformations, mutagenesis, and diseases. Trends Biochem Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Wolf A, Millar DS, Caliebe A, Horan M, Newsway V, Kumpf D, Steinmann K, Chee IS, Lee YH, Mutirangura A, Pepe G, Rickards O, Schmidtke J, Schempp W, Chuzhanova N, Kehrer-Sawatzki H, Krawczak M, Cooper DN. A gene conversion hotspot in the human growth hormone (GH1) gene promoter. Hum Mutat. 2009;30:239–247. doi: 10.1002/humu.20850. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.