Abstract
RNA silencing is a broadly conserved machinery and is involved in many biological events. Small RNAs are key molecules in RNA silencing pathway that guide sequence-specific gene regulations and chromatin modifications. The silencing machinery works as an anti-viral defense in virus-infected plants. It is generally accepted that virus-specific small interfering (si) RNAs bind to the viral genome and trigger its cleavage. Previously, we have cloned and obtained sequences of small RNAs from Arabidopsis thaliana infected or uninfected with crucifer Tobacco mosaic virus. MicroRNAs (miRNAs) accumulated to a higher percentage of total small RNAs in the virus-infected plants. This was partly because the viral replication protein binds to the miRNA/miRNA* duplexes. In the present study, we mapped the sequences of small RNAs other than virus-derived siRNAs to the Arabidopsis genome and assigned each small RNA. It was demonstrated that only miRNAs increased as a result of viral infection. Furthermore, some newly identified miRNAs and miRNA candidates were found from the virus-infected plants despite a limited number of examined sequences. We propose that it is advantageous to use virus-infected plants as a source for cloning and identifying new miRNAs.
Key words: Arabidopsis thaliana, Tobacco mosaic virus, microRNA, RNA silencing
1. Introduction
Small RNAs play important roles in RNA silencing mechanisms which are involved in many biological processes in eukaryotes. These small RNAs guide post-transcriptional gene silencing by inhibiting translation or degrading the target mRNAs, and guide transcriptional gene silencing by modifying chromatin.1–3
In Arabidopsis thaliana, functional small RNAs are mostly 21–24 nt long. The best-studied endogenous small RNAs are microRNAs (miRNAs). In Arabidopsis, miRNAs are excised from primary miRNA transcripts forming stem-loop structures by the RNase III enzyme DICER-LIKE 1 (DCL1). This enzyme works coordinately with HYL1 and SERRATE to produce miRNA/miRNA* duplexes.4–9 miRNAs are loaded into the RNA-induced silencing complex (RISC) where the complementary mRNAs are cleaved by ARGONAUTE 1 (AGO1), which has Slicer activity.10,11 Plants have some unique small RNAs in addition to miRNA. Trans-acting siRNA (tasiRNA) is produced from long double-stranded RNA (dsRNA) by DCL4 with its binding partner, DRB4.12–14 miRNA and tasiRNA are mainly involved in plant development, response to environmental stresses and so on through regulating gene expression.1–3 siRNAs derived from natural-antisense transcripts (nat-siRNAs) exist in Arabidopsis and function in the stress response and bacterial disease resistance.15,16 However, the numbers and functions of nat-siRNAs are not fully understood.
The RNA silencing pathway also serves as an anti-viral defense in plants. Viral genome-derived siRNAs are detected in plants infected with RNA viruses17,18 and DNA viruses.19,20 The lengths of these siRNAs differ with the type of viruses,18,21 suggesting the involvement of different DCLs in producing the respective viral siRNAs. It is conceived that the resulting siRNAs are loaded into the RISC as well as endogenous small RNAs, bind the viral genome through complementary sequences and direct the degradation of the viral genome. To counteract the silencing machinery of plants, many viruses have evolved genes which encode silencing suppressor proteins with distinct properties.22,23 Many suppressors have dsRNA binding activity so that virus siRNAs are trapped, resulting in inhibition of the silencing machinery.24,25 In contrast to the dsRNA binding strategy, the Cucumber mosaic virus-encoded 2b protein directly interacts with AGO1 and inhibits its activity26 and Turnip crinkle virus-encoded P38 protein suppresses DCL4 activity.18
We have examined the crucifer Tobacco mosaic virus-Cg (TMV-Cg) infection of Arabidopsis as a model system to study plant-Tobamovirus interaction.27,28 TMV-Cg is a positive-sense, single-stranded RNA virus. We have previously cloned and sequenced small RNAs from Arabidopsis infected or uninfected with TMV-Cg and demonstrated that miRNAs increased in the virus-infected plants.28 In TMV and Tomato mosaic virus (ToMV), which both belong to the Tobamovirus family, 126K replication proteins suppress RNA silencing.29,30 Thus, it is postulated that the 126K replication protein of TMV-Cg also acts as a silencing suppressor because it binds to small dsRNAs.28 Owing to this activity, 126K replication protein binds to miRNA/miRNA* duplexes, resulting in the accumulation of miRNAs. In the present study, we examined all small RNA sequences obtained from Arabidopsis infected or uninfected with TMV-Cg. We then mapped the region where each small RNA sequence was derived in the Arabidopsis genome. Among the small RNAs from virus-infected plants, we identified several newly identified miRNAs and miRNA candidates. Virus-infected plants could be an effective source to find novel miRNAs.
2. Materials and methods
2.1. Mapping of small RNAs using BLAST searches
We have analyzed small RNA sequences obtained previously; 1700 and 543 reads from the leaves of Arabidopsis infected and uninfected with TMV-Cg at 3 days post-infection (dpi), respectively.28 Each sequence of non-viral genome origin was mapped to the Arabidopsis genome by NCBI BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) and TAIR BLAST (http://www.arabidopsis.org/Blast/index.jsp) searches. During searches, we allowed one mismatch at its 3′ end, because 3′ ends of small RNAs in virus-infected plants are possibly unmethylated and unstable.31 All BLAST searches were performed automatically using BioPython script programs.
2.2. Other computational analyses
Secondary structures of miRNA precursors were predicted using the mFOLD program (http://www.bioinfo.rpi.edu/applications/mfold/).32 Targets of miRNAs were predicted using the miRU: Plant microRNA Potential Target Finder (http://bioinfo3.noble.org/miRNA/miRU.htm).33
To compare the ratio of our sequences to that of other databases, each sequence was searched against the Arabidopsis MPSS Plus Database (http://mpss.udel.edu/at/)34 and the Arabidopsis Small RNA Project (ASRP) (http://asrp.cgrb.oregonstate.edu/).35 In Table 2, miRNAs were searched against 4Co10 (Co1-0 wildtype inflorescence tissues, small RNA 454) in MPSS and Co1-0 (Leaf, Inflorescence) in ASRP. In Fig. 2, small RNA sequences were searched against all datasets in both databases.
Table 2.
Newly identified miRNAs and miRNA candidate are more abundant in TMV-Cg-infected Arabidopsis than in other databases
| miRNA | Number of read (percentage%) | |||
|---|---|---|---|---|
| TMV | MPSS | ASRP Leaf | ASRP Inflorescence | |
| miR847 (5′) | 2 (0.12%) | 0 | 0 | 0 |
| miR822 | 5 (0.29%) | 0 | 0 | 0 |
| miR823 | 1 (0.06%) | 1 (0.01%) | 7 (0.04%) | 9 (0.01%) |
| miR824 | 6 (0.35%) | 15 (0.13%) | 15 (0.09%) | 42 (0.05%) |
| Total | 1700 (100%) | 11 631 (100%) | 15 826 (100%) | 78 583 (100%) |
2.3. Plant materials and virus inoculation
The following Arabidopsis plants were used; Col-0 (wild type), dcl1-9 (BC6), dcl2-2 (SALK_123586), dcl3-1 (SALK_005512) and dcl4-1 (BC6). Dr Herve Vaucheret (INRA, France) kindly provided dcl1-9 (BC6) and dcl4-1 (BC6), which were backcrossed to Col-0 six times. All plants were grown in a growth chamber at 22°C with photoperiod of 16 h. For the virus inoculation, leaves of 4-week-old plants were dusted with carborundum and gently rubbed with 20 µg/mL of TMV-Cg solution. For mock inoculation, TMV-Cg solution was replaced with 10 mM sodium phosphate buffer.
2.4. Northern blot analysis
Total RNA was extracted at 10 dpi from the inoculated leaves, mock-inoculated leaves and inflorescences of Arabidopsis using ISOGEN reagent (Nippon gene). Aliquots of 10 µg of total RNA were loaded and resolved on a denaturing 15% polyacrylamide gel (7 M urea). To make the miR847(5′) probe, oligonucleotide (TCGGCTTCCCATTCCTCTTCA) was 5′ end-labeled with [γ-32P] ATP using T4 polynucleotide kinase (TOYOBO). Hybridization was carried out at 40°C using PerfectHyb Plus hybridization buffer (Sigma). The blots were exposed on imaging plates, and signals were visualized using BAS-2500 (Fuji).
3. Results and discussion
3.1. Increase of miRNAs in Arabidopsis infected with TMV-Cg
We have previously sequenced small RNAs from the leaves of Arabidopsis infected or uninfected with TMV-Cg at 3 dpi. We obtained 1700 sequences from the virus-infected plants and 543 sequences from mock-infected plants.28 We reported that total miRNAs were more abundant in virus-infected plants than in mock-infected plants, as shown in the northern blot results of some miRNAs. To further compare the small RNA expression profiles between virus-infected plants and mock-infected plants, we performed extensive BLAST searches to map all small RNA sequences to the Arabidopsis genome (Table 1), except for siRNAs derived from the viral genome which were 210 reads (12.4 % of total reads), as previously reported.28
Table 1.
Summary of non-viral small RNA sequences from TMV-infected or uninfected Arabidopsis
| Class | Mock | TMC-Cg | ||
|---|---|---|---|---|
| miRNA | ||||
| Known miRNA | 25 | (4.6%) | 379 | (25.4%) |
| miRNA* | 4 | (0.7%) | 37 | (2.5%) |
| Small RNA drived from miRNA precursor | 0 | (0%) | 5 | (0.3%) |
| tasiRNA | 11 | (2.0%) | 24 | (1.6%) |
| Gene | ||||
| Sense | 15 | (2.8%) | 29 | (1.9%) |
| Antisense | 3 | (0.6%) | 15 | (1.0%) |
| Sense and antisense | 0 | (0%) | 2 | (0.1%) |
| Intergenic region | 100 | (18.4%) | 94 | (6.3%) |
| rRNA | 164 | (30.2%) | 486 | (32.6%) |
| tRNA | 19 | (3.5%) | 47 | (3.2%) |
| sRNA | 2 | (0.4%) | 1 | (0.1%) |
| Transposon | 11 | (2.0%) | 9 | (0.6%) |
| Unknown | 189 | (34.8%) | 362 | (24.3%) |
| Total reads | 543 | (100%) | 1490a | (100%) |
aViral genome-derived siRNA are not included.
The percentage of total miRNAs was 5.5 times higher in TMV-infected plants (25.4%) than that in mock-infected plants (4.6%) (Table 1 and Kurihara et al.28). In contrast to the large increase of miRNAs in TMV-Cg-infected plants, the percentage of tasiRNAs was almost unchanged (2.0% in mock-infected plants and 1.6% in virus-infected plants) (Table 1). tasiRNA is another class of small RNAs in plants which is produced by the RNase III enzyme DCL4 protein.36 This suggests that miRNAs, among other characterized small RNA classes, specifically increased by the TMV infection.
We have recently demonstrated that the increase of miRNAs could be attributed to the activity of the viral-encoded 126K protein, which binds and therefore stabilizes miRNA/miRNA* duplexes.28 However, we found that there is no big difference between the ratios of miRNA to miRNA* in mock- and TMV-infected plants (6.6 times in mock-infected plants and 10 times in TMV-infected plants). This fact possibly indicates that there is still another mechanism of the miRNA increase or stabilization in the virus-infected plants, in addition to binding activity of 126K protein to miRNA/miRNA* duplexes.
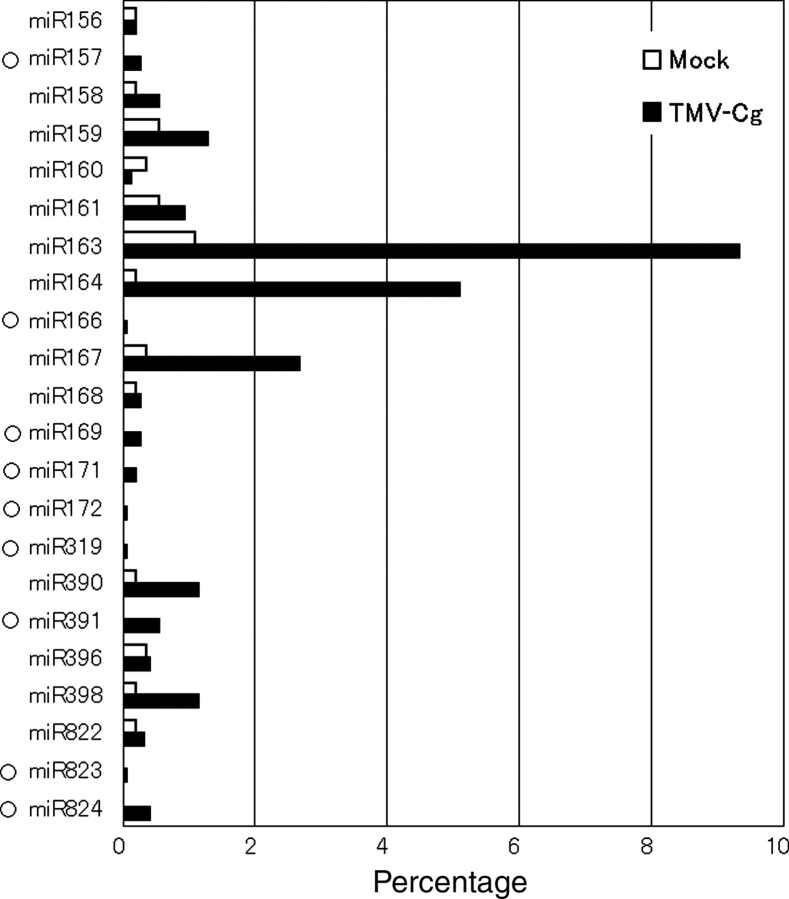
We compared the percentage of each miRNA between mock-infected and virus-infected plants (Fig. 1). The percentages of all miRNAs except for miR160 increased as a result of virus infection. Some miRNAs showed remarkable increases, especially in miR163 (from 1.1 to 9.3%), miR164 (from 0.2 to 5.1%) and miR167 (from 0.4 to 2.7%). Most others increased 2–3 times in virus-infected plants. This result indicates that binding activity of the TMV-Cg replication protein to those miRNA/miRNA* duplexes is specifically high, or that transcription of some miRNAs is also activated.
Figure 1.
Most miRNAs increased in TMV-Cg-infected plants. Percentages of each miRNA cloned in non-viral small RNAs are shown. All miRNAs except for miR160 increased in the virus-infected plants. Circle indicates miRNA that was detected only in the small RNA dataset from the virus-infected plants.
Inhibition of the miRNA pathway has been reported with many other virus-infected plants and with plants transformed with viral proteins.26,37–39 Initially we expected that miRNA up-regulation should lead the down-regulation of the target mRNAs and that might work as a defense response to virus infection. However, in the TMV-Cg-infected plants, the target mRNAs of miRNAs were not down-regulated, even though miRNAs increased (Supplementary Fig. S1). The increase of miRNA does not induce target mRNA degradation and the miRNA pathway is completely disrupted in plants infected with TMV-Cg, as is the case during infection with other plant viruses. This supports the notion that the replication protein of TMV-Cg acts as a silencing suppressor.
3.2. Unique small RNAs in gene and intergenic regions in TMV-infected and mock-infected plants
In addition to the increase of miRNA percentage in plants infected with TMV-Cg, some miRNAs were cloned and sequenced only in the virus-infected plants (Fig. 1; indicated with circles). This led us to speculate that the virus-infected plants could be a good source for finding new miRNAs.
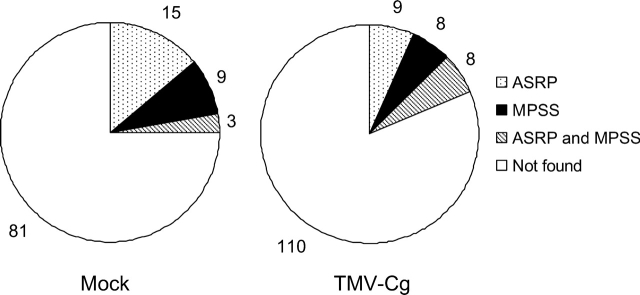
If there are new miRNAs in our small RNA sequence reads, they are likely to be mapped in the gene or intergenic region (Table 1). Therefore, we checked whether there are new sequences in the small RNAs mapped to the gene and intergenic regions compared with other small RNA databases; ASRP35 and MPSS34 (Fig. 2). Interestingly, not only in TMV-Cg-infected plants but also in mock-infected plants, most small RNAs were not detected in ASRP and MPSS databases and were unique to our reads, although there are 34 times more sequences in MPSS and 260 times more in ASRP than in our dataset from virus-infected plants. This is possibly due to the differences in RNA sources and cloning methods between ours and other groups' which performed deep sequencing using the 454 sequencing method. This suggests that many small RNAs remain to be discovered. They will be found if the cloning condition is changed even though the total reads are less than those using 454 sequencing. Thus, it is quite possible that there are new functional small RNAs in our resources of virus-infected and mock-infected plants. Especially in the TMV-infected plants in which many kinds of miRNAs were more abundant, we can expect new miRNA reads.
Figure 2.
Most sequences in this study are not found in other databases. Each small RNA sequence was searched against the ASRP and MPSS databases. Numbers of different sequences in gene and intergenic regions are shown, which are also found in ASRP and/or MPSS database, or not found in both. In both mock-infected and TMV-Cg-infected plants, most sequences are unique in this study.
3.3. miRNA candidates in plants infected with TMV-Cg
Three miRNAs (miR822, miR823, and miR824) reported very recently40 were found in our reads of virus-infected plants and, moreover, their appearances were more frequent in our total small RNAs from virus-infected plants than those in MPSS and ASRP databases (Table 2). This supports that virus-infected plants are good source for identifying novel miRNAs.
Next, we tried to find out novel miRNAs in the small RNA data of the virus-infected plants. Before confirmation and validation of new miRNAs, we performed secondary structure predictions using the mFOLD program. We used sequences ∼300 nt in length from around the matched stretch where small RNAs mapped to the gene and intergenic regions. Three different sequences formed hairpin structures, which are common structures to miRNA precursors (Fig. 3 and Supplementary Fig. S2), suggesting that these sequences are candidates of miRNAs.
Figure 3.
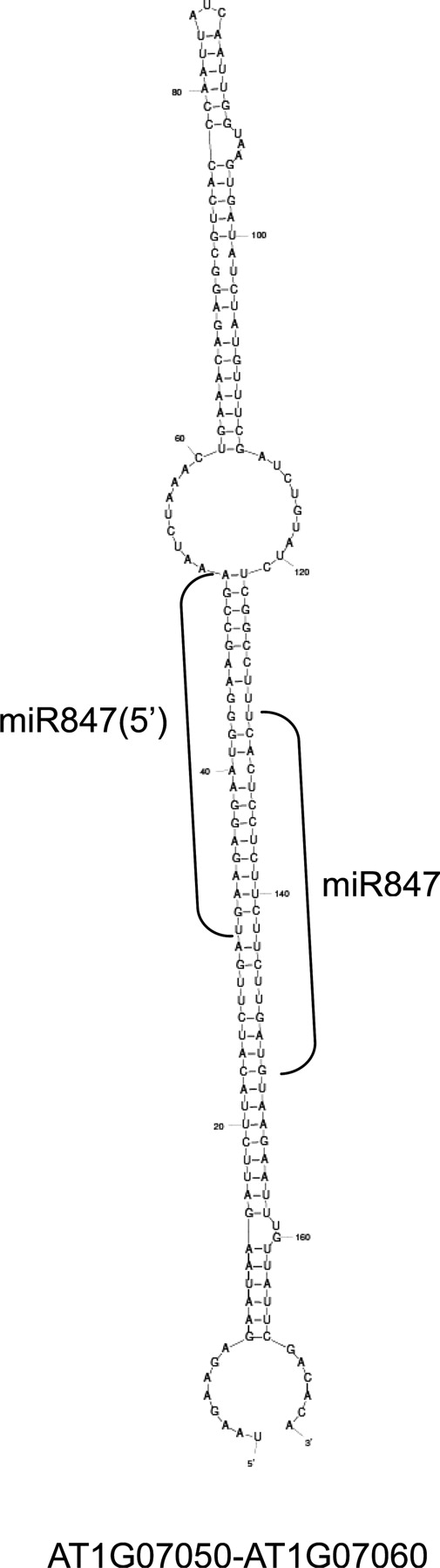
Predicted secondary structure of new miRNA candidate's precursor. Secondary structure of new miRNA candidate found in this study was predicted using the mFOLD program. miR847(5′) is derived from the 5′ arm of miR847 but they are not in miRNA/miRNA* relationship.
Out of them, we found that one candidate could be mapped to the 5′ arm of miR847 (Fig. 3). Though it may be a variant of miR847*, it is possible that the small RNA is also functional because it has an 8 nt overhang instead of 2 nt, which the DCL product generally has (Fig. 3). Thus, we named it miR847(5′) and analyzed further. In our small RNA data, there was no miR847 sequence while miR847(5′) was read twice. miR847(5′) was not cloned in ASRP and MPSS databases (Table 2). The other two sequences forming hairpin structures were named candidates A and B (Supplementary Fig. S2). Each of them was read once in the small RNA data from virus-infected plants.
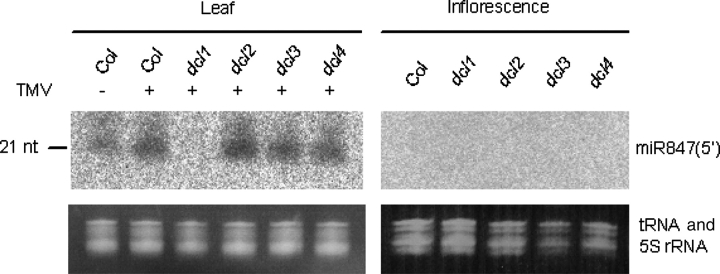
We performed northern blot analyses to detect these miRNA candidates and confirm their expression patterns. Out of three miRNA candidates, only miR847(5′) could be detected (Fig. 4). Candidates A and B were not detected.
Figure 4.
Northern blot analysis of miR847(5′). Total RNA (10 µg) from rosette leaves (mock-infected and infected with TMV-Cg) and inflorescences were used for northern blot analyses. miR847(5′) increased during TMV-Cg infection but was not detected in the dcl1mutant, indicating that miR847(5′) is miRNA. miR847(5′) was not detected in inflorescences.
miR847(5′) was not expressed in flowers but in rosette leaves. It was more abundant in TMV-Cg-infected plants than in mock-infected plants (Fig. 4). This result reflected the tendency observed in the cloning frequency discussed earlier and northern blot results of other miRNAs reported previously.28 Furthermore, miR847(5′) was expressed at a similar level in mutants except in the dcl1 mutant. In Arabidopsis, most miRNAs are produced by DCL1 in general, whereas, for example, tasiRNAs require both DCL1 and DCL4 for their productions. Therefore, if the expression of a small RNA is reduced only in dcl1 mutant plants, the small RNA is possibly a miRNA. Therefore, the northern blot result supports that miR847(5′) is an miRNA. In contrast, miR847 could not be detected by northern blot analysis neither in leaves nor in inflorescences (data not shown).
The predicted targets of these three miRNA candidates and miR847 are shown in Supplementary Table S1. However, we have not yet detected their possible cleaved fragment by the 5′ RACE method.38 On the basis of the available data, we could not conclude which of small RNAs is a functional miRNA; miR847 and miR847(5′) at present. Further analyses will be needed to make conclusion.
Recently, many results of comprehensive small RNA sequencing by 454 sequencing have been reported one after another in Arabidopsis.40–43 It has been demonstrated that miRNAs are enriched in loss-of-function mutants of RNA-dependent RNA polymerase 2 (RDR2) and RNA polymerase IV because they are required for the majority of endogenous siRNAs.41,43 By analyzing these mutant plants with the 454 sequencing method, dozens of miRNAs were discovered. In this study, we showed that plants infected with TMV-Cg are good materials for cloning and finding novel miRNAs frequently because cloning percentages of many miRNAs increase and newly identified miRNAs are found in virus-infected plants. Moreover, considering that the number of small RNA sequences read were tens to hundreds less than those of other groups, TMV-infected plants are very efficient materials to discover new miRNAs. It is anticipated that new miRNAs would be found from small RNAs in the virus-infected leaves if extensive reads were performed.
Most Arabidopsis miRNAs, expressed abundantly and conserved, have already been identified. However, there are many kinds of miRNAs which are less-expressed and non-conserved42 and it is possible that some are yet to be discovered. These miRNAs will be hereafter identified using plants under various conditions or mutant plants, which will leads to the identification of overall miRNAs and siRNAs and the understanding of how small RNAs are involved in many biological processes in Arabidopsis.
Funding
Sasakawa Scientific Research Grant from the Japan Science Society to Y.T.
Acknowledgement
We thank Dr Herve Vaucheret (INRA, France) for providing the dcl1-9 (BC6) and dcl4-1 (BC6) mutants.
Supplementary Data
Supplementary data are available online at www.dnaresearch.oxfordjournals.org.
References
- 1.Vazquez F. Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci. 2006;11:460–468. doi: 10.1016/j.tplants.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 3.Jones-Rhoades M. W., Bartel D. P., Bartel B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 4.Kurihara Y., Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurihara Y., Takashi Y., Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han M. H., Goud S., Song L., Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang L., Liu Z., Lu F., Dong A., Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- 8.Vazquez F., Gasciolli V., Crete P., Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 9.Fujioka Y., Utsumi M., Ohba Y., Watanabe Y. Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 2007;48:1243–1253. doi: 10.1093/pcp/pcm099. [DOI] [PubMed] [Google Scholar]
- 10.Qi Y., Denli A. M., Hannon G. J. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Baumberger N., Baulcombe D. C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adenot X., Elmayan T., Lauressergues D., et al. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 13.Nakazawa Y., Hiraguri A., Moriyama H., Fukuhara T. The dsRNA-binding protein DRB4 interacts with the Dicer-like protein DCL4 in vivo and functions in the trans-acting siRNA pathway. Plant Mol. Biol. 2007;63:777–785. doi: 10.1007/s11103-006-9125-8. [DOI] [PubMed] [Google Scholar]
- 14.Vaucheret H. MicroRNA-dependent trans-acting siRNA production. Sci. STKE. 2005;2005:ppe43. doi: 10.1126/stke.3002005pe43. [DOI] [PubMed] [Google Scholar]
- 15.Borsani O., Zhu J., Verslues P. E., Sunkar R., Zhu J. K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katiyar-Agarwal S., Morgan R., Dahlbeck D., et al. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molnar A., Csorba T., Lakatos L., Varallyay E., Lacomme C., Burgyan J. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deleris A., Gallego-Bartolome J., Bao J., Kasschau K. D., Carrington J. C., Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 19.Akbergenov R., Si-Ammour A., Blevins T., et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 2006;34:462–471. doi: 10.1093/nar/gkj447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moissiard G., Voinnet O. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc. Natl. Acad. Sci. USA. 2006;103:19593–19598. doi: 10.1073/pnas.0604627103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Bouche N., Lauressergues D., Gasciolli V., Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M. B., Metzlaff M. RNA silencing and antiviral defense in plants. Curr. Opin. Plant Biol. 2005;8:216–222. doi: 10.1016/j.pbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Li W. X., Ding S. W. Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 2001;12:150–154. doi: 10.1016/s0958-1669(00)00190-7. [DOI] [PubMed] [Google Scholar]
- 24.Ye K., Malinina L., Patel D. J. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakatos L., Csorba T., Pantaleo V., et al. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X., Yuan Y. R., Pei Y., et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurihara Y., Watanabe Y. A TMV-Cg mutant with a truncated coat protein induces cell death resembling the hypersensitive response in Arabidopsis. Mol. Cells. 2004;17:334–339. [PubMed] [Google Scholar]
- 28.Kurihara Y., Inaba N., Kutsuna N., Takeda A., Tagami Y., Watanabe Y. The binding of tobamovirus replication protein with small RNA duplexes. J. Gen. Virol. 2007 doi: 10.1099/vir.0.82994-0. in press. [DOI] [PubMed] [Google Scholar]
- 29.Ding X. S., Liu J., Cheng N. H., et al. The Tobacco mosaic virus 126-kDa protein associated with virus replication and movement suppresses RNA silencing. Mol. Plant Microbe. Interact. 2004;17:583–592. doi: 10.1094/MPMI.2004.17.6.583. [DOI] [PubMed] [Google Scholar]
- 30.Kubota K., Tsuda S., Tamai A., Meshi T. Tomato mosaic virus replication protein suppresses virus-targeted posttranscriptional gene silencing. J. Virol. 2003;77:11016–11026. doi: 10.1128/JVI.77.20.11016-11026.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu B., Chapman E. J., Yang Z., Carrington J. C., Chen X. Transgenically expressed viral RNA silencing suppressors interfere with microRNA methylation in Arabidopsis. FEBS Lett. 2006;580:3117–3120. doi: 10.1016/j.febslet.2006.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano M., Nobuta K., Vemaraju K., Tej S. S., Skogen J. W., Meyers B. C. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006;34:D731–D735. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gustafson A. M., Allen E., Givan S., Smith D., Carrington J. C., Kasschau K. D. ASRP: the Arabidopsis Small RNA Project Database. Nucleic Acids Res. 2005;33:D637–D640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Z., Allen E., Wilken A., Carrington J. C. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mallory A. C., Reinhart B. J., Bartel D., Vance V. B., Bowman L. H. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. USA. 2002;99:15228–15233. doi: 10.1073/pnas.232434999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kasschau K. D., Xie Z., Allen E., et al. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev. Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 39.Blevins T., Rajeswaran R., Shivaprasad P. V., et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajagopalan R., Vaucheret H., Trejo J., Bartel D. P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X., Henderson I. R., Lu C., Green P. J., Jacobsen S. E. Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. USA. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fahlgren N., Howell M. D., Kasschau K. D., et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of miRNA genes. PLoS ONE. 2007;2 doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu C., Kulkarni K., Souret F. F., et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]