Abstract
In order to understand an overview of promoter activities intrinsic to primary DNA sequences in the human genome within a particular cell type, we carried out systematic quantitative luciferase assays of DNA fragments corresponding to putative promoters for 472 human genes which are expressed in HEK (human embryonic kidney epithelial) 293 cells. We observed the promoter activities of them were distributed in a bimodal manner; putative promoters belonging to the first group (with strong promoter activities) were designated as P1 and the latter (with weak promoter activities) as P2. The frequencies of the TATA-boxes, the CpG islands, and the overall G + C-contents were significantly different between these two populations, indicating there are two separate groups of promoters. Interestingly, similar analysis using 251 randomly isolated genomic DNA fragments showed that P2-type promoter occasionally occurs within the human genome. Furthermore, 35 DNA fragments corresponding to putative promoters of non-protein-coding transcripts (ncRNAs) shared similar features with the P2 in both promoter activities and sequence compositions. At least, a part of ncRNAs, which have been massively identified by full-length cDNA projects with no functional relevance inferred, may have originated from those sporadic promoter activities of primary DNA sequences inherent to the human genome.
Key words: human genome, promoter, transcriptional start site
1. Introduction
With the unprecedented amount of data produced from both human genome1 and transcriptome2–6 projects, a new challenge of genome science is to decode how the code of the genomic DNA collectively realizes the transcriptome.7 To this end, it should be the first step to understand the code of DNA sequence to control the transcriptional initiation. Although, levels of transcripts are also controlled at post-transcription-initiation steps such as transcription elongation, splicing, nuclear export, degradation steps, and so on,8 transcription initiation is the first step in the gene expression of every gene and is supported to play fundamental roles.9,10
It is known that, in many cases, the genomic regions proximal to the transcriptional start sites (TSSs), which are called promoters, play pivotal roles in determining the rate of transcription initiation by serving as direct docking platforms for the RNA polymerase II complex.9 However, the comprehensive view is still obscure regarding which part of the human genome the RNA polymerase II is recruited to and at what frequency the transcription is initiated. There are currently only 428 genes whose transcriptional regulation is well understood (TRANSFAC7.2).10 Besides, even for those genes, the promoter activities were measured under arbitrary experimental conditions, and are therefore inappropriate for use in deciphering the transcriptional network taking place in a particular cellular context or for comprehensive quantitative analyses of promoter activities encoded by DNA sequences.
We have developed a method to construct a full-length cDNA library, which we named the oligo-capping,11 and have accumulated 1.8 million 5′-end sequences of putative full-length cDNAs.12 By utilizing the full-length cDNA information, we have been identifying exact genomic positions of the TSSs and characterizing the adjacent sequences as putative promoter regions (PPRs).13 So far we have made the collected information of the TSSs and PPRs for about 15 000 human genes available through our database, DBTSS (http://dbtss.hgc.jp/).
In the present study, we attempted to obtain quantitative experimental data about the promoter activities within a particular cell, since such a data set should serve as a firm foundation for exploring the transcriptional network of human genes. We first collected and analyzed the promoter activities of DNA fragments corresponding to PPRs of genes which are expressed in human embryonic kidney epithelial 293 cells (HEK293 cells). Then, we analyzed the promoter activities of genomic DNA fragments which were randomly isolated from non-promoter regions. By this, we wished to understand the promoter activities intrinsic to primary DNA sequences. Unexpectedly, analysis of those DNA fragments led us to hypothesis that sometimes even average human genomic sequences come to have significant promoter activities and that a class of non-protein coding RNAs should be originated from sporadically occurring promoter activities of DNA sequences, which is inherent to a long genome such as human's. Here we report our systematic experimental characterization of the promoter activities of primary DNA sequences using a uniform experimental procedure and standardized cellular conditions.
2. Material and methods
2.1. Computational procedures
Oligo-cap cDNA library was constructed from HEK293 cells (ATCC number CRL-1573) according to the procedure described previously. 12 504 one-pass sequences were produced and mapped onto the human genomic sequences (hg_17; http://genome.ucsc.edu/) as previously described.11 (We also confirmed that the update to hg_18 would make essentially no influence on the results.) The DNA sequences proximal (−1 kb to + 200 bp) to the 5′-ends of the mapped oligo-capped cDNAs were retrieved as PPRs. PPR region was defined as − 1 kb to + 200 bp, because, in many cases, TSSs are distributed over ∼100 bp wide and there are usually many TSSs located downstream of the representative TSS12 (also see Supplementary Fig. 1B). In order to cover most of those fluctuating TSSs, PCR primers were set using PRIMER3 between + 100 to + 200 bp. As a result, many of the PPR clones came to contain so-called upstream ATGs. For detailed discussion on the possible influences of those ATGs, see Supplementary Information Fig. 8. For details of the PPR sequences and the designed PCR primers, see Supplementary Tables 1–3 and 5.
For prediction of the TATA boxes,9 a matrix search program, MATCH, was run using the position weight matrices of V$TATA_C (http://www.gene-regulation.com/cgi-bin/pub/databases/transfac/getTF.cgi?AC=M00216) and V$TATA_01 (http://www.gene-regulation.com/cgi-bin/pub/databases/transfac/getTF.cgi?AC=M00252) as for the TRANSFAC7.2 database. Hits observed between − 90 and + 27 relative to the TSSs in the plus strands with cut-off value of 0.77 were scored as positives. For the statistical test, Wilcoxon test was used.
2.2. Cloning of the DNA fragments of PPRs and luciferase reporter gene assays
Using the PCR primers designed for randomly selected 1000 PPRs (out of 2170 PPRs which were identified to be active in HEK293 cells; see Supplementary Fig. 1), 50 ng of human genomic DNA (Clontech) was amplified by PCR with 10 pmol of the 5′- and 3′-PCR primers and with the 35 reaction cycles of 94°C, 1 min; 58°C, 1 min; 68°C, 2 min; using a KOD Plus PCR kit (ToYoBo). For cloning the random genomic DNAs, the human genomic DNA was amplified by a similar procedure using PCR primers solely containing attB1 and attB2 sites with the following relaxed PCR conditions: 20 reaction cycles of 94°C, 1 min; 40°C, 1 min; 72°C, 1 min; using a ExTaq PCR kit (TaKaRa). The amplified fragments were size fractionated by agarose gel electrophoresis and fragments of 1.0–1.2 kb were recovered. After the amplified fragments' lengths were confirmed by agarose gel electrophoresis, PPR and random genomic DNA fragments were cloned into a luciferase reporter gene construct using the Gateway System (Invitrogen; http://www.invitrogen.com/content.cfm?pageid=4072). We employed this multi-faceted cloning system, so that the obtained promoter clones could be used in other vector systems in future experiments. For Gateway cloning, we performed ‘one-tube reactions’ according to the supplied instructions. The insert sequences were determined from both ends.
As for ‘random’ genomic sequences, we checked their genomic positions using UCSC Genome Browser and found no explicit bias (Supplementary Table 2). We also found no difference in the distribution patterns of the G + C contents between the clones ‘random’ genomic fragments and computationally ‘randomly’-extracted genomic sequences (also see Supplementary Fig. 7). Although it is still possible that undetected bias might have been introduced by PCR amplifications, we considered the obtained clone set mostly represented ‘random’ genomic sequences.
For transient transfection of the promoter clones, the DNAs were purified using QIAwell 96 Ultra (QIAGEN). HEK293 cells were cultured in 96-well micro-titre plates with a cell density of 1.0 × 104 per well. For each well, 50 ng of the promoter clones together with the 5 ng of pTK-Renilla were transiently transfected into HEK293 cells using 0.3 µl of Fugene6 (Roche). Forty-eight hours after the transfection, dual luciferase assays were performed according to the manufacturer's instructions. The above procedures were repeated three times as independent cell culture and transfection experiments. For 86% of the data from 472 PPRs and 251 random fragments, estimated signal versus experimental noise (averaged luciferase activity versus the standard deviation of them calculated from the three experiments) ratio was less than 0.25. Identification, physical cloning, and luciferase assays of the PPRs of the ncRNAs were performed similarly with the cases of the PPRs.
3. Results and discussion
3.1. Luciferase assays of the DNA fragments corresponding to PPRs
In order to understand the overview of the promoter activities of primary DNA sequences in the human genome in a particular cell, we attempted to collect DNA fragments of PPRs which are active in HEK293 cells. For this purpose, we first collected the TSS information of the genes expressed in HEK293 cells by sequencing a 5′-end-enriched cDNA library constructed using our oligo-capping method10 (schematic representation of the workflow is shown in Supplementary Fig. 1A). The overall full-lengthness of the cDNA library was estimated to be 85% (Supplementary Fig. 2). By mapping 12504 5′-ends cDNA sequences (Genbank accessions: BP870448–BP873619; BP244227–BP249739), we identified positional information of the PPRs, which are expressed in this cell line. The genomic DNAs corresponding to − 1 kb to + 200 bp were amplified by PCR and 472 PPRs were physically cloned into the luciferase vector (for examples, see Supplementary Figs 1B and C). In the present study, we focused on − 1 kb to + 200 bp regions as PPRs (82% of the previously characterized regulatory elements were shown to be located within this range; see Supplementary Fig. 3).
We did not take the distal regulatory elements, such as enhancers and locus control regions, into consideration. Also, we did not assess the effects of chromatin structure or epigenetic modulations.8 Actually, in the reporter gene assays used in this study, influences of such factors are not reflected in exchange for its advantages in high throughput and quantitativeness. Intensive analysis focused on each gene would be indispensable for those purposes. Rather, we considered it should be prioritized to characterize the promoter activities intrinsic to the naked primary DNA sequences of the very adjacent or overlapping regions of the TSSs, providing actual docking platforms of transcriptional machineries. (In this issue, extensive analysis, comparing the activities of primary DNAs of PPRs observed using luciferase assays and the eventual mRNA expression levels observed using SAGE and microarray, is also presented in Supplementary Fig. 4.)
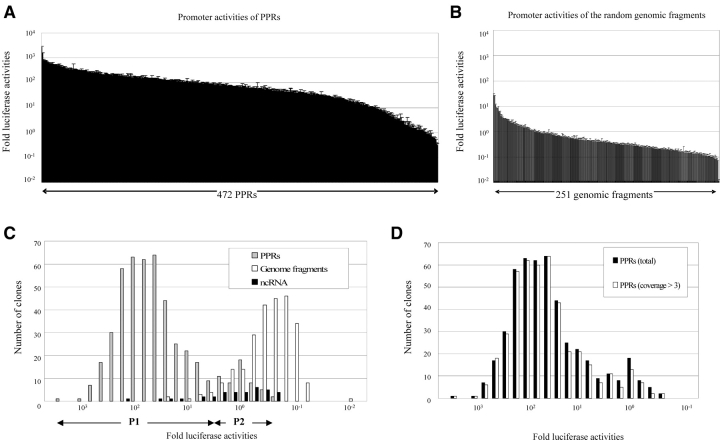
Using the 472 successfully cloned PPRs, luciferase assays were systematically carried out under standardized cell culture conditions (Fig. 1). The assay was repeated at least three times for each of the PPRs (for raw data, see Supplementary Tables 1–3). We further selected 445 PPRs that were supported by more than three independently isolated oligo-cap cDNAs. This was done to assure that the frequency of the erroneously identified PPRs due to errors of the oligo-capping would be minimized (see in what follows). For the later analyses, we used the 472 PPR data set. Essentially, the same results were obtained using the selected 445 PPR data set (Fig. 1D; for the results of each of the following analyses, see Supplementary Fig. 5).
Figure 1.
Luciferase activities of the PPRs. Luciferase activities of the PPRs (A) and the randomly isolated genomic fragments (B) Error bar indicates the standard deviation of each assay. (C) Distribution of the luciferase activities of the PPRs (gray bars), random genomic fragments (blank bars) and PPRs of the ‘ncRNAs’ (solid bars). (D) The distribution of the PPRs which are supported by more than three oligo-cap cDNAs is shown by blank bars. The average luciferase activity of the random genomic fragments was designated as 1 for all of the analyses. Details of the methods are provided as supporting information.
3.2. Two separate populations of PPRs
As shown in Fig. 1C, the observed promoter activities seemed to show a bimodal pattern of distribution. When we separated the PPRs with the luciferase activities of > 100.8 fold as P1 and those with < 100.8 fold as P2 (Table 1), we found that the bimodality of the distribution was statistically significant (Supplementary Table 4). Furthermore, sequence characterization of the PPRs revealed that there was a qualitative difference between the sequence features of P1 and P2 (Table 2).
Table 1.
Luciferase activities and the classification of the PPRs
| Luciferase activity | P | P1 | P2 | G | G1 | G2 | ncRNA |
|---|---|---|---|---|---|---|---|
| >103 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| 103–102 | 175 | 175 | 0 | 0 | 0 | 0 | 1 |
| 102–101 | 217 | 217 | 0 | 3 | 3 | 0 | 3 |
| 101–100 | 63 | 17 | 46 | 43 | 30 | 13 | 16 |
| 100–101 | 15 | 0 | 15 | 196 | 0 | 196 | 15 |
| <10−1 | 0 | 0 | 0 | 9 | 0 | 9 | 0 |
| 472 | 411 | 61 | 251 | 33 | 218 | 35 |
Statistical significances of the marked positions are shown in the margin.
Table 2.
Sequence features of the PPRs and genomic fragments
| P (%) | P1 (%) | P2 (%) | G (%) | G1 (%) | G2 (%) | |
|---|---|---|---|---|---|---|
| CpG island | 267 (57) | 259 (63)* | 8 (13) | 0 (0) | 0 (0) | 0 (0) |
| TATA box: strict | 34 (7) | 30 (7) | 4 (7) | 47 (19) | 7 (21) | 40 (18) |
| TATA box: less strict | 103 (22) | 81 (20)** | 22 (36) | 140 (56) | 20 (61) | 120 (55) |
| Average G + C content | 0.53 | 0.54*** | 0.47*** | 0.45 | 0.43 | 0.45 |
| Total | 472 | 411 | 61 | 251 | 33 | 218 |
For P1, the frequencies of strict TATA boxes (TATA[T/A][T/A]), less-strict TATA boxes (TATA-like elements; hits from the matrix search with relaxed parameters: see Material and methods) and CpG-islands were 7, 20, and 63%, respectively. The overall G + C content was 0.54, which is far more GC-rich than the average G + C content of the entire human genome (0.45). This is in good agreement with our previous statistical analysis of PPRs.13 Also, within P1, the presence of the strict TATA-containing PPRs was enriched in the PPRs whose promoter activities were in the top 25% compared with those whose activities were in the bottom 25% (18 and 3%, respectively; P < 0.001). Generally, the sequence features of P1 were consistent with the previous view that the promoters are embedded in a relatively G + C rich sequence context, often associated with CpG islands, and the view that because the presence of the canonical TATA box provides the optimal docking platform for RNA polymerase II, it drives the strongest promoter activity.8 It should be the next step analysis to further narrow down the observed promoter activities are realized by what range of the DNA sequences within the PPRs.
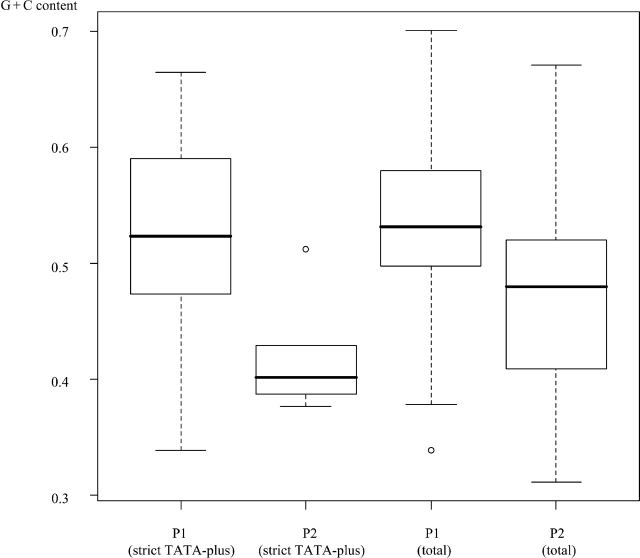
In contrast, P2 was far more AT-rich (G + C content = 0.47) than P1 (P < 6.0 × 10−10; also see Supplementary Table 6). CpG islands were far less frequent in P2 (13%; P < 1.0 × 10−6). Although the frequency of strict TATA boxes in P2 was similar to that in P1 (7%), that of the less-strict TATA-boxes was much higher (36%; P < 1.0 × 10−2). These sequence features, distinct from those of P1, were somewhat different from the features included in the classical view of promoters. Relative enrichment of the less-strict TATA boxes may indicate that sequences favourable for the binding of TATA-binding protein might be indispensable for P2, whose members otherwise meet few of the requirements of promoters as conventionally understood. Actually, G + C contents of P2 containing strict TATA boxes were lower than those of P1 similarly containing strict TATA boxes (Fig. 2). Again, this result indicates that a TATA box embedded in a relatively G + C rich sequence should be necessary for realizing strong promoter activity.
Figure 2.
G + C content of the PPRs. Box plot chart of the G + C content of the indicated population of the PPRs is shown.
We considered it unlikely that P2 consisted predominantly of erroneously identified PPRs. First, the fidelity of the identified PPRs should have increased, as the number of supporting oligo-cap cDNAs increases. As shown in Fig. 1D, among 472 PPRs, 445 (including 47 of the PPRs belonging to P2) were supported by more than three independently isolated oligo-cap cDNAs. Also, when the PPRs with more than three supporting oligo-cap cDNAs were used for all of the analyses, essentially the same results were obtained (Supplementary Fig. 5). Secondly, even for the PPRs in P2, it was not the case that the promoter activities were not observed at all. Although weak, the activities of these PPRs were clearly higher than most of the promoter activities observed for randomly isolated genomic fragments (G2: see in what follows). Lastly, supporting evidences have been reported. Although their purpose was different from ours, Trinklein et al.14 also performed luciferase assays for 152 kinds of PPRs identified from full-length cDNAs in HEK293 cells. Their results also indicated similar bimodal patterns of the promoter activities. Moreover, Versteeg et al.15 reported that human genes with high expression levels tend to be located in GC-rich regions and genes with low expression levels tend to be located in AT-rich regions according to their genome-wide ‘human transcriptome mapping’ analysis, which could be interpreted as the features of P1- and P2-driven genes, respectively.
3.3. Possible universal promoter activities of the DNA sequences in the human genome
We then attempted to examine the promoter activities of average human genomic DNA fragments in contrast to the observed PPR activities. We randomly isolated 251 non-genic genomic fragments of approximately the same length and measured their promoter activities (Fig. 1B, Supplementary Table 2; also see Material and methods). As shown in Fig. 1C, unexpectedly, we occasionally observed promoter activities comparable with those of P2. The subpopulation of the genomic fragments whose promoter activities were more than average of the P2 was designated as G1 and the others as G2. As shown in Table 2, the overall G + C content of the G1 was similar to that of P2, and CpG islands were not observed at all. Interestingly, the frequency of TATA boxes (both strict and less-strict) in G1 was the highest in all of the populations. It is possible that TATA-like sequences present among the relatively AT-rich genomic sequences may happen to possess the minimal capacity to provide a sufficient docking platform for the RNA polymerase II complex (including the TATA binding protein), allowing the corresponding genomic regions to serve as ‘pseudo-’ promoters. Among them, some might have been fixed as P2 as a result of the fact that the downstream sequences became subjected to functional constraints as genes during the course of evolution.
3.4. Possible origin of a class of ‘ncRNAs’
From the analyses of promoter activities of primary DNA sequences of PPRs and random genomic sequences, we unexpectedly observed that the randomly isolated genomic fragments occasionally resembled P2 PPRs and displayed some promoter activities. This observation led us to hypothesize that this type of promoter activity could explain the origin of a population of a novel class of transcripts, non-protein-coding RNAs (ncRNAs). Recently, both human and mouse full-length cDNA projects have demonstrated that there is an unexpectedly large number of transcript species that are unlikely to encode any proteins. For example, our FLJ project isolated 768 human full-length cDNAs that should be categorized as ‘ncRNAs’. Similarly, the FANTOM project also identified 4280 mouse ‘ncRNAs’ (‘ncRNA core’).5,16,17 The biological significance of this emerging class of ‘ncRNAs’ is currently of great interest, especially for those which are sometimes called, ‘mRNA-like long ncRNAs’ or ‘Transcripts of Unknown Functions’18 (we simply call them as ‘ncRNA’ hereafter).
We first compared those so-called ‘ncRNAs’ identified from human and mouse full-length cDNA projects with each other. We observed that they are very scarcely overlapping (4%) with any meaningful parameters.9 Since concerned that the coverage of the human/mouse cDNAs was still not adequate for a meaningful comparison, we tried to map the corresponding human/mouse cDNAs, including PPRs, against the genomes of the counter-organisms. Still, essentially in no case was a significant hit detected. Intriguingly, a recent study demonstrated that knockout mice with mega-base-scale deletions of the genomic regions, where hundreds of those putative ‘ncRNAs’ are harbored, showed no detectable phenotypic features.20 We considered, if some parts of the genomic DNA are occasionally transcribed due to the above-mentioned sporadic promoter activities, it would be natural that most of them are evolutionarily non-conserved, since such transcription events should be free from functional constraints, even though they drive deterministic transcriptions in a particular species.
In order to test this hypothesis, we analyzed the promoter activities of the PPRs of the 88 ‘ncRNAs’ that had been identified from the most intensively annotated chromosomes, 20–22, by the FLJ cDNA project. We also confirmed that none of them have any significant homology with mouse ncRNAs. We first examined the gene expression of these ‘ncRNAs’ in HEK293 cells by semi-quantitative RT–PCR analysis. Clear expression of 49 putative ‘ncRNAs’ was detected (Supplementary Table 5 and Fig. 6). Among them, we successfully cloned the PPRs of 35 ‘ncRNAs’. As shown in Fig. 1C, their promoter activities were within a similar range of the P2 or G1 groups. At the same time, the sequence features of the PPRs most resembled them (Table 3; for raw data, see Supplementary Table 3; also see Supplementary Table 6).
Table 3.
Sequence features of the PPRs of ncRNAs and the orphan cDNAs
| ncRNAcore (%) | ncRNA (293positive) (%) | Orphan cDNAs | |
|---|---|---|---|
| CpG island | 87 (11) | 3 (9) | 1872 (20) |
| TATA box: strict | 117 (15) | 6 (23) | 1411 (15) |
| TATA box: less strict | 413 (54) | 22 (63) | 4584 (49) |
| Average G + C content | 0.45 | 0.44 | 0.47 |
| Total | 768 | 35 | 9377 |
These findings suggest the possibility that at least some of the ‘ncRNAs’ may be driven by non-genic G1-like promoter activities that are evolutionarily sporadically occurring. Assumed that 33/251 (G1; 13%) of the 1 kb of the genomic DNA could possess the promoter activities, there could be 4 × 105 such promoters in the 3.0 × 109 base of human genome. Also, in our 1.8 million one-pass cDNA sequences, 9377 clusters which were supported by three or more cDNAs were located well outside of the previously annotated protein-coding genes defined by RefSeqs (‘Orphan cDNAs’). At the one-pass sequence level, most of them seemed not to correspond to protein-coding transcripts. We also analyzed the sequence features of their PPRs (Table 3). Again, we found that the PPRs of this category also resemble the features of P2 or G1. This result also supported our hypothesis that current number of orphan TSSs might be originated from sporadic promoter activities described in the present study.
4. Conclusions
In this paper, we have described promoter activities intrinsic to primary DNA sequences for PPRs of protein coding genes, random genomic regions, and PPRs of putative ncRNAs. We thereby demonstrated that there are two types of promoter activites, which are represented by P1 and P2. We also showed that average genomic DNA sequence occasionally possesses P2-type promoter activities, which may explain the origin of ncRNAs as well. Recent studies also elucidates that the genes having multiple promoters (alternative promoters) are widespread in human and mouse genes.21–25 Sporadic promoter activities emerged in the internal part of genes might explain the origin of at least some of such massively discovered alternative promoters, too. The observations and hypothesis produced by this paper should provide important viewpoint to analyze the complex nature of the transcriptome of human genes.
On the other hand, studies on alternative splicing have revealed that minor alternative splicing isoforms of transcripts tend not to be conserved between humans and mice, and it is thought that this non-conservation may possibly serve as an evolutionary reservoir for novel variants.26 Likewise, formation of ab initio promoters may take place relatively frequently among the repertory of the genomic sequences. The framework of the transcriptional modulation of human genes may have a more dynamic nature than previously thought.27 It is also possible that those sporadic promoter activities identified in the present study have been imposing an inherent problem for developing promoter prediction programs.28 Further integrative analyses of both the promoters and transcriptome of human genes will lead to a better understanding of the system architecture of the transcriptional network of human genes.
Acknowledgements
We are thankful to E. Nakajima for helpful discussion and critical reading of the manuscript. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas and by special coordination funds for promoting science and technology (SCF), both from the Ministry of Education, Culture, Sports, Science and Technology in Japan and by Institute for Bioinformatics Research and Development.
Supplementary Data
Supplementary data are available online at www.dnaresearch.oxfordjournals.org.
References
- 1.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431,:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Q. H., Ye M., Wu X. Y., Ren S. X., Zhao M., Zhao C. J., Fu G., Shen Y., Fan H. Y., Lu G., et al. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34 + hematopoietic stem/progenitor cells. Genome Res. 2000;10:1546–1560. doi: 10.1101/gr.140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiemann S., Weil B., Wellenreuther R., Gassenhuber J., Glassl S., Ansorge W., Bocher M., Blocker H., Bauersachs S., Blum H., et al. Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res. 2001;11:422–435. doi: 10.1101/gr.154701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strausberg R. L., Feingold E. A., Grouse L. H., Derge J. G., Klausner R. D., Collins F. S., Wagner L., Shenmen C. M., Schuler G. D., Altschul S. F., et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 6.Imanishi T., Itoh T., Suzuki Y., O'Donovan C., Fukuchi S., Koyanagi K. O., Barrero R. A., Tamura T., Yamaguchi-Kabata Y., Tanino M., et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser A. G., Marcotte E. M. A probabilistic view of gene function. Nat. Genet. 2004;36:559–564. doi: 10.1038/ng1370. [DOI] [PubMed] [Google Scholar]
- 8.Khodursky A. B., Bernstein J. A. Life after transcription—revisiting the fate of messenger RNA. Trends Genet. 2003;19,:113–115. doi: 10.1016/S0168-9525(02)00047-1. [DOI] [PubMed] [Google Scholar]
- 9.Roeder R. G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 10.Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A. E., Kel-Margoulis O. V., et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki Y., Sugano S. Construction of a full-length enriched and a 5′-end enriched cDNA library using the oligo-capping method. Methods Mol. Biol. 2003;221:73–91. doi: 10.1385/1-59259-359-3:73. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita R., Suzuki Y., Wakaguri H., Tsuritani K., Nakai K., Sugano S. DBTSS: DataBase of Human Transcription Start Sites, progress report 2006. Nucleic Acids Res. 2006;34,:D86–D89. doi: 10.1093/nar/gkj129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki Y., Tsunoda T., Sese J., Taira H., Mizushima-Sugano J., Hata H., Ota T., Isogai T., Tanaka T., Nakamura Y., et al. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 2001;11:677–684. doi: 10.1101/gr.164001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trinklein N. D., Aldred S. J., Saldanha A. J., Myers R. M. Identification and functional analysis of human transcriptional promoters. Genome Res. 2003;13:308–312. doi: 10.1101/gr.794803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Versteeg R., van Schaik B. D., van Batenburg M. F., Roos M., Monajemi R., Caron H., Bussemaker H. J., van Kampen A. H. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003;13:1998–2004. doi: 10.1101/gr.1649303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Numata K., Kanai A., Saito R., Kondo S., Adachi J., Wilming L. G., Hume D. A., Hayashizaki Y., Tomita M. Identification of putative noncoding RNAs among the RIKEN mouse full-length cDNA collection. Genome Res. 2003;13:1301–1306. doi: 10.1101/gr.1011603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 18.Willingham A. T., Gingeras T. R. TUF love for “junk” DNA. Cell. 2006;125,:1215–1220. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Wang J., Zhang J., Zheng H., Li J., Liu D., Li H., Samudrala R., Yu J., Wong G. K. Mouse transcriptome: neutral evolution of ‘non-coding’ complementary DNAs. Nature. 2004;431 757. [PubMed] [Google Scholar]
- 20.Nobrega M. A., Zhu Y., Plajzer-Frick I., Afzal V., Rubin E. M. Megabase deletions of gene deserts result in viable mice. Nature. 2004;431,:988–993. doi: 10.1038/nature03022. [DOI] [PubMed] [Google Scholar]
- 21.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309,:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 22.Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308,:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 23.Kim T. H., Barrera L. O., Zheng M., Qu C., Singer M. A., Richmond T. A., Wu Y., Green R. D., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436,:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura K., Wakamatsu A., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16,:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landry J. R., Mager D. L., Wilhelm B. T. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet. 2003;19,:640–648. doi: 10.1016/j.tig.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Modrek B., Lee C. J. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 27.Rockman M. V., Wray G. A. Abundant raw material for cis-regulatory evolution in humans. Mol. Biol. Evol. 2002;19:1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- 28.Bajic V. B., Tan S. L., Suzuki Y., Sugano S. Promoter prediction analysis on the whole human genome. Nat. Biotechnol. 2004;22,:1467–1473. doi: 10.1038/nbt1032. [DOI] [PubMed] [Google Scholar]