Abstract
We generated a high-density genetic linkage map of soybean using expressed sequence tag (EST)-derived microsatellite markers. A total of 6920 primer pairs (10.9%) were designed to amplify simple sequence repeats (SSRs) from 63 676 publicly available non-redundant soybean ESTs. The polymorphism of two parent plants, the Japanese cultivar ‘Misuzudaizu’ and the Chinese line ‘Moshidou Gong 503’, were examined using 10% polyacrylamide gel electrophoresis. Primer pairs showing polymorphism were then used for genotyping 94 recombinant inbred lines (RILs) derived from a cross between the parents. In addition to previously reported markers, 680 EST-derived microsatellite markers were selected and subjected to linkage analysis. As a result, 935 marker loci were mapped successfully onto 20 linkage groups, which totaled 2700.3 cM in length; 693 loci were detected using the 668 EST-derived microsatellite markers developed in this study, the other 242 loci were detected with 105 RFLP markers, 136 genome-derived microsatellite markers, and one phenotypic marker. We examined allelic variation among 23 soybean cultivars/lines and a wild soybean line using 668 mapped EST-derived microsatellite markers (corresponding to 686 marker loci), in order to determine the transferability of the markers among soybean germplasms. A limited degree of macrosynteny was observed at the segmental level between the genomes of soybean and the model legume Lotus japonicus, which suggests that considerable genome shuffling occurred after separation of the species and during establishment of the paleopolyploid soybean genome.
Key words: soybean, microsatellite markers, genetic linkage map, polymorphism information content, comparative map
1. Introduction
Soybean [Glycine max (L.) Merrill] has been an important crop throughout much of human history, and it is used not only as a food, but also as an oil source, forage, and as a raw material for a variety of industrial materials. During the last 10 years, global soybean production has increased 1.8-fold and in 2006, production levels reached 236 million tons.1 Recently, soybean has drawn particular attention as an energy source, since it can be used as the principal source of biodiesel, an alternative fuel. Thus, soybean production is expected to increase further during the coming years.
Due to its economic importance, many efforts have been made to develop genetic and genomic resources for soybean, including genetic linkage maps2–7 and expressed sequence tag (EST) collections.8 By 2006, soybean had the 6th largest EST collection,9 and in 2007, a transcriptional map comprising 1141 single nucleotide polymorphism (SNP) markers was generated using these ESTs.10 Furthermore, sequencing of the entire soybean genome is in progress [http://soybeangenome.siu.edu], and the ultimate goal of this project is acceleration of the breeding process.
With accumulating information and materials, more efficient approaches to breeding may be developed by merging genomic resources and applied genetics. An example of this new approach is the investigation of genomic diversity in soybeans, followed by identification of genes of agronomic importance. Hyten et al.11 compared genome structures in limited lengths of DNA (336–574 kb) from 120 genetically diverse soybean germplasms. The authors found a highly variable linkage disequilibrium (LD), not only among populations, but also between different regions of the genome. This finding suggests that the soybean genome comprises numerous DNA segments that exhibit various degrees of diversification, according to differing frequencies of genetic recombination. It is expected that dissection of the complex genome via evaluation of genome-wide allelic polymorphism of DNA markers, will contribute to our understanding of genomic diversity in soybean.
Comparative genomics may represent another promising means of utilizing genomic resources for applied genetics. Recently, comparisons were performed between the genome structures of two model legumes (Lotus japonicus and Medicago truncatula) and other leguminous crops.12,13 A limited degree of macrosynteny was detected between the genomes of model and crop legumes, but these observations were far from conclusive, since only partial information was available for the comparison. With the accumulation of genomic information, such as nucleotide sequences and DNA markers, the accuracy of genomic comparisons will improve and knowledge may be transferred more efficiently from model systems to soybean.
Among the various DNA markers invented thus far, EST-derived microsatellites are particularly popular because they exhibit the following characteristics: (i) they are cost-effective, especially when developed using publicly available EST information; (ii) they are informative because of their multi-allelic and co-dominant nature;14 (iii) they are useful for studying functional diversity in natural populations or germplasm collections since they are derived from transcription products;15 and (iv) they are transferable to other species and can be used for comparative mapping since most are derived from conserved coding sequences.15,16 With respect to soybean, although nearly 400 000 ESTs have been deposited in public DNA databases, only 24 polymorphic microsatellite markers have been generated using these ESTs.6
In this study, we developed microsatellite markers using publicly available EST information and generated a genetic linkage map for the entire soybean genome. In addition, we examined allele polymorphism for all the mapped EST-derived microsatellite markers and considered each marker's transferability between soybean germplasms. Moreover, we compared the genome structures of L. japonicus and soybean and discussed the transferability of structural information between them.
2. Materials and methods
2.1. Plant and DNA materials
In order to construct a genetic linkage map for soybean, we used a mapping population of 94 individual plants, which represented the F8 generation of recombinant inbred lines (RILs) derived from an F2 population generated between the parent plants ‘Misuzudaizu’ and ‘Moshidou Gong 503’.5,7 ‘Misuzudaizu’ is a Japanese cultivar and ‘Moshidou Gong 503’ is a Chinese experimental line that is an intermediate between cultivated soybean and wild soybean (G. soja). The total DNA was extracted from the leaves of each plants by CTAB method.17
2.2. Development of microsatellite markers
Microsatellite or simple sequence repeats (SSRs) ≥13 nucleotides in length, which contained all possible combinations of di-nucleotide (NN), tri-nucleotide (NNN), and tetra-nucleotide (NNNN) repeat, were identified from soybean non-redundant ESTs in public databases (Dana-Farber Center Institute; http://compbio.dfci.harvard.edu/tgi) using the a FindPatterns module from the GCG software package (Accelrys, Inc., San Diego, CA, USA). In order to amplify DNA segments containing SSRs, primer pairs were designed using the Primer3 program18 in such a way that the amplified products ranged between 90 and 300 bp in length. PCR amplifications (5 µl) were performed on 0.5 ng soybean genomic DNA in 1 × PCR buffer (BIOLINE, London, UK), 3 mM MgCl2, 0.02 U BIOTAQTM DNA Polymerase (BIOLINE), 0.2 mM dNTPs, and 0.8 µM of each primer, using the modified ‘Touchdown PCR’ protocol described by Sato et al.19 PCR products were separated by 10% polyacrylamide gel electrophoresis using TBE buffer and data collection was performed, as described previously.19
2.3. Linkage analysis
The MAPMAKER/EXP v3.0 program20 was used to analyse segregation data obtained from mapping the 94 RILs for microsatellite markers and previously reported RFLP, genomic-SSR, and phenotypic markers.7 Marker loci were analysed using the ‘group’ command with a threshold score of LOD = 3.0 and ‘ri self’ as the data type. The resulting linkage groups (LGs) were reorganized with reference to the integrated genetic map6 and a previously reported map for soybean.7 The order of marker loci and their relative genetic distances were calculated using the ‘Kosambi’ function with the ‘try’ and ‘map’ commands, respectively.
2.4. Evaluation of mapped markers and allele frequency
Polymorphism analysis was performed using a total of 24 soybean lines, including 15 Japanese cultivars and lines (‘Misuzudaizu’, ‘Nourin No.2’, ‘Ibarakimame 7 gou’, ‘Suzuyutaka’, ‘Fukuyutaka’, ‘Himeshirazu’, ‘Enrei’, ‘Toyokomachi’, ‘Yukihomare’, ‘Hayahikari’, ‘Toyomusume’, ‘Tamahomare’, ‘Saikai No. 20’, ‘Koitozairai’, and ‘Toiku No. 237’), two Japanese breeding material lines, three US cultivars (‘Jack’, ‘Adams’, and ‘Harosoy’), two Chinese lines (‘Peking’ and ‘Moshidou Gong 503’), ‘WILIS’ (Indonesia), and a wild soybean ‘Funaba 3’ (Japan). The genomic DNAs were extracted from each variety or line using DNeasy® Plant Mini Kit (QIAGEN, Germany). Following extraction of genomic DNA from each variety or line, PCR amplification was performed using the 668 primer pairs designed in this study. The number of alleles and polymorphism information content (PIC) were estimated using EST-microsatellite markers mapped onto the genetic linkage map. The PIC value was computed according to the formula described by Keim et al.:21
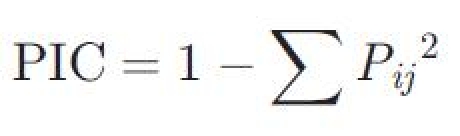 |
where, Pij is the frequency of the jth pattern for marker ith and summation extends over n patterns.
3. Results
3.1. Development of microsatellite markers derived from soybean ESTs
In order to develop microsatellite markers for soybean, we performed in silico data mining, as described in Section 2. We designed 6920 primer pairs to amplify SSRs from 63 676 non-redundant soybean ESTs obtained from public DNA databases. PCR amplification of template DNA from the parents ‘Misuzudaizu’ and ‘Moshidou Gong 503’ was achieved with 5299 (76.6%) of the primer pairs examined; 4710 pairs amplified both lines and 589 amplified only one of the parents. Clear polymorphisms between the parents were observed with 680 (12.8%) of the 5299 primer pairs; 655 detected single loci, whereas 24 and one detected double and quadruple loci, respectively. Together with 10 dominant Sequenced Tagged Site (STS) marker loci, a total of 707 marker loci were chosen for linkage mapping. SSRs of di-nucleotide repeat motifs (NN) showed polymorphisms more frequently than tri-nucleotide repeat motifs (NNN) and in fact, the ‘AT’ motifs exhibited polymorphism (23.1%; 82 of 355 primer pairs) efficiently (Supplementary Table S1). Interestingly, the largest number of primer pairs (1211) was tested for the ‘AAG’ motif and only 8.8% (107) exhibited polymorphism (Supplementary Table S1).
3.2. Construction of a genetic linkage map
In addition to 707 microsatellite markers developed in this study, we used 242 marker loci including 105 RFLP markers, 136 genome-derived microsatellite markers and one phenotypic marker that had been shown detect polymorphisms between mapping parents during construction of the genetic linkage map.7 Using the grouping module in MAPMAKER, 935 of 949 loci were assembled into 14 LGs, and these were then re-grouped into 20 LGs, according to the maps developed by Cregan et al.3 and Song et al.6 Ultimately, a total of 935 marker loci were mapped onto 20 LGs with a total genetic length of 2700.3 cM (Table 1 and Fig. 1), and 693 of the 935 loci detected were identified using the 668 EST-derived microsatellite markers developed in this study; the other 242 loci were detected using 105 RFLP markers, 136 genome-derived microsatellite markers and one phenotypic marker, as reported previously by Watanabe et al.7
Table 1.
Summary of the genetic linkage map and the mapped markers
| Linkage group | Map length (cM) | Number of markers | Average distance between two markers (cM) | Number of distorted markers (P < 0.05) | Average PIC value | ||||
|---|---|---|---|---|---|---|---|---|---|
| EST-derived microsatellite | RFLP | Genome-derived microsatellite | Phenotypic | Total | |||||
| A1 | 102.2 | 18 | 4 | 8 | 30 | 3.4 | 0 | 0.38 | |
| A2 | 194.0 | 35 | 6 | 8 | 49 | 4.0 | 2 | 0.43 | |
| B1 | 149.9 | 41 | 3 | 10 | 54 | 2.8 | 11 | 0.37 | |
| B2 | 116.7 | 27 | 5 | 2 | 34 | 3.4 | 0 | 0.43 | |
| C1 | 126.4 | 28 | 4 | 9 | 41 | 3.1 | 1 | 0.42 | |
| C2 | 191.1 | 43 | 7 | 11 | 61 | 3.1 | 1 | 0.41 | |
| D1a | 122.7 | 28 | 6 | 4 | 38 | 3.2 | 0 | 0.47 | |
| D1b | 144.0 | 48 | 5 | 7 | 60 | 2.4 | 0 | 0.43 | |
| D2 | 154.1 | 36 | 2 | 9 | 47 | 3.3 | 0 | 0.33 | |
| E | 161.3 | 34 | 9 | 8 | 51 | 3.2 | 6 | 0.45 | |
| F | 148.4 | 44 | 7 | 3 | 54 | 2.7 | 8 | 0.38 | |
| G | 130.5 | 45 | 6 | 8 | 59 | 2.2 | 3 | 0.50 | |
| H | 106.4 | 31 | 8 | 1 | 40 | 2.7 | 0 | 0.39 | |
| I | 124.8 | 28 | 3 | 6 | 1 | 38 | 3.3 | 5 | 0.42 |
| J | 75.5 | 31 | 8 | 1 | 40 | 1.9 | 5 | 0.43 | |
| K | 115.4 | 40 | 5 | 3 | 48 | 2.4 | 0 | 0.36 | |
| L | 111.6 | 35 | 6 | 9 | 50 | 2.2 | 5 | 0.38 | |
| M | 148.5 | 34 | 4 | 14 | 52 | 2.9 | 1 | 0.35 | |
| N | 132.3 | 26 | 5 | 5 | 36 | 3.7 | 0 | 0.35 | |
| O | 144.5 | 41 | 2 | 10 | 53 | 2.7 | 0 | 0.37 | |
| Total | 2700.3 | 693 | 105 | 136 | 1 | 935 | 48 | ||
| Average | 135.0 | 34.7 | 5.3 | 6.8 | 0.1 | 46.8 | 2.9 | 2.4 | 0.40 |
Figure 1.
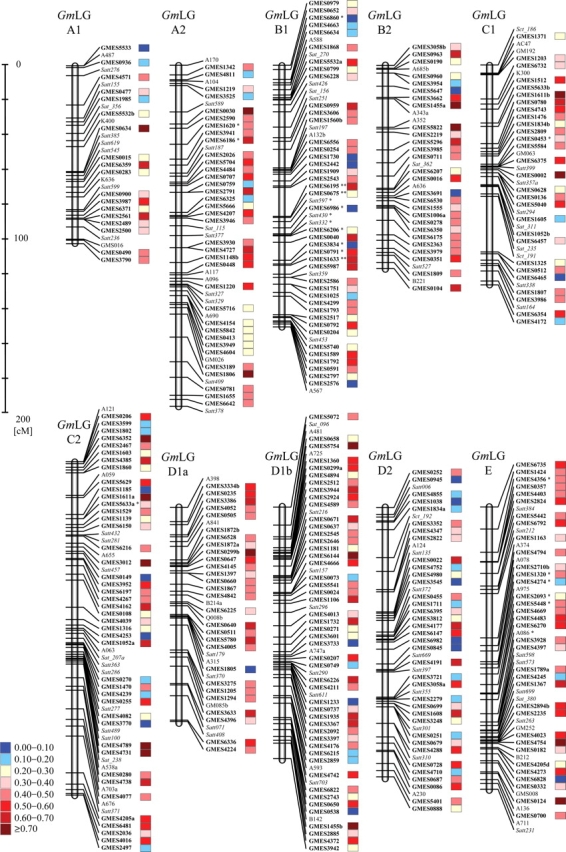
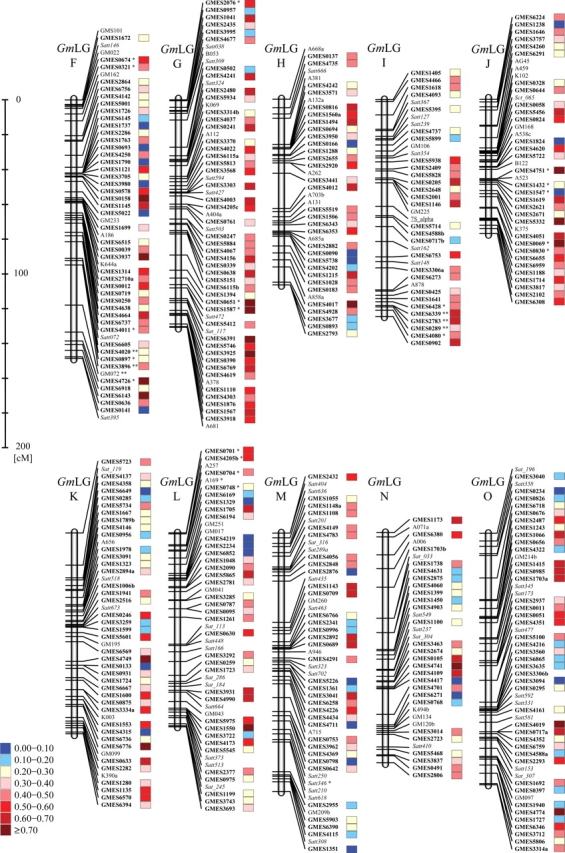
Genetic linkage map of the soybean genome. Twenty linkage groups were constructed using 105 RFLP markers (standard lettering), 136 genome-derived microsatellite markers (italic), one phenotypic marker (underlined), and 693 EST-derived microsatellite markers (bold). The marker loci indicated with an asterisk(s) show distorted loci, as determined using the Chi-square test (*P< 0.05, and **P< 0.01). Color codes inside the boxes on the right side of the marker loci indicate the level of PIC values from low (blue) to high (red), as shown on the left side of the panels. Detailed information on the EST-derived microsatellite markers is provided in Supplementary Table S2.
Although the average marker loci for each LG was 46.8, the number of EST-derived microsatellite markers ranged from 48 (LG D1b) to 18 (LG A1), with an average of 34.7 (Table 1). The average genetic length for each LG was 135.0 cM, although the sizes varied between 194.0 (LG A2) and 75.5 (LG J) cM (Table 1). The average distance between markers in each LG ranged between 1.9 and 4.0 cM (LG J and LG A2, respectively).
Most marker loci were relatively evenly distributed along the LGs, although some large gaps were observed between loci, including those between Sct_191 and GMES1325 (32.0 cM) on LG C1, and GMES1163 and A374 (25.2 cM) on LG E. The Chi-square test (P < 0.05) detected segregation distortion for 48 marker loci (5.1% of the mapped markers) and significant examples (less than P < 0.01) include the following: GMES0675, GMSE1633, and GMES6195 on LG B1; GM072, GMES3896, and GMES4020 on LG F; and GMES0289, GMES2783, and GMES6339 on LG I (Table 1).
3.3. Polymorphism of EST-derived microsatellite markers among 24 soybean lines
We examined allelic variation among 23 soybean cultivars/lines and a wild soybean line using 668 EST-derived microsatellite markers, which corresponded to 686 marker loci on the genetic linkage map (Supplementary Table S2). The number of alleles per locus ranged from 2 to 9, with a mean value of 2.8 (Fig. 2a); marker loci detecting two alleles were the most frequent. PIC values ranged from 0.08 to 0.84, with a mean value of 0.40 (Fig. 2b); marker loci with PIC values between 0.40 and 0.50 (175 markers) were most common. Although the average PIC value of the ‘AT’ motif (0.52) was one of the highest identified in this study (Supplementary Table S1), no notable correlation could be found between PIC values and SSR motifs. Marker loci with the highest and lowest average PIC values were detected in LG G (0.50) and LG D2 (0.33), respectively (Table 1). In addition, we observed clusters of marker loci with higher and lower than average PIC values such as between GMES0651 and GMES3918 on LG G (average PIC was 0.60) and between GMES2955 and GMES1351 on LG M (average PIC was 0.21; Fig. 1).
Figure 2.
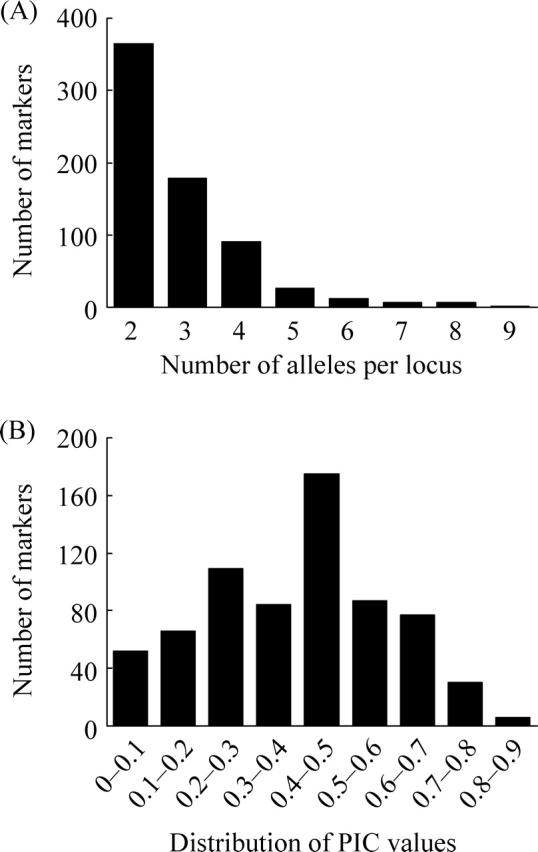
Allele frequency of mapped microsatellite markers in 23 soybean cultivars/lines and one wild soybean line. (A) The number of alleles per locus; (B) distribution of PIC values.
3.4. Comparison between the genomes of soybean and L. japonicus
We compared genome structure between soybean and the model legume L. japonicus. We performed a BLASTN search with a cut off E-value of E-20 between nucleotide sequences from soybean ESTs that corresponded to mapped microsatellite markers and L. japonicus genomic clones that had been mapped to the genetic linkage map.22 We examined 668 soybean ESTs and found 293 that showed significant sequence similarity to the 367 genomic clones comprising the L. japonicus genome (Table 2). In particular, 236 of these ESTs aligned with single loci on the L. japonicus genome, whereas the remaining 57 matched multiple loci (Supplementary Table S2). Connections could be made between the linkage maps of soybean and L. japonicus using the positional information on ESTs, as well as matched genomic clones (Fig. 3). L. japonicus chromosome 1 (hereafter referred to as LjChr) exhibited the largest number of matched soybean marker loci (97 loci), whereas LjChr 6 had the fewest (28 loci; Table 2). This variation may reflect physical lengths of the L. japonicus chromosomes.22,23
Table 2.
The number of loci with sequence similarity between the genomes of soybean and L. japonicus
| GmLG | LjChr | ||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | Total | |
| A1 | 2 | 1 | — | 5 | — | — | 8 |
| A2 | 3 | 2 | 7 | 6 | 1 | 1 | 20 |
| B1 | 3 | 5 | 5 | 1 | — | 1 | 15 |
| B2 | 2 | 7 | 1 | 2 | — | — | 12 |
| C1 | 12 | 5 | — | 4 | — | 1 | 22 |
| C2 | 11 | 1 | 1 | 2 | — | 1 | 16 |
| D1a | — | 14 | 1 | 1 | — | — | 16 |
| D1b | 2 | 12 | 1 | 3 | 6 | 3 | 27 |
| D2 | 1 | 4 | 3 | 11 | 4 | 1 | 24 |
| E | 1 | 2 | 5 | 4 | — | 6 | 18 |
| F | 2 | 1 | 4 | 8 | 2 | 2 | 19 |
| G | 11 | 1 | 1 | 5 | 3 | 4 | 25 |
| H | 5 | — | 16 | — | 1 | 1 | 23 |
| I | 1 | 1 | — | 2 | 12 | — | 16 |
| J | 5 | 4 | 2 | 6 | 2 | — | 19 |
| K | 6 | 3 | 3 | 4 | 2 | 5 | 23 |
| L | 9 | 1 | 2 | 4 | 6 | 2 | 24 |
| M | 2 | 3 | 6 | 6 | 1 | — | 18 |
| N | 11 | 1 | 2 | — | 3 | — | 17 |
| O | 8 | — | 1 | — | 14 | — | 23 |
| Total | 97 | 68 | 61 | 74 | 57 | 28 | 385* |
*The markers mapped on plural linkage groups of soybean were independently counted.
Figure 3.
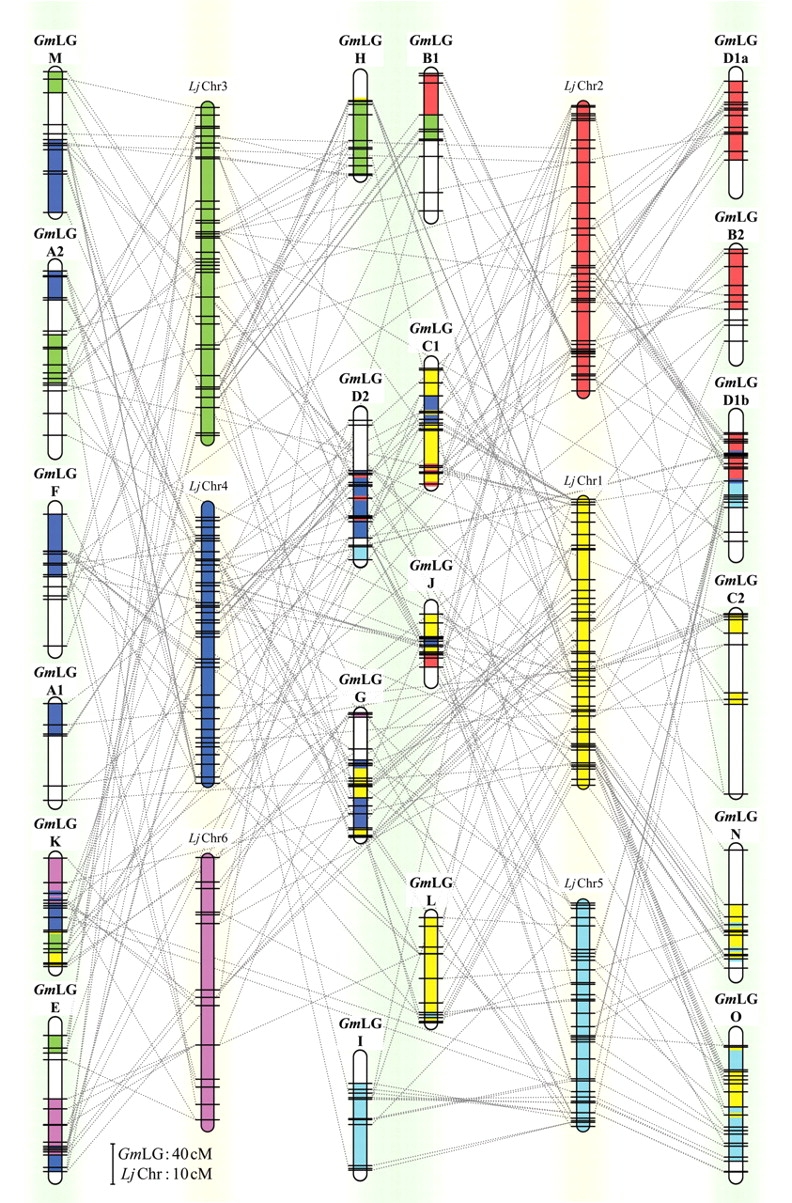
Genome alignment between soybean and L. japonicus. Twenty soybean linkage groups (GmLGs) and six L. japonicus chromosomes (LjChrs) were connected by lines based on sequences similarities, as described in Section 2. Vertical bars represent GmLGs (green background) and LjChrs (yellow background); horizontal lines indicate positions of loci showing sequence similarity between soybean and L. japonicus. The LjChrs were color-coded as follows: LjChr1; yellow, LjChr2; red, LjChr3; green, LjChr4; blue, LjChr5; light blue, LjChr6; pink. Portions of the soybean LGs were also color-coded, according to their similarity to the LjChrs.
Relationships between soybean linkage groups (hereafter referred to as GmLGs) and LjChrs appear to be rather complex (Fig. 3 and Supplementary Table S1), which suggests that considerable genome shuffling occurred after separation of the species (54 mya).24 However, to varying degrees macrosyntenic relationships were observed at the segmental level and strong co-linearity was found between the following: the bottom of LjChr 1 and the central portion of GmLG O; the central portion of LjChr 2 and the upper half of GmLG D1b, the bottom of LjChr 3 and the lower half of GmLG H; and the bottom of LjChr 5 and the lower portion of GmLG I. It is also noteworthy that a single locus on the LjChrs often corresponded to two marker loci on different GmLGs, a finding that may indicate ancestral duplication of the entire soybean genome.25
4. Discussion
In this study, a total of 6920 primer pairs (10.9%) were designed to amplify SSRs identified using a publicly available non-redundant soybean EST data set (63 676 ESTs). PCR amplification was achieved using 5299 (77%) of the 6920 primer pairs tested and 668 were located successfully onto the genetic linkage map. Altogether, ∼1% of the non-redundant ESTs could be used for development of DNA mapping markers and this study highlighted the applicability of soybean EST-SSRs for their development, although the actual efficiency of this process may differ depending upon the mapping populations.
AAG was the most frequently detected SSR motif in our marker set, and its frequency was approximately twice that of next most common motifs ATC, AAC, and AG. Shultz et al.26 reported that AT was the most frequently detected SSR motif in BAC-end sequences and that it occurred three times more frequently than the next most common motifs (AAT and AAG). These results suggest that SSR motif frequencies differ between ESTs and the whole genome in soybean. On the other hand, polymorphism frequency was higher for di-nucleotide motifs than tri-nucleotide motifs (18.3% versus 7.9%), which is consistent with observations by Shultz et al.26 The average PIC of the di-nucleotide motifs (0.46) was higher than those of tri- and tetra-nucleotide motifs (0.38 and 0.37, respectively), which also suggests that the former are more polymorphic.
The total length of the genetic linkage map generated in this study was 2700.3 cM, which was 1.1-fold greater than the previous consensus linkage map (2383.3 cM) generated by Choi et al.10 and thus, it is suggested that the present linkage map achieves a high degree of genome coverage. The genetic lengths of individual LGs in the present linkage map ranged from 0.8 (LG J) to 1.7 (LG C1) times those in the previous consensus map. When two maps are compared with respect to the common markers such as RFLP markers, the genetic distance and order on the present map fairly agreed with those on the consensus map except for LG E. In LG E, which was estimated to be 1.5-fold longer than that in the previous map, the genetic distances between any two common markers actually extended. In contrast, the genetic distances between the markers were quite similar between the two maps for LG C1, which was estimated to be 1.7-fold longer than that in the previous map, suggesting that the present map achieved longer genomic coverage.
Though there were 26 gaps of larger than 10 cM between the markers in the previous consensus linkage map of soybean,6 the EST-derived SNP markers generated recently by Choi et al.10 were mapped onto 19 of the 26 gaps, indicating usefulness of EST-derived markers to secure the uniformity of the map. The EST-derived microsatellite markers generated in the present study were successfully mapped onto 25 out of 31 gaps of larger than 20 cM in the previous linkage map constructed with RFLP and genome-microsatellite markers by Watanabe et al.7 Two large gaps on LG C1 and E still remain in both the previous and the present maps possibly because of peculiar chromosome structures that hamper marker generation.
We compared the ESTs used to develop the 1149 SNP markers identified by Choi et al.10 with the 668 non-redundant ESTs from which the mapped microsatellite markers originated in this study. Surprisingly, we found only 16 ESTs in common (data not shown), a finding that could be attributed to the different mapping populations used for detection of polymorphisms. However, it is more likely that the type of SNP and SSR polymorphism detected is a characteristic of the EST and thus, depends upon the gene from which it was derived and/or the position in the transcript. In general, the frequency of SNPs is higher than SSRs and thus in Zea mays, one SNP occurs every 28–124 bp, whereas one SSR is found every 8 kb.27 However, SNP markers are usually biallelic and less polymorphic than SSRs. Since SNP and microsatellite markers have such different characteristics, their combination should allow a larger number of EST-derived DNA markers to be mapped onto the genetic linkage map.
The allele frequency of any given marker is closely related to its transferability among germplasms, as well as the degree of variability within the marker locus. In general, nucleotide sequences are more conserved in coding regions than in other regions of the genome. Therefore, it is expected that the PIC values of EST-derived markers may be lower than for markers derived from random genomic sequences. Hossain et al.28 demonstrated that on the genetic linkage map, EST-derived markers exhibited lower average PIC values (0.40) than random genome-derived microsatellite markers (0.61). Interestingly, the 668 EST-derived microsatellite markers showed normal PIC value distributions, with a peak at 0.4–0.5; the most frequent number of alleles per marker was 2, which was the minimum number expected. The reason why two alleles per markers were most frequent was that they might include STS markers. Since the PIC calculation is based upon the number of alleles and their distribution within the examined population, different distributions of PIC and allele number suggest a uniform dispersal of most EST-derived microsatellite marker polymorphisms within soybean germplasms.
Hyten et al.11 reported that the LD is highly complex in soybean and they suggested that in addition to a large number of SNP markers, soybean would require a soybean haplotype map for whole-genome association analysis, equivalent to that of the human HapMap project.29 Although development of such a map would provide a useful information platform, it would also be very cost-intensive. In this study, we investigated the relationship between the positions of marker loci on the linkage map and their PIC, and found that there were several genomic blocks in which multiple markers exhibited higher or lower PIC values. Since the markers in our present linkage map are not all distributed evenly throughout the genome, we cannot demonstrate a comprehensive structure of diversity for multiple populations of soybean. However, if PIC values are calculated for all the markers from multiple populations, our results suggest that a conventional high-density genetic linkage map may be able to substitute for as haplotype map. This would enable identification of recombination hotspots and provide information on the block-like structures of the LD.
Comparative mapping has shown that numerous alterations contribute to genomic diversity among plants.30 Since soybean has a paleopolyploid genome, comparison with the genomes of other legume species can reveal the process of genome duplication and subsequent rearrangements during the evolution. We compared the genome structures of soybean and L. japonicus using 293 microsatellite markers, a 10-fold increase over the 22 DNA markers used in the previous comparison by Zhu et al.13 Our results indicated three types of linkage between the two genomes, i.e. soybean LGs that were related to a single Chr of L. japonicus and which exhibited co-linearity, soybean LGs that were related to two Chrs of L. japonicus and which exhibited limited degree of co-linearity, and soybean LGs that were related to three or four Chrs of L. japonicus and did not demonstrate any apparent co-linearity. In considering the evolution of the soybean genome, it is noteworthy that all soybean LGs related to three or four Chrs of L. japonicus (LGs C1, D1b, D2, E, G, J, and K) exhibited links with Chr 4. It has been demonstrated that polyoloidy is more than the sum of its gene duplications, since this process duplicates the members of entire regulatory networks.31
Shoemaker et al.25 demonstrated the segmental duplications of the soybean genome for the following pairs of the linkage groups using homeologous RFLP markers; LG A1-LG A2, LG A2-LG E, LG B1-LG H, LG B2-LG D1a/LG D1b, LG C2-LG G, LG G-LG K, and LG L-LG N. In the present study, we confirmed the possibility of segmental duplications in the previously suggested regions by inspecting the co-linearity between the genomes of soybean and L. japonicus. Furthermore, we could detect the trances of additional duplications in the following regions; a central portion of LG O-a lower half of LG L, a central portion of LG O-a lower half of LG N, and a central portion of LG I-a lower half of LG O. In this study, we have revealed relationships between the genomes of soybean and L. japonicus and this comparison, combined with information relating to regulatory networks deduced by transcriptional profiling, may provide clues toward understand the evolutionary process of the paleopolyploid soybean genome.
To date, we have accumulated a considerable amount of information and material resources for the soybean genome and the next necessary and urgent step will be to compare and combine these resources systematically. Not only will such improvements assist in our understanding of this complex genetic system, but they will provide useful information for further developments in breeding technology. In the present study, we provided new information on soybean by converting EST into DNA markers. In addition to providing positional information on the genetic linkage map, we used PIC as an index for determining genomic diversity within the species and compared genome structures between legume species. These results demonstrate the potential of using publicly available soybean ESTs for the development of valuable resources beyond microsatellite markers and genetic linkage maps.
Additional information is available online at http://www.kazusa.or.jp/soymarker/
Supplementary Data
Supplementary data are available online at www.dnaresearch.oxfordjournals.org.
Acknowledgements
We thank Dr M. Ishimoto for providing plant materials.
Funding: This work was supported by the Kazusa DNA Research Institute Foundation.
References
- 1.Production, Supply and Distribution online database. United States Department of Agriculture
- 2.Morgante M., Rafalski A., Biddle P., Tingey S., Olivieri A. M. Genetic mapping and variability of seven soybean simple sequence repeat loci. Genome. 1994;37:763–769. doi: 10.1139/g94-109. [DOI] [PubMed] [Google Scholar]
- 3.Cregan P. B., Jarvik T., Bush A. L., et al. An integrated genetic linkage map of the soybean genome. Crop Sci. 1999;39:1464–1490. [Google Scholar]
- 4.Ferreira A. R., Foutz K. R., Keim P. Soybean genetic map of RAPD markers assigned to an existing scaffold RFLP map. J. Hered. 2000;91:392–396. doi: 10.1093/jhered/91.5.392. [DOI] [PubMed] [Google Scholar]
- 5.Yamanaka N., Ninomiya S., Hoshi M., et al. An informative linkage map of soybean reveals QTLs for flowering time, leaflet morphology and regions of segregation distortion. DNA Res. 2001;8:61–72. doi: 10.1093/dnares/8.2.61. [DOI] [PubMed] [Google Scholar]
- 6.Song Q. J., Marek L. F., Shoemaker R. C., et al. A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 2004;109:122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe S., Tajuddin T., Yamanaka N., Hayashi M., Harada K. Analysis of QTLs for reproductive development and seed quality traits in soybean using recombinant inbred lines. Breeding Sci. 2004;54:399–407. [Google Scholar]
- 8.Shoemaker R., Keim P., Vodkin L., et al. A compilation of soybean ESTs: generation and analysis. Genome. 2002;45:329–338. doi: 10.1139/g01-150. [DOI] [PubMed] [Google Scholar]
- 9.Nelson R. T., Shoemaker R. Identification and analysis of gene families from the duplicated genome of soybean using EST sequences. BMC Genomics. 2006;7:204. doi: 10.1186/1471-2164-7-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi I.-Y., Hyten D. L., Matukumalli L. K., et al. A soybean transcript map: gene distribution, haplotype and single-nucleotide polymorphism analysis. Genetics. 2007;176:685–696. doi: 10.1534/genetics.107.070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyten D. L., Choi I.-Y., Song Q., et al. Highly variable patterns of linkage disequilibrium in multiple soybean populations. Genetics. 2007;175:1937–1944. doi: 10.1534/genetics.106.069740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi H.-K., Mun J.-H., Kim D.-J., et al. Estimating genome conservation between crop and model legume species. Proc. Natl. Acad. Sci. USA. 2004;101:15289–15294. doi: 10.1073/pnas.0402251101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu H., Choi H.-K., Cook D. R., Shoemaker R. C. Bridging model and crop legumes through comparative genomics. Plant Physiol. 2005;137:1189–1196. doi: 10.1104/pp.104.058891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powell W., Machray G. C., Provan J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996;1:215–222. [Google Scholar]
- 15.Varshney R. K., Graner A., Sorrells M. E. Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 2005;23:48–55. doi: 10.1016/j.tibtech.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Guo W., Cai C., Wang C., et al. A microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in Gossypium. Genetics. 2007;176:527–541. doi: 10.1534/genetics.107.070375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray M. G., Thompson W. F. Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers, In. In: Krawetz S., Misener S., editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ, USA: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 19.Sato S., Isobe S., Asamizu E., et al. Comprehensive structural analysis of the genome of red clover (Trifolium pretense L.) DNA Res. 2005;12:301–364. doi: 10.1093/dnares/dsi018. [DOI] [PubMed] [Google Scholar]
- 20.Lincoln S. E., Daly M. J., Lander E. S. Construction genetic maps with MAPMAKER/EXP 3.0. 3rd Ed. Cambridge, Massachusetts: Whitehead Institute; 1992. Whitehead Institute Technical Report. [Google Scholar]
- 21.Keim P., Beavis W., Schupp J., Freestone R. Evaluation of soybean RFLP marker diversity in adapted germ plasm. Theor. Appl. Genet. 1992;85:205–212. doi: 10.1007/BF00222861. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi M., Miyahara A., Sato S., et al. Construction of a genetic linkage map of the model legume Lotus japonicus using an intraspecific F2 population. DNA Res. 2001;8:301–310. doi: 10.1093/dnares/8.6.301. [DOI] [PubMed] [Google Scholar]
- 23.Pedrosa A., Sandal N., Stougaard J., Schweizer D., Bachmair A. Chromosomal map of the model legume Lotus japonicus. Genetics. 2002;161:1661–1672. doi: 10.1093/genetics/161.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavin M., Herendeen P. S., Wojciechowski M. F. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- 25.Shoemaker R.C., Polzin K.C., Labate J., et al. Genome duplication in soybean (Glycine subgenus soja) Genetics. 1996;144:329–338. doi: 10.1093/genetics/144.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shultz J. L., Kazi S., Bashir R., Afzal J. A., Lightfoot D. A. The development of BAC-end sequence-based microsatellite markers and placement in the physical and genetic maps of soybean. Theor. Appl. Genet. 2007;114:1081–1090. doi: 10.1007/s00122-007-0501-9. [DOI] [PubMed] [Google Scholar]
- 27.Jones E. S., Sullivan H., Bhattramakki D., Smith J. S. A comparison of simple sequence repeat and single nucleotide polymorphism marker technologies for the genotypic analysis of maize (Zea mays L.) Theor. Appl. Genet. 2007;115:361–371. doi: 10.1007/s00122-007-0570-9. [DOI] [PubMed] [Google Scholar]
- 28.Hossain K. G., Kawai H., Hayashi M., Hoshi M., Yamanaka N., Harada K. Characterization and identification of (CT)n microsatellite in soybean using sheared genomic libraries. DNA Res. 2000;7:103–110. doi: 10.1093/dnares/7.2.103. [DOI] [PubMed] [Google Scholar]
- 29.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall A. E., Fiebig A., Preuss D. Beyond the Arabidopsis genome: Opportunities for comparative genomics. Plant Physiol. 2002;129:1439–1447. doi: 10.1104/pp.004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shoemaker R. C., Schlueter J., Doyle J. J. Paleopolyploidy and gene duplication in soybean and other legumes. Curr. Opin. Plant Biol. 2006;9:104–109. doi: 10.1016/j.pbi.2006.01.007. [DOI] [PubMed] [Google Scholar]


